Abstract
After entry into the cell, the early steps of the human immunodeficiency virus type 1 (HIV-1) replication cycle are mediated by two functionally distinct nucleoprotein complexes, the reverse transcription complex (RTC) and preintegration complex (PIC). These two unique viral complexes are responsible for the conversion of the single-stranded RNA genome into double stranded DNA, transport of the DNA into the nucleus, and integration of the viral DNA into the host cell chromosome. Prior biochemical analyses suggest that these complexes are large and contain multiple undiscovered host cell factors. In this study functional HIV-1 RTCs and PICs were partially purified by velocity gradient centrifugation and fractionation, concentrated, trypsin digested, and analyzed by LC-MS/MS. A total of seven parallel infected and control biological replicates were completed. Database searches were performed with Proteome Discoverer and a comparison of the HIV-1 samples to parallel uninfected control samples was used to identify unique cellular factors. The analysis produced a total data set of 11,055 proteins. Several previously characterized HIV-1 factors were identified, including XRCC6, TFRC, and HSP70. The presence of XRCC6 was confirmed in infected fractions, and shown to be associated with HIV-1 DNA by immunoprecipitation-PCR experiments. Overall, the analysis identified 94 proteins unique in the infected fractions and 121 proteins unique to the control fractions with ≥ 2 protein assignments. An additional 54 and 52 were classified as enriched in the infected and control samples, respectively, based on a three-fold difference in total Proteome Discoverer probability score. The differential expression of several candidate proteins was validated by Western blot analysis. This study contributes additional novel candidate proteins to the growing published bioinformatic data sets of proteins that contribute to HIV-1 replication.
Keywords: Human Immunodeficiency Virus type 1, velocity gradient centrifugation, preintegration complex, reverse transcription complex, label-free MS/MS, cellular biology
INTRODUCTION
Like all retroviruses, human immunodeficiency virus type 1 (HIV-1) converts its single-stranded RNA genome into double-stranded DNA via the process of reverse transcription early after infection of the cell. The newly synthesized DNA molecule is transported via the microtubule network,1 imported into the nucleus of the cell,2 and integrated into the host cell genome via a strand transfer attack mediated by the integrase (IN) protein.3 Unlike other retroviruses, such as murine leukemia virus, which require cell division and nuclear membrane breakdown to gain access to host cell chromatin, HIV-1 and other lentiviruses can import their DNA into the nucleus of quiescent cells.2, 4 This characteristic is significant in vivo for the establishment of infection, dissemination, persistence, and disease pathogenesis.5
The critical early steps of the HIV replication cycle are mediated by two functionally defined nucleoprotein complexes (NPCs), the reverse transcription and preintegration complexes (RTC and PIC, respectively). The RTC is a filamentous structure of variable size and shape that facilitates reverse transcription of the viral RNA (vRNA) to double-stranded DNA.6 The PIC is a viral DNA (vDNA) complex that facilitates integration of the vDNA into host cell chromosome. Despite extensive research, the temporal life-span and cellular composition of both complexes is not known and it remains unresolved as to whether these complexes are biochemically distinct. The RTC is a vRNA complex reported to contain the viral reverse transcriptase (RT), integrase (IN), matrix (MA), capsid (CA), nucleocapsid (NC), Vpr, and Vif proteins.6a, 7 The presence of CA in the RTC is disputed, but a proper rate of CA uncoating from the viral core is required for efficient vDNA synthesis and PIC formation.8 Upon completion of reverse transcription, the RTC transforms into the PIC, which is operationally defined by the ability to integrate vDNA into a heterologous DNA target in vitro.3, 9 The in vitro integration reaction requires only the vDNA and IN;10 however the large estimated size of the complex11 suggests that these complexes have a complicated composition that includes a variety of viral and cellular factors which may change as the PIC travels through cytoplasm to the nuclear membrane and beyond. The PIC is a delicate complex as studies report inconsistent recovery of viral proteins from PICs, likely due to differences in the method used to purify the complexes as well as the dynamic nature of the complexes. Initially only IN was identified as a HIV-1 PIC component in complexes extracted with 0.5% TritonX-100.12 Subsequently, MA,11, 13 RT,11, 13 and Vpr14 were observed to be associated with PICs in studies which isolated the complexes with hypotonic buffers or 0.025% digitonin. NC has also been functionally demonstrated to support PIC processing and function.15
Biochemical assays have been unable to unravel the cellular interactions required for productive integration into the host cell genome. For example, the incoming complexes associate with and traverse the cell via the microtubule network,1a, 16 but the protein-protein interactions that mediate association with the dynactin complex to facilitate transport along the microtubule network remain unknown. Similarly, nuclear entry of the vDNA requires active transport once the RTC/PIC reaches the nuclear membrane,2 but the molecular events that regulate the nuclear import of the vDNA are undetermined. A central DNA flap structure formed at the end of reverse transcription17 and several viral components of the HIV PIC (MA, IN, and Vpr (HIV-1) or Vpx (HIV-2/SIV)) contain one or more karyophilic signals.18 However, many studies dispute the requirement of any single nuclear localization signal for efficient PIC nuclear import.14, 19 Recently the CA protein was shown to be the dominant factor for Transportin 3 (TNPO3) dependent nuclear transport of RTCs/PICs.20 Other cellular pathways have also been ascribed to facilitate RTC/PIC nuclear import in addition to TNPO3,21 including Rch1,22 Nup98,23 importin α3,24 importin 7,25 and tRNAs with defective ends.26 The lack of an absolute factor suggests that either the virus exploits multiple pathways for nuclear entry, or the essential or dominant pathway has yet to be discovered.
Much effort to delineate and characterize the structure of HIV-1 NPCs has focused on single protein interaction reductionistic studies. Previous studies to identify cellular NPC-interacting proteins primarily used yeast two-hybrid with IN,27 immunopurification,28 and in vitro reconstitution of salt-stripped PIC activity using purified or recombinant proteins.29 While these experiments have identified several host factors, they are biased towards IN and/or the process of integration while other NPC functions such as cytoplasmic transport have been largely unexplored. Other studies to identify host factors required for efficient infection have relied on whole genomic siRNA screens.30 Combined, these studies produced over 300 candidate proteins, but each will require further analysis to validate its importance in HIV-1 infection and determine how it interacts with HIV-1.31
Biochemical techniques that purify intact and functional complexes will likely be necessary to identify cellular proteins required specifically for the formation and transport of the RTC and PIC. Previous proteomic studies of HIV-1 have focused on virus particles,32 whole cell proteome changes during infection33 or viral protein expression,34 and biomarker discovery.35 One report examined the proteome of HIV-1 DNA complexes affinity purified using a biotinylated DNA target.36 A recent study analyzed protein interactions with 18 individual HIV-1 proteins using over-expression and affinity purification.37
The goal of the studies reported here was to screen intact RTCs and PICs to identify candidate cellular proteins that may contribute to the early steps of HIV-1 infection. To that end, functional RTCs and PICs were purified by short-length velocity gradient centrifugation. The fractions containing RTCs and PICs were concentrated and analyzed by label-free RP-HPLC MS/MS. A total of seven biological replicates were completed. The total MS data from infected and uninfected samples was compiled and unique and enriched proteins identified. Western blot detection of several of the identified proteins demonstrated the accuracy of the MS data. Furthermore, the association of XRCC6, a previously characterized PIC component, with HIV-1 DNA was confirmed. Combined, these data represent a first attempt to analyze intact, functional RTCs and PICs for associated cellular factors.
MATERIALS AND METHODS
Cell culture and viral infection
293T cells were maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal clone 3 (Hyclone, Logan, UT), 8 mM L-glutamine, 100 U/ml penicillin, and 100 ug/ml streptomycin. C8166-45 T cells were cultured in RPMI 1640 media supplemented with 10% fetal clone 3 (Hyclone), 8 mM L-glutamine, 100 U/ml penicillin, and 100 ug/ml streptomycin. PICs were produced using methods previously described.38 HIV-1 molecular clone NLX39 was produced by transient transfection of 293T cell using Polyethylenimine (PEI). Viral supernatant was harvested, filter concentrated, and stored at −80° C. Virus was treated with 100 U/ml Turbo DNase (Ambion, Austin, TX USA) for 60 min at 37° C prior to infecting cells. For each individual infection 1×108 C8166-45 cells were inoculated with 1 ml concentrated virus at an MOI ≥ 25 in a 2 ml final volume of complete media containing 16 µg/ml polybrene. The mixture was transferred to a six well plate and incubated for 10 min at room temperature to allow the cells to settle to the bottom of each well to produce an even layer of cells during spinoculation. Spinoculation was performed by centrifugation at 1050×g for 2 h and 20° C using a biosafety rotor,40 then cells were immediately placed in a tissue culture incubator for 1 h. Afterwards 2 ml of complete RPMI was added to each well, the cells transferred to 75 cm2 tissue culture flasks, and brought up to a volume of 42 ml. At the end of the infection, the cells were pelleted at 250×g for 4 min at 4° C. The media was discarded and the cells washed twice with 5 ml Buffer K(−) (20 mM HEPES (pH 7.5), 150 mM KCl, 5 mM MgCl2). For hypotonic lysis, the cells were resuspended in 1.8 ml cold hypotonic buffer (20 mM HEPES pH 7.3, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT, and 1X Protease Inhibitor Cocktail IV (EMB Biosciences, Gibbstown, NJ)), and incubated on ice for 20 min with occasional mixing by inversion. The cells were transferred to a pre-cooled Dounce homogenizer and lysed by 40 strokes with a tight pestle. The nuclei were pelleted by soft centrifugation (1000×g for 5 min), the supernatant transferred to a fresh tube, and the lysates further clarified by centrifugation at 21,000×g for 5 min. The supernatants of replicate infections were combined and separated into 550 µl aliquots that were flash frozen in liquid nitrogen and stored at −80° C until used.
