Abstract
The current study investigated transcriptional distortion in prostate cancer cells using the vitamin D receptor (VDR) as a tool to examine how epigenetic events driven by corepressor binding and CpG methylation lead to aberrant gene expression. These relationships were investigated in the non-malignant RWPE-1 cells that were 1α,25(OH)2D3 responsive (RWPE-1) and malignant cell lines that were 1α,25(OH)2D3 partially responsive (RWPE-2) and resistant (PC-3). These studies revealed that selective attenuation and repression of VDR transcriptional responses in the cancer cell lines reflected their loss of antiproliferative sensitivity. This was evident in VDR target genes including VDR, CDKN1A (encodes p21(waf1/cip1)) and GADD45A; NCOR1 knockdown alleviated this malignant transrepression. ChIP assays in RWPE-1 and PC-3 cells revealed that transrepression of CDKN1A was associated with increased NCOR1 enrichment in response to 1α,25(OH)2D3 treatment. These findings supported the concept that retained and increased NCOR1 binding, associated with loss of H3K9ac and increased H3K9me2, may act as a beacon for the initiation and recruitment of DNA methylation. Overexpressed histone methyltransferases (KMTs) were detectable in a wide panel of prostate cancer cell lines compared with RWPE-1 and suggested that generation of H3K9me2 states would be favored. Cotreatment of cells with the KMT inhibitor, chaetocin, increased 1α,25(OH)2D3-mediated induction of CDKN1A expression supporting a role for this event to disrupt CDKN1A regulation. Parallel surveys in PC-3 cells of CpG methylation around the VDR binding regions on CDKN1A revealed altered basal and VDR-regulated DNA methylation patterns that overlapped with VDR-induced recruitment of NCOR1 and gene transrepression. Taken together, these findings suggest that sustained corepressor interactions with nuclear-resident transcription factors may inappropriately transform transient-repressive histone states into more stable and repressive DNA methylation events.
Introduction
In non-malignant prostate epithelial cells control of key histone modifications during vitamin D receptor (VDR)-regulated expression of CDKN1A (encodes p21(waf1/cip1)) is spatially controlled across regulatory regions of the gene locus and dynamically regulated in time (1,2). The VDR, such as many other nuclear receptors, interacts with coactivators and corepressors during the transcriptional cycle (1–5) and these interactions combine to determine these highly choreographed distributions of histone modifications. For example, binding of NCOR1 at a specific VDR binding site was associated with the loss of H3K9ac and gain of H3K9me2 and H3K27me3 (2). In turn, these patterns of corepressor binding and changing histone modifications were associated significantly with key points of messenger RNA (mRNA) oscillation (2).
These transient histone states also act as platforms to allow effector complexes to regulate other aspects of chromatin architecture such as DNA CpG methylation. For example, gene-repressive histone modifications such as H3K9me2 associate with CpG methylation and heterochromatin. At high-density regions of CpG methylation, spanning hundreds of base pairs, these marks act as triggers to recruit heterochromatin binding protein 1 (6). The recruitment of heterochromatin binding protein 1 through interaction with the methyl-CpG binding protein MBD1 leads to recruitment of both KMT1A (called previously SUV39H1 (7)) and DNA methyltransferases (8) and thereby the entire region acquires H3K9 and H3K27 methylation and loses H3K4 methylation (reviewed in (9)). It has also emerged that in actively regulated regions, dynamic changes in DNA methylation appear to occur. For example, these have been measured in response to nuclear receptor actions (10–12). In parallel, increased corepressor binding promotes direct association with the DNA methylation-dependent transcriptional repressor ZBTB33/KAISO (13) and targets increased DNA methylation.
Despite these important roles for corepressors to regulate transcription and potentially trigger DNA methylation, they remain somewhat overlooked compared with their coactivator cousins. Ambiguity remains over how and to what extent these actions are distorted in cancer (reviewed in (14)). The sheer diversity of transcription factor and corepressor interactions contributes significantly to this uncertainty. This is in turn compounded by the fact that there are functionally different corepressor isoforms (15,16) and that corepressor actions appear specific to each phase of the cell cycle (2,17,18).
Increased NCOR1 and NCOR2/SMRT expression and localization occurs in prostate and other cancers (19–24), for example, to suppress VDR and Peroxisome proliferator-activated receptor (PPAR) responsiveness (17,20,25). In contrast, putative loss of function NCOR1 mutations have been identified contributing to breast cancer (26). The recruitment to the androgen receptor (AR) of corepressors contributes to efficacy of androgen deprivation therapy and consequently loss of expression of NCOR1 and NCOR2/SMRT is also associated with recurrent prostate cancer (27,28).
Thus, corepressor expression and interactions appear to change during the course of disease progression. These changes most probably reflect the conflicting selection pressures on corepressor expression, targeting and activity. Set against these uncertainties, dissecting corepressor actions may address the key question of separating epigenetic processes that drive cancer initiation and progression, from those that are merely a consequence of altered genomic structure. This understanding has importance for defining clinical targeting strategies, given that altered corepressor expression generates critical targets for agents that target the regulation of H3K9 acetylation and methylation states.
Therefore, the current study aimed to investigate whether corepressor binding was altered in the prostate cancer. Specifically, it focused on two inter-related questions: First, to establish whether corepressor recruitment was altered between prostate cancer (CaP) cell lines that responded differentially toward VDR activation; Second, to reveal how corepressor recruitment was associated with altered DNA methylation patterns. We exploited three prostate cell lines that displayed a range of antiproliferative responses toward 1α,25(OH)2D3. These were non-malignant RWPE-1 and its RAS-transformed variant, RWPE-2, (29), and a CaP cell line derived from a metastasis, PC-3 (30). The antiproliferative actions of 1α,25(OH)2D3 were reduced in RWPE-2 compared with RWPE-1. The cells have the same ED50 value (300nM), whereas only RWPE-1 displayed an ED75 value (900nM) ((2) and unpublished observations). PC-3 cells are well-established to be recalcitrant to the antiproliferative actions of 1α,25(OH)2D3 (17,20,31,32). Therefore, these models are attractive to study the impact of epigenetic events that may be disrupted in the initiation and progression of prostate cancer.
These studies revealed that selective corepressor recruitment was associated with altered patterns of DNA methylation and direct gene transrepression. These differences in corepressor binding and DNA methylation reflected the profound differences in VDR antiproliferative responses in different cell models.
Materials and methods
Cell lines
RWPE-1 non-malignant prostate epithelial cells and RWPE-2 cells were maintained in Keratinocyte serum-free medium supplemented with epidermal growth factor and bovine pituitary extract (Invitrogen). PC-3 cells were cultured in RPMI with 10% fetal bovine serum.
Agents
1α,25(OH)2D3 (gift of Dr Milan Uskokovic (BioXell S.p.A. Italy)) was stored as 1mM stocks in ethanol. Chaetocin (Enzo life sciences) were stored in ethanol as 10mM stock.
Live cell sorting
Cells were stained with Hoechst 33342 (Invitrogen), fractionated using a MoFlo cell sorter (Beckman-Coulter, High Wycombe, UK) and 5 × 105 cells/phase were collected.
Quantitative reverse transcription–PCR
RNA was isolated using TRizol (Invitrogen). Target gene expression was quantitated on an ABI 7900 (Applied Biosystems, http://www.appliedbiosystems.com) machine. All primers and probes were as described previously (20). Measurements were performed in technical and biological triplicate.
