SUMMARY
LDL-receptor-related protein 6 (LRP6), alongside Frizzled receptors, transduces Wnt signaling across the plasma membrane. The LRP6 ectodomain comprises four tandem β-propeller–EGF-like domain (PE) pairs which harbor binding sites for Wnt morphogens and their antagonists including Dickkopf 1 (Dkk1). To understand how these multiple interactions are integrated, we combined crystallographic analysis of the third and fourth PE pairs with electron microscopy (EM) to determine the complete ectodomain structure. An extensive inter-pair interface, conserved for the first-to-second and third-to-fourth PE interactions, contributes to a compact platform-like architecture, which is disrupted by mutations implicated in developmental diseases. EM reconstruction of the LRP6 platform bound to chaperone Mesd exemplifies a binding mode spanning PE pairs. Cellular and binding assays identify overlapping Wnt3a- and Dkk1-binding surfaces on the third PE pair, consistent with steric competition, but also suggest a model in which the platform structure supports an interplay of ligands through multiple interaction sites.
Keywords: LRP6, Wnt, Dkk1, structure, Mesd
INTRODUCTION
The Wnt family of secreted signaling molecules control animal development and homeostasis, whilst dysregulation of Wnt signaling causes many human diseases including cancer and osteoporosis (Logan and Nusse, 2004; MacDonald et al., 2009). Low-density lipoprotein receptor (LDLR)-related protein (LRP) 6 (LRP6), a member of the LRP family of type 1 transmembrane proteins, plays a central role in the canonical Wnt signaling pathway (He et al., 2004). LRP6 serves, together with seven-pass transmembrane receptors of the Frizzled (Fz) family, as a co-receptor for Wnt ligands, which induce the formation of the Fz–LRP6 complex and the subsequent activation of the Wnt downstream signaling cascade (Clevers, 2006; MacDonald et al., 2009). LRP6 is also the receptor for several Wnt antagonists, such as Dickkopf 1 (Dkk1) and Sclerostin (SOST), which prevent Wnt–LRP6 binding and thus Fz–LRP6 complex formation (Bafico et al., 2001; Li et al., 2005a; Mao et al., 2001; Semenov et al., 2005; Semenov et al., 2001). In addition, LRP6 biogenesis requires a specific, secretory pathway, chaperone Mesd (Mesoderm development), which is required for LRP6 maturation to the cell surface (Hsieh et al., 2003).
Within the LRP family, LRP6 and its close relative LRP5, plus their Drosophila homolog Arrow constitute a distinct subgroup that acts as Wnt co-receptors (Pinson et al., 2000; Tamai et al., 2000; Wehrli et al., 2000). Sequence analysis predicts that Arrow, LRP5 and LRP6 ectodomains share an architecture of four N-terminal Tyr-Trp-Thr-Asp (YWTD) β-propeller domains (P1–P4), each paired by a C-terminal epidermal growth factor (EGF)-like module (E1–E4), followed by three LDLR type A domains (L1–L3) before a transmembrane helix (Figures 1A and S1A). The four tandem YWTD β-propeller–EGF-like domain (PE) pairs (P1E1–P4E4) constitute most, if not all, of the functional recognition modules of the ectodomain, as they comprise all known Wnt- and antagonist-binding sites of LRP5 and LRP6 (He et al., 2004). A crystallographic study of a single prototypic PE pair from LDLR revealed the details of the six-bladed β-propeller structure and its tight interface with the C-terminal EGF domain (Jeon et al., 2001). However, little is known about how multiple PE pairs, which are a common feature among LRP proteins (Dieckmann et al., 2010), arrange spatially to form a functional ectodomain.
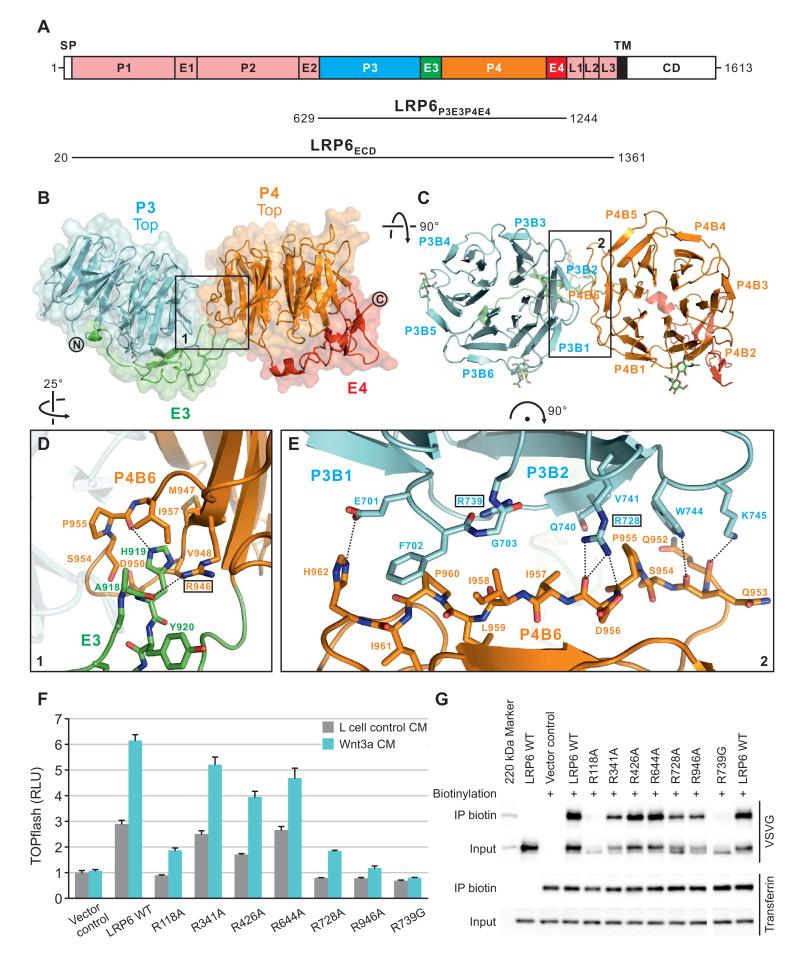
Figure 1. Crystal Structure of LRP6P3E3P4E4.
(A) Schematic domain organization of LRP6. SP, signal peptide; TM, transmembrane domain; CD, cytoplasmic domain. Constructs for crystallographic (LRP6P3E3P4E4) and electron microscopic (LRP6ECD) studies are shown. The domain color scheme is used for all figures unless otherwise stated.
(B and C) Cartoon representation of the crystal structure of LRP6P3E3P4E4 viewed from one side (B) and the top face (C) of the β-propellers. Disulfide bridges and N-linked glycans are shown in sticks. Blades of the β-propellers are numbered by convention.
(D and E) Detailed views of the boxed areas in (B) and (C) highlight contacts between E3 and P4 (D) and between P3 and P4 (E). Interface residues are shown labeled in sticks, and those chosen for mutagenesis boxed. Dashed lines indicate inter-pair hydrogen bonds or salt bridges.
(F) Mutations at the inter-pair interfaces of LRP6 affected signaling capabilities variously in Wnt-responsive TOPflash luciferase assay. RLU, relative light unit. Error bars represent standard deviations.
(G) LRP6 inter-pair interface mutants express at different levels on the cell surface as demonstrated by cell surface biotinylation. The endogenous transferrin receptor was used as a control for cell surface protein and labeling.
See also Figure S1.
Previous studies of LRP6 and LRP5 have hinted that the four tandem PE pairs could be divided into two functional units, P1E1P2E2 and P3E3P4E4. P1E1P2E2 has been identified as the primary binding domain for Wnt9b, whereas P3E3P4E4 favors Wnt3a (Bourhis et al., 2010) and Dkk1 binding (Mao et al., 2001), although there is evidence that P1E1P2E2 also contributes to Dkk1 binding (Binnerts et al., 2009; Bourhis et al., 2010; Zhang et al., 2004). Consistently experiments using antibodies that are specific to P1 and P3 of LRP6, respectively, have implied that LRP6 binding of Wnt1, Wnt9b and many other Wnts involves P1, while that of Wnt3a and Wnt3 involves P3 (Ettenberg et al., 2010; Gong et al., 2010).
In mammals, LRP5 and LRP6 have redundant, but also unique, functions during development and adult tissue homeostasis, with LRP6 playing a dominant role in embryogenesis (He et al., 2004). Mutations in LRP5 or LRP6 genes cause human diseases affecting bone accrual, eye development, cardiovascular and cerebral functions (Boyden et al., 2002; De Ferrari et al., 2007; Gong et al., 2001; Jiao et al., 2004; Little et al., 2002; Mani et al., 2007). Many of these mutations are single amino acid substitutions in the ectodomain of LRP5 or LRP6, resulting presumably in defects in receptor–ligand (agonist or antagonist) interactions and/or receptor maturation to the plasma membrane. LRP5/LRP6 antagonists are either directly involved in human diseases, as in the case of SOST (Balemans et al., 2002; Brunkow et al., 2001) or are promising therapeutic targets as in the case of both DKK1 and SOST (MacDonald et al., 2009; Mason and Williams, 2010). Therefore understanding of the LRP6/LRP5 ectodomain structure and its features determining the binding specificity of Wnts and antagonists is of profound biological and medical significance.
