Abstract
The chemopreventive agent curcumin has anti-proliferative effects in many tumour types, but characterization of cell cycle arrest, particularly with physiologically relevant concentrations, is still incomplete. Following oral ingestion, the highest concentrations of curcumin are achievable in the gut. Although it has been established that curcumin induces arrest at the G2/M stage of the cell cycle in colorectal cancer lines, it is not clear whether arrest occurs at the G2/M transition or in mitosis. To elucidate the precise stage of arrest, we performed a direct comparison of the levels of curcumin-induced G2/M boundary and mitotic arrest in eight colorectal cancer lines (Caco-2, DLD-1, HCA-7, HCT116p53+/+, HCT116p53–/–, HCT116p21–/–, HT-29 and SW480). Flow cytometry confirmed that these lines underwent G2/M arrest following treatment for 12h with clinically relevant concentrations of curcumin (5–10 μM). In all eight lines, the majority of this arrest occurred at the G2/M transition, with a proportion of cells arresting in mitosis. Examination of the mitotic index using fluorescence microscopy showed that the HCT116 and Caco-2 lines exhibited the highest levels of curcumin-induced mitotic arrest. Image analysis revealed impaired mitotic progression in all lines, exemplified by mitotic spindle abnormalities and defects in chromosomal congression. Pre-treatment with inhibitors of the DNA damage signalling pathway abrogated curcumin-induced mitotic arrest, but had little effect at the G2/M boundary. Moreover, pH2A.X staining seen in mitotic, but not interphase, cells suggests that this aberrant mitosis results in DNA damage.
Introduction
Many cancers have a long interval between initiation and development of metastatic disease leading to mortality. This presents a significant window of opportunity for preventing or slowing tumour growth. Many prevention strategies are under investigation, including the usefulness of dietary agents. Among these, curcumin has been found to be biologically active against a variety of tumour types, both in vitro and in vivo. However, a more detailed understanding of its mechanism of action at various stages of the carcinogenic process is required to fully exploit its potential. Curcumin treatment in preclinical models of colorectal cancer (CRC) generally induces G2/M cell cycle arrest as assessed by flow cytometry (1–4). However, this method does not differentiate between cells arresting at the G2/M transition, and those blocked within M phase.
Aberrant mitosis following curcumin treatment was first described by Holy, who discovered an accumulation of cells in M phase and an abnormal distribution of the microtubule bundling protein NuMA (5). Curcumin was later shown to act as an anti-mitotic agent via inhibition of microtubule assembly (6). Incorporation into microtubule polymers occurred in a concentration-dependent fashion, staining tubulin polymers yellow. Binding resulted in conformational changes in tubulin and a reduced GTPase activity of microtubules. This can lead to altered microtubule assembly dynamics, highlighted by the mislocalization of the mitotic kinesin Eg5 in breast cancer cells (7).
Two common mitotic abnormalities noted in curcumin-treated cells are monopolar spindles and abnormal chromatin structures (5,7–9). Further analysis of the kinases involved in the regulation of mitosis has revealed reduced levels of Aurora A (10). Impairment of the chromosomal passenger complex via down-regulation of survivin may contribute to the observed mislocalization of Aurora B (9–11). Increased levels of Mad2 and BubR1 at the kinetochores indicate the involvement of the spindle-assembly checkpoint in curcumin-induced mitotic arrest (7). Activation of this checkpoint during mitosis can inhibit the activity of the anaphase promoting complex/cyclosome (APC/C). For cells to progress from metaphase to anaphase, the APC/C is required to target securin for destruction, which in turn activates separase, a protease necessary for the separation of sister chromatids. Metaphase to anaphase progression in curcumin-treated cells is likely to be hampered by the direct binding of curcumin to the APC/C member CDC27, which has been shown to result in reduced APC/C activity (12).
Curcumin-mediated cell cycle arrest has been reported to involve DNA damage signalling (13–15). However, the exact nature of the trigger and the mechanisms involved are unclear. Curcumin has been associated with the production of reactive oxygen species (ROS), topoisomerase poisoning and chromosomal alterations (16–18). In a variety of curcumin-treated cell types, including HCT116 cells exposed to 50 µM, a significant increase in mitochondrial and nuclear DNA damage was measured by the comet assay (13,19–21). In K562 leukaemia cells, curcumin acted as a topoisomerase poison, but the resulting damage was prevented by the antioxidant N-acetyl cysteine (NAC) (18). However, some of the above effects were only observed using non-physiological concentrations of curcumin.
The ability of curcumin to modulate DNA damage signalling depends on cell type. In primary leukaemia and HTLV-1-infected T cells it reduced expression of cyclin D1, CDC25C, CDK1 and survivin, inducing G2/M and G1 arrest (15). However, in human bladder cancer cells, there was no effect on expression or phosphorylation status of CDC25C, CDK1 or Wee1, but p21 was up-regulated (14). A study in pancreatic cells found that activation of ATM and Chk1 resulted in inhibition of the CDC25C phosphatase activity, a decrease in expression of CDK1 and cyclin B, and an increase in phosphorylation of H2A.X (22).
The status of mismatch repair genes appears to influence the response to curcumin in HCT116 cells because in mismatch repair-proficient cells, treatment resulted in much greater formation of pH2A.X foci and Chk1-mediated arrest than in mismatch repair-deficient cells (17). The authors proposed that ROS induced double-strand breaks and showed that phosphorylation of H2A.X, ATM and Chk1 and levels of arrest were reduced when curcumin was combined with the ROS inhibitor, NAC.
In this study, we used a panel of CRC cell lines to further characterize curcumin-induced G2/M transition and mitotic arrest. Curcumin treatment generally resulted in higher levels of arrest at the G2/M transition than in mitosis. We found that curcumin disrupted early events in mitosis, such as centrosomal separation, chromosomal congression and bipolar metaphase spindle formation. Following treatment, cells rarely progressed beyond the prophase/prometaphase stages of mitosis. Pre-treatment with inhibitors, caffeine or debromohymenialdisine (DBH) reduced the mitotic index, implicating the DNA damage network in mitotic arrest. In accordance with this, image analysis revealed higher levels of pH2A.X staining in curcumin-treated mitotic cells.
Materials and methods
Cell lines and treatments
The Caco-2 line (ATCC) was cultured in modified Eagle’s medium with 20% fetal calf serum (FCS), 1% glutamax and 1% non-essential amino acid solution (NEAA) (Sigma–Aldrich). HCA-7 Colony 29 cells (HPA Culture Collections) were maintained in Dulbecco’s modified Eagle’s medium with 10% FCS and 1% glutamax. The HCT116 lines (p53+/+, p53–/– and p21–/–), a kind gift from Dr B.Vogelstein, were cultured in McCoy’s medium with 10% FCS and 1% glutamax. The HCT116p21–/– line was generated by homozygous deletion of the p21 gene using homologous recombination (23). The genotype control HCT116p53+/+ cells are the parental HCT116Wt strain used to construct the HCT116p53–/– cell line via targeted homologous recombination of the p53 gene (24). HT-29 (from ATCC) was cultured in Dulbecco’s modified Eagle’s medium and the DLD-1 and SW480 lines (ATCC) in RPMI, with 10% FCS. Curcumin (Sigma–Aldrich), nocodozole (Sigma–Aldrich) and DBH (Santa Cruz) were prepared as stocks in dimethyl sulfoxide (DMSO), whereas caffeine (Sigma–Aldrich) was prepared in H2O.