Multitarget microfluidic quantitative reverse transcription–PCRM
Measurement of multiple gene transcripts was undertaken on custom-designed TaqMan® Low Density Array (ABI 7900HT Fast Real-Time PCR System) as described previously (22) and included known and putative VDR target genes and control genes (31,33–36). mRNA from cell-cycle sorted cells was quantified in triplicate samples measured in duplicate as described previously (17,22).
Fold changes were calculated for single target gene expression and statistical analyses were carried out using the TIGR MultiExperiment Viewer 4.0, MeV (www.tm4.org). A one-sample t-test analysis based on permutation (Westfall Young stepdown (37) – MaxT correction) was used to identify genes significantly expressed in each phase of the cell cycle comparing PC-3 to RWPE-1 cells. Vectors containing gene expression values were tested against the mean of 18S fold changes. One-way analysis of variance was used to identify genes that were expressed differentially across the three phases of the cell cycle.
ChIP protocols
X-ChIP was used to measure the association of NCOR1 binding as described previously (2,4). Briefly, chromatin from 1.5 × 106 mid-exponential cells was cross-linked. Precleared inputs were immunoprecipited with NCOR1 (Abcam ab24552). Complexes were recovered using magnetic beads, washed, cross-linking was reversed and further cleared DNA was recovered by standard precipitation approaches. 25ng DNA was used per quantitative PCR using SYBRgreen with preoptimized primers as described previously (2).
DNA methylation assays in cell lines
CpG dinucleotide methylation was measured around VDR binding regions and the transcription start site (TSS) of the CDKN1A locus. Specifically, CpG regions in an approximately 300bp region centered on the VDR binding region were used to undertake MassArray Quantitative Methylation Analysis (MAQMA) on the Sequenome platform in the RPCI Genomics Core Facility as described previously (38–40). This approach is high-throughput, with 384 assays performed simultaneously. DNA was isolated from the cells at the indicated time points following treatment. CpG dinucleotide methylation appears to be strand-specific (11) and therefore bisulfite PCR primers specific to each strand were designed for each region of interest.
Results
Suppressed VDR target gene regulation in 1α,25(OH)2D3- recalcitrant cells
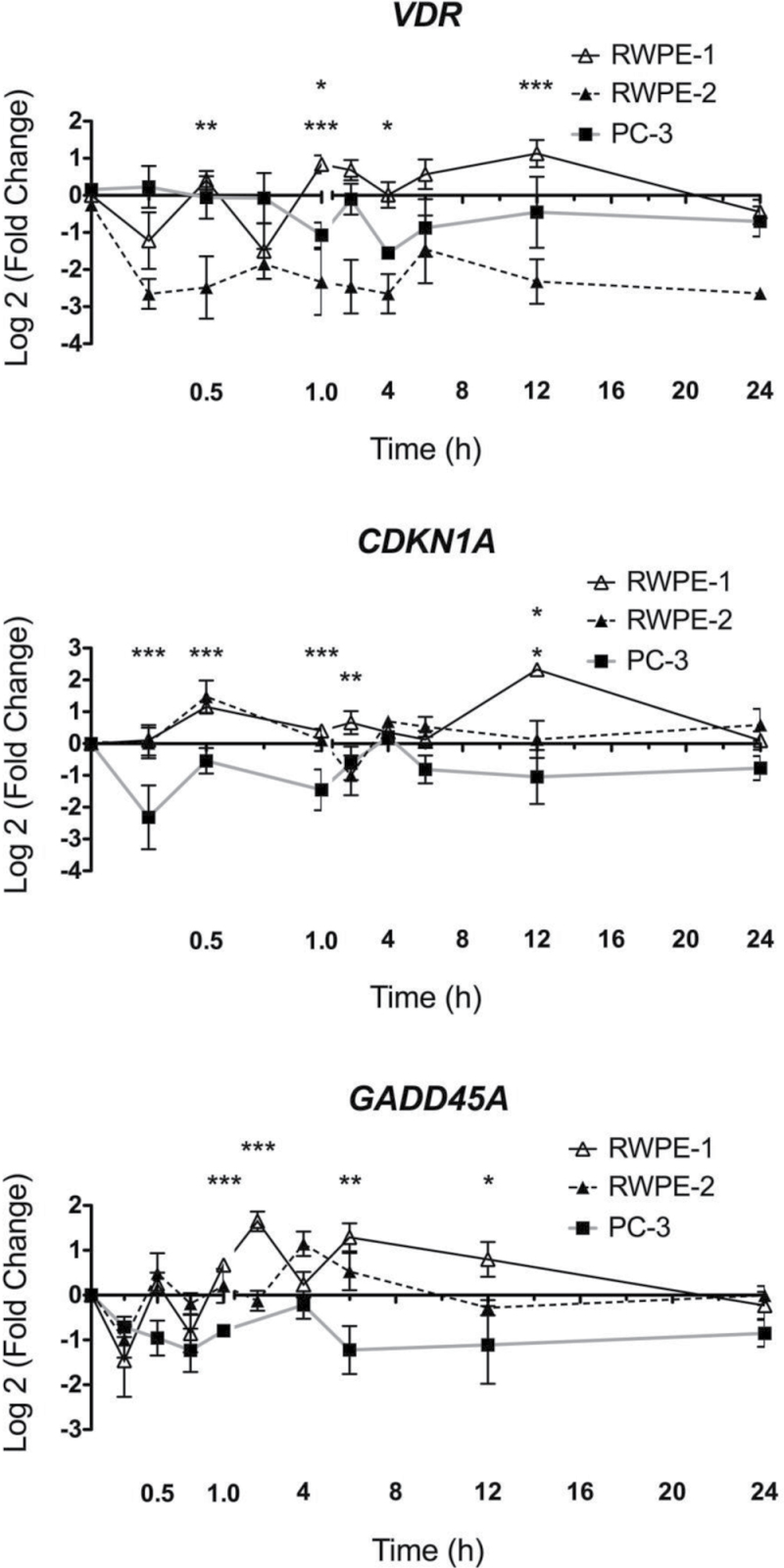
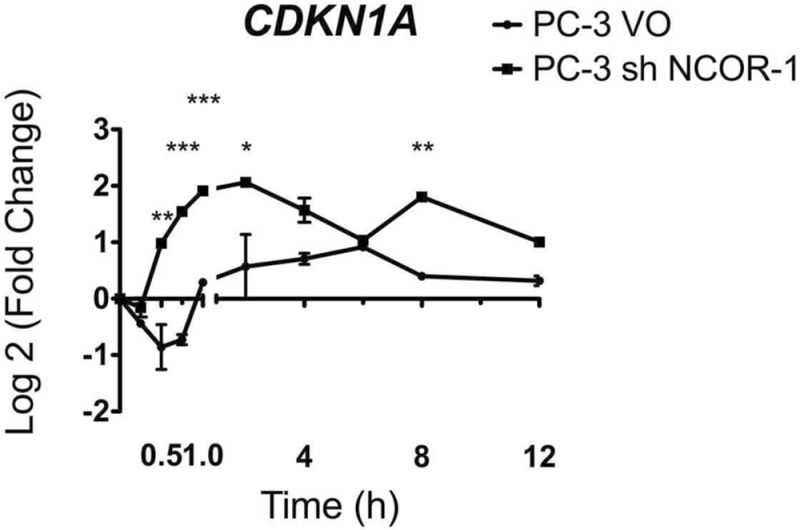
As a functional indicator of 1α,25(OH)2D3 actions, VDR-mediated gene regulatory actions were examined in RWPE-1, RWPE-2 and PC-3 cells. Time-resolved regulation studies were undertaken with three established VDR target genes (VDR, CDKN1A, GADD45A (1,20,41)). The patterns of VDR-mediated gene regulation were selectively distorted in the RWPE-2 and/or PC-3 cells compared with RWPE-1 cells. VDR regulation was distorted most clearly in RWPE-2, being profoundly repressed compared with RWPE-1 at multiple time points (Figure 1). The kinetics of CDKN1A mRNA regulation in RWPE-1 cells reflected previous findings (2), whereas the regulation in RWPE-2 was repressed, for example, at 12h. Transrepression was evident in PC-3 at multiple time points. In RWPE-1 and RWPE-2 cells, GADD45A also displayed rapid accumulation at 0.5h and 2h (RWPE-1 only). Again the fold induction was attenuated significantly in PC-3 cells, for example, at 0.5h and 6h (Figure 1). Using a clone of PC-3 cells, we established previously to have stable knock down of NCOR1 (17) and we examined CDKN1A induction following 1α,25(OH)2D3 treatment. In this case, we found that the regulation was significantly enhanced with a loss of the transrepression observed in the parental cells. Interestingly, and probably reflecting some aspect of stable selection, the levels of CDKN1A induction in the vector controls were also beyond the levels seen in RWPE-1 cells (Figure 2).