Here we report a high-resolution crystal structure of a portion of the human LRP6 ectodomain comprising the third and fourth PE pairs, P3E3P4E4, which are the principal Wnt3a- and Dkk1-binding region, and a negative stain electron microscopy reconstruction of the entire human LRP6 ectodomain. We define key elements of the interface between P3E3 and P4E4, which appear to be conserved for the interface between P1E1 and P2E2, and demonstrate how these two pairs of tandem PE pairs are arranged spatially to form a compact ectodomain. We also find that this LRP6 ectodomain architecture permits Mesd to engage two PE pairs simultaneously in a pH-dependent manner. We combined structure-guided mutagenesis with Wnt3a and Dkk1 binding assays to map common binding surfaces on P3 and flanking contributions from P4 resulting in an integrated model for LRP5/LRP6 binding to agonists and antagonists. These results provide a framework in which to interpret the functional consequences of LRP5/LRP6 mutations associated with human diseases.
RESULTS
Crystal Structure of LRP6P3E3P4E4
A construct comprising the membrane-proximal two YWTD β-propeller–EGF domain pairs of human LRP6 (LRP6P3E3P4E4; Figure 1A) was co-expressed in human embryonic kidney (HEK) 293 cells with the LRP-specific chaperone Mesd. Immobilized metal affinity chromatography yielded purified LRP6P3E3P4E4 (Figure S1B). The isolated protein inhibited Wnt3a activity in the standard Wnt-responsive TOPflash luciferase reporter assay (which measures transcriptional activation of the luciferase reporter via multimerized Wnt-responsive elements), presumably by competing with endogenous LRP6 for Wnt3a binding (Figure S1C). We determined the crystal structure of LRP6P3E3P4E4 to a resolution of 1.9 Å (Table 1) and revealed the two pairs, P3E3 and P4E4, abut side by side sharing an extensive interface (Figures 1B and 1C). The ‘top’ faces of the YWTD β-propeller domains are juxtaposed to provide a continuous, curved surface while the EGF-like domains pack against the ‘bottom’ faces of the β-propellers (Figures 1B and 1C).
Table 1.
Data Collection and Refinement Statistics
| Data Collection | |
|---|---|
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 169.8, 42.7, 119.9 |
| α, β, γ (°) | 90.0, 97.5, 90.0 |
| Resolution (Å) | 30.0–1.9 (2.0–1.9) |
| R merge | 0.098 (0.555) |
| I/σI | 9.4 (2.0) |
| Completeness (%) | 98.0 (88.4) |
| Redundancy | 4.1 (3.0) |
|
| |
| Refinement | |
|
| |
| Resolution (Å) | 30.0–1.9 (1.95–1.90) |
| Number of reflections | 66,493 (3924) |
| Rwork/Rfree | 0.181/0.211 (0.306/0.309) |
| Number of atoms | |
| Protein | 4848 |
| Ligand/glycan/ion | 8/84/6 |
| Water | 437 |
| B-factors (Å2) | |
| Protein | 29.2 |
| Ligand/glycan/ion | 28.1/54.7/22.5 |
| Water | 38.5 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.099 |
The highest-resolution shell is shown in parentheses.
The structures of the individual β-propellers (P3 and P4) are typical of the YWTD class (Figure S1D). Six blades of four-stranded antiparallel β-sheets are symmetrically arranged around a central channel. The electron density suggests the presence of three Ca2+ ions, one bound in the central channel of each of the β-propellers plus a third on the surface of P3 (Figure S1E). P3 and P4 are structurally closely related (root mean square deviation (r.m.s.d.) of 1.19 Å for 221 Cα pairs), as is P3 to the YWTD β-propeller domains of LDLR (Jeon et al., 2001) (r.m.s.d. of 1.13 Å for 223 matching Cα atoms) and nidogen (Takagi et al., 2003) (r.m.s.d. of 1.13 Å for 221 equivalent Cα atoms). P4 superposes less well on these other examples of YWTD β-propellers (LDLR: r.m.s.d. of 1.35 Å for 211 Cα pairs; nidogen: r.m.s.d. of 1.34 Å for 203 Cα pairs) suggesting that P4 may have evolved from P3. The structures of E3 and E4 conform to the EGF-type fold. The overall architecture of the PE pair in both P3E3 and P4E4 is similar to that previously reported for LDLR (Figures 1B and S1F); the linker between β-propeller and EGF spans the base of the β-propeller, positioning the EGF domain to form a predominantly hydrophobic contact with the second and third blades (Figure S1G). This β-propeller–EGF contact features conserved aliphatic residues on the EGF domain (E3: Leu903 and Leu905, E4: Ile1217 and Leu1219, and LDLR: Leu659 and Leu661), however, the linker between P4 and E4 differs slightly in conformation from those in P3E3 and LDLR, forming an α-helix extending from the β-propeller so that the entrance to the central channel is exposed (Figure S1H).
The two PE pairs of LRP6P3E3P4E4 bury ~1200 Å2 of total solvent-accessible area at the inter-pair, P3E3–P4E4, interface. The shape complementarity (Sc) of the interface surfaces is high (0.714) (Figures 1B–1E). The majority of the buried area (~720 Å2) is contributed by a side-to-side contact between the β-propellers, blades 1 and 2 of P3 (P3B1 and P3B2) abutting the outermost β-strand of P4 blade 6 (P4B6) (Figures 1C and 1E). This elongated interface comprises a network of hydrophobic stacking interactions, hydrogen bonds and salt bridges, involving nine residues from P3 and eleven residues from P4. Notably, Arg728 (P3B2) and Asp956 (P4B6) form hydrogen bonds and a salt bridge at the core of the interface. A second arginine residue, Arg739 on P3B2, is buried at the interface, and stabilizes the local structure of P3 through a salt bridge and multiple hydrogen bonds (Figure 1E). The EGF–β-propeller interaction of E3–P4 also contributes significantly to the inter-pair interface (~500 Å2 buried; Sc 0.757), with a loop from E3 docking into a pocket between the outer two β-strands of P4B6: His919 at the apex of the loop participating in a network of hydrogen bonds and hydrophobic interactions that involve Arg946 on P4 (Figure 1D). In total the interactions between P3E3 and P4E4 suggest a relatively rigid and distinctive inter-pair arrangement.
To address the functional significance of the P3E3–P4E4 interface, we introduced single point mutations R728A, R739G (P3–P4 interaction) (Figure 1E) and R946A (E3–P4 interaction) (Figure 1D) into full-length LRP6. As detailed above, all three arginine residues are embedded within the interface, and are involved in multiple hydrogen bonds as well as hydrophobic stacking interactions. In the Wnt-responsive reporter assay, the wild-type (WT) LRP6 transfected alone activated the TOPflash reporter, likely due in part, to its activation by the endogenous Wnt proteins produced in these HEK293T cells, and LRP6 could be further stimulated by the exogenously added Wnt3a conditioned medium (CM) (Figure 1F), as has been documented extensively. Consistent with the structural roles of these residues we found that all three mutant receptors showed loss-of-function when expressed alone or stimulated by Wnt3a (Figure 1F). This was fully attributable to severely reduced levels of these LRP6 mutant proteins on the cell surface likely due to misfolding of the receptor, as demonstrated by cell surface protein biotinylation (Figure 1G). We note that WT LRP6 and mutants migrated in gel electrophoresis as two discrete bands, with the predominant upper slower migrating one being the biotinylated LRP6 species on the cell surface (Figure 1G), as we have previously observed (Semenov and He, 2006).
We extended our mutagenesis study to assess whether the architecture of the P3E3–P4E4 interface was conserved among other tandem PE pairs. Sequence alignments indicate that Arg728 (which our results show has a central structural role in the P3–P4 interface) is invariably conserved at the equivalent residue position in all tandem PE pairs found in Arrow and each vertebrate LRP5 and LRP6 (Figure S1A). In addition, Arg946, which we find makes crucial contributions to the E3–P4 interface, is also well conserved (Figure S1A). A simple modeling exercise in which the side chains of the P3E3–P4E4 interface are replaced with those for the equivalent residues in LRP6 P1E1–P2E2 suggests that Arg118 may play a similar role to Arg728 in stabilizing the propeller–propeller interface (Figure S1I). Indeed, we found that the R118A mutation prevented LRP6 cell surface expression and Wnt signaling (Figures 1F and 1G). Generally, the residues of the predicted P1E1–P2E2 interface generate a broadly compatible match in charge and hydrophobicity when substituted into the architecture of the P3E3–P4E4 interface. Interestingly, in the context of the predicted P1E1–P2E2 interface Arg341 (the equivalent of Arg946) no longer plays a pivotal role in a hydrogen bond network (Figure S1I), and, indeed, the R341A mutation exhibited a relatively minor effect on cell surface expression and Wnt signaling (Figures 1F and 1G). In contrast, the residues of P2E2 and P3E3 do not generate a convincing interface when simply juxtaposed in the manner of the P3E3–P4E4 structure and the R426A mutation (which is analogous to the R728A mutation that affected the P3–P4 interface) had a milder effect on the folding/trafficking of LRP6 to the plasma membrane and Wnt signaling (Figures 1F and 1G). These findings are consistent with the possibility of a different mode of interaction between P2E2 and P3E3. The conclusion that the characteristics of the P2E2–P3E3 interface differ substantially from those of P3E3–P4E4 and P1E1–P2E2 is reinforced by quasi-normal levels of surface expression and Wnt signaling exhibited by the R644A mutant (equivalent to the E3–P4 mutation R946A), which behaved similarly to the WT LRP6 (Figures 1F and 1G).