Tandem mass spectrometry measurement of DNA oxidative damage
DNA was extracted for liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis from HCT116 cells using the Wako sodium iodide extractor kit (Wako Chemicals GmbH Neuss, Germany) as instructed, but with modifications (25). Next, 100 µg DNA per sample was dried down with 5 µl of each stable internal standard, [15N5]8-oxodG and [15N5]8-oxodA (each 1 pmol/µl). DNA was then enzymically digested, centrifuged and the supernatant analysed by online column switching LC-MS/MS. MS conditions were as described (25), with the exception that a Waters Micromass Quattro Ultima tandem quadrupole mass spectrometer was used for the analysis (Micromass, Waters Ltd, Manchester, UK). Levels of 8-oxodG and 8-oxodA were determined from the ratio of peak areas of internal standards to those of the analytes.
Immunocytochemistry. Mitotic index was evaluated by staining DNA with Hoechst 33258 (Invitrogen) and identifying mitotic cells with an anti-phospho-H3Ser10 antibody (9706, mouse, Cell Signaling) and scoring under an Axioskop2 Plus fluorescence microscope using a ×40 lens. Further immunofluorescence experiments were conducted with antibodies to α-tubulin (T5168; Sigma–Aldrich), Aurora B (ab2254; Abcam), centrin (sc-27793-R; Santa Cruz), phospho-histone H3Ser10 (3377, rabbit; Cell Signaling), phospho-histone H2A.XSer139 (05-636; Merck Millipore) with secondary fluorescent antibodies (A11001, A11012; Invitrogen). Brinkley 1980 buffer was used as a pre-extraction buffer when staining cells for α-tubulin to help visualize microtubules. Chromatin was stained with 4′,6-diamidino-2-phenylindole (DAPI) dilactate (Sigma–Aldrich). Centrosomes were defined as separated if more than 2 µm apart. All images were captured from a Nikon microscope (×100 lens) with an Xcite Illumination system, using a Hamamatsu ORCA-R2 digital camera connected to a Mac running Openlab software (Improvision).
Flow cytometry
DNA content.
Cells were seeded onto 6-well plates at a density of 1×105 per well and left to adhere for ~24h prior to treatment. Samples were prepared for cell cycle analysis by staining with propidium iodide as described previously (3). Analysis of DNA content was performed using a Becton Dickinson FACScan Flow Cytometer, and data were captured using the Cell Quest program. At least 5000 cells were analysed per sample and gating on forward scatter versus side scatter was used to exclude doublets and debris. Cell cycle data analysis was performed using Modfit LT software.
Mitotic cells.
Cells were seeded at 1×106 in 9cm dishes, left to adhere for ~24h and then treated with curcumin or nocodazole for 12h. Floating cells were retained and added to the adherent cell mixture for analysis. To assess mitosis, cells were resuspended in solution of 2% formaldehyde and 1% bovine serum albumin (BSA) for 10–15min, centrifuged and resuspended in ice cold methanol for 10–15min. Cells were then centrifuged, washed with 1% BSA in phosphate-buffered saline and incubated with mouse anti-phospho-H3Ser10 antibodies (9706; Cell Signaling) or mouse IgG1 isotype control (Sigma–Aldrich) diluted in 1% BSA for 1h at room temperature. After washing with 1% BSA, they were incubated for a further 1h with anti-mouse Alexa Fluor680-conjugated antibodies (A10038; Invitrogen), resuspended in 1ml DAPI solution (0.1% Triton-X 100 and 250 μg/ml DAPI dilactate in phosphate-buffered saline), syringed to remove clumps and analysed. At least 10 000 counts per sample were analysed. Evaluation of the mitotic index was done on a Becton Dickinson FACS Aria II, using the FACS Diva 6 and WinMDI programmes. The near-UV 355nm laser with a 450/50 filter was used to detect DAPI and the 640nm laser with a 670/14 filter to detect Alexa Fluor680.
Western blotting.
Following electrophoresis of cell lysates, proteins were transferred to nitrocellulose membranes and incubated with primary antibodies to α-tubulin (Sc-8035; Santa Cruz), actin (Sc-1616-R; Santa Cruz), CDC25C (Sc-13138; Santa Cruz) or phospho-Chk1Ser345 (2341; Cell Signaling), followed by secondary anti-mouse (926–32210; LI-COR) or anti-rabbit antibodies (926–32210; LI-COR). The Odyssey imaging system was used to analyse protein expression.
Results
Sensitivity of CRC cells to curcumin-induced arrest
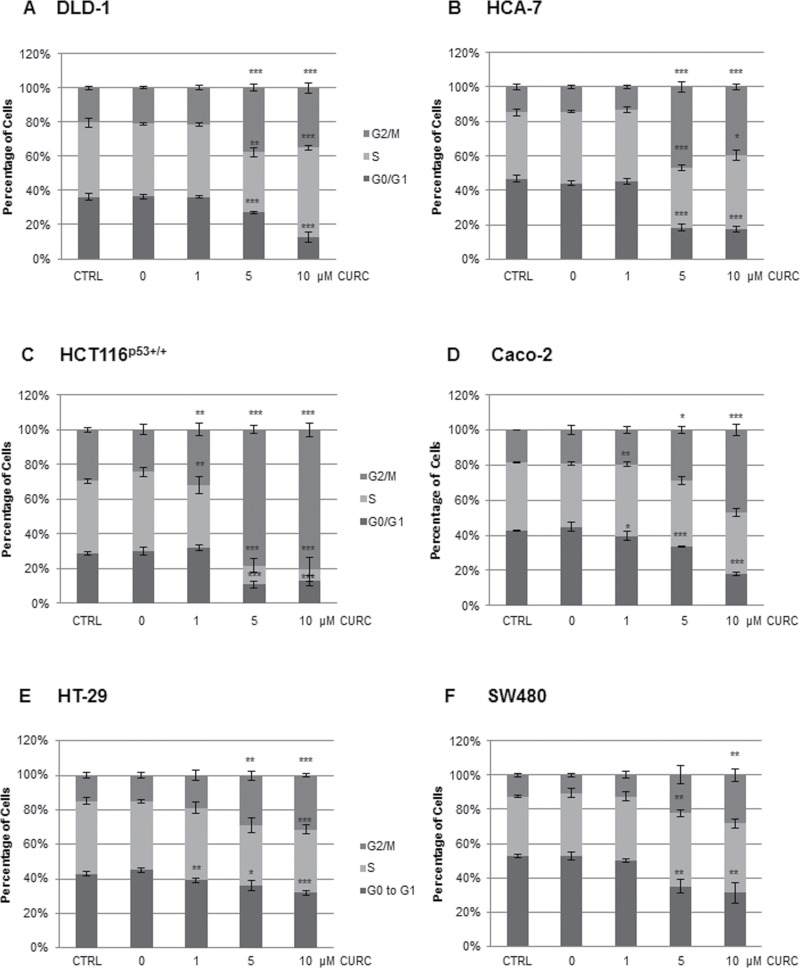
In agreement with our own and other published data, flow cytometry analysis of DNA content showed that curcumin induced G2/M arrest in MIN DLD-1, HCA-7, HCT116p53+/+, HCT116p21–/–, and HCT116p53–/– and CIN Caco-2, SW480 and HT-29 CRC cells (1–4). Overall, levels of G2/M arrest were highest in the HCT116 lines (Figure 1, Supplementary Figures 1 and 2, available at Carcinogenesis Online). In four of the lines (DLD-1, HCA-7, HCT116p53–/– and HCT116p21–/–), treatment with 5 µM resulted in the greatest G2/M arrest with 10 µM causing an increase in S phase arrest. It is well established in the literature that the response to curcumin can be dose dependent.