Velocity gradient centrifugation and fractionation
Gradients were made in 14×89 mm open-top polyclear centrifuge tubes (Seton Scientific, Las Gatos, CA) which were pre-rinsed thrice with millipure water and dried. Gradients were formed utilizing the Gradient Master Station as directed by the manufacturer (BioComp, Fredericton, NB, Canada). After formation the gradients were kept on ice. A 500 µl volume was removed from the top of each gradient and 500 µl of lysate applied drop-wise. Ultracentrifugation was performed with an LE-80 Ultracentrifuge (Beckman Coulter, Fullerton, CA USA) using slow acceleration and slow deceleration. The fractions were collected using the piston fractionator on the Gradient Master Station using cold Buffer K(−) as a wash buffer. The fractions were collected at the speed of 0.5 mm/sec, distance 0.395 mm, for a total distance of 82.95 mm. The collection line was rinsed with Buffer K(−) between each fraction. Fractions were collected on ice during fractionation and stored at 4° C. The linearity of gradients was monitored by measuring the refractive index of a 15 µl sample of each fraction using a refractometer. Gradient fractions were concentrated using 3,000 MWCO column concentrating filters at 4°C as directed by the manufacturer (Millipore). A combined 10.0 ml of sample plus 1.0 ml of PBS was added at a time to each column. After centrifugation the flow through discarded, the column refilled with lysate/PBS and re-centrifuged. The fractions were concentrated to a volume less than or equal to 500 µl. The concentrated sample was transferred to a 1.6 ml tube and the filter rinsed with 100 to 200 µl PBS to recover residual protein. Each sample was brought up to a final volume of ~650 µl with PBS. The refractive index of each sample was measured and if the concentration was > 5% (w/v) sucrose the samples were returned to the column, diluted with PBS, and re-centrifuged. This process was repeated until each sample contained < 5% (w/v) sucrose.
Viral DNA quantification and integration assays
DNA was isolated from samples (lysates or gradient fractions) using PureLink PCR Purification Kit (Invitrogen) with a vacuum manifold (Qiagen, Valencia, CA USA) according to manufacturer’s protocol except that the DNA was eluted using 50 µl of Tris-EDTA pre-warmed to 65° C. DNA samples were stored at −20° C. Ten-fold dilutions (100,000 - 10 fg/µl) of pNLX plasmid in 3 ng/µl tRNA were used as a PCR standard. Therefore the presented values are relative to the amplification of the plasmid standard. Real-time PCR reactions contained 1X IQ SybrGreen Supermix (Bio-Rad, Hercules, CA USA), 250 nM of primers, and 2 µl of DNA sample or standard. The primers used to detect late reverse transcription (LRT) product vDNA were NL919 (5’-TTCGCAGTTAATCCTGGCCTT-3’) and cNL1054 (5’-GCACACAATAGAGGACTGCTATTGTA-3’) corresponding to n.t. 919-1054 of NL4-3 (Genbank # M19921); and the primers used to detect early reverse transcription (ERT) product vDNA were NL497 (5’-GCTAACTAGGGAACCCACTGCTT-3’) and cNL-574 (5’-ACAACAGACGGGCACACACTAC-3’) corresponding to n.t. 497-574 of NL4-3. Real-time PCR was performed using an iQ5 Multicolor real-time PCR detection system (Bio-Rad). The PCR parameters were: an initial denaturization for 6 min at 95° C, then 38 cycles of denaturation at 95° C for 20 sec, annealing at 55° C for 20 sec, and extension at 72° C for 30 sec. The real time data capture occurred during the extension step. DNA extraction efficiency was monitored by quantifying mitochondrial DNA levels using primers and amplification parameters described previously.41 DNA extraction efficiency was monitored by quantifying mitochondrial DNA levels using primers and amplification parameters described previously.41
In-solution tryptic digest
Protein levels from concentrated fractions were measured using NanoDrop ND-1000 (Thermo Fisher Scientific, Inc., Waltham, MA). Approximately 10 µg of protein were added to 15 µl of digestion buffer (50 mM ammonium bicarbonate), 1.5 µL of reducing buffer (100 mM DTT), and 0.5 µL water. The samples were heated to 95° C for 5 minutes and allowed to cool. Next 3 µL of alkylation buffer (100 mM Iodoacetamide) was added and the sample was incubated for 20 minutes. Finally, 1 µL trypsin (1 µg/µL) (Promega, Madison, WI) was added and incubated at 37° C for 3 hours. An additional 1 µL trypsin was added and the sample was incubated overnight at 30° C. After tryptic digest samples were purified using C18 Zip TipsR (Millipore, Billerica, MA) according to manufacturer's procedure. Samples were resuspended in 0.1% formic acid in water prior to LC-MS/MS analysis.
Nano-LC-MS/MS
Mass spectrometric analyses were performed based on previously published work.42 Specifically, samples were analyzed with a LTQ Orbitrap XL (Thermo Scientific, Inc., San Jose, CA) with Eksigent nano-LC system equipped with two alternating peptide traps and a PicoFrit C18 column-emitter from New Objectives (Woburn, MA). Samples were loaded onto the peptide trap with 2% acetonitrile (ACN) + 1% formic acid and eluted using a 60 minute linear gradient of 2–60% ACN + 1% formic acid. The instrument was tuned by direct infusion of angiotension and calibrated every 2–3 days using standards provided by the manufacturer. The acquisition method was created in data-dependant mode with one precursor scan in the Orbitrap, followed by fragmentation of the 5 most abundant peaks in the CID, detected in the LTQ. Resolution of the precursor scan was set to 60,000, scanning from 300–2000 m/z. Precursor peaks with a minimum signal count of 50,000 were dynamically excluded after two selections for 60 seconds within a range +/− 10 ppm. Monoisotopic Precursor selection (MIPS) was enabled. No charge state rejection was used but previously found background peaks were included in a mass rejection list. Collision energy was set to 35 using an isolation width of 2 and an activation Q of 0.250.
The peak list generating software used was Proteome Discoverer 1.2. The outline of the method used in the spectrum selector node of the Proteome Discoverer software that included the following scan event filters: fragmentation method, ionization source, and Unrecognized Activation Type Replacements parameters. All were set to default settings to generate MS/MS spectra. The precursor charge state (high/low), retention time, minimum peak count, total intensity threshold value was all set to default settings. The Max and Min precursor mass settings were 5000 Da and 350 Da, respectively.
The NCBI.fasta database from http://ftp.ncbi.nih.gov was created on April 2010 with a file size of 118783 kB and was restricted to Homo sapiens. The search parameters included in this study were: Two missed cleavages allowed, fully tryptic peptides only, fixed modification of carboxymethyl cysteine (+58.00Da), and variable modifications of oxidized methionine (+15.995Da). The spectra were searched using Sequest™ algorithm in Proteome Discoverer 1.2 software (Thermo Scientific Inc., San Jose, CA) using the following parameters: threshold for Dta generation = 10000, precursor ion mass tolerance = 10 ppm, peptide tolerance for ion trap ms/ms was 1.80Da. Database NCBI.fasta from http://ftp.ncbi.nih.gov was used with two missed cleavage sites allowed and at least two unique peptides were required for protein identification. The criteria used for acceptance of peptide assignments are as follows: Minimal Xcorr value for each charged state ranging from 1 to 7 was 1.50, 2.0, 2.25, 1.0, 1.0, 1.0 and 1.0 respectively and the minimal X-corr value for charge state > 7 was 1.0. Two target values for a decoy database search were applied: strict FDR of 0.01 and a relaxed FDR of 0.05.