Fig. 1.
Dynamic regulation of VDR target genes. RWPE-1, RWPE-2 and PC-3 cells were treated with 1α,25(OH)2D3 (100nM) or ethanol control and mRNA was extracted at the indicated time points, and accumulation of indicated genes was measured using TaqMan Q-RT–PCR. Accumulation of each target is given as log2 (fold change). Each data point represented the mean of triplicate experiments in triplicate wells ± standard error mean (*P < 0.05, **P < 0.01, ***P < 0.001).
Fig. 2.
ShRNA to NCOR1 changes the regulation of CDKN1A. Stable transfectants PC-3 VO (vector only) and PC-3 shNCOR1 cells were treated with 1α,25(OH)2D3 (100nM), mRNA extracted at the indicated time points, and accumulation of CDKN1A measured using TaqMan Q-RT–PCR. Accumulation is given as log2 (fold change). Each data point represents the mean of triplicate experiments in triplicate wells ± standard error mean (*P < 0.05, **P < 0.01, ***P < 0.001).
Repression of the VDR mRNA regulation response was also observed when controlling for the impact of the different distributions of cells through the cell cycle in RWPE-1 and PC-3 cells. We noted that in RWPE-1 and PC-3 cells, the regulation of CDKN1A and GADD45A appeared to return to basal levels at 4h but differed at all time points. Therefore, we selected this time point to examine regulation of genes across the cell cycle. Specifically, a microfluidic quantitative reverse transcription (Q-RT)–PCRM approach (22) was applied to reveal 1α,25(OH)2D3-regulated expression patterns in cells in each phase of the cell cycle (Table 1). Cells in G1 displayed the greatest differential response between RWPE-1 and PC-3 cells. In G1-sorted cells, all VDR targets were regulated positively in RWPE-1 cells but these effects were mostly either reduced or repressed in PC-3 cells in the same phase. This was less pronounced in the other phases with only CYP24A1 and IGFBP3 significantly different in their regulation in all phases. Interestingly, CDKN1A regulation differed significantly in only G2/M cells. NCOR2/SMRT, a previously established VDR target gene, was differentially regulated but NCOR1 was not and is not included in the table. Control genes B2M and GAPDH were not regulated differentially among cell lines.
Table I.
Significantly distorted regulation of VDR target genes in each phase of the cell cycle
| Target gene | Log2 fold change in VDR target genes | |||||
|---|---|---|---|---|---|---|
| G1 | S | G2/M | ||||
| RWPE-1 | PC-3 | RWPE-1 | PC-3 | RWPE-1 | PC-3 | |
| CYP24A1 | 9.2 | 4.2 | 8.4 | 3.9 | 7.2 | 1.7 |
| IGFBP3 | 2.7 | −0.1 | 1.6 | 0.2 | 0.5 | −0.5 |
| TGFB2 | 2.5 | −0.2 | ||||
| G0S2 | 2.0 | 0.3 | ||||
| GADD45A | 1.8 | −0.4 | ||||
| CDH1 | 1.7 | −0.1 | ||||
| CDKN1A | −1.6 | −0.5 | ||||
| CDKN1B | 1.4 | −0.5 | ||||
| NCOR2 | 1.2 | −0.4 | ||||
| ABCG2 | 1.0 | −0.07 | ||||
| VDR | 0.9 | 0.7 | ||||
| BAX | 0.9 | −0.02 | ||||
| TP53 | 0.5 | 0.05 | −1.3 | −0.2 | ||
| B2M | ||||||
| GAPDH | ||||||
Exponentially growing RWPE-1 and PC-3 cells were treated with either 1α,25(OH)2D3 (100nM) or ethanol (control) for 4h and then fractionated into each phase of the cell cycle prior to mRNA extraction and Q-RT-PCRM to measure regulation of multiple VDR target genes compared with 18S RNA (B2M and GAPDH were included as further controls) and the fold change was transformed into log2 values. The table is organized to highlight, at the top of the list, genes that changed the most and specifically only those that were significantly modulated. Levels of gene expression are indicated in log2 fold changes; significance was calculated with a one-sample t-test comparing the expression levels with the value of 0, corresponding to no differences in gene expression between the two cell lines; shaded cell indicates not significant.
Together these data indicate that gene regulation by 1α,25(OH)2D3 was most dynamic in cells that were most responsive to the antiproliferative effects (RWPE-1 cells). These dynamic patterns included both positive and negative mRNA regulation. Furthermore, cells in G1 of the cell cycle were the most responsive. By comparison in RWPE-2 and PC-3 cells, the mRNA regulation profiles were increasingly and selectively attenuated. For CDKN1A, at least, NCOR1 appeared to play a significant role in suppressing the accumulation.