Fig. 1.
Curcumin induces G2/M arrest in CRC cell lines. Levels of G2/M arrest were assessed by flow cytometry following curcumin treatment for 12h. At least 5000 cells were analysed per datapoint. n = at least 3 ±SD. *P < 0.05, **P < 0.01, ***P < 0.001 compared with the DMSO control.
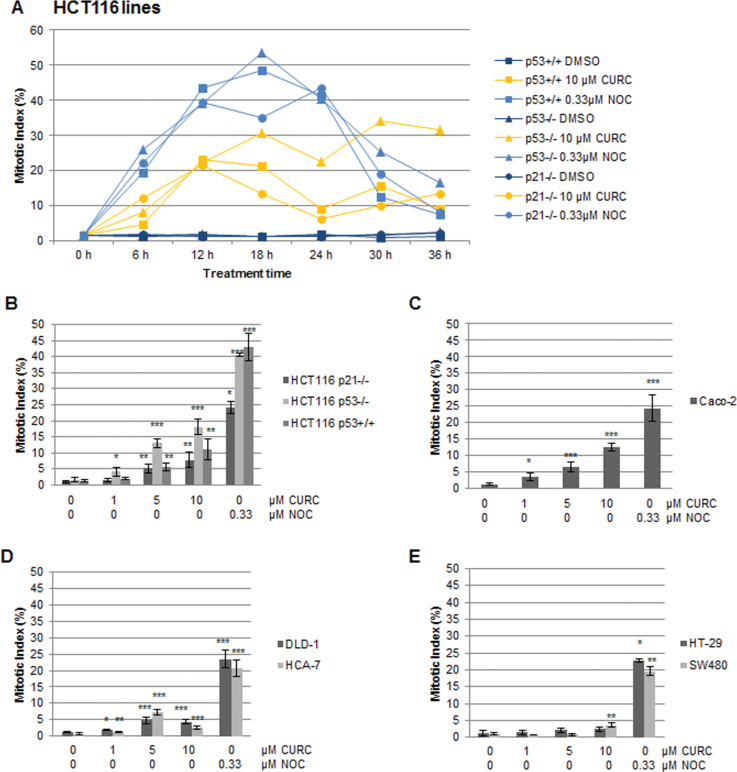
Mitotic index was analysed to differentiate between cells arresting at the G2/M boundary and in M phase. Results in the HCT116 cells suggested that mitotic counts were influenced by the status of the p53 gene, peaking at around 12h in the HCT116p53+/+ and HCT116p21–/– lines and somewhat later in the HCT116p53–/– line (Figure 2A). Detailed analysis showed a dose response and indicated that the HCT116p53–/– line was the most sensitive to curcumin-mediated mitotic arrest (Figure 2B). The Caco-2 cell line was also sensitive, with an average mitotic index of 13% following treatment with 10 µM curcumin for 12h (Figure 2C). In the DLD-1, HCA-7, HT-29 and SW480 cell lines, mitotic arrest was much less apparent or absent, in concordance with overall lower levels of G2/M arrest (Figure 2D and E). From the comparison of flow cytometry and mitotic index data in all CRC lines, it appeared that the majority of cells arrested at the G2/M boundary rather than in M phase.
Fig. 2.
Curcumin-induced mitotic arrest in CRC cell lines. Cells treated as shown with curcumin or nocodazole were stained with anti-phospho-H3 antibodies and Hoescht and assessed by fluorescence microscopy. (A) Analysis of levels of mitotic arrest in HCT116 lines over a period of 36h. For each datapoint 1000+ cells were analysed (n = 1). (B–E) Dose response analysis of mitotic arrest following treatment for 12h in (B and D) MIN lines, and (C and E) CIN lines. At least 500 cells were scored per datapoint. n = at least 3 ±SD. *P < 0.05, **P < 0.01, ***P < 0.001 compared with the DMSO control.
Curcumin disrupts early events in mitosis, arresting cells in prophase/prometaphase
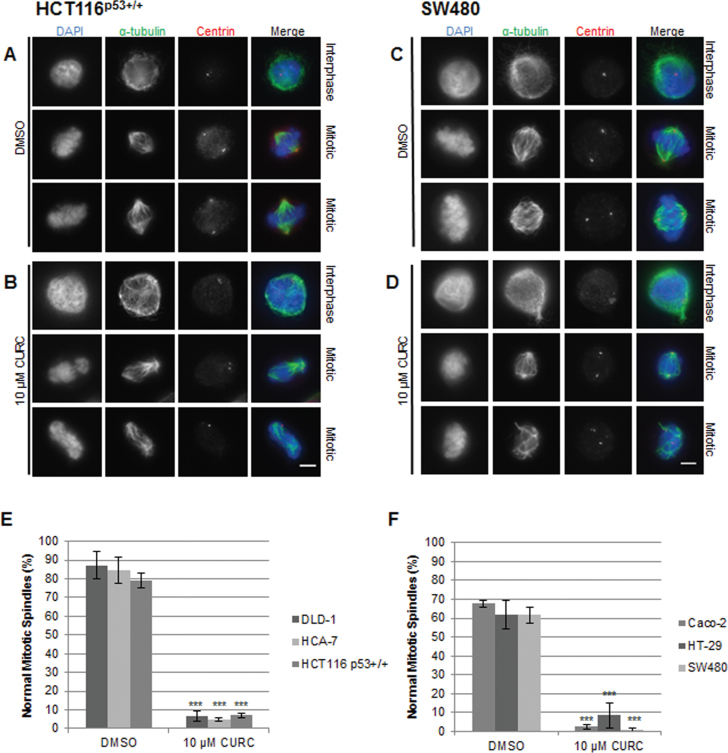
The effect of curcumin on the formation of mitotic spindles was examined in metaphase cells by immunofluorescence microscopy. In comparison with the bipolar mitotic spindle formation and alignment of chromosomes on the metaphase plate in untreated cells, abnormalities were readily apparent in treated cells (Figure 3, Supplementary Figure 3, available at Carcinogenesis Online). Where spindle pole separation had occurred, chromosomes appeared entangled and incorrectly aligned on the spindle equator and the spindles themselves were asymmetric. We noted that curcumin consistently increased the levels of spindle abnormalities in all eight lines (Figure 3).