Specific interactions have been previously demonstrated between Mesd (FLAG-tagged) and LRP5/6 (Hsieh et al., 2003). In particular, some mutations in P1 of LRP5 have been shown to exhibit compromised Mesd binding (Ai et al., 2005; Zhang et al., 2004). We therefore analyzed whether our LRP6 mutations affected its interaction with the chaperone Mesd using coimmunoprecipitation. WT LRP6, but not LRP6ΔN (which lacks the extracellular domain of the receptor), was precipitated by FLAG-tagged Mesd. All the LRP6 mutants showed Mesd binding comparable to the WT receptor, except for the one carrying R118A (the only P1 mutation of this panel) which did not co-precipitate with Mesd (Figure S1J). This is consistent with the previously reported effects of P1 mutations on Mesd binding (Ai et al., 2005; Zhang et al., 2004).
Thus the combined structural and functional analyses suggest that the LRP6 P3E3–P4E4 interface revealed by the crystal structure defines a generic architecture for units comprising tandem PE pairs (i.e. P1E1–P2E2 and P3E3–P4E4), but that the pair-pair interfaces between these units, as in P2E2–P3E3, may have different characteristics.
Negative Stain Electron Microscopy of the LRP6 Ectodomain
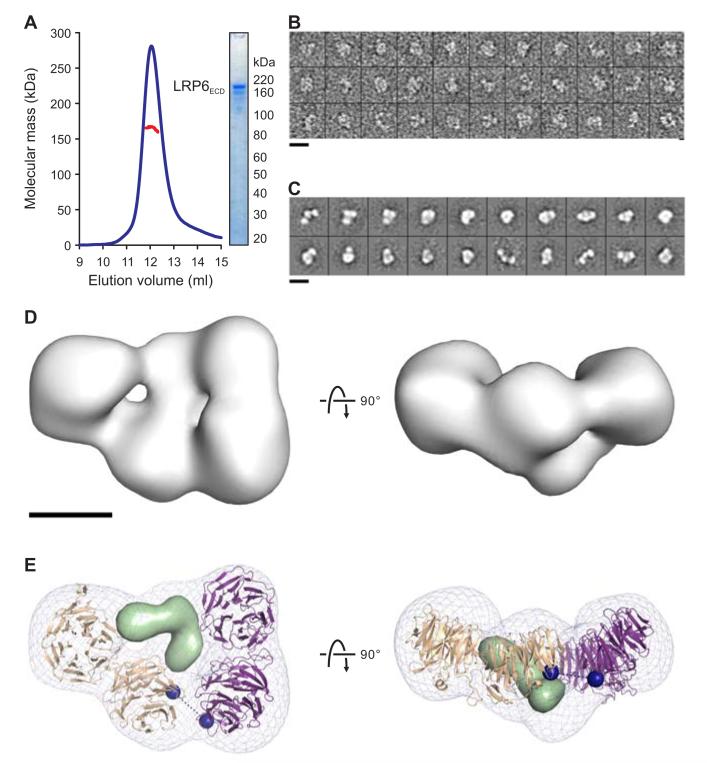
To understand how multiple tandem PE pairs arrange to provide the functional context for agonist and antagonist binding, we set out to generate insight into the structure of the entire LRP6 ectodomain (LRP6ECD; Figure 1A). Low yields of pure LRP6ECD made its structure determination by X-ray crystallography challenging. Therefore, we adopted a negative stain electron microscopic approach, which required significantly less material. Multi-angle light scattering (MALS) coupled with gel filtration chromatography indicated that LRP6ECD is monomeric (Figure 2A). LRP6ECD raw images (Figure 2B) were aligned using reference-free alignment and classified into groups. Two-dimensional (2D) class averages showed that the receptor is a compact horseshoe-like structure with less density in the middle (Figure 2C). To reduce the potential for model bias, a featureless Gaussian blob of dimensions similar to the 2D class averages was used as an initial reference for the ab initio structure determination of LRP6ECD. The refinement converged on a reconstruction whose resolution was 25 Å, determined by the Fourier shell correlation cut-off of 0.5 (Figures 2D and S2A). The averages of the data obtained by angular refinement were identical to the reference-free averages, supporting the consistency of the structure (Figure S2C). The raw images, 2D reference-free aligned class averages and the reconstruction of LRP6ECD are all consistent with a skewed planar horseshoe-like structure with three sides, each approximately 100-Å long and 50-Å wide (Figure 2D). Four globular domains comprise the perimeter and less density occupies the central cavity. Three of the lobes are of similar dimensions (50 Å in diameter), while the fourth is slightly larger (60 Å).
Figure 2. LRP6ECD Is a Monomer With a Compact Fold.
(A) Multi-angle light scattering (MALS) indicates an experimental molecular mass (red line) of 165.2 ± 2.0 kDa for LRP6ECD (blue line; elution profile in 50 mM NaAc (pH 5.0), 150 mM NaCl, axis not shown) as observed by SDS–PAGE (inset) and in agreement with the calculated molecular mass for a monomer (169.9 kDa).
(B) Representative raw images of negatively stained LRP6ECD.
(C) Reference-free 2D class averages reveal a skewed horseshoe structure.
(D) Two orthogonal views of the 3D reconstruction of LRP6ECD (grey surface) at 25-Å resolution (Figure S2).
(E) A pseudo-atomic model for LRP6ECD consisting of the LRP6P3E3P4E4 crystal structure and a homology model for LRP6P1E1P2E2 were fit into the reconstruction (grey mesh), and refined as two rigid bodies. Two possible placements of the model, related by a 180° rotation interchanging the N- and C-termini were scored. Having equivalent correlation coefficients, the one shown here corresponds to the model that minimizes the distance linking the two rigid bodies. Black dashed line connects the two termini (blue spheres). Tandem PE pairs are shown as tan and purple ribbons. Density corresponding to a homology model of the three LDLR type A domains of LRP6 is shown as a light green surface. Scale bars for (B) and (C) (140 Å), and (D) and (E) (50 Å) are indicated.
See also Figure S2.
The high sequence identity between the PE pairs of LRP6, short linkers between consecutive domains and the extensive interface between successive PE pairs observed in our crystal structure informed an initial model for LRP6ECD. A homology model was constructed in which each inter-pair orientation duplicated that of P3E3–P4E4. We found that this simple exercise to create a model for the four PE pairs automatically generated a horseshoe shape. The initial square horseshoe model was docked into the LRP6ECD reconstruction, and refined as a single rigid body. The fit matched the contour of three out of the four PE pairs, leaving one pair out of density. Our mutational analysis, investigating the similarities between inter-pair interfaces, indicated that P1E1–P2E2, but not P2E2–P3E3, shares a similar architecture to P3E3–P4E4 in the crystal structure. Based on this experimental constraint, the docked horseshoe model was split and subjected to further real-space refinement as two rigid bodies (P1E1P2E2 and P3E3P4E4). Two rotations of the initial model differing by 180° were placed into both hands of the reconstruction. Given the pseudo-twofold symmetric domain organization of LRP6, we were unable to assign specific β-propellers to individual lobes of density. All models matched the EM density equally well and had real-space correlation coefficients of 0.87 (Figure 2E).
In addition to the density occupied by the four PE pairs, our map contains density at the center of the horseshoe that contacts multiple β-propellers and contributes to the overall compact architecture of the LRP6ECD structure. The final PE pair (P4E4) is succeeded by three C-terminal LDLR type A domains in LRP6ECD (Figure 1A). Due to the small size of these LDLR type A domains and the resolution of the reconstruction, we have not included them in our refinement; however, it is noteworthy that the size and shape of this additional density is consistent with the structure of three LDLR type A domains from LDLR (Rudenko et al., 2002). In the case of the LDLR, these domains are thought to act as a surrogate for substrates at low pH and we cannot preclude that our structure (determined at pH 5.0, see Experimental Procedures) may reflect an inactive conformation.