Spatial-temporal distribution of NCOR1 to CDKN1A is altered in 1α,25(OH)2D3-recalcitrant cells
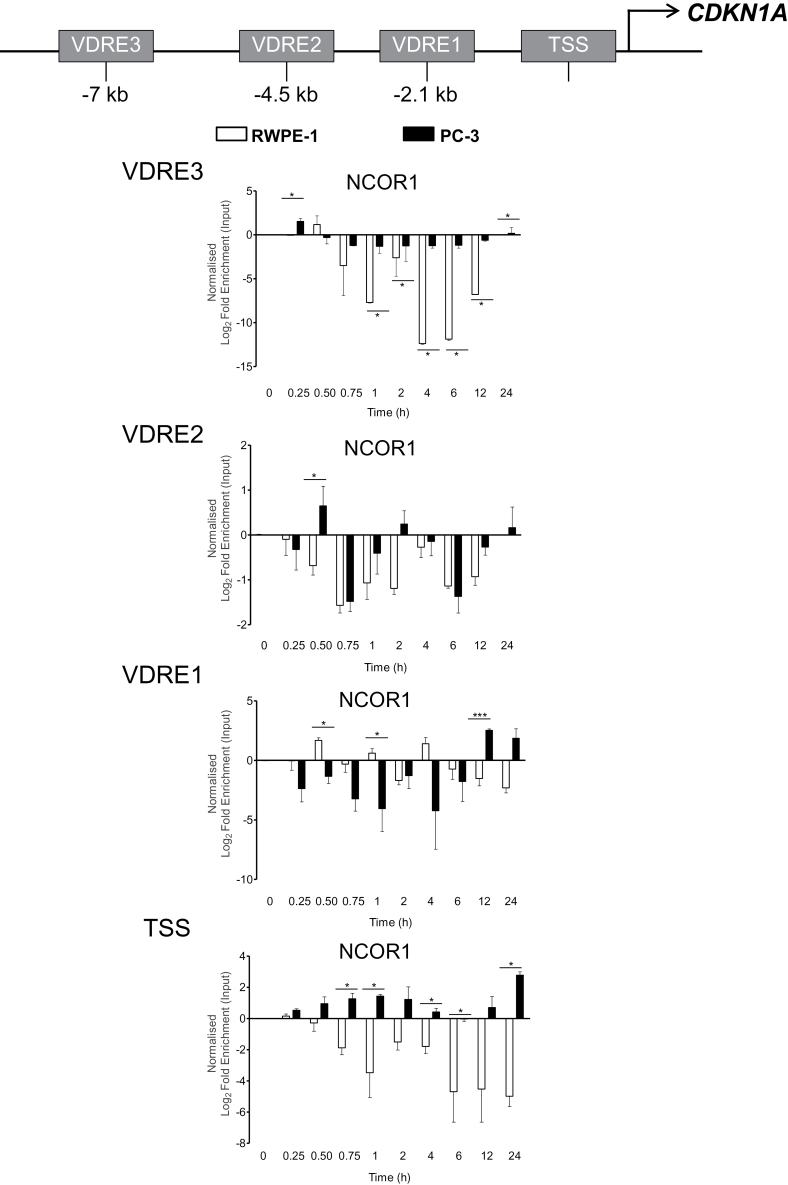
A fine-resolution X-ChIP time course was undertaken to examine NCOR1 recruitment to CDKN1A in cells that were 1α,25(OH)2D3-sensitive (RWPE-1) and 1α,25(OH)2D3-recalcitrant (PC-3). 1α,25(OH)2D3-regulated binding was measured at three VDR binding regions (VDREs 1, 2 and 3) and the TSS on CDKN1A (1) (Figure 3).
Fig. 3.
NCOR1 differentially associates with CDKN1A regulatory regions. RWPE-1 and PC-3 cells were treated with 1α,25(OH)2D3 (100nM) or ethanol control for indicated time points. Association of NCOR1 was measured at each region using X-ChIP with ChIP grade antibodies and normalized and given as fold enrichment over input (2). Enrichment was measured using Q-PCR with primers specific to these regions that amplified products <150bp. All measurements were performed in technical duplicate and biological triplicate and are given as log2 fold enrichment (*P < 0.05, **P < 0.01, ***P < 0.001).
In the basal state, there were no significant differences between the basal binding of NCOR1 between the two cell models (data not shown). In contrast, following 1α,25(OH)2D3 treatment, the recruitment of NCOR1 differed significantly between the two cell lines. In RWPE-1 cells, there was a pronounced loss of NCOR1 at VDRE3, VDRE2 and the TSS. This trend was significantly reduced in PC-3 cells where the loss of NCOR1 at VDRE3 did not occur to any significant extent. At specific time points at VDRE2 and VDRE1, NCOR1 enrichment was reciprocal in the two models. Notably at VDRE2 at 0.5h and at VDRE1 at 12h and 24h, NCOR1 was positively enriched in PC-3 cells and lost at the same time point in RWPE-1 cells (Figure 3). The most extreme inversion of the kinetics of NCOR1 recruitment occurred at the TSS where NCOR1 was lost from this region in RWPE-1 cells but exclusively recruited and enriched in PC-3 cells. These suggest that NCOR1 was recruited differentially to the promoter of CDKN1A, following 1α,25(OH)2D3 activation in the responsive RWPE-1 cell line as compared with the 1α,25(OH)2D3-recalcitrant PC-3 cell line.
Altered histone methyltransferase expression in prostate cancer cells
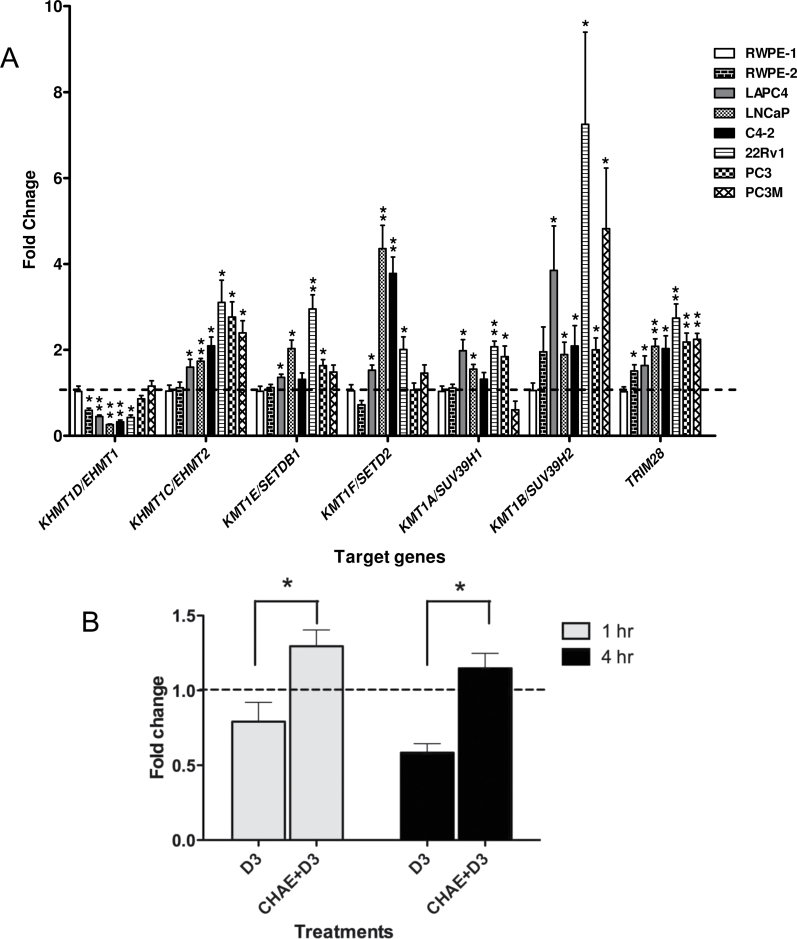
We next addressed the question as to whether the association of NCOR1 could result in altered DNA methylation at regions adjacent to the VDR binding elements. As a prelude to these studies, we examined the basal expression of several enzymes known to be H3K9 methyltransferases; specifically KHMT1D/EHMT1, KHMT1C/EHMT2, KMT1E/SETDB1, KMT1F/SETD2, KMT1A/SUV39H1, KMT1B/SUV39H2 and also TRIM28 and ZBTB33/KAISO. Expression was examined in a wide panel of prostate cancer cell lines compared with RWPE-1 cells. Five of the KMT enzymes were elevated across the cancer cell lines including KHMT1C/EHMT2 and KMT1B/SUV39H2. Interestingly KHMT1D/EHMT1 was repressed in the cancer cell lines suggesting specificities of action (Figure 4A). ZBTB33/KAISO was unaltered in the cell line panel.
Fig. 4.
