Abstract
Osteoporosis in patients with systemic lupus erythematosus (SLE) is thought to be the result of accelerated osteoclastogenesis induced by pro-inflammatory cytokines such as tumor necrosis factor (TNF). However, the molecular mechanisms involved in the osteoblastogenesis in SLE patients are not fully understood. We investigated the bone morphogenetic protein-2 (BMP-2)-induced osteoblastic capacity of bone marrow-derived mesenchymal stem cells (BMMSCs) from SLE patients and the TNF signaling system in determining BMP-2-induced regulatory pathways. It showed that the capacity of osteogenic differentiation of BMMSCs from SLE patients was reduced compared with that from healthy controls. The nuclear factor κB (NF-κB) signaling was activated while the BMP/Smad pathway was repressed in BMMSCs from SLE patients. TNF activated NF-κB pathway and inhibited the phosphorylation of Smad 1/5/8 and BMP-2-induced osteoblastic differentiation in BMMSCs from normal controls, while addition of pyrollidine dithiocarbamate (PDTC), an NF-κB inhibitor, to SLE-BMMSCs could partially reverse these effects. Thus, our findings have shown that the activated NF-κB pathway in SLE-BMMSCs inhibits the BMP-2-induced osteoblastic differentiation through BMP/Smad signaling pathway, suggesting that the impaired osteoblastic differentiation may participate in the pathology of osteoporosis in SLE patients.
Introduction
Systemic lupus erythematosus (SLE) is a chronic, multisystemic autoimmune disease that involves multiple organs including renal, cardiovascular, neural, musculoskeletal, and cutaneous systems. Osteoporosis (OP), a metabolic bone disease characterized by decreased bone mineralization, increased bone fragility, and increased risk of fracture, has been found to be prevalent in patients with SLE. Cross-sectional studies have shown an increased prevalence of OP in SLE patients, which is estimated to be between 4% and 22% [1–3].
The mechanisms of OP in SLE patients remain incompletely understood and may include the inflammatory disease itself, disease-related comorbidity, and the treatment of the disease including the corticosteroids (CS). Although exposure to CS is generally considered to be a major factor contributing to the development of OP, the systemic inflammation itself is recognized to be the most important risk factor of OP in SLE patients. Tumor necrosis factor (TNF) is one of the most potent pro-inflammatory cytokines and is known to be a catabolic factor in the inflammatory reaction of diseases such as rheumatoid arthritis (RA) [4]. SLE patients are reported to have increased TNF in its serum [5,6]. TNF induces osteoclastogenesis either by promoting the proliferation of osteoclast precursor cells or by causing the activation of the differentiated osteoclast through RANK/RANKL signaling pathway [7–9]. TNF stimulates osteoclasts to express RANK to interact with its ligand RANKL to accelerate the activity of osteoclasts.
Indeed, in physiologic remodeling, activation of bone resorption requires contact between cells of the osteoblast and osteoclast lineages. Osteoblasts play an essential role in the pathogenesis of OP. Osteoblasts produce RANKL, which activates the differentiation of osteoclasts and maintains their bone resorptive function. Osteoblasts also produce and secrete osteoprotegerin, a decoy receptor that can block RANKL/RANK interactions [10,11]. Since bone mass loss in SLE is believed to be associated with TNF system, it has also been hypothesized that TNF directly controls osteoblast survival and/or its function [12]. However, the role of TNF signaling in modulating osteoblast's function and differentiation remains controversial. Some groups reported that treatment of human mesenchymal stem cells (MSCs) with TNF during differentiation resulted in enhanced expression of osteogenic markers [13,14]. On the contrary, other studies have demonstrated that TNF blocked osteoblast differentiation in multiple model systems such as fetal calvaria, bone marrow (BM) stromal cells, and MC3T3-E1 cells [15–17]. Moreover, both spontaneous and bone morphogenetic protein (BMP)-induced osteoblast differentiation could be inhibited by TNF in vitro [18,19].
Osteoblasts are derived from MSCs, which can differentiate into osteoblasts both in vitro and in vivo [20,21]. We have previously reported that bone marrow-derived mesenchymal stem cells (BMMSCs) from SLE patients showed significantly decreased bone-forming capacity and impaired reconstruction of BM osteoblastic niche in vivo [22]; however, the cellular mechanism underlying this deficiency has not yet been elucidated.
BMPs, members of the transforming growth factor (TGF)-β superfamily, have been shown to play critical roles in governing various aspects of embryological development, including brain, heart, kidney, and eye [23]. BMPs are also known to promote the differentiation of MSCs into chondrocytes and osteoblasts and the differentiation of osteoprogenitor cells into osteoblasts through the Smad signaling transduction pathway via 2 transmembrane serine-threonine kinase receptors, BMP receptor (BMPR) type I and BMPR type II [24]. The activated receptor kinases, in turn, phosphorylate the transcription factors Smad 1, 5, and 8. The phosphorylated Smads then form a heterodimeric complex with Smad 4 in the nucleus and activate the expression of target genes. In this study, we investigated the nuclear factor κB (NF-κB) and BMP/Smad signaling pathways in the BMMSCs from SLE patients. We demonstrated the role of TNF activated NF-κB pathway in regulating osteoblastic differentiation and found that the activated NF-κB pathway of BMMSCs from SLE patients inhibited BMP-2-induced osteogenic differentiation through downregulating Smad signaling pathway. Together, our results have uncovered a novel mechanism that TNF participates in the pathology of OP in SLE patients.
Materials and Methods
Patients and controls
BM cells for cDNA microarray were obtained from 4 patients with SLE, according to the American College of Rheumatology [25], (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd). All the patients were female with the mean age of 37±11 years (range 20–44). The normal controls were 1 male and 3 females, with the mean age of 39±7 years (range 29–45). MSCs from 10 SLE patients (mean 37±13 years, range 15–57 years) and 10 age- and sex-matched normal controls were examined for various osteoblastic markers by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis. Three to five of these 10 samples mentioned above were used for western blot analysis. All the SLE patients above had active disease with a Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) [26] score of more than 8 at the time of BM aspiration. Serum from 16 patients (15 females and 1 male, mean age of 33±14 years, range 13–57, SLEDAI: 2–21) and 15 female normal controls (mean age of 32±13 years, range 16–57) were tested for BMP-2 levels (Supplementary Table S2). All participants gave written consent to the study, which was approved by the Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing University Medical School [27].
Cell culture and flow cytometry
BM samples were taken from the iliac crest of SLE patients and healthy subjects. The BM mononuclear cells were plated at 105/mL density in low glucose Dulbecco modified Eagle medium (L-DMEM; Gibco) supplemented with 10% heat inactivated fetal bovine serum (Invitrogen) and 1% antibiotic-antimycotic and incubated at 37°C in a 5% (vol/vol) humidified CO2 chamber as described previously (27). Markers CD29, CD44, C105, CD34, CD45, and HLA-DR on the cells at passage 2 were consequently detected by flow cytometry [27].
Cells at passage 3–5 were plated on a 6-cm dish (2.5×104/mL) and were grown to confluence for 3 days at 37°C in 5% CO2 and then stimulated with osteogenic medium including 10 mM β-glycerophosphate (GP; Sigma), 50 μg/mL ascorbic acid (AA; Sigma), and 300 ng/mL BMP-2 (Miltenyi Biotec) for 2, 14, or 21 days.