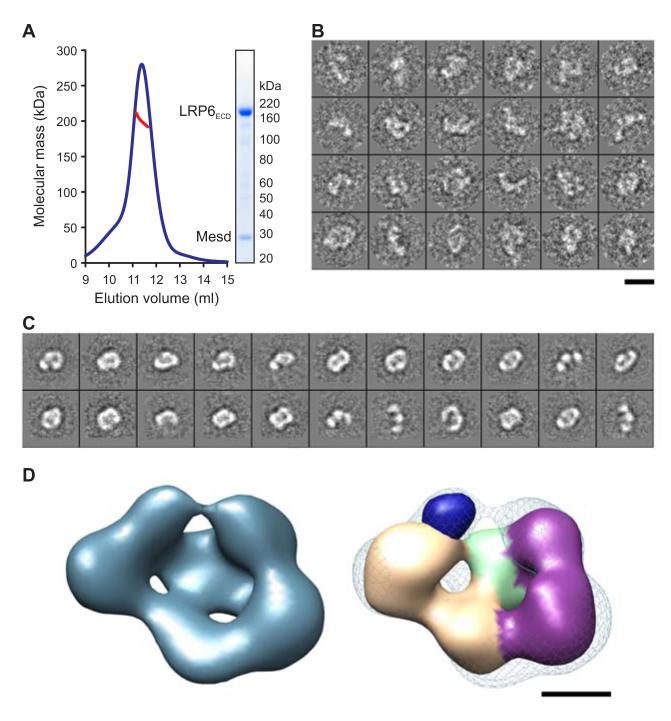
Structure of the LRP6ECD–Mesd Complex
Mesd binding to LRP6 antagonizes ligand binding and matures LRP6 during biogenesis (Hsieh et al., 2003; Li et al., 2005b). Interestingly, co-expression of LRP6ECD with Mesd resulted in a secreted complex (Figure 3A). MALS analysis indicates the LRP6ECD–Mesd complex to be a 1:1 heterodimer (Figure 3A). The structure of the LRP6ECD–Mesd complex was visualized by negative stain electron microscopy. Reference-free aligned class averages reveal a structure similar to that of the apo LRP6ECD (Figure 3B). The Gaussian blob used to initiate the refinement of the unbound LRP6 also served as a reference for ab initio structure determination of the complex. The resulting reconstruction converged at 26-Å resolution (Figures 3C and S3A) and as a ring-like structure with a less-dense interior. A comparison with the unbound LRP6ECD revealed an overall similar architecture. However, in addition to the four globular domains present around the perimeter of the apo structure, the complex possesses a fifth domain between the two ends of the horseshoe (Figure 3D). The size and shape of this additional density is consistent with recent structures of the compact core domain of Mesd (Collins and Hendrickson, 2011; Kohler et al., 2011). In agreement with our assignment of this density, Mesd has been shown to interact in isolation with tandem PE pairs P1E1P2E2 (Koduri and Blacklow, 2007) and P3E3P4E4 (Liu et al., 2009).
Figure 3. LRP6ECD–Mesd Complex.
(A) MALS analysis indicates an experimental molecular mass (red line) of 199.8 ± 5.4 kDa for the LRP6ECD–Mesd complex (blue line; elution profile in 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, axis not shown) as observed by SDS–PAGE (inset) and in agreement with the calculated molecular mass for a 1:1 LRP6ECD–Mesd complex (195.4 kDa).
(B) Raw images of the negatively stained LRP6ECD–Mesd complex.
(C) Representative class averages of the complex show a skewed ring-like structure of similar dimensions to LRP6ECD alone.
(D) 3D reconstruction of the LRP6ECD–Mesd complex at 26 Å resolution (blue surface, left panel) (Figure S3). The apo LRP6ECD reconstruction (density corresponding to tandem PE pairs colored tan and purple while density representing LDLR type A domains in light green) was real-space refined into density for the complex (blue mesh, right panel). The crystal structure of the mouse Mesd core domain (dark blue surface) was rendered at 25 Å resolution and manually placed into the remaining density of the reconstruction. Scale bars for (B) and (C) (140 Å), and (D) and (E) (50 Å) are indicated.
See also Figure S3.
Wnt3a Contacts the Top Face of β-propeller 3 of LRP6
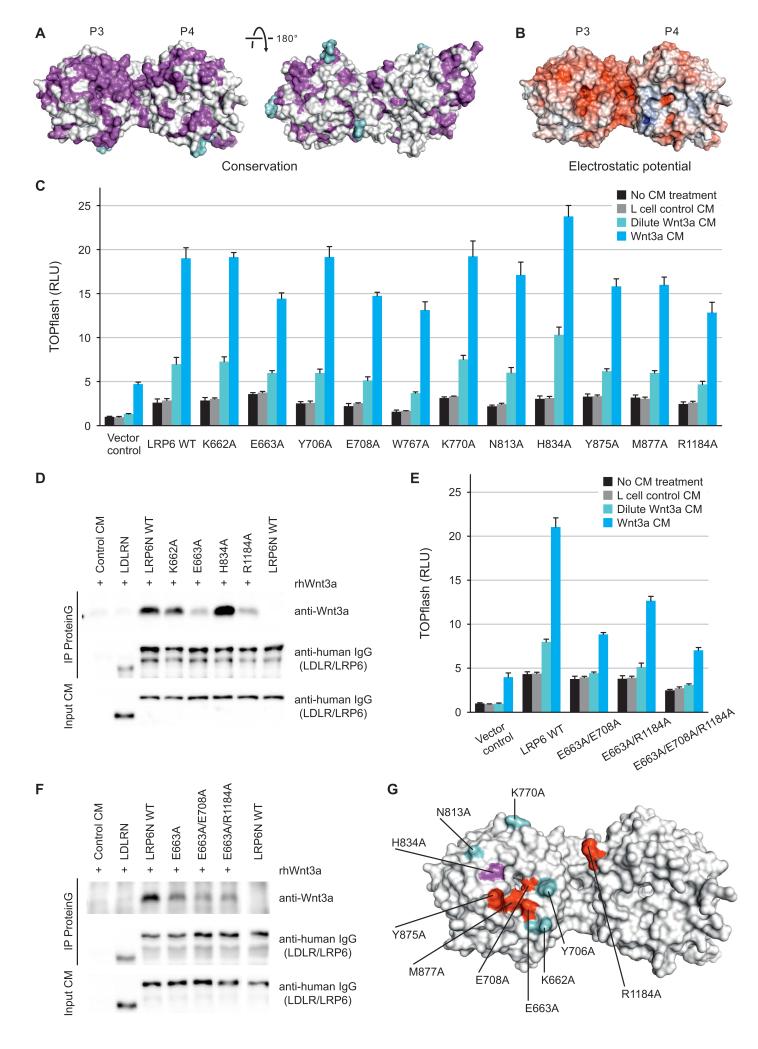
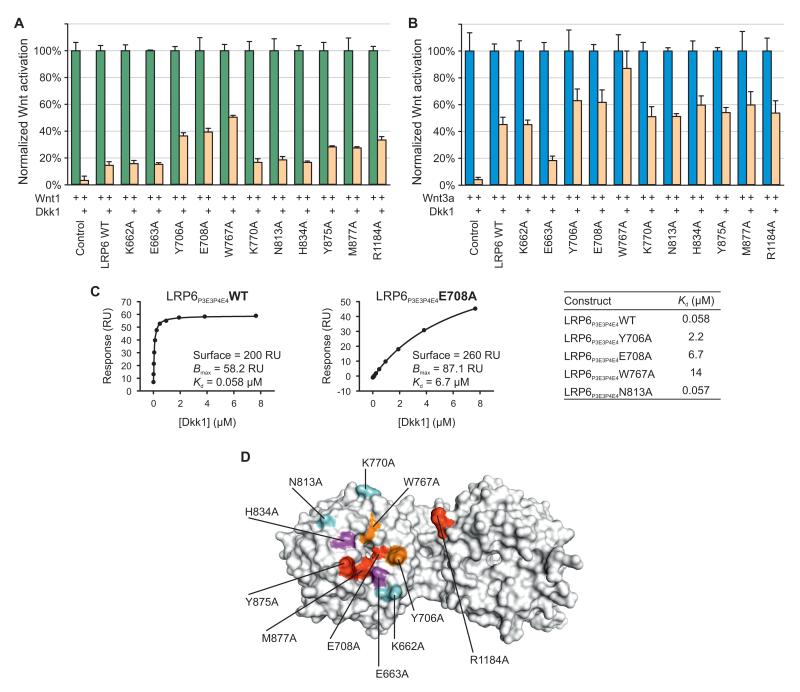
P3E3P4E4 harbors the primary binding site within the LRP6 ectodomain for Wnt3a (Bourhis et al., 2010), and consistent with this, we have demonstrated that purified LRP6P3E3P4E4 counteracts Wnt3a in the TOPflash reporter assay (Figure S1C). However, LRP6ECD has much stronger binding to Wnt3a than LRP6P3E3P4E4, implying that LRP6P1E1P2E2 influences Wnt3a-binding, directly or indirectly (Bourhis et al., 2010). Our crystal structure of LRP6P3E3P4E4 displays a cluster of conserved residues on the top face of P3 and an adjacent area on P4 (Figure 4A). This extensive surface is free from glycosylation (Figure 4A), and shows a pronounced acidic character (Figure 4B). It has previously been proposed that the surface of a YWTD β-propeller is a favored site for ligand interactions (Takagi et al., 2003), and the equivalent surface in LRP5 P3 has been implicated in Dkk1 inhibition (Zhang et al., 2004). How LRP6 engages its ligands, in particular Wnt proteins, has been unresolved. To test whether the top face of LRP6 P3 is important for interaction with Wnt3a, we generated a panel of LRP6 mutants, each containing a single alanine substitution, and examined their ability to respond to Wnt3a. We selected residues covering the P3 top face. In addition, we included in our panel Arg1184, a residue on P4, which our crystal structure shows exposed proximal to the top of P3.