Fig. 3.
Curcumin treatment results in mitotic abnormalities in CRC cells. Cells were treated with DMSO (A and C) or curcumin (B and D) for 12h and stained with antibodies against α-tubulin (microtubules, green) and centrin (centrosomes, red). DNA was stained with DAPI. Interphase and mitotic images are representative of three separate experiments. Scale bar, 5 µm. Quantitation of mitotic spindle abnormalities in (E) MIN lines and (F) CIN lines (treated for 12h). Normal metaphase was defined as a bipolar mitotic spindle with chromosomes clearly aligned on the metaphase plate, and at least 50 metaphase cells were counted per slide (n = 3).
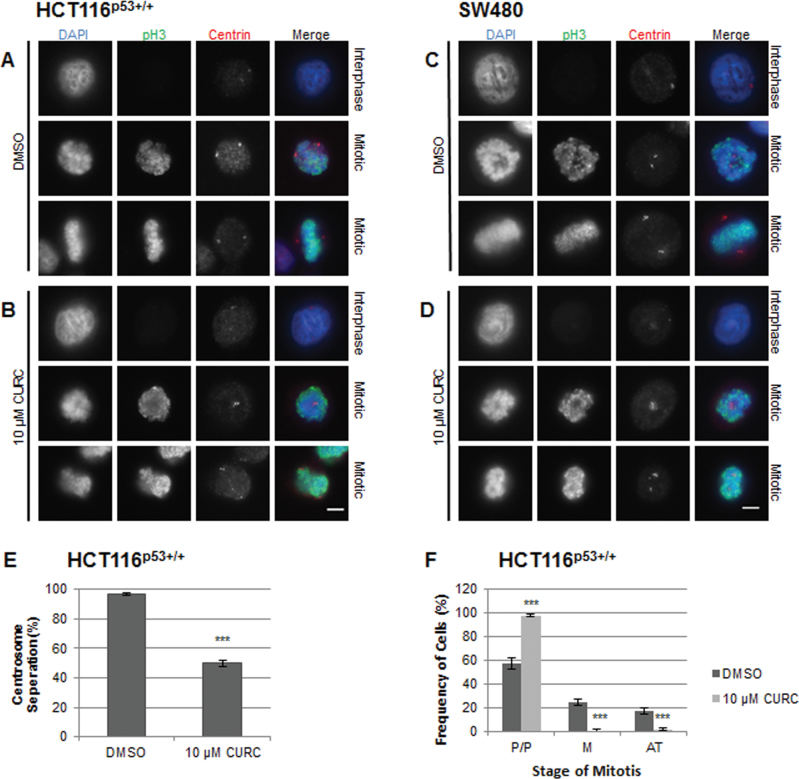
As curcumin has been shown to induce chromatin abnormalities during mitosis (5,9), we used fluorescence microscopy to investigate the effects of treatment on chromosomal alignment in mitotic cells (Figure 4, Supplementary Figure 4, available at Carcinogenesis Online). Some prophase/prometaphase cells appeared to have an indent in their chromatin, over which unseparated centrosomes could be seen. The chromosomes of curcumin-treated mitotic cells failed to congress to form a normal metaphase plate. We next conducted experiments to quantify the effect of curcumin on centrosomal separation, as monopolar spindles have been identified as a key feature of curcumin-induced mitotic arrest (5,7,8). Centrosomal separation was reduced by about 50% in HCT116p53+/+ cells (Figure 4E, Supplementary Figure 5, available at Carcinogenesis Online). Curcumin consistently arrested cells at the prophase/prometaphase stage of mitosis and reduced the number of cells in the later stages of mitosis (Figure 4F).
Fig. 4.
Curcumin impairs chromosomal congression and centrosomal separation in mitotic CRC cells. Cells were treated with DMSO (A and C) or curcumin (B and D) for 12h, stained with antibodies against phospho-histone H3 (green) and centrin (centrosomes, red) and analysed by immunofluorescence microscopy. DNA was stained with DAPI (blue). Representative interphase and mitotic cells are shown for each treatment. n = ≥3 separate experiments. Scale bar, 5 μm. (E) Centrosomal separation (>2 µm apart) in HCT116 mitotic cells. At least 50 cells were counted per slide (n = 3). Mean ±SD, ***P < 0.001 compared with DMSO control, following a two-tailed t-test assuming equal variance. (F) Stage of curcumin-induced mitotic arrest. To more clearly identify the stage of mitosis, cells were stained with antibodies against Aurora B and phosphoH3. DNA was stained with DAPI. Prophase/prometaphase cells (P/P) were defined by condensed chromosomes. Metaphase cells (M) had a bipolar mitotic spindle with chromosomes clearly aligned on the metaphase plate. Anaphase and telophase (AT) cells were counted together. In anaphase sister chromatid separation was visible, and in telophase separation has progressed with contraction of the cleavage furrow. At least 50 mitotic cells were counted per slide (n = 3).