RNA extraction and quantitative real-time RT-PCR analysis
BMMSCs from SLE patients were cultured in a 6-well plate (2.0×105 viable cells), and treated with indicated concentrations of TNF (R&D), BMP-2, pyrollidine dithiocarbamate (PDTC), or both of them for indicated times; BMMSCs in the presence or absence of stimulation were placed in TRIzol (Invitrogen) and total cellular RNA was extracted. cDNA was synthesized using PrimerScript RT reagent Kit (TaKaRa). qRT-PCR was performed on an ABI 7500 FAST real-time PCR detection system (Applied Biosystems) using SYBR Green detection mix [25]. The following primers were used in this study: alkaline phosphatase (ALP) Sense 5′-GCACCGTCAAGGCTGAG AAC-3′, Antisense 5′-TGGTGAAGACGCCA GTGGA-3′; Runx2 Sense 5′, CACTGGCGCTGCAACAAGA-3′, Antisense 5′-CATTCCGGAGCTCAGCAG AATAA-3′; osteocalcin (OCN) Sense 5′-CGGTGCAGAGT CCAGCAAAG-3′, Antisense 5′-TACAGGTAGCGCCTGGGTCTCT-3′; COLIA2 Sense 5′-GAGGGCAACAGC AGGTTCACTTA-3′, Antisense 5′-TCA GCACCAC CGATGTCCA-3′; and (GAPDH) sense: 5′-TGACTTCAACAGCGACACCCA-3′, antisense: 5′-CACCCTGTTG CTGTAG CCAAA-3′.
The expression of the target genes in SLE samples as compared with that in controls was examined using 2−ΔΔCt method. Briefly, for each sample, a value for the cycle threshold (Ct) was determined, defined as the mean cycle at which the fluorescence curve reached an arbitrary threshold. The ΔCt for each sample was then calculated according to the formula Ct target gene−—Ct GAPDH; ΔΔCt values were then obtained by subtracting the ΔCt of a reference sample (average ΔCt of the control group) from the ΔCt of the studied samples. Finally, the levels of expression of the target genes in the studied samples as compared with the reference sample were calculated as 2-ΔΔCt.
cDNA microarray analysis
BMMSCs at passage 3 from 4 patients and 4 normal controls were placed in Trizol (Invitrogen) and processed for RNA extraction using the RNeasy kit according to the instructions of the manufacturer (Qiagen). The universal human reference RNA samples (Stratagene Corporation) were used as a common reference in the 2 channel microarrays. Total RNA was reverse-transcribed, and the cDNA of BMMSCs from SLE patients and controls were added with Cy3-dCTP while the cDNA of human reference were added with Cy5-dCTP in the presence of Klenow enzyme (GE Healthcare Cat. No. PA 55021/PA 53021) [28].
Microarray analysis was performed in CapitalBio Corp using 22K Human Genome Array. Labeled samples were quantitatively adjusted based on the efficiency of Cy-dye incorporation and mixed into 80 μL hybridization solution [3×saline sodium citrate (SSC), 0.2% sodium dodecyl sulfate (SDS), 25% formamide, and 5× Denhart's]. DNA in hybridization solution was denatured at 95°C for 3 min prior loading on a microarray. The array was hybridized at 42°C overnight and washed with 2 consecutive washing solutions (0.2% SDS, 2× SSC at 42°C for 5 min and 0.2% SSC) for 5 min at room temperature. Finally, arrays were scanned with a confocal LuxScan 10 KA scanner (CapitalBio).
The data of obtained images were extracted with LuxScan 3.0 software (CapitalBio). Genes with the signal intensity more than 800 (Cy3 or Cy5) were regarded as the expressed genes. In every 2 channel slides, the intensity ratio of the Cy3 to Cy5 of each spot was calculated after normalization with LOWESS regression. Statistical data and differential analysis files were generated by using SAM software 3.0 (Stanford University). The significantly changed genes were selected based on P-value <0.05 and >2-fold as criteria. All the differentially expressed genes were analyzed using a free web-based Molecular Annotation System 2.0 (MAS 2.0, http://bioinfo.capitalbio.com/mas).
All data are MIAME compliant and that the raw data have been deposited in a MIAME compliant database (GEO). The raw data can be seen at www.ncbi.nlm.nih.gov/geo/query/acc.cgi acc, =GSE 21649 and the accession number is GSE 21649.
ALP staining
Fourteen days after stimulation, BMMSCs were washed twice with phosphate-buffered saline, fixed with 0.5 mL/well formalin/methanol/H2O (1:1:1.5) for 15 min at room temperature, and washed thrice with distilled water. For staining, 1 FAST5-bromo-4-chloro-3-indolylphosphate/nitro blue tetrazolium tablet (Sigma) was dissolved in 10 mL of water, and 0.5 mL of substrate solution was added to the fixed cultures for 15 min at room temperature. After staining, cultures were washed thrice with distilled water and air-dried.
Alizarin Red S staining for mineralization
Twenty-one days after stimulation, BMMSCs were washed twice in phosphate-buffered saline, fixed with 0.5 mL/well formalin/methanol/H2O (1:1:1.5) for 15 min at room temperature, and washed thrice. Saturated Alizarin Red S solution (pH 4.1) was filtered, and 1.5 mL/well was added and incubated for 5 min at room temperature. Cells were then washed 4–5 times and air-dried.
Western blotting analysis
Cells (2×105 viable cells) were precultured in 6-well plates in DMEM containing 10% fetal calf serum for 48 h. After preculture, the medium was replaced with serum-free fresh medium, and then indicated concentrations of BMP-2, TNF, or PDTC were added to the culture medium for different time intervals. After stimulation, cells were lysed with SDS-sample buffer containing 20 mM Tris-HCl (pH 7.6), 250 mM NaCl, 0.5% NP-40, 3 mM ethylenediaminetetraacetic acid, and 1.5 mM ethyleneglycoltetraacetic acid with 10 mg/mL Aprotinin, 10 mg/mL leupeptin, 1 mM DTT, 1 mM paranitrophenylphosphate, and 0.1 mM Na3VO4 as protease and phosphatase inhibitor. Cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore). Blots were probed by anti-phospho-Smad1/5/8 antibodies (Cell Signaling Technology, Inc.), anti-total-Smad1/5/8 antibodies (Santa Cruz Biotechnology, Inc.), anti-Sma4 antibodies (Cell Signaling Technology, Inc.), anti-inhibitor κB (IκB) antibody (Cell Signaling Technology, Inc.), and anti-GAPDH antibody (Millipore) before visualizing with horseradish peroxidase-conjugated secondary antibodies followed by development with FluorChem FC2 System (Alpha Innotech Corporation).
Immunoprecipitation analysis
BMMSCs from patients and healthy controls were starved in serum-free fresh medium for 24 h and stimulated with BMP-2 for 30 min, then lysed with (SDS)-sample buffer, and diluted to 1 μg/μL. Anti-Smad1/5/8 antibodies were added to 1 mL cell lysates at 4°C overnight. The immunocomplex was captured by adding 100 μL Protein A Agarose, Fast Flow, bead slurry (Millipore). After incubation at 4°C for 2 h, the carrier beads were washed 4 times in buffer. The samples were boiled in Laemmli loading buffer for 10 min, applied to 10% SDS-PAGE gels, and electroblotted onto PVDF western blot membranes (Roche Diagnostics). The Smad4 protein was identified by western blot analysis.
Enzyme-linked immunosorbent assay analysis
Serum samples from 16 SLE patients and 15 healthy controls were collected, and the concentrations of BMP-2 of each individual were measured using commercial ELISA kit (R&D) according to the manufactory introduction.
Statistical analysis
All results are shown as mean±standard error of the mean of data from at least 3 separate experiments, each performed with triplicate samples. Differences between groups were analyzed for statistical significance using analysis of variance. Differences between 2 independent groups were analyzed for student's t-test (SPSS 16.0 software). P values <0.05 were accepted as statistically significant.