Histone methyltransferases expression in prostate cell lines. (A) Differences in expression levels of the indicated histone methyltrasnferase, compared with 18S expression, measured by TaqMan Q-RT–PCR in the indicated prostate cancer cells compared with RWPE-1 cells. Each data point represents the mean of three separate experiments amplified in triplicate wells ± standard error mean (*P < 0.05, **P < 0.01). (B) PC-3 cells were pretreated with chaetocin (200nM) or ethanol control for 24h then treated with either 1α,25(OH)2D3 or ethanol control for a further 1h & 4h and mRNA was extracted, and accumulation of CDKN1A (encodes p21(waf1/cip1)) was measured using TaqMan Q-RT–PCR. For 1α,25(OH)2D3 only treatment (no chaetocin pretreatment), expression levels were compared with ethanol control and presented as fold change. For chaetocin pretreatments cells, the fold change in CDKN1A in response to 1α,25(OH)2D3 was compared with ethanol control. Therefore, fold expression changes were calculated for 1α,25(OH)2D3 compared with ethanol control with no chaetocin pretreatment (D3) or 1α,25(OH)2D3 compared with ethanol with chaetocin pretreatment (CHAE+D3). Each data point represented the mean of biological and technical triplicates ±standard error mean.
The upregulation of five of the six tested KMTs including KHMT1C/EHMT2 and KMT1B/SUV39H2 in PC-3 and the other CaP cells may co-operate with elevation of NCOR1 to either induce and/or sustain inappropriate levels of H3K9me2. In turn, this repressive histone modification attracts the HP-1 complex and leads to elevated local DNA methylation associated with stable gene silencing and/or transrepression in response to 1α,25(OH)2D3 treatment. We examined this possibility by examining gene regulation in response to the cotreatment of 1α,25(OH)2D3 and the KMT inhibitor chaetocin, compared with inhibitor alone (42,43). Supporting a role for deregulated KMT activity to distort transcription, this cotreatment with chaetocin reduced the 1α,25(OH)2D3-transrepression of CDKN1A in PC-3 cells (Figure 4B).
Changes in DNA methylation on the CDKN1A promoter in response to 1α,25(OH)2D3
To examine how the basal and regulated patterns of CpG methylation differed between the two cell models, we used MAQMA assays. This approach allowed measurement of the DNA methylation at multiple points along the CDKN1A promoter including regions that were within 300 nucleotides of the center of the three VDR binding regions and the TSS.
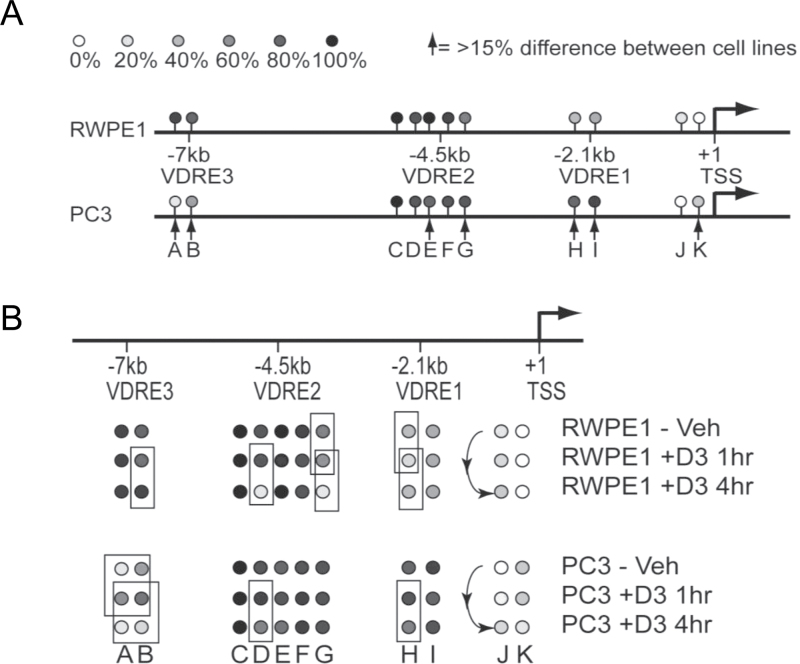
These studies revealed that the basal methylation patterns differed significantly and very specifically between the two cell models. There is a large CpG island in the area surrounding the TSS and there were 19 CpGs that were informative with the MAQMA. Most showed very low levels of methylation (0–10%) with no difference between the cell lines. However, at CpG position K that is in close proximity to the TSS (−104 relative to TSS), shown in Figure 5A, we observed only 2% methylation in RWPE-1 but 28% in the non-responsive PC-3 cells. The three VDREs are outside the context of a CpG island but do contain individual CpG positions. At VDRE2 in both cell lines, the basal level of methylation was rather high with position E showing more methylation in RWPE-1 (99%) compared with PC-3 (77%), whereas this was reversed at position G (66% in RWPE-1 and 86% in PC-3). VDREs 1 and 3 have only two CpG positions each, but they show strong differences at specific methylation. Methylation at VDRE1 was found to be higher in the non-responsive PC-3 cells (positions H and I; ~35% in RWPE-1 and ~85% in PC-3) but the opposite was true for VDRE3 (positions A and B; ~90% in RWPE-1 and ~25% in PC-3).
Fig. 5.
Altered DNA methylation patterns on the CDKN1A regulatory regions. RWPE-1 and PC-3 cells were treated with 1α,25(OH)2D3 (100nM) or ethanol control for indicated time points and DNA and RNA were extracted. DNA methylation was measured by quantitative bisulfite sequencing using MAQMA and the percentage of methylation at each CpG position is indicated by the gray scale shown at the top of the figure. (A) Comparison of basal methylation levels between RWPE-1 and PC-3 cells. Arrows indicate positions where there is a difference in percentage of methylation greater than 15%. (B) 1α,25(OH)2D3 induced changes in DNA methylation for both cell lines. Boxes indicate comparisons where the 1α,25(OH)2D3 treatment led to a change in percentage of methylation greater than 15%.
In RWPE-1, following 1α,25(OH)2D3 treatment, there were regions near the TSS and the VDREs where DNA methylation levels changed. In particular, VDRE2 showed marked and progressive loss of methylation 1 and/or 4h after treatment in positions D and G, going from 85% to 14% at positions D after 4h and from 66% to 11% at position G. In contrast, in the non-responsive PC-3 cells, there was little to no methylation change in VDRE2. Position D went from 89% to 63%, which is a much smaller reduction than seen in RWPE-1, whereas there was no change at position G (Figure 5B). VDRE1 showed a small reduction of methylation in RWPE-1 cells after 1h of treatment at position H that reverted back by 4h (34%, to 17%, to 34%). Whereas in PC-3 cells, at position H the higher basal level of methylation remained with only a slight reduction at 4h of treatment from 80% to 65%. Regardless, the relatively high level of methylation in PC-3 at VDRE1 remained in comparison with RWPE-1. Very little change in methylation patterns were observed at VDRE3 in RWPE-1 cells, but in the relatively undermethylated PC-3 cells, we observed an increase in methylation at 1h at both positions A and B, followed by a reduction of methylation at 4h. The TSS at position J also showed increased methylation driven by 1α,25(OH)2D3 treatment in PC-3 cells.