Activation of the DNA damage signalling pathway by curcumin
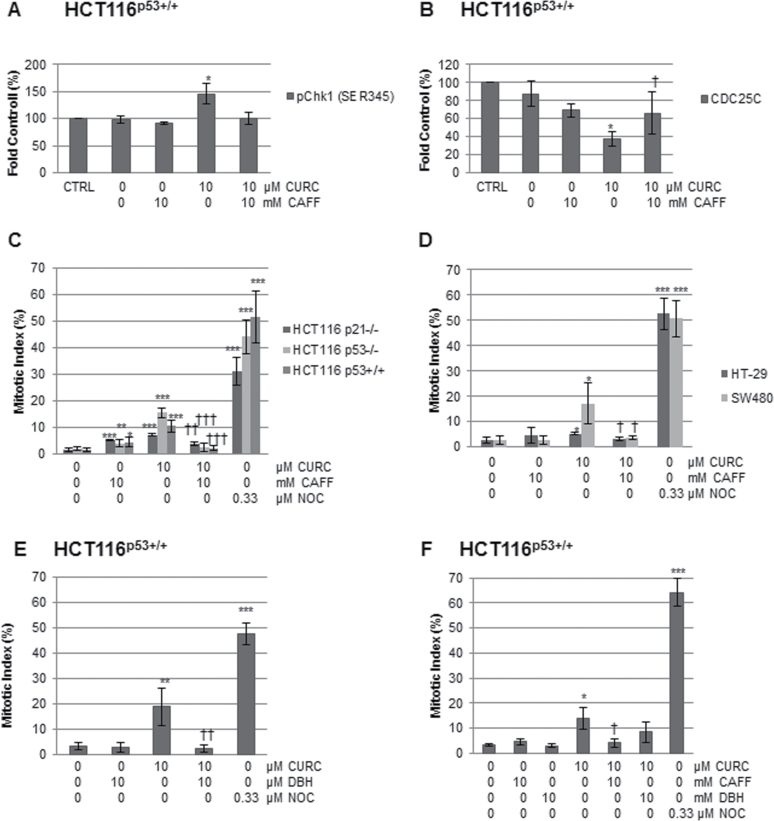
To investigate whether curcumin-mediated ROS production triggered DNA damage signalling, LC-MS/MS analysis of DNA from curcumin-treated HCT116 cells was performed. Activation of signalling appeared to be independent of ROS production, as no significant increase in the levels of 8-oxodG or 8-oxodA adducts was found (Supplementary Figure 6A, available at Carcinogenesis Online). Caffeine has been shown to inhibit the activity of the DNA damage network signalling kinases, ATM, ATR, DNA-PK and Chk1 at IC50 values of 0.2mM, 1.1mM, 10mM and 5mM, respectively (26). To investigate the involvement of the DNA damage network further, the phosphorylation status of Chk1 (Ser345) was examined by western blotting. Curcumin increased phosphorylation of Chk1 about 1.4-fold, an effect inhibited by caffeine (Figure 5A, Supplementary Figure 6B, available at Carcinogenesis Online). Chk1 and Chk2 in turn phosphorylate and inactivate CDC25C, targeting it for proteasomal degradation. Curcumin reduced total CDC25C protein levels by around 60% in the HCT116p53+/+ line. Although the average levels of CDC25C appeared to be partially restored by caffeine pre-treatment, the reversal was not statistically significant (Figure 5B, Supplementary Figure 6C, available at Carcinogenesis Online).
Fig. 5.
The effect of inhibitors on DNA damage signalling and mitotic arrest. Cell lysates, treated as indicated for 12h, were analysed by western blotting for (A) pChk1S345 (B) CDC25C and levels normalized to loading controls. *P < 0.05 and **P < 0.01 between DMSO and curcumin, following a two-tailed t-test assuming equal variance. Mean ± SD (n = 3). †P < 0.05 between cells treated with curcumin and those pre-treated with caffeine followed by curcumin. (C and D) Cells were pre-treated with caffeine for 1h then curcumin for 12h and assessed using fluorescence microscopy. At least 500 cells were scored per datapoint. n = at least 3 ±SD. *P < 0.05, **P < 0.01, ***P < 0.001 compared with the DMSO control, and †P < 0.05, ††P < 0.01, †††P < 0.001 in cells pre-treated with caffeine then curcumin compared with curcumin alone. (E) Cells were pre-treated with DBH for 1h followed by curcumin for 12h, and assessed as above. At least 100 cells were scored per datapoint. Mean (±SD) n = 3. **P < 0.01, ***P < 0.001 compared with DMSO control. ††P < 0.01 between curcumin alone and DBH plus curcumin. (F) Cells were pre-treated with DBH or caffeine for 1h, followed by curcumin for 12h and mitotic index assessed by flow cytometry using antibodies to phospho-histone H3. At least 10 000 cells were analysed per datapoint. Mean (±SD) of at least 3 independent experiments. *P < 0.05, ***P < 0.001 difference from the DMSO control. † indicates a significant difference (P < 0.05) between cells treated with curcumin alone and cells pre-treated with caffeine or DBH followed by curcumin.
DNA damage signalling and mitotic arrest
We next investigated the effects of caffeine on curcumin-induced mitotic arrest using fluorescence microscopy. In all lines, pre-treatment with caffeine abrogated curcumin-induced mitotic arrest (Figure 5C and 5D, Supplementary Figure 6D and E, available at Carcinogenesis Online). Overall, G2/M arrest, measured by FACS analysis of DNA content, was partially inhibited by caffeine (10mM) in the Caco-2 and HCT116 cells, but no effect was apparent in the DLD-1, HCA-7, HT-29 or SW480 cells (data not shown). These data suggest that DNA damage checkpoint signalling could be involved in mitotic arrest in response to curcumin, but not in G2/M boundary arrest. Lu et al. (13) also reported caffeine-insensitive curcumin-induced G2/M arrest in HCT116 cells, but did not look specifically at mitotic arrest.
The involvement of checkpoint kinases in curcumin-induced mitotic arrest
Following induction of DNA damage signalling, Chk1/2 effector kinases are activated downstream of proximal kinases, ATM and ATR. To investigate the involvement of checkpoint kinases in curcumin-induced mitotic arrest, HCT116p53+/+ cells were pre-treated with the inhibitor, DBH(27). Using immunofluorescence microscopy, we found a significant decrease in the mitotic index of curcumin-treated cells (Figure 5E). However, although flow cytometry analysis of cells stained for phospho-histone H3 (pH3) confirmed the reduction in mitotic cells following caffeine treatment, the decrease observed in response to DBH did not reach statistical significance (Figure 5F).