Results
Characterization of osteoblastic differentiation in BMMSCs from SLE patients
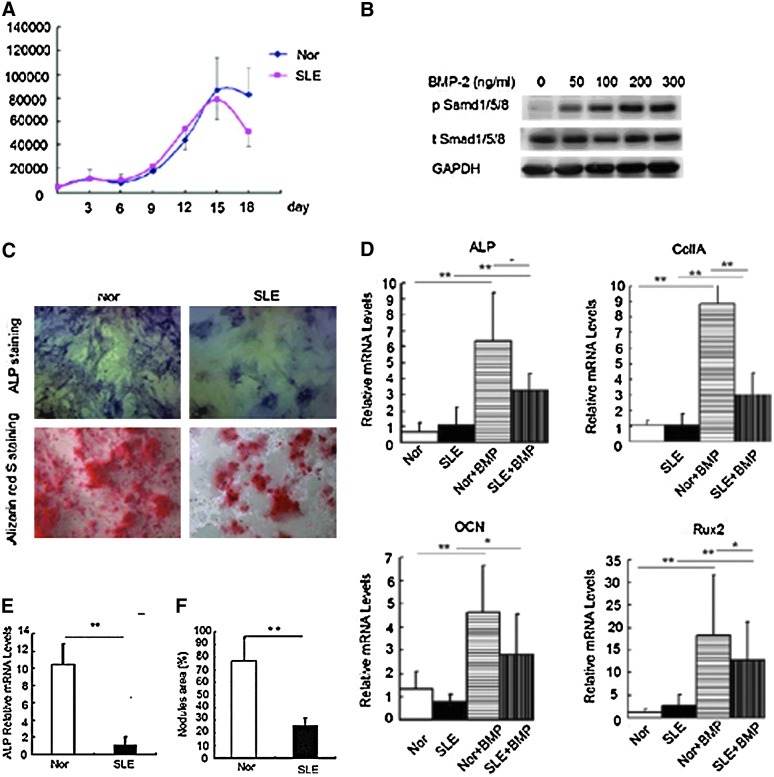
Consistent with our previous findings, BMMSCs from both SLE patients and normal controls were positive for CD29, CD44, and CD105, and negative for CD14, CD34, CD45, and HLA-DR. There was no significant difference between the phenotypic features of MSCs from SLE patients and normal controls [27]. To exclude the effect of senescence on the osteogenic capacity of BMMSCs, we compared the proliferation of BMMSCs during the culturing of osteogenic medium with BMP-2 (OMB) (AA, GP, and BMP-2). The growth curves showed that both the 2 groups rapidly grew from about the 9th day and reached the highest level at the 14th day. Cell amount decreased after 14 days during the osteogenic medium culture. There was no difference between the cell numbers of normal controls (n=3) and SLE patients (n=3) at every point (all P>0.05; Fig. 1A). And then, we evaluated the osteogenic capacity of BMMSCs from SLE patients induced by OMB. As BMP-2 activates Smad signaling pathway via the phosphorylation of Smad1/5/8, we firstly treated BMMSCs with different concentrations of BMP-2 to find that BMP-2 induced the pSmad1/5/8 in a dose-dependent manner, so we selected 100 ng/mL BMP-2 in the following experiments (Fig. 1B). Before stimulation, BMMSCs from both SLE and control subjects showed a low expression of ALP, Runx2, OCN, and collagen II A2 (CoIIA2) at the transcriptional level. In response to OMB treatment for 2 days, BMMSCs from both groups showed a significant increase in mRNA levels of ALP, CoIIA2, Runx2, and OCN. Of note, BMMSCs from SLE patients showed lower expression of ALP, CoIIA, and Runx2 post-stimulation as compared with that from the normal controls, while no statistical difference could be found in OCN mRNA level between these 2 groups (Fig. 1C). In addition, after a longer exposure to osteogenic stimulus, the ALP activity and the area of the mineralized nodules were lower in SLE-BMMSCs as detected by ALP and Alizarin Red S staining on day 14 and 21, respectively (Fig. 1D). These results suggested BMMSCs from SLE patients display impaired capacity of osteoblastic differentiation.
FIG. 1.
Characterization of osteoblastic differentiation in BMMSCs from SLE patients. (A) The growth curves of BMMSCs during osteogenic differentiation from SLE patients and normal controls. Two groups of cells were cultured in the osteogenic medium with BMP-2 (OMB) for 0∼18 days. (B) Stimulation with different doses of BMP-2 on the BMMSCs. Cells were starved for 24 h, and stimulated for 30 min with different concentrations of BMP-2. The phosphorylated Smad1/5/8 was detected by western blotting analysis. (C) Alkaline phosphatase (ALP) staining of cells for 14 days and Alizarin Red S staining for 21 days after treatment with OMB. *P<0.05 and **P<0.01 versus control groups. ALP-positive cells are stained as blue, and mineralization is visible as red spots on the photos shown. (D) qRT-PCR analysis of osteoblastic markers after osteogenic stimulus with BMP-2 for 2 days. The relative mRNA levels were analyzed by comparing with normal groups. (E) qRT-PCR analysis of ALP activity on day 14. **P<0.01 versus indicated groups. (F) The area of mineralized nodules of BMMSCs from SLE patients and healthy controls. **P<0.01 versus normal control groups (n=3). Nor, normal controls; SLE, systemic lupus erythematosus, BMMSCS, bone marrow-derived mesenchymal stem cells; BMP-2, bone morphogenetic protein-2; qRT-PCR, quantitative reverse transcriptase-polymerase chain reaction.