These findings were consistent with the changes in the binding of NCOR1 following 1α,25(OH)2D3 treatment in PC-3 versus RWPE-1. The basal levels of DNA methylation may represent the probability of NCOR1 association. Thus, at position K at the TSS, there is elevated basal CpG methylation in PC-3 cells and was accompanied by the ligand-induced enrichment of NCOR1 at the TSS. Furthermore in PC-3 cells, ligand-induced enrichment of NCOR1 at VDRE2 at 0.5h and 2h was accompanied with sustained DNA methylation at positions D and G. Also in PC-3 cells, immediate ligand-induced enrichment of NCOR1 at VDRE3 was accompanied by increased DNA methylation at regions A and B at 1h. Finally, NCOR1 ligand-induced enrichment was apparent in PC-3 cells at multiple time points at the TSS and was accompanied by increased DNA methylation at position J.
Discussion
The current study was undertaken to investigate epigenetic mechanisms that distort transcriptional responses in cancer using the VDR as a model transcription factor. The VDR governs and influences antimitotic and prodifferentiation transcriptional programs, and these actions are distorted in prostate cancer cells (44). Therefore, dissecting the 1α,25(OH)2D3-recalcitrant phenotype is also of potential clinical significance. To address this aim, we considered two components of epigenetic regulation. First, we examined whether the corepressor protein NCOR1 was differentially recruited to target genes that are known to regulate these antimitotic transcriptional programs, in particular we focused on CDKN1A (encodes p21(waf1/cip1)). Second, we investigated to what extent the altered regulation of genes was also reflected by differential basal and regulated patterns of DNA methylation.
As a starting point to these questions, the current study undertook a comprehensive time-resolved approach to reveal differential mRNA regulation of a panel of VDR target genes in three different prostate cell models. These studies revealed that 1α,25(OH)2D3-regulated expression was attenuated and repressed in models with reduced and recalcitrant responses to the antimitotic actions of 1α,25(OH)2D3. Compared with non-malignant RWPE-1 cells, in most cases, the magnitude of 1α,25(OH)2D3-stimulated gene regulation in the isogenic transformed RWPE-2 cells was reduced. These patterns in mRNA expression in RWPE-1 cells were comparable with those reported previously that associated with cyclical patterns of protein expression (2). In PC-3 cells, the cyclical mRNA transactivation was abolished and in many cases replaced by transrepression.
Gene targets were also regulated selectively through the cell cycle in RWPE-1 cells, with G1 being the most transcriptionally permissive phase, and this was also suppressed in PC-3 cells. Previously, we established in RWPE-1 that epigenetic mechanisms significantly favored VDR regulation in G1 (2), an event that has also been demonstrated for AR signaling (18). We have also established altered expression of corepressors through the cell cycle between RWPE-1 and PC-3 cells (17). Thus, the regulation of genes observed in bulk culture may well represent a subset of cells, in G1, that are maximally responsive. The suppressed gene regulation in 1α,25(OH)2D3-recalcitrant cells may reflect the actions of different corepressor components within separate phases of the cell cycle. Similarly, the small magnitudes of regulation (and dysregulation) of mRNA are probably to translate to a greater collective impact on cell-cycle regulatory networks and therefore phenotypes.
Building on these studies, we examined the binding of NCOR1 following VDR activation and revealed that 1α,25(OH)2D3 induced greater NCOR1 association on the CDKN1A promoter in PC-3 cells, compared with RWPE-1 cells. Thus, NCOR1 was sustained and enriched at all three VDR binding sites to different extents and at different time points. Probably, reflecting looping events, the TSS showed sustained NCOR1 enrichment throughout the time course (45).
We reasoned that the consequences of this enhanced and sustained recruitment would be a critical loss of H3K9ac at, and around, the VDRE binding regions and may in turn allow KMT enzymes to modify this lysine and sustain H3K9me2 levels. Supportively, in other non-cancer systems, increased targeting of KMT1A/SUV39H1 to the CDKN1A promoter sustained H3K9me2 (43,46). We therefore undertook a survey of multiple KMTs in a broad panel of prostate cell lines and revealed that five out of six KMTs were commonly overexpressed, including KHMT1C/EHMT2 and KMT1B/SUV39H2. Cotreatment with the KMT inhibitor, chaetocin, reversed the gene transrepression in PC-3 cells supporting a role for these enzymes to alter the patterns of CDKN1A regulation.
Given that H3K9me2 levels can attract the machinery that drives DNA CpG methylation, we examined the basal and regulated DNA methylation patterns on the CDKN1A promoter in RWPE-1 and PC-3 cells. Significantly, both the basal and 1α,25(OH)2D3-regulated CpG methylation differed in a position-specific manner. Basal differences were evident, but more surprisingly 1α,25(OH)2D3 treatment resulted in clear changes in the site-specific methylation in both models. For example, VDRE2 and the TSS contained CpG regions that were either de-methylated or unchanged in RWPE-1 cells but in PC-3 cells they either remained highly methylated or displayed increased methylation. Again, these findings also reflected our earlier work in RWPE-1 cells that identified VDRE2 as a critical to the activation of CDKN1A (2). In parallel, Carlberg et al. revealed that VDRE2 was a key responsive element involved in chromatin looping and gene activation (45). Taken together, these findings supported the concept that inappropriate NCOR1 recruitment coupled with elevated levels of key KMTs such as KMT1B/SUV39H2 can sustain H3K9me2 levels that in turn attract the DNA methylation machinery, for example through HP-1, and sustain transcriptional silencing by inducing DNA methylation (6,13)(reviewed in (9)). A parallel inference may also be that this mechanism could contribute to the transrepression by the VDR of targets such as c-MYC (47) and therefore it is tempting to speculate that gene regulation behavior reflects how sustained the interactions are with corepressors such as NCOR1 and NCOR2/SMRT.
It is now over 30 years since the initial reports demonstrated the anticancer actions of 1α,25(OH)2D3 (48–50). Following these studies, antiproliferative effects were demonstrated in a wide variety of cancer cell lines, including those from prostate (51–54), as well as xenograft and transgenic CaP models (55,56). As the anticancer effects of the ligand emerged, large-scale epidemiological studies found inverse associations between circulating 25OHD3 and cancer risk and advanced disease (57–65). However, although in vitro, in vivo and epidemiological data support links between replete VDR signaling, growth restraint and broad anticancer activities, clinical exploitation of this receptor has been limited. A significant impediment to translation remains the inability to predict accurately which patients will respond to either chemoprevention or chemotherapy strategies centered on vitamin D compounds. The mechanisms that drive this resistant phenotype are often illusive and probably involve multiple aspects of disruption. Key mechanisms include gene amplification of the 1α,25(OH)2D3 metabolizing enzyme CYP24A1 (66) and repression of the VDR by more general repressors such as SNAIL (67). The process of inappropriate corepressor recruitment leading to stable gene silencing also contributes to this phenotype and in particular may shed light on why the VDR and other nuclear receptors are often expressed in non-malignant and retained in malignant prostate epithelial cells (17).
The differential recruitment of corepressors also addresses another ambiguity in their cancer biology. Increased NCOR1 and NCOR2/SMRT expression occurs in breast and bladder cancer associated with suppressed responsiveness of nuclear receptors that exert mitotic restraint, such as VDR and PPARα/γ (17,19–25). In contrast, other studies have shown that downregulated NCOR1 and NCOR2/SMRT enhanced AR transcriptional programs in CaP (27,28). Thus, in CaP, there appears to be conflicting pressures on the expression of corepressor expression. Instead, we propose that gene-specific recruitment may be at least as significant as changes in expression.