Higher levels of pH2A.X foci in curcumin-treated cells
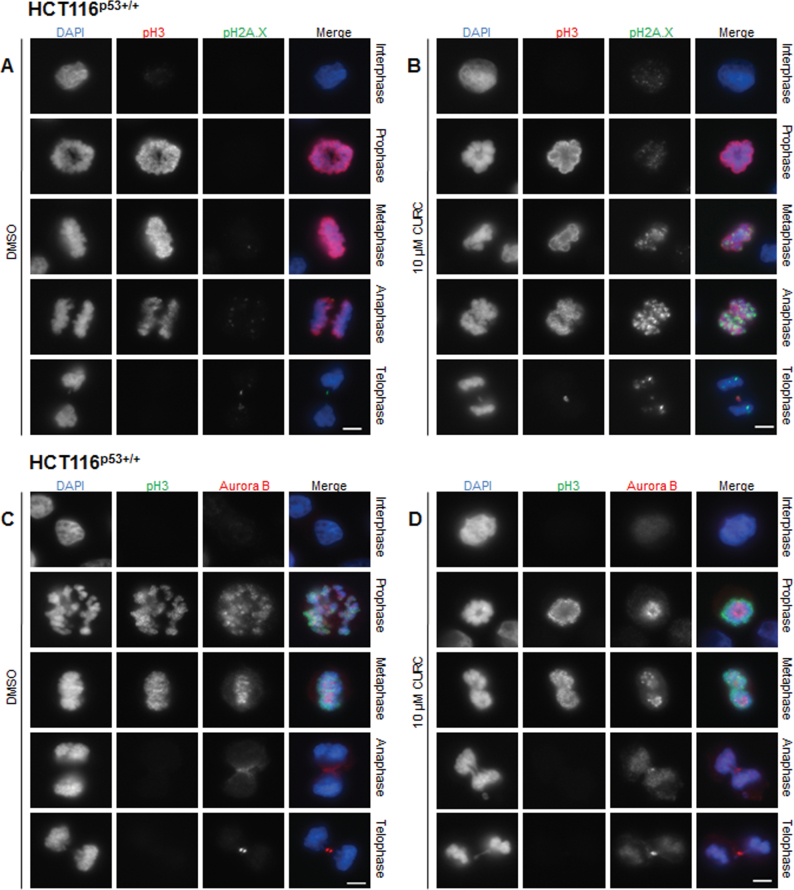
As DNA damage signalling was shown to be linked to curcumin-induced mitotic arrest, experiments to elucidate the source of damage were conducted. An increase in pH2A.X staining had been observed in HCT116 cells treated with spindle poisons such as nocodazole, paclitaxel or monastrol due to aberrant mitosis (28). As curcumin can act as a spindle poison, we used immunofluorescent microscopy to investigate whether pH2A.X foci formed in curcumin-treated mitotic cells. Our experiments revealed increased pH2A.X staining, most notably in metaphase, anaphase and telophase (Figure 6A and 6B). During interphase and prophase, levels were similar in control and treated cells. The highest levels of pH2A.X staining occurred at anaphase. Such cells appear to have tangled chromosomes that had failed to separate. Much less frequent foci of pH2A.X staining were also visible in the few curcumin-treated cells that reached telophase and cytokinesis. No increase in pH2A.X in response to curcumin was observed in unsynchronized HCT116 whole cell lysates by western blotting (data not shown), to be expected if the increase only occurred in mitotic cells that were in the minority.
Fig. 6.
Curcumin increased pH2A.X staining and Aurora B mislocalization in mitotic cells. Cells were treated for 12h as indicated and stained with antibodies against pH3 (red) and pH2A.X (green) (A and B) or against pH3 (green) and Aurora B (red) (C and D). DNA was stained with DAPI (blue). Representative images (from n = ≥3 experiments) are shown in interphase prophase, metaphase, anaphase and telophase. Exposure and gain were kept constant for pH2A.X staining. Scale bar, 5 µm.
The effects of curcumin treatment on the localization of Aurora B during mitosis
In mouse progenitor cells, curcumin was reported to cause mislocalization of Aurora B during mitosis (9). To investigate whether this occurred in human CRC cells, HCT116p53+/+ cells were co-labelled using antibodies to Aurora B and phospho-histone H3. In untreated prophase/prometaphase cells, Aurora B was localized at the centromeres and chromosome arms, but following treatment it was concentrated at the centre of the condensed chromosomes (Figure 6C and 6D). During normal metaphase Aurora B coincided with aligned chromosomes on the metaphase plate, but in curcumin-treated cells staining was much less uniform, with foci of bright stain. In anaphase cells, instead of being localized solely at the midzone and cortex, it was partially retained on chromatin. Finally, in late telophase and during cytokinesis when Aurora B was normally confined to the cleavage furrow, in response to treatment, it was still visible at the midzone and on chromatin. In curcumin-treated cells, contraction of the spindle midbody could be seen while chromatin was still present in the cleavage plane.
Discussion
Curcumin treatment resulted in increased levels of overall G2/M arrest in all cell lines tested, with HCT116 lines being the most sensitive. Using a combination of flow cytometric analysis of DNA content and analysis of the mitotic index with fluorescence microscopy, we have shown that the majority of cells arrest at the G2/M boundary. However, in several lines (HCT116 and Caco-2) significant numbers arrest in mitosis.
In all treated cell lines, a significant increase in abnormal mitotic spindles was observed. Mitotic progression was impaired and cells frequently failed to form a normal metaphase plate. Curcumin exerts anti-mitotic effects by depolymerizing microtubules and impairing microtubule assembly (6). It has previously been shown to disturb the localization of NuMA, a protein involved in organizing microtubules into asters during mitosis, as well as the kinesin protein Eg5, crucial for bipolar mitotic spindle morphogenesis (7,29–32). Moreover, curcumin can decrease the levels of Aurora A, a mitotic kinase that controls centrosome maturation and spindle formation (10). Disruption of these key regulators of early events in mitosis may contribute to the spindle abnormalities, failed centrosomal separation and prometaphase/prophase arrest that we observed in curcumin-treated mitotic cells. In addition, perturbed microtubule dynamics probably affect the microtubule capture of chromosomes at kinetochores, a process that takes longer in CIN lines compared with MIN lines due to an increased chromosomal content relative to diploid lines (33). This may account for the high levels of spindle abnormalities observed in the HT-29 and SW480 lines, which occur in 99% of SW480 cells following treatment.
Defects in chromosome segregation and cytokinesis in curcumin-treated mouse progenitor cells transfected with the Bcr-Abl gene were reported in a model of chronic myeloid leukaemia (9). However, a high number of cells still reached metaphase and the later stages of mitosis, albeit with many abnormalities in chromosome segregation. In contrast, we found that very few HCT116p53+/+ cells reached the later stages of mitosis, but those that did, had similar abnormalities. Curcumin impaired chromosomal congression and segregation, and inhibition of chromosome condensation and decatenation may contribute to this phenomenon. Curcumin has previously been shown to negatively regulate SMC2 (structural maintenance of chromosomes 2) and this may impair chromosome condensation during mitosis (34). Curcumin can act as a topoisomerase II inhibitor that may prevent the removal of any residual catenanes between sister chromatids prior to the onset of anaphase (18). We found that chromosomes in treated anaphase cells failed to segregate, and that these cells resembled the cut (chromosomes untimely torn) phenotype observed in fission yeast cells expressing a non-degradable version of Cut2, and human cells expressing a non-degradable version of securin (35–37). They also resembled the anaphase chromosomes of Caenorhabditis elegans cells that have been depleted of SMC-4 (structural maintenance of chromosomes 4 ) (38). Intriguingly, new data indicate that curcumin can target the APC/C protein CDC27 in HCT116 cells (12). If the activity of the APC/C was impaired, thus reducing the activation of separase, this may account for the failure of sister chromatids to separate correctly as observed in our study.
Curcumin activates DNA damage signalling in a variety of cell types, and the source of this damage has been linked to the production of ROS. In this study, we found that curcumin-activated DNA damage network signalling in the absence of oxidative DNA damage. This resulted in activatory phosphorylation of Chk1 and loss of the protein phosphatase CDC25C, which removes inhibitory phosphate residues on CDK1 during the G2/M transition and mitosis. Our data show that pre-treatment with caffeine inhibited curcumin-induced mitotic arrest, implicating the involvement of the DNA damage signalling in this type of arrest for the first time. Activation of the spindle checkpoint via a Chk1-pathway has also been shown to occur in HCT116 cells undergoing mitotic arrest following treatment with another chemopreventive agent, diallyl trisulfide (39).