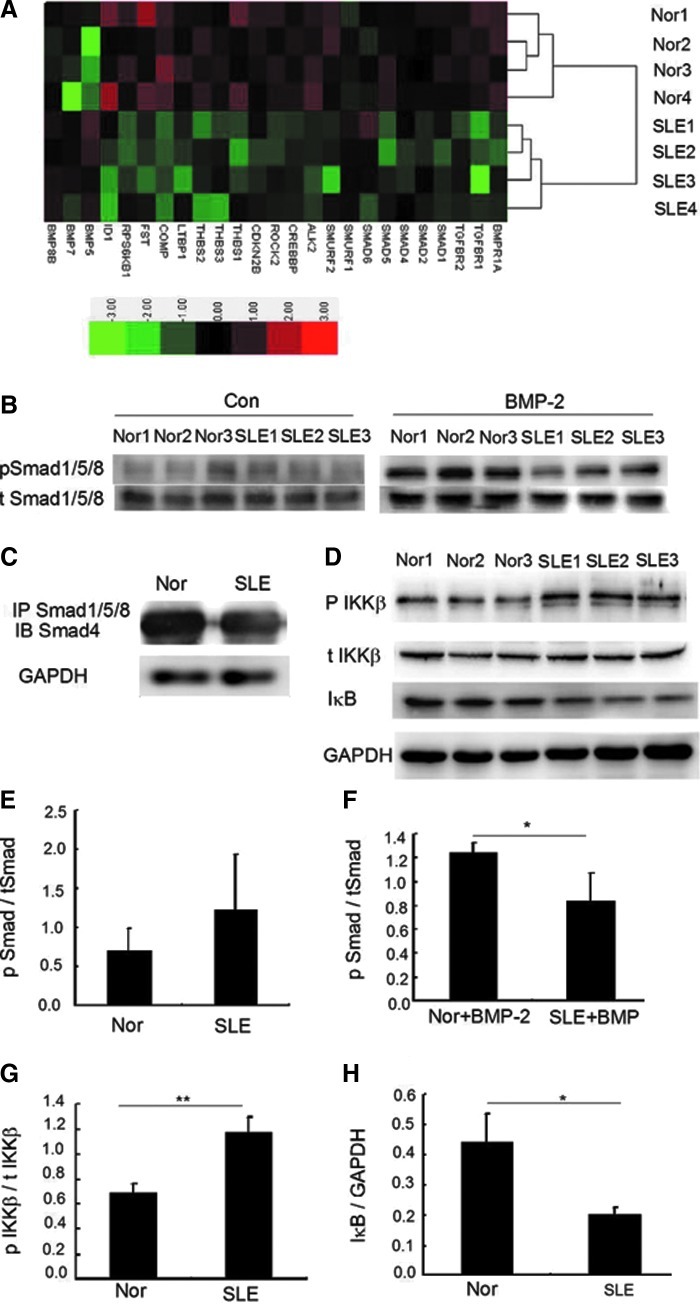
Repressed BMP/Smad and activated NF-κB signaling pathways in BMMSCs from SLE patients
As many signaling pathways like Wnt, Notch, fibroblast growth factors, and BMP/Smad pathways [29–32] are involved in the osteoblastic differentiation of MSCs, we compared the gene expression profiles of BMMSCs between SLE patients and normal controls, and found that all the differentially expressed genes in BMP/TGF-β signaling pathway except for BMP5 were downregulated in the patients' cells (Fig. 2A). Western blot analysis revealed that the Smad1/5/8 phosphorylation was higher in normal BMMSCs 30 min after BMP-2 stimulation (Fig. 2B). Moreover, the complex of Smad1/5/8 and Smad4, which was translocated into nucleus for the initiation of the mRNA transcription of target genes such as Runx2, was also reduced in SLE-BMMSCs compared with normal BMMSCs by immunoprecipitation analysis (Fig. 2C). These results revealed a suppressed BMP/Smad signaling pathway in SLE-BMMSCs. It was previously reported that TNF could inhibit bone formation and osteoblastic differentiation induced by BMPs [15,16] through NF-κB signaling pathway [33], besides, higher level of TNF was found in the serum of SLE patients compared with normal controls [5,6]. We next examined the state of NF-κB signaling pathway in BMMSCs from SLE patients, and found that the phosphorylated inhibitor κB kinase β (IKKβ) was higher. As the target of the IKK complex, the IκB was phosphorylated and subsequently degraded and decreased. We further found that the level of IκB was decreased in SLE-BMMSCs (Fig. 2D), which suggested an activated status of the NF-κB pathway in the patients' cells.
FIG. 2.
Repressed BMP/Smad and activated NF-κB signaling pathways in BMMSCs from SLE patients. (A) Clustering analysis of genes in BMP/TGF-β signaling pathway in BMMSCs from SLE patients and healthy controls. (B) Western blotting analysis of phosphorylated Smad1/5/8 of BMMSCs from SLE patients and healthy controls. Cells were starved (the left) and incubated with BMP-2 (100 ng/mL) for 30 min (the right). (C) Immunoprecipitation (IP) analysis of BMMSCs from SLE patients and healthy controls. (D) Western blotting analysis of P IKKβ, t IKKβ, and IκB in BMMSCs from SLE patients and healthy controls. (E–H) Quantity analysis of western blotting analysis. *P<0.05 and **P<0.01 versus normal control groups. Nor, normal controls, SLE, systemic lupus erythematosus, Nor+BMP-2, normal BMMSCs stimulated with BMP-2; SLE+BMP-2, SLE-BMMSCs stimulated with BMP-2; TGF-β, transforming growth factor-β; NF-κB, nuclear factor κB; IκB, inhibitor κB; IKKβ, inhibitor κB kinase β.
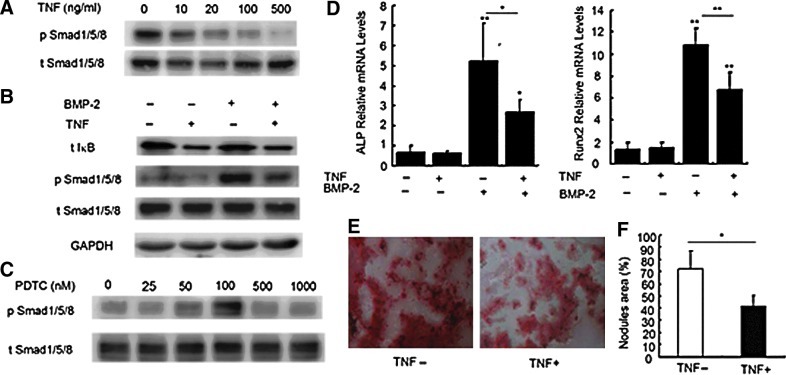
Activation of NF-κB inhibited BMP-2-induced expression of osteogenic markers in BMMSCs from normal controls
Next, we investigated the effect of TNF on the phosphorylation of Smad1/5/8 through NF-κB signaling pathway in BMMSCs from normal controls. Following 2-h pretreatment with TNF, BMMSCs were stimulated with BMP-2 for 30 min. As shown in Fig. 3A, TNF inhibited the phosphorylation of Smad1/5/8 induced by BMP-2 in a dose-dependent manner. TNF (20 ng/mL) alone effectively activated the NF-κB pathway as detected by the decreased IκB, but it had no effect on the activation of Smad1/5/8; nevertheless it could inhibit the Smad1/5/8 phosphorylation induced by BMP-2 (Fig. 3B). To further observe whether TNF inhibited the Smad1/5/8 phosphorylation through NF-κB pathway, PDTC, an NF-κB inhibitor was added to the BMP-2-TNF system. As shown in Fig. 3C, PDTC, at the concentration of 50 and 100 nM, reversed the inactivation of Smad1/5/8 caused by TNF. These data collectively indicate that the activation of NF-κB inhibits the BMP/Smad signaling pathway.
FIG. 3.
Effect of activating NF-κB signaling pathway on BMP-2-induced osteogenic markers in normal BMMSCs. (A) Effect of different concentrations of TNF on the Smad1/5/8 phosphorylation in BMMSCs revealed by western blotting analysis. BMMSCs were starved for 24 h and subsequently stimulated with TNF for 2 h and BMP-2 for 30 min. (B) Effect of TNF (20 ng/mL) on the levels of IκB and phosphorylation of Smad1/5/8 induced by BMP-2. (C) Effect of PDTC on the Smad1/5/8 phosphorylation in the presence of BMP-2 (100 ng/mL) and TNF (20 ng/mL). Starved cells were treated with TNF and PDTC for 2 h, followed by BMP-2 for 30 min. (D) qRT-PCR analysis of ALP and Runx2 mRNA levels. Cells were treated with TNF or BMP-2 for 48 h. *P<0.05 and **P<0.01 versus controls or between the indicated groups (n=4). (E) Alizarin Red S staining of cells cultured in the OMB for 21 days in the presence or absence of TNF. (F) The quantity analysis of mineralized nodules of normal BMMSCs in the OMB for 21 days in the presence or absence of TNF. *P<0.05 versus normal control groups (n=3). TNF, tumor necrosis factor; PDTC, pyrollidine dithiocarbamate.
For further investigation of the mechanism by which NF-κB affects the BMP-induced osteblastic differentiation, we examined the mRNA levels of ALP and Runx2 and the extracellular mineralization in normal BMMSCs treated with BMP-2 and/or TNF. Consistent with the western blot results, TNF inhibited ALP activity and Runx2 mRNA expression induced by BMP-2 for 48 h (Fig. 3D). Moreover, TNF inhibited BMP-2 induced extracellular mineralization on day 21 (Fig. 3E).