Nuclear receptors display a range of distributions between the cytoplasm and the nucleus. In the absence of ligand, steroidal receptors such as the AR are cytoplasmic, whereas others including the VDR are resident more predominantly in the nucleus. The conflicting pressures on corepressor function in prostate cancer maybe resolved by considering their location within the cell, and on chromatin. By being inappropriately retained on nuclear receptors resident in the nucleus, they may distort a transient epigenetic process, the control of H3K9 methylation status, to favor a more stable epigenetic event, namely DNA methylation. Thus, the increased recruitment of corepressor association may convert a transient epigenetic silencing process, which is part of the normal nuclear receptor-transcriptional cycle, into a stable and heritable epigenetic event. The consequences of this could therefore be the targeted methylation of genes where NCOR1 and other corepressors are recruited by the VDR, or other nuclear resident transcription factors.
Therefore, these receptors may provide a route for the silencing of critical transcriptional programs by selective NCOR1 recruitment and thereby allow CaP cells to escape mitotic restraint. Given the role for corepressors to sequestrate and direct histone deacetylases and methyltransferases, these findings have important implications for the effective targeting of epigenetic therapies during CaP progression with current and next-generation epigenetic drugs.
Funding
NucSys, a European Community FP6- Marie Curie Research Training Network, the Biotechnology and Biological Sciences Research Council; National Institute of Health (R01 CA095367-06 and 2R01-CA-095045-06) to M.J.C.; NCI Cancer Center Support Grant to the Roswell Park Cancer Institute (CA016056) to M.J.C.; Cancer Research UK (C1015/A9077) to B.M.T.
Acknowledgement
1α,25(OH)2D3 was a gift from Dr. Milan Uskokovic (BioXell S.p.A., Italy).
Conflict of interest: The authors declare no conflict of interest.
Glossary
Abbreviations:
- AR
androgen receptor
- MAQMA
MassArray Quantitative Methylation Analysis
- mRNA
messenger RNA
- Q-RT–PCR
quantitative reverse transcription–PCR
- TSS
transcription start site
- VDR
vitamin D receptor.
References
- 1. Saramäki A., et al. (2006). Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res., 34, 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thorne J.L., et al. (2011). Epigenetic control of a VDR-governed feed-forward loop that regulates p21(waf1/cip1) expression and function in non-malignant prostate cells. Nucleic Acids Res., 39, 2045–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Métivier R., et al. (2003). Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell, 115, 751–763 [DOI] [PubMed] [Google Scholar]
- 4. Saramäki A., et al. (2009). Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1alpha,25-dihydroxyvitamin D3. J. Biol. Chem., 284, 8073–8082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kang Z., et al. (2004). Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol. Endocrinol., 18, 2633–2648 [DOI] [PubMed] [Google Scholar]
- 6. Mohn F., et al. (2009). Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet., 25, 129–136 [DOI] [PubMed] [Google Scholar]
- 7. Fujita N., et al. (2003). Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J. Biol. Chem., 278, 24132–24138 [DOI] [PubMed] [Google Scholar]
- 8. Estève P.O., et al. (2006). Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev., 20, 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng X., et al. (2010). Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry, 49, 2999–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Le May N., et al. (2010). NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol. Cell, 38, 54–66 [DOI] [PubMed] [Google Scholar]
- 11. Métivier R., et al. (2008). Cyclical DNA methylation of a transcriptionally active promoter. Nature, 452, 45–50 [DOI] [PubMed] [Google Scholar]
- 12. Kangaspeska S., et al. (2008). Transient cyclical methylation of promoter DNA. Nature, 452, 112–115 [DOI] [PubMed] [Google Scholar]
- 13. Yoon H.G., et al. (2003). N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell, 12, 723–734 [DOI] [PubMed] [Google Scholar]
- 14. Battaglia S., et al. (2010). Transcription factor co-repressors in cancer biology: roles and targeting. Int. J. Cancer, 126, 2511–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J.D., et al. (1996). SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc. Natl. Acad. Sci. U.S.A., 93, 7567–7571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodson M.L., et al. (2005). Alternative mRNA splicing of SMRT creates functional diversity by generating corepressor isoforms with different affinities for different nuclear receptors. J. Biol. Chem., 280, 7493–7503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Battaglia S., et al. (2010). Elevated NCOR1 disrupts PPARalpha/gamma signaling in prostate cancer and forms a targetable epigenetic lesion. Carcinogenesis, 31, 1650–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Altintas D.M., et al. (2011). Cell cycle regulated expression of NCoR might control cyclic expression of androgen responsive genes in an immortalized prostate cell line. Mol. Cell. Endocrinol., 332, 149–162 [DOI] [PubMed] [Google Scholar]
- 19. Girault I., et al. (2003). Expression analysis of estrogen receptor alpha coregulators in breast carcinoma: evidence that NCOR1 expression is predictive of the response to tamoxifen. Clin. Cancer Res., 9, 1259–1266 [PubMed] [Google Scholar]
- 20. Khanim F.L., et al. (2004). Altered SMRT levels disrupt vitamin D3 receptor signalling in prostate cancer cells. Oncogene, 23, 6712–6725 [DOI] [PubMed] [Google Scholar]
- 21. Banwell C.M., et al. (2006). Altered nuclear receptor corepressor expression attenuates vitamin D receptor signaling in breast cancer cells. Clin. Cancer Res., 12(7 Pt 1)2004–2013 [DOI] [PubMed] [Google Scholar]
- 22. Abedin S.A., et al. (2009). Elevated NCOR1 disrupts a network of dietary-sensing nuclear receptors in bladder cancer cells. Carcinogenesis, 30, 449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim J.Y., et al. (2009). Involvement of SMRT corepressor in transcriptional repression by the vitamin D receptor. Mol. Endocrinol., 23, 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Z., et al. (2006). NCOR1 mRNA is an independent prognostic factor for breast cancer. Cancer Lett., 237, 123–129 [DOI] [PubMed] [Google Scholar]
- 25. Chang T.H., et al. (2002). Enhanced growth inhibition by combination differentiation therapy with ligands of peroxisome proliferator-activated receptor-gamma and inhibitors of histone deacetylase in adenocarcinoma of the lung. Clin. Cancer Res., 8, 1206–1212 [PubMed] [Google Scholar]
- 26. Stephens P.J., et al. (2012). The landscape of cancer genes and mutational processes in breast cancer. Nature, 486, 400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liao G., et al. (2003). Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J. Biol. Chem., 278, 5052–5061 [DOI] [PubMed] [Google Scholar]
- 28. Hodgson M.C., et al. (2005). The androgen receptor recruits nuclear receptor CoRepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J. Biol. Chem., 280, 6511–6519 [DOI] [PubMed] [Google Scholar]