We found pH2A.X foci in curcumin-treated, but not control, mitotic cells, suggesting that aberrant mitosis and not ROS may contribute to DNA damage. Other spindle poisons such as nocodazole, and K5I, an inhibitor of the kinesin Eg5, can also induce phosphorylation of histone H2A.X (28,40). In MCF-7 breast cancer cells treated with K5I, a prolonged mitotic arrest resulted in activation of caspase-activated DNAase followed by phosphorylation of histone H2A.X by ATM (41). In that study, activation of caspase 9 preceded activation of caspase 7, followed by cleavage of inhibitor of caspase-activated DNase (ICAD). The authors proposed that cell stress, as a result of prolonged mitotic arrest, initiated pro-apoptotic signalling leading to DNA damage. We have shown previously that curcumin can activate caspases 7, 8 and 9 in HT-29 and HCT116 cells (3).
It has generally been considered that ROS are the main source of DNA damage in curcumin-treated cells, as when treatment is combined with the antioxidant, NAC, the formation of pH2A.X foci, DNA damage signalling, and induction of apoptosis are significantly reduced (17,18,42,43). Interestingly, because NAC is an enzymatic inhibitor of topo II, it may also impair the ability of curcumin to act as a topo II poison, and thus generate dsDNA breaks (44). Because curcumin can act as both an anti-mitotic agent and a topoisomerase poison, DNA damage incurred from pro-apoptotic signalling and topo-mediated strand breaks may contribute to the formation of pH2A.X foci in mitotic cells. Moreover, we found that treatment with physiological levels of curcumin did not induce significant levels of oxidative DNA damage in the HCT116 line, indicating that in this instance, ROS are unlikely to be the source of damage.
These results support the role of curcumin as an anti-mitotic agent in CRC cells, which acts by disrupting microtubule dynamics and hampering the bi-orientation of chromosomes between spindle poles. Depolymerization of microtubules and inhibition of the mitotic kinesin Eg5 may impair kinetochore capture by microtubules during mitosis, and the effects of curcumin on chromosome condensation and congression may contribute to the deregulation of this process. In the HCT116 and Caco-2 lines, this resulted in activation of the spindle-assembly checkpoint and significant M phase arrest. Recently, concerns have been raised regarding the safety of high doses of curcumin on a long-term basis in a clinical setting (45). These data show that impaired mitotic spindle morphogenesis and defects in chromosomal segregation may result in the replication of cells with non-lethal levels of DNA damage acquired during aberrant mitosis. Further research is required to examine the possibility of increased chromosomal aberrations following treatment in healthy colorectal cells in vivo. It is possible that damaged cells may acquire a senescent phenotype similar to those that undergo prolonged mitotic arrest following treatment with KI5 (46). Indeed, a recent study indicates that HCT116p53+/+ and HCT116p53–/– cells undergo growth arrest and begin to display markers of senescence after treatment with 10 µM curcumin for 24h (47). If a link between aberrant mitosis and senescence was to be established, this could be an important step in alleviating concerns regarding the potential mutagenic properties of this agent.
Supplementary material
Supplementary Figures 1–6 can be found at http://carcin.oxfordjournals.org/
Funding
L.M.B. was in receipt of a Biotechnology and Biological Sciences Research Council studentship and this study was partly funded by Cancer Research-UK (C6048/A10896 to M.M.M.); Cancer Prevention Research Trust to M.M.M.; The FACSAria II cell sorter was funded by a grant from the Medical Research Council (G0802524). C.B.’s salary is funded by Cancer Research UK (C325/A13101).
Supplementary Material
Acknowledgements
The authors wish to thank Prof. Andrew Fry and Dr Raj Patel (University of Leicester) for much useful discussion and Dr Suzy Prosser for her assistance with immunofluorescence microscopy.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- APC/C
anaphase promoting complex/cyclosome
- BSA
bovine serum albumin
- CRC
colorectal cancer
- DAPI
4′,6-diamidino-2-phenylindole
- DBH
debromohymenialdisine
- DMSO
dimethyl sulfoxide
- FCS
fetal calf serum
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- NAC
N-acetyl cysteine
- ROS
reactive oxygen species.
References
- 1. Chen H, et al. (1999). Curcumin inhibits cell proliferation by interfering with the cell cycle and inducing apoptosis in colon carcinoma cells. Anticancer Res., 19, 3675–3680 [PubMed] [Google Scholar]
- 2. Hanif R, et al. (1997). Curcumin, a natural plant phenolic food additive, inhibits cell proliferation and induces cell cycle changes in colon adenocarcinoma cell lines by a prostaglandin-independent pathway. J. Lab. Clin. Med., 130, 576–584 [DOI] [PubMed] [Google Scholar]
- 3. Howells L.M, et al. (2007). Comparison of oxaliplatin- and curcumin-mediated antiproliferative effects in colorectal cell lines. Int. J. Cancer, 121, 175–183 [DOI] [PubMed] [Google Scholar]
- 4. Jaiswal A.S, et al. (2002). Beta-catenin-mediated transactivation and cell-cell adhesion pathways are important in curcumin (diferuylmethane)-induced growth arrest and apoptosis in colon cancer cells. Oncogene, 21, 8414–8427 [DOI] [PubMed] [Google Scholar]
- 5. Holy J.M. (2002). Curcumin disrupts mitotic spindle structure and induces micronucleation in MCF-7 breast cancer cells. Mutat. Res., 518, 71–84 [DOI] [PubMed] [Google Scholar]
- 6. Gupta K.K, et al. (2006). Dietary antioxidant curcumin inhibits microtubule assembly through tubulin binding. FEBS J., 273, 5320–5332 [DOI] [PubMed] [Google Scholar]
- 7. Banerjee M, et al. (2010). Curcumin suppresses the dynamic instability of microtubules, activates the mitotic checkpoint and induces apoptosis in MCF-7 cells. FEBS J., 277, 3437–3448 [DOI] [PubMed] [Google Scholar]
- 8. Basile V, et al. (2009). Curcumin derivatives: molecular basis of their anti-cancer activity. Biochem. Pharmacol., 78, 1305–1315 [DOI] [PubMed] [Google Scholar]
- 9. Wolanin K, et al. (2006). Curcumin affects components of the chromosomal passenger complex and induces mitotic catastrophe in apoptosis-resistant Bcr-Abl-expressing cells. Mol. Cancer Res., 4, 457–469 [DOI] [PubMed] [Google Scholar]
- 10. Liu H.S, et al. (2011). Curcumin-induced mitotic spindle defect and cell cycle arrest in human bladder cancer cells occurs partly through inhibition of aurora A. Mol. Pharmacol., 80, 638–646 [DOI] [PubMed] [Google Scholar]
- 11. Magalska A, et al. (2006). Resistance to apoptosis of HCW-2 cells can be overcome by curcumin- or vincristine-induced mitotic catastrophe. Int. J. Cancer, 119, 1811–1818 [DOI] [PubMed] [Google Scholar]