Figure 4. Wnt3a Contacts the Top Face of β-propeller 3 of LRP6.
(A) LRP6P3E3P4E4 is color-coded by conservation from white (not conserved) to magenta (conserved) based on a sequence alignment across LRP5 and LRP6 in vertebrates with glycans shown in cyan. The left panel has the same view as Figure 1C.
(B) The solvent-accessible surface of LRP6P3E3P4E4 is color-coded by electrostatic potential from red (−9 kbT/ec; negatively charged) to blue (9 kbT/ec; positively charged).
(C) Responsiveness of LRP6 mutants to Wnt3a in luciferase assays. Error bars represent standard deviations.
(D) Binding between representative LRP6 ectodomain mutants and Wnt3a, as assayed by co-immunoprecipitations.
(E and F) Luciferase reporter (E) and co-immunoprecipitation (F) assays of LRP6 double or triple mutants corroborated the top face of LRP6 P3 as a Wnt3a-interacting interface. Error bars represent standard deviations.
(G) A Wnt3a-binding surface on LRP6 deduced from cellular and binding assays. Residues are colored according to mutational effects. Hypoactive mutations are in red whereas the hyperactive one in magenta. Residues in cyan when mutated had little or no effect on LRP6 signaling capacity.
See also Figure S4.
Compared to the WT LRP6, which when expressed alone or in response to two concentrations of Wnt3a CM showed dose-dependent activation of the TOPflash reporter, a number of LRP6 mutants exhibited diminished Wnt signalling activity, either alone or upon Wnt3a stimulation (Figure 4C). E663A, E708A, Y875A, M877A and R1184A mutants showed reduced capability to respond to Wnt3a despite similar cell surface expression compared to the WT LRP6 (Figure S4G). Glu663 is conserved across vertebrate LRP5 and LRP6 sequences and is surface exposed, forming a single hydrogen bond to Ser682 (Figure S4A), whereas Glu708 lies exposed on the rim of the central channel of the P3 β-propeller and hydrogen bonds to Arg751 (Figure S4B). Met877 also lies on the rim, making hydrophobic interactions with Phe836 and Trp850 (Figure S4C). Tyr875 protrudes particularly prominently from the P3 surface and forms a face-to-edge ring stacking interaction with Trp850 (Figure S4C). The effects of these four mutations are therefore consistent with a reduction of functional interaction with Wnt3a. W767A also showed a compromised Wnt3a response, but it also showed reduced cell-surface expression (Figures 4C and S4G). The aromatic side chain of Trp767 is sandwiched between a pair of arginine side chains (Arg751 and Arg792) (Figure S4D). Loss of these stacking interactions could somewhat destabilize the structure as well as change the potential interaction properties of P3, making interpretation of the W767A mutation result more complicated. In contrast to the above mutations, the H834A mutant showed enhanced Wnt3a responsiveness while its surface expression was comparable to the WT LRP6 (Figures 4C and S4G). From our LRP6P3E3P4E4 crystal structure, mutation of His834 to alanine will add to the already substantial hydrophobicity of a cluster of adjacent surface-exposed residues including Trp767, Leu810, Phe836 and Trp850, providing a potential enhancement of hydrophobic interactions with Wnt3a (Figure S4E). In this context it is noteworthy that Wnt3a has particularly hydrophobic characteristics due to its lipid modification (Willert et al., 2003), which in fact appears to be required for Wnt3a binding to LRP6 (Cong et al., 2004; Komekado et al., 2007). Thus His834, and possibly adjacent hydrophobic residues, seem to be involved in Wnt3a interaction in addition to Glu663, Glu708, Tyr875 and Met877. R1184A, the mutation in P4 (Figure S4F), also resulted in compromised LRP6 response to Wnt3a (Figure 4C), further suggesting the involvement of a relatively extensive surface in Wnt3a binding.
We then examined binding between several representative LRP6 mutants and Wnt3a using co-immunoprecipitation. We employed secreted LRP6N–IgG (Tamai et al., 2000), which is a tagged version of LRP6ECD, and its mutant derivatives. Wnt3a was specifically precipitated by WT LRP6N–IgG, but not LDLRN–IgG, a secreted and equivalently tagged extracellular domain of LDLR (Figure 4D). Wnt3a was precipitated equivalently by the K662A mutant, which behaved indistinguishably in signaling assays to the WT LRP6 (Figure 4C). However, E663A and R1184A mutants showed diminished binding to Wnt3a, whereas the H834A mutant showed enhanced binding (Figure 4D). Therefore these binding results show strong concordance with the functional assay for LRP6 activity.
We sought further evidence that the residues we identified on the top face of P3 are a part of a Wnt3a-interacting surface by combining representative single point mutations together. Double mutants E663A/R1184A, and E663A/E708A in particular, and a triple mutant E663A/E708A/R1184A showed significantly weaker Wnt signaling activity than the respective single mutants (Figure 4C and 4E), consistent with more pronounced defects in Wnt3a engagement. The residual LRP6 signaling of these double/triple mutants (Figure 4E) may be due to the contribution of either other extracellular regions including P1E1P2E2 (Bourhis et al., 2010; Gong et al., 2010) or the endogenous LRP6 in these cells to Wnt3a signaling. The two double mutants exhibited further diminished Wnt3a-binding capacity than the E663A single mutant (Figure 4F). Therefore our functional and binding assays implicate the top face of LRP6 P3 and the immediately neighboring P4 residue (Arg1184) in Wnt3a binding (Figure 4G).
Mesd binding to LRP6 has been reported to antagonize ligand binding to the receptor under certain experimental conditions (Li et al., 2005b). We therefore tested if the mutations on this β-propeller surface altered the interaction between LRP6 and Mesd. All LRP6 mutants tested showed behavior similar to the WT LRP6 in co-immunoprecipitation with Mesd (Figures S4G and S4H). Thus our data suggest that the top face of P3 contributes minimally to Mesd binding.
We next examined whether these P3 and P4 mutations affect Wnt1–LRP6 signaling. Wnt1 is less soluble and hardly available in CM compared to Wnt3a, we therefore performed co-transfection of Wnt1 with WT LRP6 or LRP6 mutants. We note that the WT LRP6 and LRP6 mutants behaved similarly regardless of whether Wnt3a was provided via co-transfection or as CM (data not shown). LRP6 mutants that diminished Wnt3a responsiveness, including single mutants E663A, E708A, and R1184A, and double mutants E663A/E708A and E663A/R1184A (Figures 4C and 4E), or the H834A mutant that enhanced Wnt3a signaling (Figure 4C), each had minimal effect on LRP6 responsiveness to Wnt1 (Figures S4I and S4J). The E663A/E708A/R1184A triple mutant, which exhibited drastically reduced Wnt3a responsiveness (Figure 4E), affected Wnt1 signaling only mildly (Figure S4J). These results are consistent with the suggestion that Wnt1 and Wnt3a engage LRP6 primarily via P1E1P2E2 and P3E3P4E4, respectively (Bourhis et al., 2010; Ettenberg et al., 2010; Gong et al., 2010), and mutations in P3 therefore mostly affect signaling by Wnt3a, but not Wnt1.
Dkk1 Binding to β-propeller 3 of LRP6 Is Involved in Inhibition of Wnt1 and Wnt3a
Alanine substitutions of several residues on the top face of LRP5 P3, via a homology model based on the LDLR β-propeller structure, have previously been examined, and reported to attenuate Dkk1 inhibition of Wnt1 through LRP5 (Zhang et al., 2004). This and other studies led to the suggestion that Dkk1 binds primarily to P3E3 (plus P4E4), but also to P1E1P2E2, of LRP5 and LRP6 (Bourhis et al., 2010; Zhang et al., 2004). We therefore examined how our panel of LRP6 single residue mutations in LRP6 P3 (and P4) affect Dkk1 binding and inhibition of LRP6. Importantly, because P3 mutations primarily affected Wnt3a, but not Wnt1 signaling through LRP6, we investigated whether Dkk1 inhibits Wnt3a and Wnt1 signaling through LRP6 via a common or different mechanism(s). Intriguingly, we found that although Dkk1 inhibited Wnt1 and Wnt3a with different efficacy, an identical set of LRP6 mutations were identified that reduced Dkk1 inhibition of both Wnt1 and Wnt3a, in particular Y706A, E708A and W767A (Figures 5A and 5B). In the LRP6P3E3P4E4 structure the aromatic residue Tyr706 is prominently exposed on the top face of P3. Glu708 and Trp767 participate in a network of interactions: Glu708 hydrogen bonds to Arg751 which provides one of a pair of arginine side chains which sandwich Trp767, as previously described. We note that because Dkk1 interacts with and inhibits LRP6 in a stoichiometric fashion, Dkk1 inhibition of Wnt signaling through overexpressed LRP6 or its mutants was less effective than that through the endogenous LRP6 (Figures 5A and 5B), as previously observed (Bafico et al., 2001; Mao et al., 2001; Semenov et al., 2001; Zhang et al., 2004).