- 29. Webber M.M., et al. (1997). Acinar differentiation by non-malignant immortalized human prostatic epithelial cells and its loss by malignant cells. Carcinogenesis, 18, 1225–1231 [DOI] [PubMed] [Google Scholar]
- 30. Kaighn M.E., et al. (1979). Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest. Urol., 17, 16–23 [PubMed] [Google Scholar]
- 31. Campbell M.J., et al. (1997). Inhibition of proliferation of prostate cancer cells by a 19-nor-hexafluoride vitamin D3 analogue involves the induction of p21waf1, p27kip1 and E-cadherin. J. Mol. Endocrinol., 19, 15–27 [DOI] [PubMed] [Google Scholar]
- 32. Campbell M.J., et al. (2000). The anti-proliferative effects of 1alpha,25(OH)2D3 on breast and prostate cancer cells are associated with induction of BRCA1 gene expression. Oncogene, 19, 5091–5097 [DOI] [PubMed] [Google Scholar]
- 33. Matilainen M., et al. (2005). Regulation of multiple insulin-like growth factor binding protein genes by 1alpha,25-dihydroxyvitamin D3. Nucleic Acids Res., 33, 5521–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Towsend K., et al. (2006). Identification of VDR-responsive gene signatures in breast cancer cells. Oncology, 71, 111–123 [DOI] [PubMed] [Google Scholar]
- 35. Chêne G., et al. (2007). n-3 and n-6 polyunsaturated fatty acids induce the expression of COX-2 via PPARgamma activation in human keratinocyte HaCaT cells. Biochim. Biophys. Acta, 1771, 576–589 [DOI] [PubMed] [Google Scholar]
- 36. Seuter S., et al. (2007). Functional characterization of vitamin D responding regions in the human 5-Lipoxygenase gene. Biochim. Biophys. Acta, 1771, 864–872 [DOI] [PubMed] [Google Scholar]
- 37. Qiu X., et al. (2006). Assessing stability of gene selection in microarray data analysis. BMC Bioinformatics, 7, 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Camoriano M., et al. (2008). Phenotype-specific CpG island methylation events in a murine model of prostate cancer. Cancer Res., 68, 4173–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coolen M.W., et al. (2007). Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res., 35, e119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ehrich M., et al. (2005). Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A., 102, 15785–15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zella L.A., et al. (2007). Enhancers located in the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. J. Steroid Biochem. Mol. Biol., 103, 435–439 [DOI] [PubMed] [Google Scholar]
- 42. Lakshmikuttyamma A., et al. (2010). Reexpression of epigenetically silenced AML tumor suppressor genes by SUV39H1 inhibition. Oncogene, 29, 576–588 [DOI] [PubMed] [Google Scholar]
- 43. Cherrier T., et al. (2009). p21(WAF1) gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene, 28, 3380–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thorne J., et al. (2008). The vitamin D receptor in cancer. Proc. Nutr. Soc., 67, 115–127 [DOI] [PubMed] [Google Scholar]
- 45. Saramaki A., et al. (2009). Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1alpha,25-dihydroxyvitamin D3. J. Biol. Chem., 284, 8073–8082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pollard P.J., et al. (2008). Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem. J., 416, 387–394 [DOI] [PubMed] [Google Scholar]
- 47. Toropainen S., et al. (2010). The down-regulation of the human MYC gene by the nuclear hormone 1alpha,25-dihydroxyvitamin D3 is associated with cycling of corepressors and histone deacetylases. J. Mol. Biol., 400, 284–294 [DOI] [PubMed] [Google Scholar]
- 48. Colston K., et al. (1981). 1,25-dihydroxyvitamin D3 and malignant melanoma: the presence of receptors and inhibition of cell growth in culture. Endocrinology, 108, 1083–1086 [DOI] [PubMed] [Google Scholar]
- 49. Miyaura C., et al. (1981). 1 alpha,25-Dihydroxyvitamin D3 induces differentiation of human myeloid leukemia cells. Biochem. Biophys. Res. Commun., 102, 937–943 [DOI] [PubMed] [Google Scholar]
- 50. Abe E., et al. (1981). Differentiation of mouse myeloid leukemia cells induced by 1 alpha,25-dihydroxyvitamin D3. Proc. Natl. Acad. Sci. U.S.A., 78, 4990–4994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Campbell M.J., et al. (1997). Inhibition of proliferation of prostate cancer cells by a 19-nor-hexafluoride vitamin D3 analogue involves the induction of p21waf1, p27kip1 and E-cadherin. J. Mol. Endocrinol., 19, 15–27 [DOI] [PubMed] [Google Scholar]
- 52. Elstner E., et al. (1999). Novel 20-epi-vitamin D3 analog combined with 9-cis-retinoic acid markedly inhibits colony growth of prostate cancer cells. Prostate, 40, 141–149 [DOI] [PubMed] [Google Scholar]
- 53. Peehl D.M., et al. (1994). Antiproliferative effects of 1,25-dihydroxyvitamin D3 on primary cultures of human prostatic cells. Cancer Res., 54, 805–810 [PubMed] [Google Scholar]
- 54. Colston K., et al. (1982). 1,25-dihydroxyvitamin D3 receptors in human epithelial cancer cell lines. Cancer Res., 42, 856–859 [PubMed] [Google Scholar]
- 55. Blutt S.E., et al. (2000). A calcitriol analogue, EB1089, inhibits the growth of LNCaP tumors in nude mice. Cancer Res., 60, 779–782 [PubMed] [Google Scholar]
- 56. Banach-Petrosky W., et al. (2006). Vitamin D inhibits the formation of prostatic intraepithelial neoplasia in Nkx3.1;Pten mutant mice. Clin. Cancer Res., 12, 5895–5901 [DOI] [PubMed] [Google Scholar]
- 57. Garland C.F., et al. (1980). Do sunlight and vitamin D reduce the likelihood of colon cancer? Int. J. Epidemiol., 9, 227–231 [DOI] [PubMed] [Google Scholar]
- 58. Garland F.C., et al. (1990). Geographic variation in breast cancer mortality in the United States: a hypothesis involving exposure to solar radiation. Prev. Med., 19, 614–622 [DOI] [PubMed] [Google Scholar]
- 59. Luscombe C.J., et al. (2001). Prostate cancer risk: associations with ultraviolet radiation, tyrosinase and melanocortin-1 receptor genotypes. Br. J. Cancer, 85, 1504–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Giovannucci E. (2005). The epidemiology of vitamin D and cancer incidence and mortality: a review (United States). Cancer Causes Control, 16, 83–95 [DOI] [PubMed] [Google Scholar]
- 61. Schwartz G.G., et al. (1990). Is vitamin D deficiency a risk factor for prostate cancer? (Hypothesis). Anticancer Res., 10(5A)1307–1311 [PubMed] [Google Scholar]
- 62. John E.M., et al. (2005). Sun exposure, vitamin D receptor gene polymorphisms, and risk of advanced prostate cancer. Cancer Res., 65, 5470–5479 [DOI] [PubMed] [Google Scholar]
- 63. Chen T.C., et al. (2003). Prostatic 25-hydroxyvitamin D-1alpha-hydroxylase and its implication in prostate cancer. J. Cell. Biochem., 88, 315–322 [DOI] [PubMed] [Google Scholar]
- 64. Hsu J.Y., et al. (2001). Reduced 1alpha-hydroxylase activity in human prostate cancer cells correlates with decreased susceptibility to 25-hydroxyvitamin D3-induced growth inhibition. Cancer Res., 61, 2852–2856 [PubMed] [Google Scholar]
- 65. Ahonen M.H., et al. (2000). Prostate cancer risk and prediagnostic serum 25-hydroxyvitamin D levels (Finland). Cancer Causes Control, 11, 847–852 [DOI] [PubMed] [Google Scholar]
- 66. Albertson D.G., et al. (2000). Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat. Genet., 25, 144–146 [DOI] [PubMed] [Google Scholar]
- 67. Pálmer H.G., et al. (2004). The transcription factor SNAIL represses vitamin D receptor expression and responsiveness in human colon cancer. Nat. Med., 10, 917–919 [DOI] [PubMed] [Google Scholar]