Western Blots
Concentrated protein fractions were TCA precipitated, washed twice with 100% acetone, and resuspended in sample buffer. Samples were separated by SDS-PAGE and transferred to PVDF using a semi-dry transfer cell (Bio-Rad). XRCC6/Ku70, RPS6, hnRNP L, ILF2, ILF3, PRKRA, and Annexin A6 were detected using anti-Ku70 (E-5) antibody, anti-RPS6 antibody (C-8), anti-hnRNP L antibody (4D11), anti-ILF2 antibody (A-8) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-ILF3 antibody (AJ1402a; Abgent, San Diego, CA), anti-PRKRA antibody (10771-1; Proteintech Group Inc, Chicago, IL), and anti-Annexin A6 antibody (31026; Abcam, Cambridge, MA) followed by HRP conjugated anti-rabbit or anti-mouse IgG secondary antibody (GE Healthcare, Piscataway, NJ) and visualization by chemiluminescence (Pierce Biotechnology, Rockford, IL).
Immunoprecipitation-PCR
C8166-45 cells were infected with equivalent amounts of NLX + VSVg by spinoculation. The virus pellet was resuspended in 300 µl of IP lysis buffer (Pierce) and pre-cleared with 50 µl protein G agarose beads (GE Healthcare) pre-equilibrated in lysis buffer for 1 h at 4°C. The beads were removed and the samples incubated with antibody at 1/100 dilution overnight at 4° C. The following day 50 µl of protein G agarose beads were added and incubated for an additional 4 h at 4° C. The immune complexes were washed three times for 15 min in lysis buffer and digested for 30 min at 37°C with 100 µg proteinase K in 200 µl of 50 mM Tris (pH 8.0)/1% SDS/10 mM EDTA. Viral DNA was extracted with one round each of saturated phenol, acid phenol-chloroform extraction, and precipitated by 100% ethanol with 1µl LPA. DNA was purified by centrifugation for 10 min at 13,000×g, washed with 70% ethanol, and eluted in 50 µl Tris-EDTA. Viral DNA was detected using primers NL919 and cNL1054.
RESULTS AND DISCUSSION
Optimal infection parameters
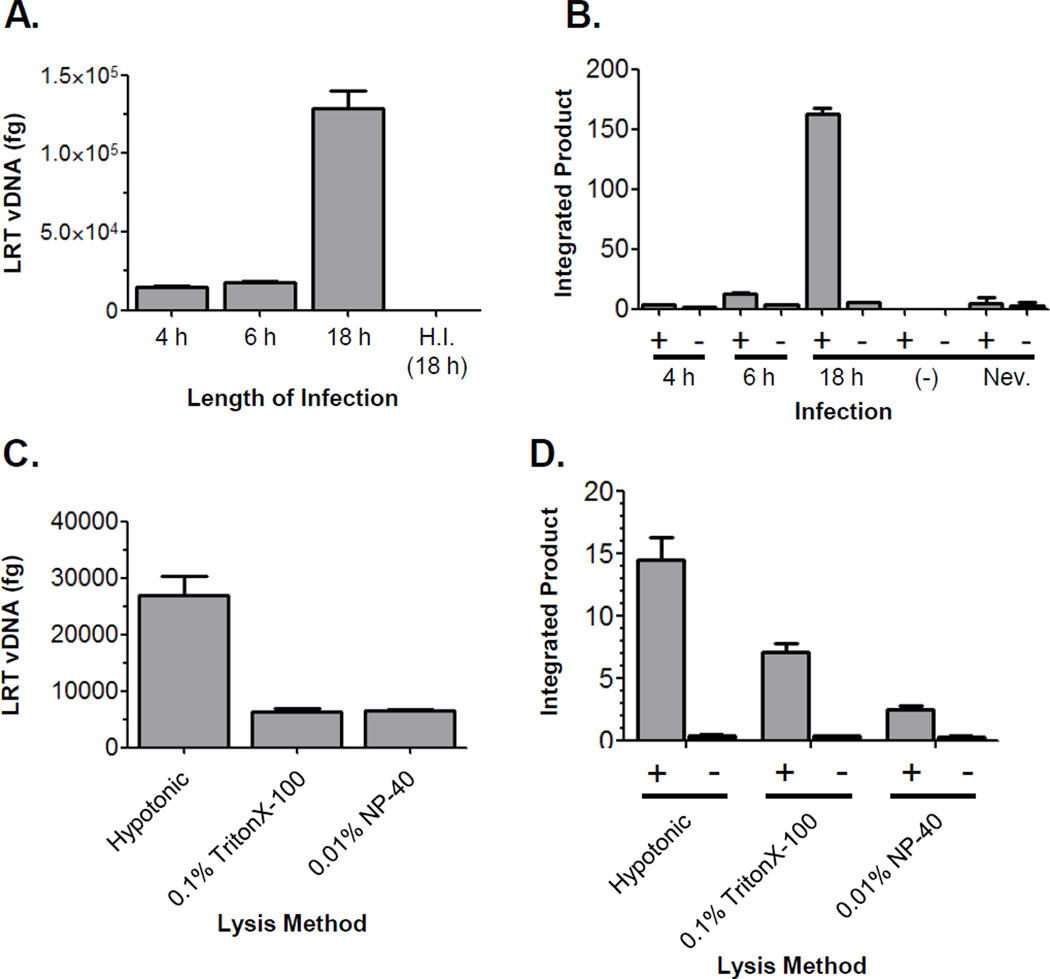
To identify cellular co-factors of HIV-1 infection it was critical to isolate the maximal quantity of functional viral complexes while avoiding any disruption. To determine the time point at which the greatest number of PICs were present in cells, we infected C8166-45 cells with cell-free, VSVg-pseudotyped, replication competent HIV-1NLX and measured the level of late reverse transcription (LRT) product DNA and PIC activity. Infected cells were harvested at 4, 6, and 18 hours post infection (hpi). LRT product DNA was quantified by real-time PCR using primers specific to the MA region of the gag gene. The absence of plasmid DNA contamination was monitored by control infections with heat-inactivated virus (H.I.). Compared with the 4 or 6 h time points, the highest level of viral DNA (vDNA) was detected at 18 hpi (Figure 1A). PIC activity was measured using an in vitro integration assay that measures vDNA integration into a plasmid target DNA by nested PCR.38a, 43 As a control, the PIC activity in each sample was measured (+) or (−) target DNA. Specific integration activity was detected at every time point (Figure 1B). Consistent with the viral DNA levels, greater than ten-fold higher levels of integration activity were observed in lysates harvested at 18 hpi compared to those harvested at 4 or 6 hpi. Control reactions with uninfected lysates and lysates generated from cells treated with the reverse transcriptase inhibitor nevirapine confirmed the specific extraction of PIC activity from samples. Combined, the vDNA and PIC activity data indicated that the maximal levels of PICs were recovered at 18 hpi. Previous studies demonstrated that entry and reverse transcription occur within 4–6 hpi.7b, 44 Thus, the higher levels of PIC activity obtained at 18 hpi was likely due to a burst of cell-cell spread of HIV-1 after the initial robust infection, which is more efficient than cell-free virus infection.45
Figure 1. Assessment of infection length and extraction method.
(A) Recovery of LRT vDNA at various times post infection. DNA was isolated from cell lysates obtained by hypotonic swelling and Dounce homogenization at the indicated times post infection. DNA extractions were normalized by quantification of mitochondrial DNA (data not shown) and viral DNA levels quantified by real-time PCR using gag-specific primers. H.I.= control infection with virus heat inactivated at 65°C for 30 min. Data is representative of two independent experiments. (B) Total level of PIC activity recovered by hypotonic swelling and Dounce homogenization at various times post-infection. The levels (fg) of integrated products were determined by real-time PCR for reactions performed in the presence (+) and absence (−) of target DNA. PICs were harvested from infected C8166-45 cells at the times indicated; (−)= control reaction with uninfected lysates; Nev.= reaction with lysate from cells treated with 20 mM nevirapine. Data shown is representative of three independent experiments. (C) Recovery of vDNA from lysates extracted using the indicated methods. A single infection was split into equivalent amounts, DNA isolated, and LRT vDNA quantified by real-time PCR. Plot is representative of three independent experiments. (D) Total integration activity recovered from lysates extracted by the indicated methods. Data is representative of three independent experiments. In all experiments the error bars denote the standard deviation of the representative experiment.