Figure 5. Dkk1 Binding to β-propeller 3 of LRP6 Is Involved in Inhibition of Wnt1 and Wnt3a.
(A and B) Mutations on the top face of P3 affect Dkk1-mediated inhibition of Wnt1 (A) and Wnt3a (B) in luciferase reporter assays. RLU was set at 100% in the absence of Dkk1. Error bars represent standard deviations.
(C) SPR binding results between Dkk1 and LRP6P3E3P4E4 or its mutants are consistent with those from luciferase reporter assays. RU, response units.
(D) Wnt3a and Dkk1 have overlapping binding surfaces on LRP6. Residues important for the activity and binding of both Wnt3a and Dkk1 (red), of Wnt3a only (magenta), of Dkk1 only (orange) and of neither (cyan) are labeled.
See also Figure S5.
To assess directly the Dkk1-binding properties of the LRP6 mutants we purified a selected set of secreted LRP6P3E3P4E4 mutants and performed surface plasmon resonance (SPR) binding experiments to measure their Dkk1-binding affinities. WT LRP6P3E3P4E4 exhibited a dissociation constant (Kd) of 58 nM for Dkk1 (Figure 5C), generally consistent with a reported value (Bourhis et al., 2010). The N813A mutant showed essentially WT binding to Dkk1, whereas Y706A, E708A and W767A mutants displayed most diminished binding to Dkk1 (Figure 5C), demonstrating strong correlation between Dkk1 binding to P3E3P4E4 and Dkk1 inhibition of LRP6 signaling (Figures 5A and 5B). Although reduced cell surface expression of LRP6 W767A complicated somewhat the interpretation of this particular mutant, the less effective inhibition of LRP6 Y706A, E708A and W767A by Dkk1 correlates with reduced Dkk1 binding by these mutants in SPR. In combination, these results suggest these residues directly participate in (or are in the proximity of) the Dkk1 interaction surface.
The E663A mutation appeared to show enhanced Dkk1 inhibition of Wnt3a but not Wnt1 through LRP6 (Figures 5A and 5B). This likely reflects diminished Wnt3a binding to this particular mutant, making Dkk1 more effective for competing with Wnt3a for LRP6. Thus despite Wnt1 and Wnt3a binding primarily to different β-propeller domains, Dkk1 binding to a common top surface of P3 of LRP6 is important for its inhibition of both Wnt1 and Wnt3a. The shared Dkk1- and Wnt3a-binding surfaces on LRP6 P3 suggest antagonism involves direct steric inhibition (Figure 5D). We note however, that it remains possible that our mutations induce conformational changes in LRP6 that indirectly affect Wnt3a and/or Dkk1 binding.
Mapping Disease-Associated LRP5/6 Mutations
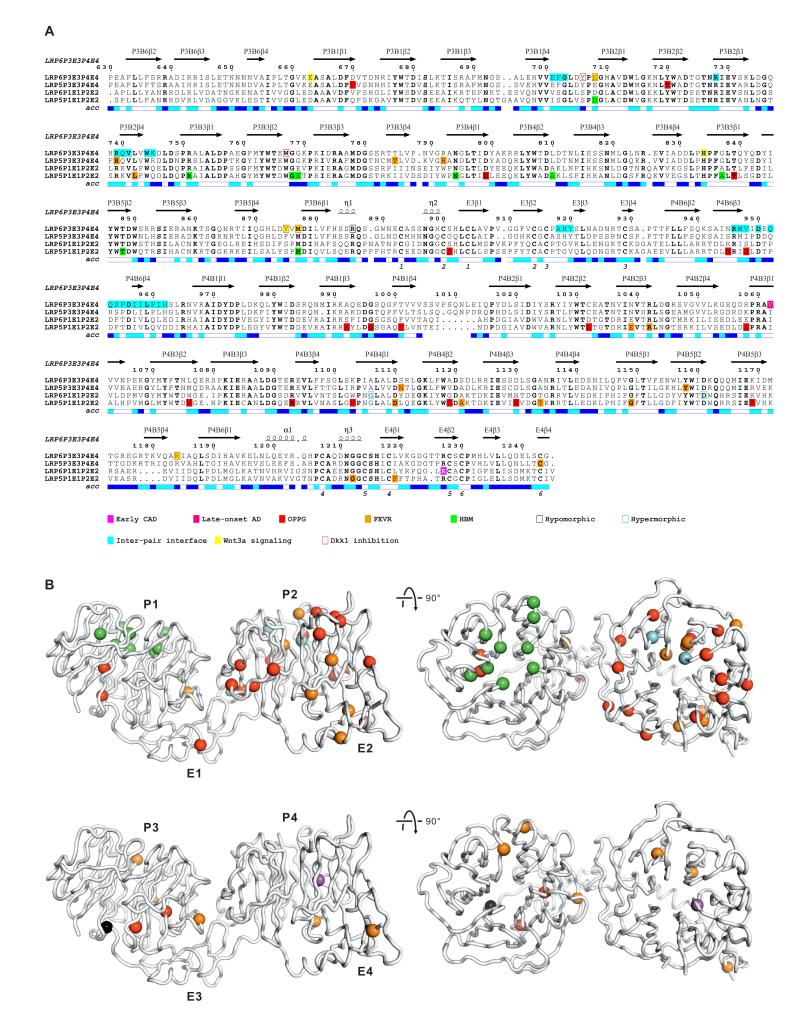
Mutations in LRP5 and LRP6 ectodomains have been implicated in a number of human diseases (Boyden et al., 2002; De Ferrari et al., 2007; Gong et al., 2001; Jiao et al., 2004; Little et al., 2002; Mani et al., 2007). To assess the molecular basis for these pathological effects, we used a sequence alignment of tandem PE pairs to relate human mutations, and mutations from mouse models, to our structural and functional results (Figure 6A). Thus we could map the combined data for human diseases (early coronary artery disease, early CAD; late-onset Alzheimer’s disease, late-onset AD; osteoporosis pseudoglioma syndrome, OPPG; familial exudative vitreoretinopathy, FEVR; high bone mass, HBM) and mouse models (Table S1) into the context of the LRP6P3E3P4E4 structure (Figure 6B).
Figure 6. Disease-Associated LRP Mutations.
(A) The amino acid sequence alignment of tandem PE pairs from human LRP6 (P1E1P2E2, residues 20–629; P3E3P4E4, residues 630–1244), LRP5 (P1E1P2E2, residues 32–642; P3E3P4E4, residues 643–1254). LRP5 and LRP6 residues implicated in diseases (Table S1) are indicated. Secondary structure assignments derived from LRP6P3E3P4E4 are displayed above the alignment. The relative accessibility of each residue is indicated below each block: white for buried (<10% exposed); cyan for moderately exposed (10–40% exposed); blue for exposed (>40% exposed). Residues at the inter-PE pair interface observed in the crystal structure and those important for Wnt3a and Dkk1 binding are shown.
(B) Cα traces of LRP6P3E3P4E4. Residues implicated in human diseases and mouse models from P1E1P2E2 and P3E3P4E4 are respectively mapped to the equivalent Cα atom positions of two copies of the crystal structure of LRP6P3E3P4E4, shown as spheres and color-coded as in the sequence alignment (A).
See also Table S1.
Many of the loss-of-function mutations affect residues that we predict are essential for the structural integrity of the individual domains or a single PE pair. These include residues that contribute to the hydrophobic cores of the β-propeller blades as well as cysteines involved in EGF domain disulfide bonds, or residues in their close proximity. Some mutations map to interpair interfaces, consistent with a precisely defined juxtaposition of the four PE pairs being a necessary requirement for LRP5/6 function. One such example is a R752G mutation in the third β-propeller of LRP5 attributed to the human hereditary eye disorder, FEVR (Jiao et al., 2004). Arg752 in LRP5 is equivalent to Arg739 in LRP6. Our LRP6P3E3P4E4 crystal structure positions this residue in a key stabilizing role at the P3E3–P4E4 interface, and our functional data for a R739G mutant in LRP6 indicate loss of Wnt signaling due to a reduction in cell surface expression, consistent with the disease mutation in LRP5 impacting on function by destabilizing the compact structure of the LRP5 ectodomain (see above; Figures 1F and 1G). In contrast to the loss-of-function mutations, which have been found in each of the LRP5 PE pairs, the LRP5 HBM mutations reported to interfere with antagonist binding (Table S1) map only to residue positions that are exposed on the top surfaces of β-propellers. This parallels our results that associate a Wnt3a- and Dkk1-binding site with the top surface of LRP6 P3 (Figure 4G and 5D). We note that the paralogy between LRP5 and LRP6 with regard to these HBM mutations is not only based on homology but has also been demonstrated by the transplantation of one of these mutations, G171V, into LRP6, which similarly reduced affinity to Dkk1 (Ai et al., 2005). In total, the available structural and functional data point to the LRP5/6 ectodomain serving as a compact platform for ligand binding.