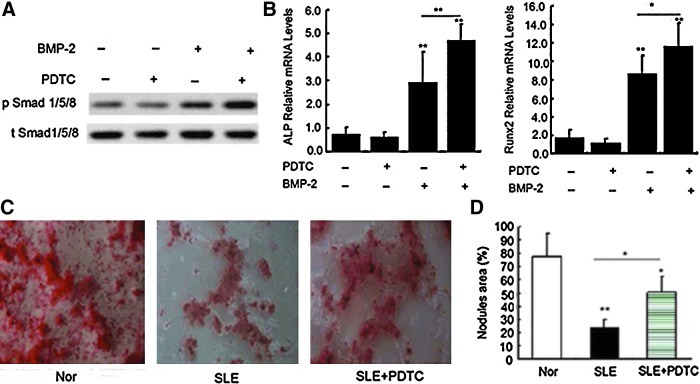
PDTC restored BMP-2-induced osteoblastic differentiation of BMMSCs from SLE patients
Next, we investigated whether inhibiting the activation of NF-κB could restore the osteoblastic differentiation of BMMSCs in SLE patients. Although PDTC alone had no effect on the phosphorylation of Smad1/5/8, it increased the phosphorylation of Smad1/5/8 induced by BMP-2 in the absence of exogenous TNF (Fig. 4A). Similarly, PDTC enhanced the levels of Runx2 and ALP mRNA treated with BMP-2 for 48 h (Fig. 4B). After exposure to osteogenic stimulus for 21 days, PDTC increased the mineralization of BMMSCs from SLE patients, although it did not restore it to the level of normal cells (Fig. 4D).
FIG. 4.
Effect of PDTC on BMP-2 induced osteogenic differentiation in SLE-BMMSCs. (A) Western blotting analysis of phosphorylated Smad1/5/8 stimulated with PDTC (100 nM) and/or BMP-2 (100 ng/mL). Cells were starved for 24 h and treated with PDTC for 2 h, followed by BMP-2 for 30 min. (B) qRT-PCR analysis of ALP and Runx2 mRNA levels. Cells were treated with PDTC and/or BMP-2 for 48 h. *P<0.05 and **P<0.01 versus controls or between the indicated groups (n=3). (C) Alizarin Red S staining of SLE-BMMSCs cultured in the OMB for 21 days in the presence or absence of PDTC. (D) Quantity analysis of mineralization area. *P<0.05 and **P<0.01 versus normals or between the indicated groups (n=3). Nor, normal controls; SLE, systemic lupus erythematosus.
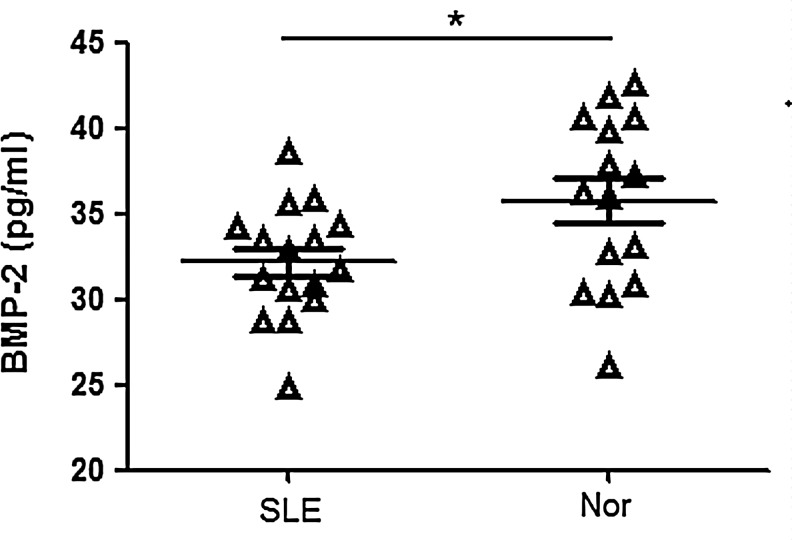
The circulating levels of BMP-2 in patients with SLE
BMP-2 is secreted by osteoblasts to bone matrix and also is one of the markers of osteoblastogenesis bone formation. Although we hardly detected BMP-2 in the BM of SLE patients, the level of BMP-2 in the serum of the patients (n=16) was lower than normal controls (n=15), suggesting a decreased osteoblastogenesis in SLE patients (Fig. 5).
FIG. 5.
The circulating levels of BMP-2 in patients with SLE. *P<0.05 SLE (n=16) versus normal controls (n=15). Nor, normal controls; SLE, systemic lupus erythematosus.
Discussion
BMMSCs from SLE patients demonstrated early signs of senescence [34,35]. SLE-BMMSCs were also observed to grow slower than the normal controls in ordinary cultural medium [27,34]. However, in this study, we did not find the difference in the proliferation rates between the 2 groups in the OMB. Different from the ordinary cultural medium, the numbers of the BMMSCs in OMB decreased after about 14 days, suggesting an enhanced trend to differentiation. The growth curve excluded the effect of senescence on the osteoblastic differentiation.
Both patients and normal BMMSCs cultured in ordinary medium showed extremely low levels of all the osteogenic markers, confirming that BMMSCs, even for a long period of time in culture, cannot differentiate into osteoblasts without osteogenic stimulation [36]. After stimulating with BMP-2 for 2 days, the mRNA levels of ALP, ColIIA2, OCN, and Runx2 in BMMSCs were increased. Besides, the mRNA levels of aforementioned genes in BMMSCs from SLE patients were lower than that from normal controls except for OCN. The ALP activity (at day 14) and the mineralized level (at day 21) were also lower in SLE patients compared with normal subjects. All the results mentioned above indicated there was a defective capacity of SLE-BMMSCs to differentiate into osteoblasts. OCN, one of the later markers of osteoblast differentiation compared with ALP and Runx2, was reported to be induced at the third day on MC3T3 cells [37] or at the sixth day on RD-C6 cells [38] induced by BMP-2. Therefore, the difference in OCN expression between BMMSCs from SLE patients and normal controls may not be detected at day 2 of culture.
Our results differ from findings by Nie et al., which suggested a similar ability of MSCs differentiating into osteoblasts between SLE patients and healthy subjects [34]. The underlying reasons responsible for this discrepancy remains unclear, but several possibilities might exist: since MSCs in vitro have been recognized to represent heterogeneous group of progenitors with different self-renewal properties [39], MSCs of different passages may have distinct capacity of differentiation [40]; moreover [41], the osteoblastic differentiation may change with the alternations of cytokines in the osteoblastic medium [42]; some medications, such as glucocorticoid (GC) [43,44], methotrexate, and warfarin may affect the osteoblastic capacity of MSCs [45]; at last, in the study of Nie et al. [34] the quantitative analysis of Alizarin Red S staining had not been measured.
In this study, cDNA microarray analysis revealed 19 differentially expressed genes in BMP/TGF-β signaling pathway. Among them, 18 genes were downregulated, including Smad1, Smad5, BMPRIA, and Id1 (inhibitor of differentiation or inhibitor of DNA binding-1). Moreover, the lower level of Smad1/5/8 phosphorylation and the decreased complex of Smad1/5/8 with Smad4 confirmed the decreased BMP/smad signaling pathway in SLE-BMMSCs.