Identification of candidate interacting proteins in NPCs which are functionally active but not biochemically characterized is a challenging task. Detergents and/or chaotropic agents typically used for sample preparation may disrupt non-specific interactions, yet they can also lead to separation of critical components which are bound with low affinity. However, using too mild of conditions may result in infusing false negative results due to non-specific interactions. HIV-1 PICs are sensitive to low levels of detergents,46 and the recovery of viral proteins from the complexes varies by the extraction method used. The sensitivity of PICs represented a major challenge in our effort to recover intact complexes for MS analysis. Mechanical disruption seemed to be the ideal method to extract complexes, but it was unclear how efficient this method would be compared to detergent lysis. To investigate this, we compared the recovery of PICs harvested by hypotonic lysis/Dounce homogenization with two mild detergent buffers (0.1 % TritonX-100 or 0.01% NP-40). Infected C8166-45 cells were harvested at 18 hpi, split into three samples for each lysis method, and the recovery of PICs measured by viral DNA and in vitro integration activity as described above. Four-fold higher levels of LRT vDNA were recovered consistently from cells harvested by hypotonic swelling/Dounce homogenization compared to the detergent methods (Figure 1C). In parallel, the lysates were measured for total PIC activity (Figure 1D). Analogous to the LRT vDNA levels, two- to four-fold higher levels of PIC activity were recovered from the cells lysed by hypotonic swelling/Dounce homogenization as compared to the cells lysed with mild detergent. These data demonstrated that mechanical disruption isolated a larger amount of functional PICs compared to mild detergent lysis. Notably, calculation of the levels of PIC activity to the amount of vDNA in each sample indicated no statistical difference in PIC activity/fg vDNA between the different lysis methods, indicating the increase in the hypotonic samples resulted from recovery of a higher quantity of PICs instead of differences in activity among the extraction methods. Combined, these data validate the method of PIC isolation using 18 h infections and lysis by hypotonic swelling/Dounce homogenization.
Gradient purification of NPCs
Previous studies have purified RTCs and PICs using both density/equilibrium (isopycnic)13, 47 and velocity (rate-zonal) centrifugation,6b, 47 size-exclusion chromatography,11–12, 29a and recently affinity purification.36 Only the affinity purification study was coupled to MS analysis. Velocity gradient centrifugation is a useful method to purify viruses, cellular organelles, and NPCs. It utilizes a short centrifugation time that separates macromolecular complexes by size. Numerous types of sucrose gradients (5–20%, 5–30%, 5–45%, 10–30%, 20–60% (w/v) in buffer K(−)) and lengths of centrifugation time (30 min. – 16 h) were investigated (results not shown). Strikingly different profiles of viral factors were observed for different gradients and centrifugation parameters. Notably, the amount of recoverable PIC activity was reduced in gradients run for ≥ 4 h.
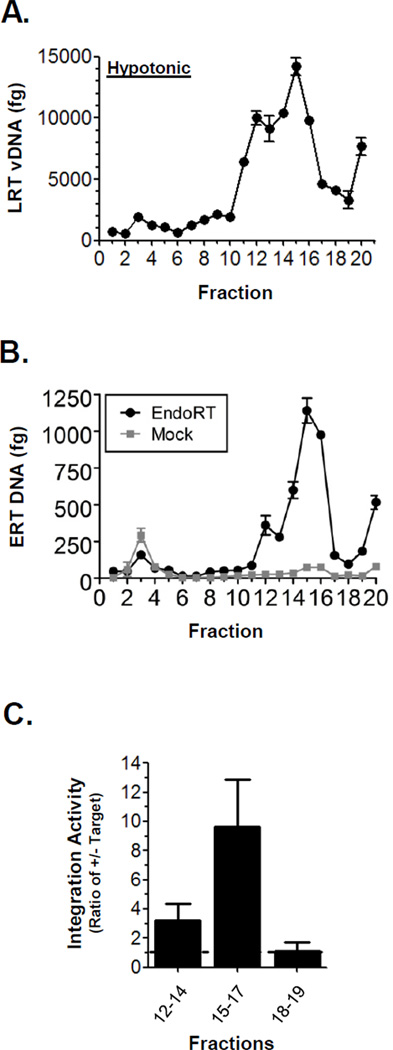
In this study, the HIV-1 NPCs were purified using 5–45% sucrose gradients centrifuged for 1 h at 207,570×g. Gradients were fractionated from the top to the bottom by positive displacement. To ascertain the sedimentation pattern of PICs, both the level of LRT vDNA and PIC activity were measured for each fraction. First, DNA was isolated from each fraction and vDNA levels quantified by real-time PCR using the LRT primers (Figure 2A). A major peak of vDNA was identified in fractions 12–17. Next the sedimentation of RTCs in gradients was determined. This was accomplished by measuring the level of endogenous RT activity in each fraction. These assays were performed using a previously described method that monitors the synthesis of the first product of reverse transcription, the strong stop or early RT (ERT) vDNA product.6b The assay was performed as described previously, but adapted to utilize real-time PCR to quantify ERT synthesis. Also, for each experiment parallel mock reactions were assembled and incubated on ice as a negative control to confirm ERT synthesis (Figure 2B, gray plot). ERT vDNA synthesis was detected in fractions 12–16, indicating the presence of functional RTCs. Both MA and CA were also detected in these fractions by Western blot (data not shown).
Figure 2. Isolation of functional HIV-1 preintegration complexes and reverse transcription complexes.
(A) Sedimentation of HIV-1 vDNA complexes in 5–45% (w/v) sucrose gradients run for 1 h. Fractions were obtained by piston displacement from the top (fraction 1) to the bottom of the tube. The fraction number is indicated at bottom of each graph and fraction 20 represents the irretrievable portion in the round bottom of the tube. DNA was isolated from each fraction and the vDNA quantified by real-time PCR. Plot represents greater than 3 independent infections and fractionations. The error bars denote the standard deviation of the single real-time PCR. (B) Endogenous RT (EndoRT) activity was measured by in vitro strong stop DNA synthesis as quantified by real-time PCR using ERT vDNA primers. Result from a parallel mock reaction held on ice is show by gray plot. (C) Integration activity recovered from the fractions of 1 h 5–45% sucrose gradients. For each experiment six replicate lysates were centrifuged, fractionated, combined, and concentrated. The concentrated fractions were individually assayed for PIC activity. Integration activity was determined by calculating the integrated product in the presence and absence of target DNA. The bars in the graph present the combined mean PIC activity for the fractions indicated. A ratio > 1 indicates IN activity (dashed line). Error bars denote the combined standard error of the mean for the data of least three independent experiments. Endogenous RT (EndoRT) activity was measured by in vitro strong stop DNA synthesis as quantified by real-time PCR using ERT vDNA primers. Result from a parallel mock reaction held on ice is show by gray plot.
To determine if the vDNA containing fractions contained PICs we assayed the fractions for in vitro integration activity. PIC assays were performed using concentrated, replicate fractions that were buffer exchanged to < 5% sucrose. For these experiments the level of specific integration activity was calculated for each fraction. This represented the ratio of the activity in the (+) target and (−) target reactions. Thus, a value > 1 indicated the presence of specific integration activity and a value ≤ 1 indicated no specific activity. The mean specific activity for the fractions is shown in Figure 2C. Fractions 15–17 contained the highest level of PIC activity. A lower level of PICs were recovered from fractions 12–14, however no PIC activity was detected in fractions 18–19. These results demonstrated the presence of functional PICs in fractions 12–17 of 1 h 5–45% sucrose gradients. Combined, the data indicated that PICs and RTCs co-sediment in 1 h 5–45% sucrose gradients. In addition, the data suggested that this purification method did not dissociate proteins critical for proper function of these complexes. Additional studies determined that even small amounts of detergent (0.1%) led to a significant decrease in recoverable integration activity and a loss of associated viral proteins MA and CA (data not shown). Thus, these results demonstrated the partial purification of functional RTCs and PICs using 5–45% sucrose gradients.
MS Analysis of HIV-1 RTCs and PICs
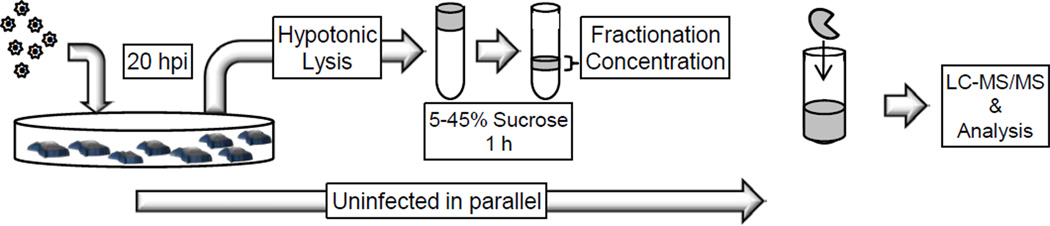
A summary of the overall experimental approach is presented in Figure 3. For each biological replicate infected and control uninfected samples were prepared in parallel. A total of ~ 1.6×109 C8166-45 cells were infected using the spinoculation technique.40 The lysates were obtained by hypotonic lysis/Dounce homogenization, clarified by centrifugation, snap frozen in liquid nitrogen, and stored at −80°C until gradient purification. After velocity gradient centrifugation the sedimentation pattern of each replicate was monitored by quantifying the levels of LRT viral DNA in each fraction. All replicates produced a single broad peak of vDNA spanning fractions 11–19 similar to Figure 2A (data not shown). The infections and NPC purifications for proteomic analysis were performed a total of seven times (seven independent cultures and infections), which were defined as “biological replicates.” The level of infection and yield of complexes was consistent across all replicates. For each experimental replicates, sucrose gradient fractions 11 to 19 were digested individually with trypsin and analyzed separately by nano-RP-LC-MS/MS. The MS spectra of each fraction were submitted separately for database searches using Proteome Discoverer (Thermo Scientific). In total 63 samples (9 fractions×7 biological replicates) were processed for both the infected and control (uninfected) groups. Results of database searches were exported to a .xlsx file. After completion of all seven biological replicates, the data were combined into control and infected groups.