DISCUSSION
Tandem YWTD β-propeller–EGF domain pairs are a feature of many members of the LRP family (Dieckmann et al., 2010; He et al., 2004; Springer, 1998). In the LRP6 ectodomain, four consecutive pairs interact with several classes of secreted ligands (agonists and antagonists of Wnt signaling), serving as a nexus for the integration of these disparate interactions into activation or inhibition of canonical Wnt signaling. Our crystal structure of the third and fourth PE pairs of human LRP6 defined a spatial architecture of tandem PE pairs and a critical interface between them. Mutational analysis of the inter-PE pair interfaces showed that this form of interface is shared between the first and second PE pairs and is required to stabilize tandem PE pairs. Intriguingly, however, the interface between P2E2 and P3E3 appeared to be governed in a distinct manner. Single-particle EM reconstructions supported this view and further revealed a compact LRP6 ectodomain, whether alone as a monomer or in complex with its chaperone Mesd. In combination these crystallographic, mutational and EM analyses are consistent with a platform-like LRP6 ectodomain constructed from tandem PE pairs. Although Wnt proteins may interact primarily with different PE pairs in LRP6, we speculate that a tray-like platform might be suited to permit various Wnt–LRP6 complexes to have similar spatial relationships to the plasma membrane and to Fz receptors, thereby mediating signaling by distinct Wnt proteins through a common mechanism of Wnt–Fz–LRP6 complexes. This platform model provides a template in which to explore the architecture of other members of the LRP family.
Our structure-guided mutagenesis studies map overlapping binding sites for Wnt3a and Dkk1 on the top face of LRP6 β-propeller 3. The binding sites for both ligands involve residues contributed from an extensive area and no single mutation completely eliminates binding. This demonstration that Wnt3a and Dkk1 share, at least in part, the same or overlapping binding sites, is consistent with a mode of inhibition through direct competition. Though some experiments suggest a model for Dkk1-mediated inhibition of Wnt signaling by LRP6 endocytosis (Mao et al., 2002), subsequent biochemical and genetic data indicate that promotion of LRP6 endocytosis is unlikely to be the primary mechanism for Dkk1 action (Ellwanger et al., 2008; Semenov et al., 2008; Wang et al., 2008). Our data support the direct competition model (Semenov et al., 2001). However, our results also suggest an interplay of interactions that extends beyond steric inhibition at a single, contiguous binding surface on one β-propeller. Consistent with the observations of others on the mapping of the primary binding sites for Wnt3a and Wnt1 to different PE pairs in the LRP6 ectodomain (Bourhis et al., 2010; Ettenberg et al., 2010; Gong et al., 2010), we find that mutations on the top surface of P3 affect Wnt3a, but not Wnt1, signaling, yet mutations at this site affect Dkk1-mediated inhibition of both Wnt ligands. Dkk1 has been reported to bind also to P1E1P2E2 in LRP6 (Ai et al., 2005; Binnerts et al., 2009; Bourhis et al., 2010; Bourhis et al., in press; Zhang et al., 2004). Our results are consistent with these findings, and further uncover interplay between Dkk1 binding at P3E3P4E4 and its inhibition of Wnt1 signaling through P1E1P2E2. Notably, the compact architecture of LRP6ECD revealed by our EM analyses provides a platform-like structure well suited to interactions involving more than one PE pair, and indeed the structure of the LRP6ECD–Mesd complex demonstrates how a single ligand could simultaneously interact with multiple PE pairs. Therefore one scenario is that Dkk1 binding to P3E3P4E4 facilitates its additional interaction with P1E1P2E2, either via Dkk1 bipartite interaction with LRP6 or Dkk1 oligomerization, thereby inhibiting Wnt1 binding at P1E1P2E2. Consistent with this possibility, Kd of Dkk1 binding to LRP6ECD is over an order of magnitude higher than that to P3E3P4E4 (Bourhis et al., 2010); this study). During revision of this manuscript a crystal structure of LRP6 P1E1 in complex with a peptide derived from the N-terminal domain of Dkk1 has been solved (Bourhis et al., in press). This study implicates the top surface of LRP6 P1 in providing a Dkk1-binding site that is separate from LRP6 P3, which is believed to bind to the cysteine-rich domain (CRD) 2 of Dkk1 (Brott and Sokol, 2002; Chen et al., 2008; Wang et al., 2008; Zhang et al., 2004). Thus Dkk1 appears to be able to engage two separate LRP6 β-propeller domains, in line with our findings reported here. However we cannot rule out the possibility that the apparent interplay of binding sites could be indirect, resulting from conformational changes in the LRP6 ectodomain induced by Dkk1 binding.
The effects of mutations implicated in human diseases support our model for the LRP6/5 ectodomain functioning as a compact platform. These single residue mutations impact globally on the structural integrity of the ectodomain or locally on ligand binding sites. For example our results for the R739G mutation in LRP6 are consistent with the equivalent, disease-associated mutation in LRP5 destabilizing the inter-pair interfaces of the LRP5 signaling platform. Conversely, all ‘gain-of-function’ HBM mutations in the first PE pair of LRP5, which are associated with reduced Dkk1 and SOST binding (Ai et al., 2005; Semenov and He, 2006), map to residues that our structural results predict are exposed on the top surface of the β-propeller. Indeed modeled on the structure of LRP6 P1E1 in complex with a Dkk1 N-terminal peptide, these LRP5 HBM mutations appear to occur predominantly at or near the ligand-binding interface (Bourhis et al., in press). In combination with our mutagenesis studies, which implicated the equivalent surface in LRP6 P3E3 in Wnt3a- and Dkk1-binding, these LRP5 disease mutations reinforce our model that a platform-like architecture serves to present the β-propellers as multiple ligand-binding sites.
In summary, our findings provide insights into the structure and function of the LRP6 ectodomain, revealing the characteristics of a signaling platform comprising a compact arrangement of four β-propeller–EGF domain pairs. This platform construction supports an interplay of interactions between LRP6 and its agonists or antagonists that play a fundamental role in Wnt signaling, and as such provides a framework for elucidation of aberrant Wnt signaling in human diseases and development of structure-based therapeutics for related disorders.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human LRP6P3E3P4E4 and LRP6ECD (NP_002327.2; residues 629–1244 and 20–1361, respectively) were cloned into the pHLsec vector (Aricescu et al., 2006) in-frame with a C-terminal His6 tag. Transient expression in HEK293 cell derivatives HEK293S GnTI– (for crystallization) and HEK293T (for other analyses) was carried out as co-expression with human Mesd (NP_055969.1; cloned into the same vector with a Flag tag introduced between residues 35 and 36) to increase levels of secreted protein. Human Dkk1 (NP_036374.1) was cloned into the same vector with a C-terminal His6 tag and transiently expressed in HEK293T cells. Secreted recombinant proteins were isolated from CM by immobilized metal affinity chromatography and further purified by gel filtration chromatography. Subsequent to purification in 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, the LRP6ECD–Mesd complex was buffer-exchanged into 50 mM NaAc (pH 5.0), 150 mM NaCl in which it dissociated allowing LRP6ECD to be purified separately by gel filtration. Dissociation was verified by MALS (see Supplemental Experimental Procedures), and polyacrylamide gels visualized by Coomassie staining as well as Western blotting.
Crystal Structure Determination
LRP6P3E3P4E4 was concentrated to ~5.3 mg/ml in 10 mM Tris-HCl (pH 8.0), 150 mM NaCl for crystallization trials (see Supplemental Experimental Procedures). Crystals grew at 21 °C as needle clusters in 20% (w/v) polyethylene glycol 3350, 0.2 M calcium chloride; optimization yielded single long, thin plates. Crystals were briefly immersed in mother liquor supplemented with 25% ethylene glycol before flash freezing in liquid nitrogen. Data were collected at 100 K from multiple positions along a single crystal using the PILATUS 6M detector on beamline I24 of the Diamond Light Source. X-ray diffraction data were processed, phased by molecular replacement and the structure refined using standard protocols (See Supplemental Experimental Procedures).
Assays and Binding Studies
Luciferase Wnt reporter assays in HEK293T cells were performed as described (MacDonald et al., 2008). Firefly luciferase activity was normalized against that of Renilla luciferase. Site-directed mutagenesis, cell surface biotinylation and co-immunoprecipitation assays were carried out according to standard protocols as were binding studies using surface plasmon resonance (see Supplemental Experimental Procedures).
Electron Microscopy and Image Processing
LRP6ECD and LRP6ECD-Mesd, imaged by negative stain electron microscopy, were sorted into classes using reference-free alignment. 3D reconstructions were calculated using angular reconstitution and projection matching. A homology model for LRP6, based on the LRP6P3E3P4E4 crystal structure, was fitted into the reconstruction and rigid-body refined. (See also Supplemental Experimental Procedures).
Supplementary Material
HIGHLIGHTS.