- 12. Lee S.J, et al. (2012). Anaphase-promoting complex/cyclosome protein Cdc27 is a target for curcumin-induced cell cycle arrest and apoptosis. BMC Cancer, 12,: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu J.J, et al. (2011). Curcumin induces DNA damage and caffeine-insensitive cell cycle arrest in colorectal carcinoma HCT116 cells. Mol. Cell. Biochem., 354, 247–252 [DOI] [PubMed] [Google Scholar]
- 14. Park C, et al. (2006). Induction of G2/M arrest and inhibition of cyclooxygenase-2 activity by curcumin in human bladder cancer T24 cells. Oncol. Rep., 15, 1225–1231 [PubMed] [Google Scholar]
- 15. Tomita M, et al. (2006). Curcumin (diferuloylmethane) inhibits constitutive active NF-kappaB, leading to suppression of cell growth of human T-cell leukemia virus type I-infected T-cell lines and primary adult T-cell leukemia cells. Int. J. Cancer, 118, 765–772 [DOI] [PubMed] [Google Scholar]
- 16. Giri A.K, et al. (1990). Sister chromatid exchange and chromosome aberrations induced by curcumin and tartrazine on mammalian cells in vivo . Cytobios, 62, 111–117 [PubMed] [Google Scholar]
- 17. Jiang Z, et al. (2010). The mismatch repair system modulates curcumin sensitivity through induction of DNA strand breaks and activation of G2-M checkpoint. Mol. Cancer Ther., 9, 558–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. López-Lázaro M, et al. (2007). Curcumin induces high levels of topoisomerase I- and II-DNA complexes in K562 leukemia cells. J. Nat. Prod., 70, 1884–1888 [DOI] [PubMed] [Google Scholar]
- 19. Blasiak J, et al. (1999). Curcumin damages DNA in human gastric mucosa cells and lymphocytes. J. Environ. Pathol. Toxicol. Oncol., 18, 271–276 [PubMed] [Google Scholar]
- 20. Cao J, et al. (2006). Mitochondrial and nuclear DNA damage induced by curcumin in human hepatoma G2 cells. Toxicol. Sci., 91, 476–483 [DOI] [PubMed] [Google Scholar]
- 21. Kelly M.R, et al. (2001). Disparate effects of similar phenolic phytochemicals as inhibitors of oxidative damage to cellular DNA. Mutat. Res., 485, 309–318 [DOI] [PubMed] [Google Scholar]
- 22. Sahu R.P, et al. (2009). Activation of ATM/Chk1 by curcumin causes cell cycle arrest and apoptosis in human pancreatic cancer cells. Br. J. Cancer, 100, 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Waldman T, et al. (1995). p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res., 55, 5187–5190 [PubMed] [Google Scholar]
- 24. Bunz F, et al. (1998). Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science, 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 25. Singh R, et al. (2009). Simultaneous determination of 8-oxo-2’-deoxyguanosine and 8-oxo-2’-deoxyadenosine in DNA using online column-switching liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom., 23, 151–160 [DOI] [PubMed] [Google Scholar]
- 26. Sarkaria J.N, et al. (1999). Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res., 59, 4375–4382 [PubMed] [Google Scholar]
- 27. Curman D, et al. (2001). Inhibition of the G2 DNA damage checkpoint and of protein kinases Chk1 and Chk2 by the marine sponge alkaloid debromohymenialdisine. J. Biol. Chem., 276, 17914–17919 [DOI] [PubMed] [Google Scholar]
- 28. Dalton W.B, et al. (2007). Human cancer cells commonly acquire DNA damage during mitotic arrest. Cancer Res., 67, 11487–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blangy A, et al. (1995). Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo . Cell, 83, 1159–1169 [DOI] [PubMed] [Google Scholar]
- 30. Gaglio T, et al. (1995). NUMA is required for the organization of microtubules into aster-like radial arrays. J. Cell. Biol, 131, 693–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holy J. (2004). Curcumin inhibits cell motility and alters microfilament organization and function in prostate cancer cells. Cell Motil. Cytoskeleton, 58, 253–268 [DOI] [PubMed] [Google Scholar]
- 32. Kang D, et al. (2002). The checkpoint protein Chfr is a ligase that ubiquitinates Plk1 and inhibits Cdc2 at the G2 to M transition. J. Cell Biol., 156, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Silkworth W.T, et al. (2009). Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE, 4, e6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen G, et al. (2006). Modulation of nuclear factor E2-related factor 2-mediated gene expression in mice liver and small intestine by cancer chemopreventive agent curcumin. Mol. Cancer Ther., 5, 39–51 [DOI] [PubMed] [Google Scholar]
- 35. Funabiki H, et al. (1996). Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature, 381, 438–441 [DOI] [PubMed] [Google Scholar]
- 36. Hagting A, et al. (2002). Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J. Cell Biol., 157, 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hirano T, et al. (1986). Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J., 5, 2973–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hagstrom K.A, et al. (2002). C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev., 16, 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiao D, et al. (2009). Diallyl trisulfide-induced apoptosis in human cancer cells is linked to checkpoint kinase 1-mediated mitotic arrest. Mol. Carcinogenesis, 48, 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shi J, et al. (2008). Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Res., 68, 3269–3276 [DOI] [PubMed] [Google Scholar]
- 41. Orth J.D, et al. (2012). Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol. Biol. Cell, 23, 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khar A, et al. (2001). Induction of stress response renders human tumor cell lines resistant to curcumin-mediated apoptosis: role of reactive oxygen intermediates. Cell Stress Chaperones, 6, 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scott D.W, et al. (2004). Curcumin-induced GADD153 gene up-regulation in human colon cancer cells. Carcinogenesis, 25, 2155–2164 [DOI] [PubMed] [Google Scholar]
- 44. Grdina D.J, et al. (1998). Effects of thiols on topoisomerase-II alpha activity and cell cycle progression. Cell Prolif., 31, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burgos-Moron E. (2010). The dark side of curcumin. Int. J. Cancer, 126, 1771–1775 [DOI] [PubMed] [Google Scholar]
- 46. Nakai R, et al. (2009). K858, a novel inhibitor of mitotic kinesin Eg5 and antitumor agent, induces cell death in cancer cells. Cancer Res., 69, 3901–3909 [DOI] [PubMed] [Google Scholar]
- 47. Mosieniak G, et al. (2012). Curcumin induces permanent growth arrest of human colon cancer cells: link between senescence and autophagy. Mech. Ageing Dev., 133, 444–455 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.