Figure 3. Schematic depiction of the methods used in this analysis.
C8166 T-cells were infected with HIV-1 + VSVg pseudotyped virus for 20 hours. Cells were lysed by hypotonic swelling and Dounce homogenization. Lysates were loaded on a 5–45% sucrose gradient and subjected to ultracentrifugation for 1 hour at 207,570×g. Gradients were fractionated and digested in solution with trypsin prior to MS/MS analysis. For each infected replicate, a parallel uninfected sample was subjected to the same procedure.
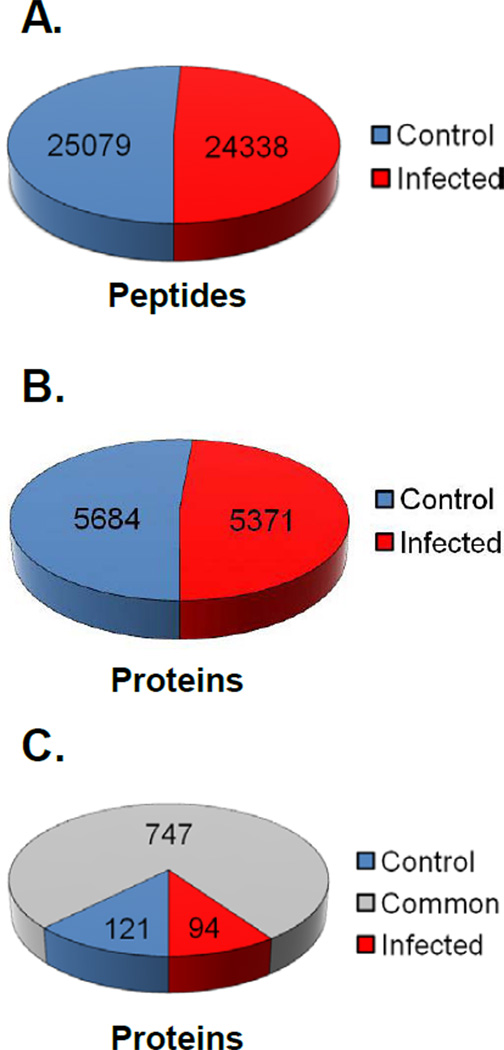
In total, database searches across all experiments yielded 49,417 high confidence peptide identifications, 25,079 from control fractions and 24,338 from infected fractions (Figure 4A). This resulted in the identification of 11,055 total proteins from the peptide pools-5684 (51.42%) and 5371 (48.58%) proteins in the control and infected groups, respectively (Figure 4B). There were 974 protein assignments based on single peptide identifications. Removal of these lowered the number of protein identifications to 5186 (51.44%) and 4895 (48.56%) in the control and infected groups, respectively. The comparison of the total number of peptides and protein identifications in the control and infected samples over the seven biological replicates indicated a near 1:1 ratio with a minute bias toward the control fractions. To further judge whether there was overall parity between the control and infected samples, we assessed if there was a difference between the control and infected groups by comparing three metrics using the data of the most abundant proteins common to both control and infected samples across all replicates. The dataset included 31 proteins with ≥ 25 independent protein assignments. First, we compared the number of high confidence peptide identifications per individual protein assignment (Supplemental Figure 1A). Second we compared the number of individual protein assignments for each NCBI GI number across the seven biological replicates (Supplemental Figure 1B). Finally we calculated and compared the coverage of each protein across the seven replicates (Supplemental Figure 1C). For all three metrics there was no statistical difference between the infected and control groups. These data provide good evidence that there was an absence of bias in the comparison of control and infected MS data due to differences in the sample quality between the infected and control groups.
Figure 4. Distribution of peptides and proteins identified in the MS analysis.
The total Proteome Discoverer data was organized and analyzed manually using Microsoft Excel. (A) Representation of the total number of peptides identified with high confidence across all seven biological replicates for both the control and infected samples. (B) A chart of the number of unique NCBI GI numbers identified across all seven replicates (including duplicates) for both the infected and control MS data. (C) Graphic illustration of the number of common and unique proteins in each data set after compilation of all seven replicates and the removal of protein assignments based on single peptide identifications.
To identify the proteins composing RTCs/PICs we assumed that proteins unique or significantly enriched to the HIV-1 infected cells would represent viral interacting proteins. Therefore, the proteins common to both the control and infected data sets were excluded to identify candidate RTC/PIC proteins. Due to the high frequency in every sample, Ribosomal proteins were also excluded from our analyses. Despite the large pool of proteins identified, there were less than 100 unique proteins in the infected samples (Figure 4C). These were ranked by total Proteome Discoverer Score and the top ten candidates are presented in Table 1. The protein unique to the infected samples with the highest total peptide identifications was XRCC6 (P12956, XRCC6_HUMAN) which belongs to Ku70 family (UniProt KB). This protein functions in the non-homologous end joining pathway and was described previously as a HIV-1 interacting protein.48 XRCC6 is incorporated into HIV-1 particles and protects integrase from degradation upon entry into target cells.49 Other HIV-1 interacting proteins identified in the infected dataset included: HSP70,50 TFRC,51 NONO,36 and TAF652 (Table 1). The presence of these known HIV-1 factors affirmed our experimental approach.
Table 1.
Top ten proteins unique to infected samples across 7 biological replicates1
| NCBI GI # | Gene name2 | Total Peptide Identifications3 |
Total PD Score4 |
|---|---|---|---|
| 194385986 | XRCC6 (Ku70) | 16 | 66.73 |
| 49425327 | FCRL6 Fc receptor-like 6 | 9 | 50.15 |
| 119609608 | glutamate receptor, ionotropic, N-methyl D-aspartate 2C | 9 | 46.21 |
| 483355 | NONO non-POU domain containing, octamer-binding; p54(nrb) | 10 | 43.07 |
| 33877895 | MARCKS protein | 8 | 42.69 |
| 119581245 | MAX-like protein X | 9 | 38.11 |
| 239750920 | Golgi matrix protein 130 (GM130) | 8 | 35.03 |
| 110238585 | FAM167B family with sequence similarity 167, member B | 14 | 34.99 |
| 52545770 | Serine/threonine kinase 33 (STK33) | 10 | 31.95 |
| 194375674 | PDIA3 protein disulfide isomerase family A, member 3 | 9 | 30.38 |
Ranked by Proteome Discoverer Score. Ribosomal proteins were omitted due to the high frequency in both infected and control samples.
NCBI Gene name. Unknown protein hits were identified by BLAST homology search.
The total number of peptides identified across all seven replicates that referenced the indicated GI number.
The sum total of Proteome Discoverer (PD) score for all protein identifications across the study.