Crystal structure of LRP6 tandem repeats defines domain interfaces
Electron microscopy reconstruction of LRP6 reveals a compact platform-like shape
Wnt3a and Dkk1 share overlapping binding surfaces on the top face of LRP6 P3
Our platform model provides a structural basis for disease-associated LRP mutations
ACKNOWLEDGEMENTS
We thank Karl Harlos, Jing-Shan Ren, Tom Walter, and staff of Diamond Light Source for assistance; Xin-Jun Zhang for advice on Wnt3a binding assays; Xiao-Fan Wang for advice and discussions. This work was funded by Cancer Research UK, the Spanish Ministry of Science and Innovation (SAF2008-00451, SAF2011-22988), the “Red Temática de Investigación Cooperativa en Cáncer (RD06/0020/1001), and the NIH (RO1 GM074241). S.C. is a recipient of the Chinese Ministry of Education–University of Oxford scholarship; D.B. is supported by EMBO; A.R.A. is a UK Medical Research Council Career Development Award Fellow; C.S. is a Wellcome Trust Research Career Development Fellow; X.H. is a Leukemia and Lymphoma Society Scholar; and E.Y.J. is a Cancer Research UK Principal Research Fellow.
Footnotes
ACCESSION CODES Coordinates and structure factors for the crystal structure of LRP6P3E3P4E4 have been deposited in the Protein Data Bank under accession code 4A0P. EM density maps for LRP6ECD and LRP6ECD–Mesd complex have been deposited to Electron Microscopy Data Base under accession codes EMD-1964 and EMD-1965, respectively.
SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures, Supplemental References, five figures and one table and can be found with this article online at XXX.
REFERENCES
- Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Molecular and cellular biology. 2005;25:4946–4955. doi: 10.1128/MCB.25.12.4946-4955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- Balemans W, Patel N, Ebeling M, Van Hul E, Wuyts W, Lacza C, Dioszegi M, Dikkers FG, Hildering P, Willems PJ, et al. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet. 2002;39:91–97. doi: 10.1136/jmg.39.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnerts ME, Tomasevic N, Bright JM, Leung J, Ahn VE, Kim KA, Zhan X, Liu S, Yonkovich S, Williams J, et al. The first propeller domain of LRP6 regulates sensitivity to DKK1. Mol Biol Cell. 2009;20:3552–3560. doi: 10.1091/mbc.E08-12-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis E, Tam C, Franke Y, Bazan JF, Ernst J, Hwang J, Costa M, Cochran AG, Hannoush RN. Reconstitution of a frizzled8.Wnt3a.LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J Biol Chem. 2010;285:9172–9179. doi: 10.1074/jbc.M109.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis E, Wang W, Tam C, Hwang J, Zhang Y, Spittler D, Huang OW, Gong Y, Estevez A, Zilberleyb I, et al. Wnt antagonists bind through a short peptide to the first β-propeller domain of LRP5/6. Structure. doi: 10.1016/j.str.2011.07.005. (in press) [DOI] [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- Brott BK, Sokol SY. Regulation of Wnt/LRP signaling by distinct domains of Dickkopf proteins. Molecular and cellular biology. 2002;22:6100–6110. doi: 10.1128/MCB.22.17.6100-6110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. American journal of human genetics. 2001;68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang K, Shao Y, Huang J, Li X, Shan J, Wu D, Zheng JJ. Structural insight into the mechanisms of Wnt signaling antagonism by Dkk. J Biol Chem. 2008;283:23364–23370. doi: 10.1074/jbc.M802375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Collins MN, Hendrickson WA. Structural characterization of the Boca/Mesd maturation factors for LDL-receptor-type beta propeller domains. Structure. 2011;19:324–336. doi: 10.1016/j.str.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong F, Schweizer L, Varmus H. Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, Frizzled and LRP. Development. 2004;131:5103–5115. doi: 10.1242/dev.01318. [DOI] [PubMed] [Google Scholar]
- De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, Major MB, Myers A, Saez K, Henriquez JP, Zhao A, et al. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer’s disease. Proc Natl Acad Sci U S A. 2007;104:9434–9439. doi: 10.1073/pnas.0603523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann M, Dietrich MF, Herz J. Lipoprotein receptors--an evolutionarily ancient multifunctional receptor family. Biol Chem. 2010;391:1341–1363. doi: 10.1515/BC.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwanger K, Saito H, Clement-Lacroix P, Maltry N, Niedermeyer J, Lee WK, Baron R, Rawadi G, Westphal H, Niehrs C. Targeted disruption of the Wnt regulator Kremen induces limb defects and high bone density. Molecular and cellular biology. 2008;28:4875–4882. doi: 10.1128/MCB.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettenberg SA, Charlat O, Daley MP, Liu S, Vincent KJ, Stuart DD, Schuller AG, Yuan J, Ospina B, Green J, et al. Inhibition of tumorigenesis driven by different Wnt proteins requires blockade of distinct ligand-binding regions by LRP6 antibodies. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1007428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Bourhis E, Chiu C, Stawicki S, DeAlmeida VI, Liu BY, Phamluong K, Cao TC, Carano RA, Ernst JA, et al. Wnt isoform-specific interactions with coreceptor specify inhibition or potentiation of signaling by LRP6 antibodies. PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Lee L, Zhang L, Wefer S, Brown K, DeRossi C, Wines ME, Rosenquist T, Holdener BC. Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell. 2003;112:355–367. doi: 10.1016/s0092-8674(03)00045-x. [DOI] [PubMed] [Google Scholar]
- Jeon H, Meng W, Takagi J, Eck MJ, Springer TA, Blacklow SC. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat Struct Biol. 2001;8:499–504. doi: 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- Jiao X, Ventruto V, Trese MT, Shastry BS, Hejtmancik JF. Autosomal recessive familial exudative vitreoretinopathy is associated with mutations in LRP5. Am J Hum Genet. 2004;75:878–884. doi: 10.1086/425080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koduri V, Blacklow SC. Requirement for natively unstructured regions of mesoderm development candidate 2 in promoting low-density lipoprotein receptor-related protein 6 maturation. Biochemistry. 2007;46:6570–6577. doi: 10.1021/bi700049g. [DOI] [PubMed] [Google Scholar]
- Kohler C, Lighthouse JK, Werther T, Andersen OM, Diehl A, Schmieder P, Du J, Holdener BC, Oschkinat H. The structure of MESD45-184 brings light into the mechanism of LDLR family folding. Structure. 2011;19:337–348. doi: 10.1016/j.str.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komekado H, Yamamoto H, Chiba T, Kikuchi A. Glycosylation and palmitoylation of Wnt-3a are coupled to produce an active form of Wnt-3a. Genes Cells. 2007;12:521–534. doi: 10.1111/j.1365-2443.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. The Journal of biological chemistry. 2005a;280:19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen J, Lu W, McCormick LM, Wang J, Bu G. Mesd binds to mature LDL-receptor-related protein-6 and antagonizes ligand binding. J Cell Sci. 2005b;118:5305–5314. doi: 10.1242/jcs.02651. [DOI] [PubMed] [Google Scholar]
- Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. American journal of human genetics. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Pearson C, Bu G. Cooperative folding and ligand-binding properties of LRP6 beta-propeller domains. The Journal of biological chemistry. 2009;284:15299–15307. doi: 10.1074/jbc.M807285200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Yokota C, Tamai K, Zeng X, He X. Wnt signal amplification via activity, cooperativity, and regulation of multiple intracellular PPPSP motifs in the Wnt coreceptor LRP6. The Journal of biological chemistry. 2008;283:16115–16123. doi: 10.1074/jbc.M800327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani A, Radhakrishnan J, Wang H, Mani MA, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D, Lifton RP. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- Mason JJ, Williams BO. SOST and DKK: Antagonists of LRP Family Signaling as Targets for Treating Bone Disease. J Osteoporos. 2010;2010 doi: 10.4061/2010/460120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- Semenov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. The Journal of biological chemistry. 2005;280:26770–26775. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- Semenov MV, He X. LRP5 mutations linked to high bone mass diseases cause reduced LRP5 binding and inhibition by SOST. J Biol Chem. 2006;281:38276–38284. doi: 10.1074/jbc.M609509200. [DOI] [PubMed] [Google Scholar]
- Semenov MV, Tamai K, Brott BK, Kuhl M, Sokol S, He X. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr Biol. 2001;11:951–961. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- Semenov MV, Zhang X, He X. DKK1 antagonizes Wnt signaling without promotion of LRP6 internalization and degradation. J Biol Chem. 2008;283:21427–21432. doi: 10.1074/jbc.M800014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer TA. An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. Journal of molecular biology. 1998;283:837–862. doi: 10.1006/jmbi.1998.2115. [DOI] [PubMed] [Google Scholar]
- Takagi J, Yang Y, Liu JH, Wang JH, Springer TA. Complex between nidogen and laminin fragments reveals a paradigmatic beta-propeller interface. Nature. 2003;424:969–974. doi: 10.1038/nature01873. [DOI] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang Y, Li X, Chen L, Wang H, Wu J, Zheng J, Wu D. Characterization of the Kremen-binding site on Dkk1 and elucidation of the role of Kremen in Dkk-mediated Wnt antagonism. J Biol Chem. 2008;283:23371–23375. doi: 10.1074/jbc.M802376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli M, Dougan ST, Caldwell K, O’Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, 3rd, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Li X, Zhang J, Mao J, Li Z, Zheng J, Li L, Harris S, Wu D. The LRP5 high-bone-mass G171V mutation disrupts LRP5 interaction with Mesd. Mol Cell Biol. 2004;24:4677–4684. doi: 10.1128/MCB.24.11.4677-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.