Several possible mechanisms may be responsible for the reduction of Smad1/5/8 phosphorylation: First, the inactive status of BMPR (BMPR I and BMPR II) because of the weak stimulation of BMPs in BM or the intrinsic defect of the receptors; second, the regulation of cytokines or growth factors that directly inhibits the Smad1/5/8 phosphorylation. We found in this study that the protein level of BMPR-IA did not change as analyzed by western blot (Supplementary Fig. S1), although we found the level of BMP-2 in the serum of SLE patients was lower than normal controls. It has been shown that many BMPs, including BMP2, BMP6, BMP7, and BMP9 may induce osteoblast lineage-specific differentiation of MSCs [46–48], and the total amount of BMP activity is more important than the activity of a specific BMP [49]; moreover, other BMPs may be able to compensate for the loss of one BMP [50]. Therefore, with the normal BMPR-IA, the total amount of BMPs in SLE-BMMSCs might remain unchanged. Thus, our data suggest that the change in BMP/Smad signaling pathway likely results from the function of other cytokines or growth factors in the BM.
TNF is a potent inflammatory cytokine that contributes to local and systemic bone loss in inflammatory bone diseases such as RA, periodontitis, and multiple myeloma, and in estrogen deficiency [51–54]. It is now well known that TNF can induce osteoclastogenesis both in vivo and in vitro. However, the effect of TNF on the differentiation or maturation of osteoblast and their mechanisms remain unresolved: TNF may enhance the osteoblastic differentiation through NF-κB pathway [13,14], or it activates SAPK/JNK, ERK1/2, and Ras/Rho-MAPK signaling pathway to inhibit the spontaneous or BMP-2nduced osteoblastic differentiation [18,19]. We found here that TNF inhibited BMP-2-induced osteoblastic differentiation of BMMSCs from normal controls. We further found that TNF inhibited the osteoblastic differentiation of BMMSCs by inactivating Smad1/5/8, and NF-κB inhibition restored the activation of Smad1/5/8. These data collectively indicate that the inhibitory effect of TNF is attributed to, at least in great part, the activation of NF-κB.
In the current study, we found that the NF-κB activity was increased while the BMP signaling induced by BMP-2 was inactive in BMMSCs from SLE patients. Importantly, NF-κB inhibitor, PDTC, restored the activation of BMP signaling and subsequent ostoblastic markers and mineralization of BMMSCs from SLE patients. Similar results were obtained in the addition of another NF-κB inhibitor, SN50 (Supplementary Fig. S2). It is interesting that PDTC failed to repair the osteogenic capacity of SLE-BMMSCs stimulated with dexamethasone (Dex), GP, and AA (Supplementary Fig. S3). These results suggest that an enhanced NF-κB activity by high level of TNF suppressed the BMP/Smad, rather than other osteogenesis-related pathways in SLE patients.
GC is used extensively for the treatment of SLE. In this study, we cannot eliminate the effect of GC received by these patients. It is reported that GC alters bone metabolism at the cellular and molecular levels not only by increasing osteoclastic action [55] but also by inhibiting osteoblastic growth and differentiation [56,57]. Dex suppressed the BMP-2 induced Smad1/5/8 activation in mouse myoblastic C2C12 cells in vitro [58]. We show here in the Supplementary Data that Dex, at the concentration of 0.1 and 1 μM, reduced the phosphorylation of Smad1/5/8 induced by BMP-2, but it had no any effect on the NF-κB activation (Supplementary Fig. S4). Therefore, theoretically, the NF-κB inhibitory treatment cannot restore the inactivation of pSmad1/5/8 induced by GC. From above, we indicate that in SLE patients, with the treatment of GC, the suppressed BMP signaling pathway is at least partially attributed to the activation of NF-κB.
The inhibitory mechanism of NF-κB to Smad1/5/8 is not fully elucidated. Inconsistent with our results, Yamazaki et al. did not observe an inhibitory effect of TNF activated NF-κB on the phosphorylation of Smad1, Smad5, and Smad8 or on the nuclear translocation of the Smad1-Smad4 complex. NF-κB inhibited Smad pathway by interfering with the DNA binding of Smad proteins to its target gene in MC3T3-E1 [33]. Although we could not explain this discrepancy, the discrepancy might depend on the stage of the osteoblast differentiation, the type of the cell, and the methods of the experiments and needed further investigation.
In conclusion, we have demonstrated an activated NF-κB activity and NF-κB inhibited BMP/Smad signaling pathways in BMMSCs derived from SLE patients. The activated NF-κB pathway inhibits BMP-2-induced osteogenic differentiation through downregulating Smad signaling. Thus, our results may suggest a new approach to increasing osteoblastic differentiation for the treatment of OP in SLE patients.
Supplementary Material
Acknowledgments
The authors thank Prof. Wanjun Chen, Mucosal Immunology Unit, Oral Infection and Immunity Branch, National Institute of Dental and Craniofacial Research, NIH, MD 20892, for his critical review of the article. The study was supported by the Major International (Regional) Joint Research Project (no. 81120108021), National Natural Science Foundation of China (no. 30972736, no. 81202358); Jiangsu Province Natural Science Foundation (BK2009034); and Jiangsu Province Kejiao Xingwei Program, Chinese National 115 Supporting Program (2008BAI 59B02).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Sinigaglia L. Varenna M. Binelli L. Zucchi F. Ghiringhella D. Gallazzi M. Limonta M. Zeni S. Fantini F. Determinants of bone mass in systemic lupus erythematosus: a cross sectional study on premenopausal women. J Rheumatol. 1999;26:1280–1284. [PubMed] [Google Scholar]
- 2.Kalla AA. Fataar AB. Jessop SJ. Bewerunge L. Loss of trabecular bone mineral density in systemic lupus erythematosus. Arthritis Rheum. 1993;36:1726–1734. doi: 10.1002/art.1780361212. [DOI] [PubMed] [Google Scholar]
- 3.Petri M. Musculoskeletal complications of systemic lupus erythematosus in the Hopkins Lupus Cohort: an update. Arthritis Care Res. 1995;8:137–145. doi: 10.1002/art.1790080305. [DOI] [PubMed] [Google Scholar]
- 4.Feldmann M. Brennan FM. Maini RN. Role of cytokines in rheumatoid arthritis. Annu Rev Immunol. 1996;14:397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- 5.Studnicka-Benke A. Steiner G. Petera P. Smolen JS. Tumour necrosis factor alpha and its soluble receptors parallel clinical disease and autoimmune activity in systemic lupus erythematosus. Br J Rheumatol. 1996;35:1067–1074. doi: 10.1093/rheumatology/35.11.1067. [DOI] [PubMed] [Google Scholar]