The Presence of XRCC6 in PICs Validates the Extraction Method
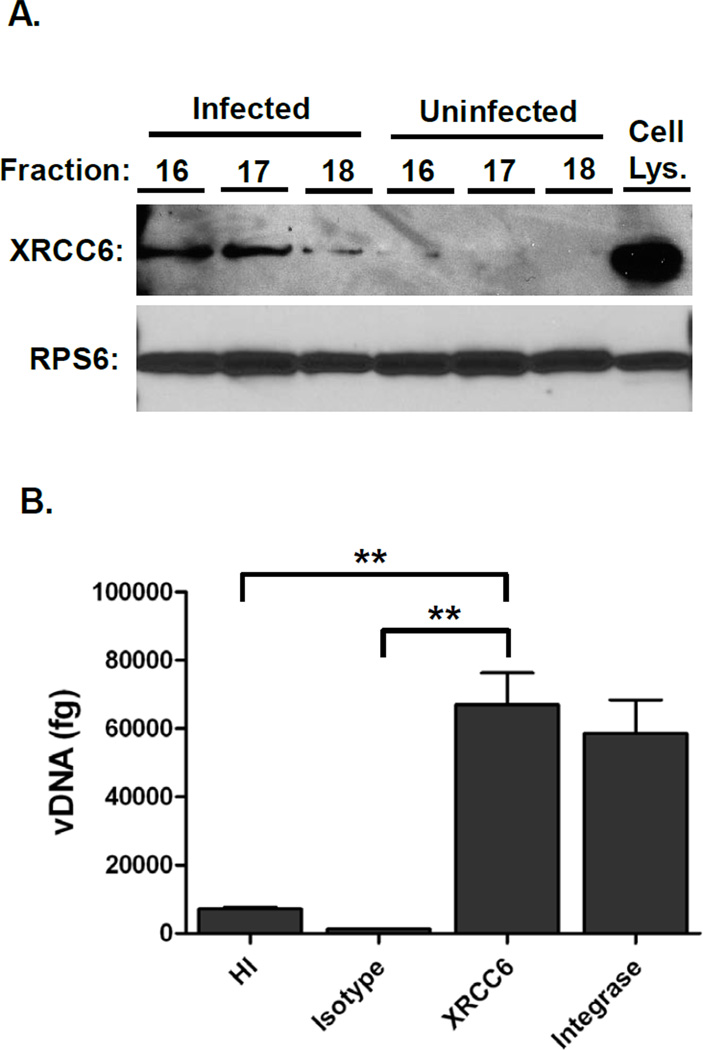
The mechanical lysis and complex composition of the gradient fractions posed a risk of producing non-specific interactions. To confirm the integrity of the partially purified complexes we tested whether XRCC6 (Ku70), which was unique in the infected samples, was associated with PICs. To confirm the presence of XRCC6 in the proteomic samples, Western blots were performed on the concentrated fraction samples (Figure 5A, top panel). Ribosomal protein S6 (RPS6), identified at similar levels in both the infected and control fractions by MS (total Proteome Discover Scores of 415.95 (infected) and 392.20 (control)), was used as a control in these assays (Figure 5A, lower panel). Consistent with the MS data, RPS6 was detected abundantly in all fractions in both the infected and control samples. In agreement with the Proteome Discoverer data, XRCC6 was observed in fractions 16–18 of the infected samples, but nearly undetectable in the parallel control fractions. Moreover, these results were consistent with previous studies that demonstrate XRCC6 is associated with PICs.48–49 To confirm that XRCC6 associated directly with HIV-1 NPCs immunoprecipitation-PCR assays were performed. Infected C8166-45 cells were lysed and imunoprecipitated with isotype control, anti-XRCC6, or anti-integrase antibody as a positive control. As an additional control cells were infected with heat-inactivated virus. After extensive washing, the immunoprecipitates were digested with proteinase K and DNA isolated by phenol:chloroform extraction. Viral DNA was detected by real-time PCR using HIV-1 gag-specific primers (Figure 5B). Importantly, only a small amount of DNA was detected in samples infected with heat-inactivated virus and immunoprecipitated with anti-XRCC6 antibody, and infected lysates immunoprecipitated with isotype control antibody. Anti-XRCC6 antibody immunoprecipitated HIV-1 DNA in infected cells at levels comparable to the anti-HIV-1 integrase positive control antibody, confirming the presence of XRCC6 with PICs early after HIV-1 infection. These data validate the capability of the experimental design to isolate and identify a factor known to interact with HIV-1 NPCs.
Figure 5. XRCC6 is present in the infected samples and is associated with HIV-1 DNA.
(A) Proteomic fractions 16–18 were TCA precipitated, resuspended in sample buffer, and separated by SDS-PAGE. Western blot was performed using the antibodies to XRCC6 and RPS6. The Cell Lys. sample denotes a C8166 whole cell lysate. (B) C8166 cells were infected with NLX + VSVg for 20h, lysed, and immunoprecipitated with the indicated antibodies. DNA was isolated and viral DNA detected by real-time PCR using gag-specific primers. HI denotes heat948 inactivated virus. ** denotes p < 0.01 as calculated by two-tailed T test. Data represents three independent replicates
Candidate HIV Factors
The goal of the study was to identify novel proteins that may contribute to HIV-1 replication. Proteins identified in the analysis that were not reported previously as components of HIV complexes included: FCRL6, GRIN2C, MARCKS, MAX-like protein X, GM130, FAM167B, STK33, and PDIA3. Intriguing among these proteins is STK33. It is a serine/threonine kinase and member of the Ca2+/calmodulin-dependent kinase family.53 STK33 autophosphorylates and interacts with and phosphorylates Vimentin. Vimentin, an intermediate filament protein, reportedly interacts with HIV-1 Vif,54 is cleaved by HIV-protease,55 and its phosphorylation can be inhibited by the HIV-1 envelope protein, gp120.56 It was suggested previously that Vimentin played a role the early steps of HIV-1 replication,57 but the results were not corroborated. Moreover, intermediate filament inhibitors can block the early steps of HIV replication.58 Combined, these findings support the hypothesis that HIV-1 usurps the function of STK33 to facilitate the early steps of virus infection.
The search results also uncovered numerous proteins that appeared to be enriched in the infected fractions. As there is no precedent in this type of modified unlabelled proteomic experiment, we arbitrarily defined any protein that had 3-fold or greater total Proteome Discoverer Score in the infected data set compared to the uninfected set as “enriched.” This provided a reasonable list of 32 candidate proteins, the top ten of which are listed in Table 2 with their respective Proteome Discoverer scores. These may represent proteins which are normally present in particular fractions, but actively migrate to the HIV-1 nucleoprotein complexes upon infection. One protein that caught our attention in this dataset was Annexin A6 (ANXA6). ANXA6 is a member of the annexin protein family which are dynamic and multifunctional calcium and membrane-binding proteins.59 ANXA6 is the largest member of the annexin family and research suggests it functions in cholesterol homeostasis, membrane trafficking/structure, actin-cytoskeleton, and signal transduction (see 60 and references therein). Another annexin family member, A2, was implicated in HIV-1 infection as a factor required for viral production in monocyte-derived macrophages (MDM).61 A subsequent study confirmed the importance of A2 in particle production in MDMs but this role was not recapitulated in other cell types.62 Therefore, A2 may be a macrophage-specific host cell factor that regulates HIV-1 infectivity. Published data suggest that ANXA6 expression is closely linked to caveolae formation in various cell types.63 Of note, the major caveolae protein caveolin-1 can inhibit HIV-1 infection through transcriptional repression mediated by NF-κB.64 Thus it is tempting to theorize that HIV-1 may alter caveolae homeostasis via ANXA6 to achieve productive infection.
Table 2.
Top ten proteins enriched in infected samples across 7 biological replicates1
| NCBI GI # | Gene name2 | Total PD Score (Inf/Cont)3 |
|---|---|---|
| 179976, 34526818, 34533483, 221042282 | Annexin A6 | 234.15 / 74.99 |
| 18105045 | HIST1H2AH histone cluster 1, H2ah | 284.96 / 17.86 |
| 119584828 | CKLF-like MARVEL transmembrane domain containing 7 | 60.17 / 8.58 |
| 119624259 | SFRS protein kinase 1 | 153.09 / 21.97 |
| 2224547 | MAST4 | 18.09 / 3.88 |
| 194375063 | Lung cancer oncogene 7 | 51.4 / 12.18 |
| 119626383 | AR4/FMR2 family, member 1 | 31.61 / 8.21 |
| 4098637 | MAP1A | 51.01 / 14.83 |
| 194377490 | RAPGEF2 Rap guanine nucleotide exchange factor (GEF) 2 | 29.48 / 9.41 |
| 30795231 | Brain abundant, membrane attached signal protein 1 | 348.49 / 102.9 |
Ranked by fold enrichment as measured by the ratio of total Proteome Discover (PD) score in the infected versus uninfected samples. Ribosomal proteins were omitted due to the high frequency in both infected and control samples.
NCBI Gene name. Unknown protein hits were identified by BLAST homology search.
The sum total of PD score for all protein identifications across the seven biological replicates.
The next dataset generated in the analysis were proteins that were unique to the fractions of the uninfected control cells. A total of 121 proteins were specific to the control cells (Figure 4C). These proteins could represent cellular factors that are mislocalized or down-regulated by HIV-1 infection. A list of the ten proteins unique to control samples with the highest Proteome Discoverer Scores is presented in Table 3. The highest scored protein was PABPC1. The interaction of PABPC1 with HIV-1 is inconclusive. It was shown to bind HIV-1 RNA in the presence of the viral Rev protein,65 but was also found to be degraded by the HIV-1 protease in a separate study.66 The unique presence of PABPC1 in control samples in our studies could be consistent with the latter finding, but additional studies will be needed to test this hypothesis.
Table 3.
Top ten proteins unique to control samples across 7 biological replicates1
| NCBI GI # | Gene name2 | Total Peptide Identifications5 |
Total PD Score6 |
|---|---|---|---|
| 194386544 | Poly(A) binding protein, cytoplasmic 1 (PABPC1) | 67 | 215.65 |
| 4503483 | Eukaryotic translation elongation factor 2 | 34 | 174.96 |
| 194381632 | BASP1 | 21 | 79.72 |
| 55958544 | hnRNP K | 14 | 73.63 |
| 18375673 | UPF1 | 20 | 59.42 |
| 194382312 | SRSF10 | 6 | 51.65 |
| 157412270 | hnRNP M | 11 | 48.65 |
| 55959640 | Poly(A) binding protein, cytoplasmic 4 (PABPC4) | 9 | 45.04 |
| 13544110 | Calmodulin 3 | 6 | 39.01 |
| 119577230 | hnRNP L | 4 | 30.71 |
Ranked by Proteome Discoverer score. Ribosomal proteins were omitted due to the high frequency in both infected and control samples.