- 6.Aringer M. Smolen JS. SLE—complex cytokine effects in a complex autoimmune disease: tumor necrosis factor in systemic lupus erythematosus. Arthritis Res Ther. 2003;5:172–177. doi: 10.1186/ar770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li P. Schwarz EM. O'Keefe RJ. Ma L. Boyce BF. Xing L. RANK signaling is not required for TNFalpha-mediated increase in CD11(hi) osteoclast precursors but is essential for mature osteoclast formation in TNFalpha-mediated inflammatory arthritis. J Bone Miner Res. 2004;19:207–213. doi: 10.1359/JBMR.0301233. [DOI] [PubMed] [Google Scholar]
- 8.Takayanagi H. Ogasawara K. Hida S. Chiba T. Murata S. Sato K. Takaoka A. Yokochi T. Oda H, et al. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature. 2000;408:600–605. doi: 10.1038/35046102. [DOI] [PubMed] [Google Scholar]
- 9.Theill LE. Boyle WJ. Penninger JM. RANK-L and RANK: T cells, bone loss, and mammalian evolution. Annu Rev Immunol. 2002;20:795–823. doi: 10.1146/annurev.immunol.20.100301.064753. [DOI] [PubMed] [Google Scholar]
- 10.Suda T. Takahashi N. Udagawa N. Jimi E. Gillespie MT. Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999;20:345–357. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- 11.Rodan GA. Martin TJ. Role of osteoblasts in hormonal control of bone resorption—a hypothesis. Calcif Tissue Int. 1982;34:311. doi: 10.1007/BF02411258. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert LC. Rubin J. Nanes MS. The p55 TNF receptor mediates TNF inhibition of osteoblast differentiation independently of apoptosis. Am J Physiol Endocrinol Metab. 2005;288:E1011–E1018. doi: 10.1152/ajpendo.00534.2004. [DOI] [PubMed] [Google Scholar]
- 13.Hess K. Ushmorov A. Fiedler J. Brenner RE. Wirth T. TNFalpha promotes osteogenic differentiation of human mesenchymal stem cells by triggering the NF-kappaB signaling pathway. Bone. 2009;45:367–376. doi: 10.1016/j.bone.2009.04.252. [DOI] [PubMed] [Google Scholar]
- 14.Cho HH. Shin KK. Kim YJ. Song JS. Kim JM. Bae YC. Kim CD. Jung JS. NF-kappaB activation stimulates osteogenic differentiation of mesenchymal stem cells derived from human adipose tissue by increasing TAZ expression. J Cell Physiol. 2010;223:168–177. doi: 10.1002/jcp.22024. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert L. He X. Farmer P. Boden S. Kozlowski M. Rubin J. Nanes MS. Inhibition of osteoblast differentiation by tumor necrosis factor-alpha. Endocrinology. 2000;141:3956–3964. doi: 10.1210/endo.141.11.7739. [DOI] [PubMed] [Google Scholar]
- 16.Nanes MS. Tumor necrosis factor-alpha: molecular and cellular mechanisms in skeletal pathology. Gene. 2003;321:1–15. doi: 10.1016/s0378-1119(03)00841-2. [DOI] [PubMed] [Google Scholar]
- 17.Nakase T. Takaoka K. Masuhara K. Shimizu K. Yoshikawa H. Ochi T. Interleukin-1 beta enhances and tumor necrosis factor-alpha inhibits bone morphogenetic protein-2-induced alkaline phosphatase activity in MC3T3-E1 osteoblastic cells. Bone. 1997;21:17–21. doi: 10.1016/s8756-3282(97)00038-0. [DOI] [PubMed] [Google Scholar]
- 18.Mukai T. Otsuka F. Otani H. Yamashita M. Takasugi K. Inagaki K. Yamamura M. Makino H. TNF-alpha inhibits BMP-induced osteoblast differentiation through activating SAPK/JNK signaling. Biochem Biophys Res Commun. 2007;356:1004–1010. doi: 10.1016/j.bbrc.2007.03.099. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita M. Otsuka F. Mukai T. Otani H. Inagaki K. Miyoshi T. Goto J. Yamamura M. Makino H. Simvastatin antagonizes tumor necrosis factor-alpha inhibition of bone morphogenetic proteins-2-induced osteoblast differentiation by regulating Smad signaling and Ras/Rho-mitogen-activated protein kinase pathway. J Endocrinol. 2008;196:601–613. doi: 10.1677/JOE-07-0532. [DOI] [PubMed] [Google Scholar]
- 20.Prockop DJ. Marrow stromal cells as steam cells for nonhematopoietic tissues. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 21.Kaewsrichan J. Wongwitwichot P. Chandarajoti K. Chua KH. Ruszymah BH. Sequential induction of marrow stromal cells by FGF2 and BMP2 improves their growth and differentiation potential in vivo. Arch Oral Biol. 2011;56:90–101. doi: 10.1016/j.archoralbio.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Sun L. Akiyama K. Zhang H. Yamaza T. Hou Y. Zhao S. Xu T. Le A. Shi S. Mesenchymal stem cell transplantation reverses multiorgan dysfunction in systemic lupus erythematosus mice and humans. Stem Cells. 2009;27:1421–1432. doi: 10.1002/stem.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddi AH. Bone morphogenetic proteins: an unconventional approach to isolation of first mammalian morphogens. Cytokine Growth Factor Rev. 1997;8:11–20. doi: 10.1016/s1359-6101(96)00049-4. [DOI] [PubMed] [Google Scholar]
- 24.Lieberman JR. Daluiski A. Einhorn TA. The role of growth factors in the repair of bone. Biology and clinical applications. J Bone Joint Surg Am. 2002;84-A:1032–1044. doi: 10.2106/00004623-200206000-00022. [DOI] [PubMed] [Google Scholar]
- 25.Tan EM. Cohen AS. Fries JF. Masi AT. McShane DJ. Rothfield NF. Schaller JG. Talal N. Winchester RJ. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 26.Schned ES. Glickstein SL. Doyle MA. Derivation of the SLEDAI. Arthritis Rheum. 1993;36:877–878. doi: 10.1002/art.1780360623. [DOI] [PubMed] [Google Scholar]
- 27.Sun LY. Zhang HY. Feng XB. Hou YY. Lu LW. Fan LM. Abnormality of bone marrow-derived mesenchymal stem cells in patients with systemic lupus erythematosus. Lupus. 2007;16:121–128. doi: 10.1177/0961203306075793. [DOI] [PubMed] [Google Scholar]
- 28.Guo Y. Guo H. Zhang L. Xie H. Zhao X. Wang F. Li Z. Wang Y. Ma S, et al. Genomic analysis of anti-hepatitis B virus (HBV) activity by small interfering RNA and lamivudine in stable HBV-producing cells. J Virol. 2005;79:14392–14403. doi: 10.1128/JVI.79.22.14392-14403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Day TF. Guo X. Garrett-Beal L. Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 30.Bai S. Kopan R. Zou W. Hilton MJ. Ong CT. Long F. Ross FP. Teitelbaum SL. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem. 2008;283:6509–6518. doi: 10.1074/jbc.M707000200. [DOI] [PubMed] [Google Scholar]
- 31.Mishina Y. Starbuck MW. Gentile MA. Fukuda T. Kasparcova V. Seedor JG. Hanks MC. Amling M. Pinero GJ. Harada S. Behringer RR. Bone morphogenetic protein type IA receptor signaling regulates postnatal osteoblast function and bone remodeling. J Biol Chem. 2004;279:27560–27566. doi: 10.1074/jbc.M404222200. [DOI] [PubMed] [Google Scholar]
- 32.Yu K. Xu J. Liu Z. Sosic D. Shao J. Olson EN. Towler DA. Ornitz DM. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 33.Yamazaki M. Fukushima H. Shin M. Katagiri T. Doi T. Takahashi T. Jimi E. Tumor necrosis factor alpha represses bone morphogenetic protein (BMP) signaling by interfering with the DNA binding of Smads through the activation of NF-kappaB. J Biol Chem. 2009;284:35987–35995. doi: 10.1074/jbc.M109.070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nie Y. Lau CS. Lie AKW. Chan GCF. Mok MY. Defective phenotype of mesenchymal stem cells in patients with systemic lupus erythematosus. Lupus. 2010;19:850–859. doi: 10.1177/0961203309361482. [DOI] [PubMed] [Google Scholar]