NCBI Gene name. Unknown protein hits were identified by BLAST homology search.
The total number of peptides identified across all seven biological replicates that reference the indicated GI number.
The sum total of Protoeome Discoverer (PD) score for all seven bioloigical replicates.
Finally, we compiled a list of protein assignments enriched in control fractions (Table 4). Identical to the infected dataset, 32 proteins had a three-fold or higher Proteome Discoverer Score compared to the infected samples. The top ten candidate proteins are listed in Table 4. The top protein in this category was PRKRA, with a 15.6 fold enrichment over infected samples. PRKRA (also known as PACT and DYT16) is a double-stranded RNA binding protein and activator of the protein kinase PKR, part of the interferon induced anti-viral response. However PRKRA activation of PKR can occur a dsRNA independent manner.19e, 67 PRKRA shares some homology with another PKR interacting protein and HIV-1 factor, the transactivation response (TAR) RNA binding protein (TRBP). TRBP was discovered due to its affinity for the HIV-1 TAR RNA, and can inactivate the activity of PKR.68 The same study demonstrated that PRKRA can relieve PRK mediated transcription inhibition, including HIV-1 gene expression. These data suggest that PRKRA would be a positive regulator of HIV-1 replication, however recent in vivo studies found that PRKRA was negatively regulated in lymphatic tissue of HIV-1 infected individuals.69 Our data is consistent with the in vivo data and indicate further studies are needed to define the role of PRKRA in HIV-1 replication.
Table 4.
Top ten proteins enriched in control samples across 7 biological replicates1
| NCBI GI # | Gene name2 | Total PD Score (Inf/Cont)3 |
|---|---|---|
| 213417919 | PRKRA protein kinase, interferon-inducible double stranded RNA dependent activator | 4.9 / 76.28 |
| 33875035 | STAU1 protein | 2.23 / 22.22 |
| 181486 | YBX1 Y box binding protein 1 | 45.86 / 348.68 |
| 9714266, 212549553 | Interleukin enhancer binding factor 3, 90kDa | 6.92 / 226.94 |
| 55962124 | Interleukin enhancer binding factor 2, 45kDa | 16.49 / 89.63 |
| 194391252 | PRPF40A PRP40 pre-mRNA processing factor 40 homolog A | 8.54 / 47.22 |
| 194383864 | Heterogeneous nuclear ribonucleoprotein F | 22.04 / 71.15 |
| 194387074 | Heterogeneous nuclear ribonucleoprotein C | 21.18 / 152.18 |
| 7242140 | ClpX caseinolytic peptidase X homolog | 8.52 / 40.08 |
| 3005744 | Family with sequence similarity 120A | 9.15 / 54.44 |
Ranked by fold enrichment as measured by the ratio of total Proteome Discover (PD) score in the infected versus uninfected samples. Ribosomal proteins were omitted due to high frequency in both infected and control samples.
NCBI Gene name. Unknown protein hits were identified by BLAST homology search.
The sum total of PD score for all protein identifications across the seven biological replicates in both the infected (inf) and control (cont) groups.
Two related proteins were also ranked within the top ten candidates identified as enriched in the control samples, ILF2 and ILF3. These two proteins form a heterodimer that functions in gene expression and the stabilization of mRNAs.70 ILF3 controls the expression of Cyclin T1, a component of the P-TEFb transcription factor complex that is recruited by HIV Tat for HIV transcription. Modulation of ILF3 expression consequently affects HIV replication: RNAi knockdown of ILF3 reduces HIV transduction, and over-expression of ILF3 increases HIV gene expression. It has also been shown that a C-terminal variant of ILF3 can interact with HIV-1 Rev and inhibit HIV-1 mRNA export by altering the intracellular localization of Rev.71 Our proteomic data suggested that HIV down modulates the cytoplasmic localization of ILF2 and ILF3 during the early steps of virus replication.
Validation of Candidate Proteins
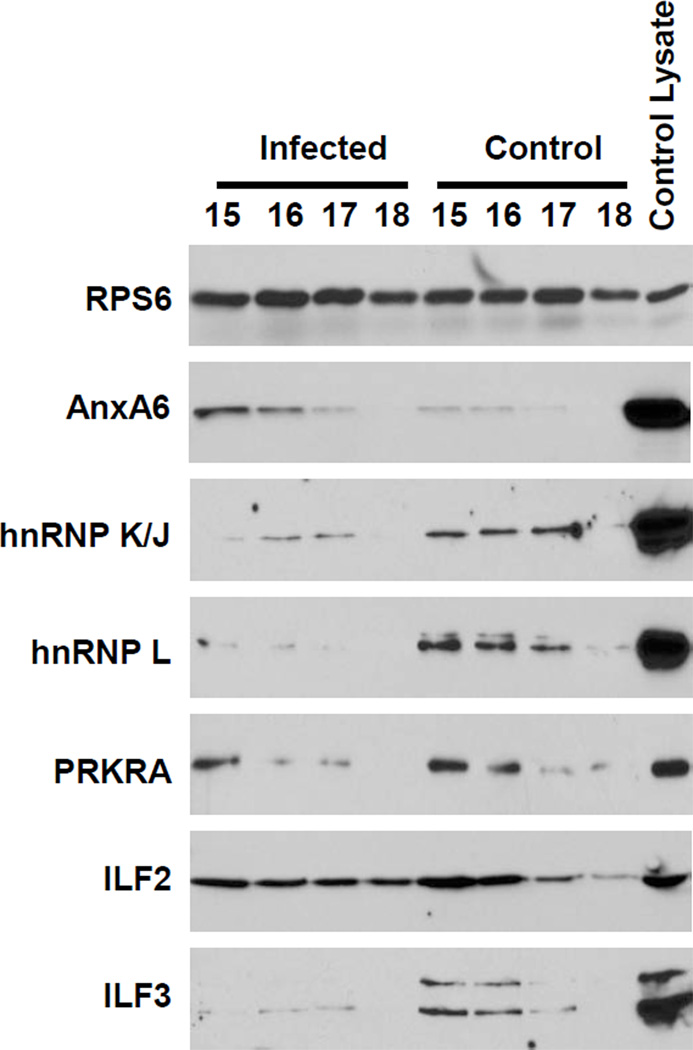
As with XRCC6, Western blot analysis was performed on fractions 15–18 to validate the differential presence of candidate proteins between the control and infected fractions (Figure 6). The detection of RPS6 was again used as a control for these studies. RPS6 was readily detected at similar levels in all fractions indicating equivalent levels of proteins were loaded in each lane (top panel). The candidate proteins that were screened are listed on the left side of Figure 6. As shown in the second panel of Figure 6, higher levels of ANXA6 were detected in the infected fractions compared to the control fractions. This confirmed the MS results that ANXA6 was enriched in the HIV-1 NPC-containing fractions. Next, we assessed the presence of two the proteins found to be unique to the control data sets: heterogeneous nuclear ribonucleoproteins J/K and L (hnRNP K/J and hnRNP L; panels 3 and 4). Both hnRNPs were readily detected in the fractions of the control samples; however in both cases a small amount of protein was found in the fractions of the infected samples. This was similar to the results with XRCC6 and suggested that there was an insufficient level of both hnRNPs in the infected samples to be detected by MS. Finally we investigated the presence of three of the proteins found to be enriched in the control samples: PRKRA, ILF2, and ILF3. All three of the proteins were detected in both the infected and control samples. But as predicted by the MS data, all three proteins were detected at higher levels in the control compared to the infected fractions. Taken together, these Western blot data validated the MS results and demonstrated the efficacy of the experimental approach. Therefore these data provide numerous candidate proteins that are differentially regulated during early HIV infection and may possibly be associated with RTCs and/or PICs.
Figure 6. Validation of candidate proteins identified by MS.
Hypotonic gradient fractions were TCA precipitated, resuspended in sample buffer, and separated by SDS-PAGE. Several candidate proteins were detected using protein-specific antibodies. A whole C8166 cell lysate was used as a positive control to ensure proper detection of proteins.
SUMMARY
Determination of the cellular proteins associated with HIV-1 RTCs and PICs may identify cellular factors and pathways critical for virus replication. Isolation of these complexes is a challenging task because of the small number of complexes per cell, their complex composition, and their inherent instability and sensitivity to detergent. Our experimental approach to perform MS analysis on partially purified intact and functional NPCs was novel, and produced data sets identifying candidate factors both up- and down-regulated in the fractions containing RTCs and PICs. The presence of a number of proteins in the data sets previously reported to be associated with HIV NPCs, and the validation of the differential expression of several candidate proteins demonstrated the efficacy of the experimental approach. More importantly, these results identified a panel of novel cellular proteins that are potentially associated within these structures. Further studies focused on role of these candidate proteins in HIV replication are needed to determine their importance in HIV-1 pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
These studies were funded by the National Institutes of Health (R01A1080348) (M.B.) and (R01DA030962) (P.C.).