- 35.Li X. Liu L. Meng D. Wang D. Zhang J. Shi D. Liu H. Xu H. Lu L. Sun L. Enhanced apoptosis and senescence of bone-marrow-derived mesenchymal stem cells in patients with systemic lupus erythematosus. Stem Cells Dev. 2012;21:2387–2394. doi: 10.1089/scd.2011.0447. [DOI] [PubMed] [Google Scholar]
- 36.Kulterer B. Friedl G. Jandrositz A. Sanchez-Cabo F. Prokesch A. Paar C. Scheideler M. Windhager R. Preisegger KH. Trajanoski Z. Gene expression profiling of human mesenchymal stem cells derived from bone marrow during expansion and osteoblast differentiation. BMC Genomics. 2007;8:70. doi: 10.1186/1471-2164-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamurovic N. Cappellen D. Rohner D. Susa M. Coordinated activation of Notch, Wnt, and transforming growth factor-beta signaling pathways in bone morphogenic protein 2-induced osteogenesis Notch-Target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J Biol Chem. 2004;279:37704–37715. doi: 10.1074/jbc.M403813200. [DOI] [PubMed] [Google Scholar]
- 38.Liu T. Gao Y. Sakamoto K. Minamizato T. Furukawa K. Tsukazaki T. Shibata Y. Bessho K. Komori T. Yamaguchi A. BMP-2 promotes differentiation of osteoblasts and chondroblasts in Runx2-deficient cell lines. J Cell Physiol. 2007;211:728–735. doi: 10.1002/jcp.20988. [DOI] [PubMed] [Google Scholar]
- 39.Dominici M. Paolucci P. Conte P. Horwitz EM. Heterogeneity of multipotent mesenchymal stromal cells: from stromal cells to stem cells and vice versa. Transplantation. 2009;87:S36–S42. doi: 10.1097/TP.0b013e3181a283ee. [DOI] [PubMed] [Google Scholar]
- 40.Kasper G. Mao L. Geissler S. Draycheva A. Trippens J. Kuhnisch J. Tschirschmann M. Kaspar K. Perka C. Duda GN. Klose J. Insights into mesenchymal stem cell aging: involvement of antioxidant defense and actin cytoskeleton. Stem Cells. 2009;27:1288–1297. doi: 10.1002/stem.49. [DOI] [PubMed] [Google Scholar]
- 41.Uehara R. Suzuli Y. Ichikawa Y. Methotrexate (MTX) inhibis osteoblastic differentiation in vitro: possible mechanisim of MTX osteopathy. J Rheumatol. 2001;28:251–256. [PubMed] [Google Scholar]
- 42.Maegawa N. Kawamura K. Hirose M. Yajima H. Takakura Y. Ohgushi H. Enhancement of osteoblastic differentiation of mesenchymal stromal cells cultured by selective combination of bone morphogenetic protein-2 (BMP-2) and fibroblast growth factor-2 (FGF-2) J Tissue Eng Regen Med. 2007;1:306–313. doi: 10.1002/term.41. [DOI] [PubMed] [Google Scholar]
- 43.Leclerc N. Luppen CA. Ho VV. Nagpal S. Hacia JG. Smith E. Frenkel B. Gene expression profiling of glucocorticoid-inhibited osteoblasts. J Mol Endocrinol. 2004;33:175–193. doi: 10.1677/jme.0.0330175. [DOI] [PubMed] [Google Scholar]
- 44.Vidal NO. Brandstrom H. Jonsson KB. Ohlsson C. Osteoprotegerin mRNA is expressed in primary human osteoblast-like cells: down-regulation by glucocorticoids. J Endocrinol. 1998;159:191–195. doi: 10.1677/joe.0.1590191. [DOI] [PubMed] [Google Scholar]
- 45.Menon RK. Gill DS. Thomas M. Kernoff PB. Dandona P. Impaired carboxylation of osteocalcin in warfarin-treated patients. J Clin Endocrinol Metab. 1987;64:59–61. doi: 10.1210/jcem-64-1-59. [DOI] [PubMed] [Google Scholar]
- 46.Cheng H. Jiang W. Phillips FM. Haydon RC. Peng Y. Zhou L. Luu HH. An N. Breyer B, et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs) J Bone Joint Surg Am. 2003;85-A:1544–1552. doi: 10.2106/00004623-200308000-00017. [DOI] [PubMed] [Google Scholar]
- 47.Lavery K. Swain P. Falb D. Alaoui-Ismaili MH. BMP-2/4 and BMP-6/7 differentially utilize cell surface receptors to induce osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. J Biol Chem. 2008;283:20948–20958. doi: 10.1074/jbc.M800850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo J. Tang M. Huang J. He BC. Gao JL. Chen L. Zuo GW. Zhang W. Luo Q, et al. TGFbeta/BMP type I receptors ALK1 and ALK2 are essential for BMP9-induced osteogenic signaling in mesenchymal stem cells. J Biol Chem. 2010;285:29588–29598. doi: 10.1074/jbc.M110.130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ten Dijke P. Bone morphogenetic protein signal transduction in bone. Curr Med Res Opin. 2006;1(22 Suppl):S7–S11. doi: 10.1185/030079906X80576. [DOI] [PubMed] [Google Scholar]
- 50.Solloway MJ. Dudley AT. Bikoff EK. Lyons KM. Hogan BL. Robertson EJ. Mice lacking Bmp6 function. Dev Genet. 1998;22:321–339. doi: 10.1002/(SICI)1520-6408(1998)22:4<321::AID-DVG3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 51.Ralston SH. Russell RG. Gowen M. Estrogen inhibits release of tumor necrosis factor from peripheral blood mononuclear cells in postmenopausal women. J Bone Miner Res. 1990;5:983–988. doi: 10.1002/jbmr.5650050912. [DOI] [PubMed] [Google Scholar]
- 52.Pacifici R. Brown C. Puscheck E. Friedrich E. Slatopolsky E. Maggio D. McCracken R. Avioli LV. Effect of surgical menopause and estrogen replacement on cytokine release from human blood mononuclear cells. Proc Natl Acad Sci U S A. 1991;88:5134–5138. doi: 10.1073/pnas.88.12.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ammann P. Rizzoli R. Bonjour JP. Bourrin S. Meyer JM. Vassalli P. Garcia I. Transgenic mice expressing soluble tumor necrosis factor-receptor are protected against bone loss caused by estrogen deficiency. J Clin Invest. 1997;99:1699–16703. doi: 10.1172/JCI119333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li B. Shi M. Li J. Zhang H. Chen B. Chen L. Gao W. Giuliani N. Zhao RC. Elevated tumor necrosis factor-α suppresses TAZ expression and impairs osteogenic potential of Flk-1+ mesenchymal stem cells in patients with multiple myeloma. Stem Cells Dev. 2007;16:921–930. doi: 10.1089/scd.2007.0074. [DOI] [PubMed] [Google Scholar]
- 55.Kondo T. Kitazawa R. Yamaguchi A. Kitazawa S. Dexamethasone promotes osteoclastogenesis by inhibiting osteoprotegerin through multiple levels. J Cell Biochem. 2008;103:335–345. doi: 10.1002/jcb.21414. [DOI] [PubMed] [Google Scholar]
- 56.Rubin MR. Bilezikian JP. Clinical review 151: The role of parathyroid hormone in the pathogenesis of glucocorticoid-induced osteoporosis: a re-examination of the evidence. J Clin Endocrinol Metab. 2002;87:4033–4041. doi: 10.1210/jc.2002-012101. [DOI] [PubMed] [Google Scholar]
- 57.Weinstein RS. Jilka RL. Parfitt AM. Manolagas SC. Inhibition of osteoblastogenesis and promotion of apoptosis of osteoblasts and osteocytes by glucocorticoids. Potential mechanisms of their deleterious effects on bone. J Clin Invest. 1998;102:274–282. doi: 10.1172/JCI2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsumoto Y. Otsuka F. Takano M. Mukai T. Yamanaka R. Takeda M. Miyoshi T. Inagaki K. Sada KE. Makino H. Estrogen and glucocorticoid regulate osteoblast differentiation through the interaction of bone morphogenetic protein-2 and tumor necrosis factor-alpha in C2C12 cells. Mol Cell Endocrinol. 2010;325:118–127. doi: 10.1016/j.mce.2010.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.