ABBREVIATIONS
- HIV
human immunodeficiency virus
- RTC
reverse transcription complex
- PIC
preintegration complex
- LRT
late reverse transcription
- ERT
early reverse transcription
- vDNA
viral DNA
- vRNA
viral RNA
- IN
integrase
- RT
reverse transcriptase
- MA
matrix
- CA
capsid
- NC
nucleocapsid
Contributor Information
Cameron J. Schweitzer, Email: cameronschweitzer@creighton.edu.
Teena Jagadish, Email: tjagadish@unmc.edu.
Nicole Haverland, Email: nhaverland@unmc.edu.
Pawel Ciborowski, Email: pciborowski@unmc.edu.
References
- 1.(a) McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002;159(3):441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Naghavi MH, Valente S, Hatziioannou T, de Los Santos K, Wen Y, Mott C, Gundersen GG, Goff SP. Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells. Embo J. 2007;26(1):41–52. doi: 10.1038/sj.emboj.7601475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukrinsky MI, Sharova N, Dempsey MP, Stanwick TL, Bukrinskaya AG, Haggerty S, Stevenson M. Active Nuclear Import of Human Immunodeficiency Virus Type 1 Preintegration Complexes. PNAS. 1992;89(14):6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farnet CM, Haseltine WA. Integration of Human Immunodeficiency Virus Type 1 DNA in vitro. PNAS. 1990;87(11):4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukrinsky MI, Stanwick TL, Dempsey MP, Stevenson M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 1991;254(5030):423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blankson JN, Persaud D, Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med. 2002;53:557–593. doi: 10.1146/annurev.med.53.082901.104024. [DOI] [PubMed] [Google Scholar]
- 6.(a) Chen M, Garon CF, Papas TS. Native ribonucleoprotein is an efficient transcriptional complex of avian myeloblastosis virus. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(3):1296–1300. doi: 10.1073/pnas.77.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nermut MV, Fassati A. Structural Analyses of Purified Human Immunodeficiency Virus Type 1 Intracellular Reverse Transcription Complexes. J. Virol. 2003;77(15):8196–8206. doi: 10.1128/JVI.77.15.8196-8206.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Carr JM, Coolen C, Davis AJ, Burrell CJ, Li P. Human immunodeficiency virus 1 (HIV-1) virion infectivity factor (Vif) is part of reverse transcription complexes and acts as an accessory factor for reverse transcription. Virology. 2008;372(1):147–156. doi: 10.1016/j.virol.2007.10.041. [DOI] [PubMed] [Google Scholar]; (b) Fassati A, Goff SP. Characterization of Intracellular Reverse Transcription Complexes of Human Immunodeficiency Virus Type 1. J. Virol. 2001;75(8):3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Dismuke DJ, Aiken C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol. 2006;80(8):3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Yamashita M, Perez O, Hope T, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3:1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellison V, Abrams H, Roe T, Lifson J, Brown P. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64(6):2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Bushman FD, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249(4976):1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]; (b) Craigie R, Mizuuchi k, Bushman FD, Engelman A. A rapid in vitro assay for HIV DNA integration. Nucl. Acids Res. 1991;19(10):2729–2734. doi: 10.1093/nar/19.10.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 1997;71(7):5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnet CM, Haseltine WA. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 1991;65(4):1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley WG, Stevenson M. Association of Integrase, Matrix, and Reverse Transcriptase Antigens of Human Immunodeficiency Virus Type 1 with Viral Nucleic Acids Following Acute Infection. PNAS. 1993;90(13):6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinzinger NK, Bukrinsky MI, Haggerty SA, Ragland AM, Kewalramani V, Lee M, Gendelman HE, Ratner L, Stevenson M, Emerman M. The Vpr Protein of Human Immunodeficiency Virus Type 1 Influences Nuclear Localization of Viral Nucleic Acids in Nondividing Host Cells. PNAS. 1994;91(15):7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Buckman JS, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J Virol. 2003;77(2):1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix JL. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21(4):831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arhel N, Genovesio A, Kim K-A, Miko S, Perret E, Olivo-Marin J-C, Shorte S, Charneau P. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat Meth. 2006;3(10):817–824. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- 17.(a) Ao Z, Yao X, Cohen EA. Assessment of the role of the central DNA flap in human immunodeficiency virus type 1 replication by using a single-cycle replication system. J Virol. 2004;78(6):3170–3177. doi: 10.1128/JVI.78.6.3170-3177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Arhel N, Munier S, Souque P, Mollier K, Charneau P. Nuclear Import Defect of Human Immunodeficiency Virus Type 1 DNA Flap Mutants Is Not Dependent on the Viral Strain or Target Cell Type. J. Virol. 2006;80(20):10262–10269. doi: 10.1128/JVI.00974-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 18.(a) Bouyac-Bertoia M, Dvorin JD, Fouchier RAM, Jenkins Y, Meyer BE, Wu LI, Emerman M, Malim MH. HIV-1 Infection Requires a Functional Integrase NLS. Molecular Cell. 2001;7(5):1025–1035. doi: 10.1016/s1097-2765(01)00240-4. [DOI] [PubMed] [Google Scholar]; (b) Bukrinsky MI, Haggerty S, Dempsey MP, Sharova N, Adzhubei A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365(6447):666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Belshan M, Ratner L. Identification of the nuclear localization signal of human immunodeficiency virus type 2 Vpx. Virology. 2003;311(1):7–15. doi: 10.1016/s0042-6822(03)00093-x. [DOI] [PubMed] [Google Scholar]; (d) Jenkins Y, McEntee M, Weis K, Greene WC. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J Cell Biol. 1998;143(4):875–885. doi: 10.1083/jcb.143.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Kamata M, Aida Y. Two putative alpha-helical domains of human immunodeficiency virus type 1 Vpr mediate nuclear localization by at least two mechanisms. J Virol. 2000;74(15):7179–7186. doi: 10.1128/jvi.74.15.7179-7186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Haffar OK, Popov S, Dubrovsky L, Agostini I, Tang H, Pushkarsky T, Nadler SG, Bukrinsky M. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. Journal of Molecular Biology. 2000;299(2):359–368. doi: 10.1006/jmbi.2000.3768. [DOI] [PubMed] [Google Scholar]
- 19.(a) Devroe E, Engelman A, Silver PA. Intracellular transport of human immunodeficiency virus type 1 integrase. J Cell Sci. 2003;116(Pt 21):4401–4408. doi: 10.1242/jcs.00747. [DOI] [PubMed] [Google Scholar]; (b) Fouchier RA, Meyer BE, Simon JH, Fischer U, Malim MH. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. Embo J. 1997;16(15):4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Limon A, Devroe E, Lu R, Ghory HZ, Silver PA, Engelman A. Nuclear Localization of Human Immunodeficiency Virus Type 1 Preintegration Complexes (PICs): V165A and R166A Are Pleiotropic Integrase Mutants Primarily Defective for Integration, Not PIC Nuclear Import. J. Virol. 2002;76(21):10598–10607. doi: 10.1128/JVI.76.21.10598-10607.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Mannioui A, Nelson E, Schiffer C, Felix N, Le Rouzic E, Benichou S, Gluckman JC, Canque B. Human immunodeficiency virus type 1 KK26-27 matrix mutants display impaired infectivity, circularization and integration but not nuclear import. Virology. 2005;339(1):21–31. doi: 10.1016/j.virol.2005.05.023. [DOI] [PubMed] [Google Scholar]; (e) Petit C, Schwartz O, Mammano F. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J Virol. 2000;74(15):7119–7126. doi: 10.1128/jvi.74.15.7119-7126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Yamashita M, Emerman M. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 2005;1(3):e18. doi: 10.1371/journal.ppat.0010018. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Dvorin J, Bell P, Maul G, Yamashita M, Emerman M, Malim M. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J Virol. 2002;76:12087–12096. doi: 10.1128/JVI.76.23.12087-12096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Limon A, Nakajima N, Lu R, Ghory HZ, Engelman A. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J Virol. 2002;76(23):12078–12086. doi: 10.1128/JVI.76.23.12078-12086.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Riviere L, Darlix JL, Cimarelli A. Analysis of the viral elements required in the nuclear import of HIV-1 DNA. J Virol. 2010;84(2):729–739. doi: 10.1128/JVI.01952-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnan L, Matreyek KA, Oztop I, Lee K, Tipper CH, Li X, Dar MJ, Kewalramani VN, Engelman A. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J Virol. 2010;84(1):397–406. doi: 10.1128/JVI.01899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christ F, Thys W, De Rijck J, Gijsbers R, Albanese A, Arosio D, Emiliani S, Rain JC, Benarous R, Cereseto A, Debyser Z. Transportin-SR2 imports HIV into the nucleus. Curr Biol. 2008;18(16):1192–1202. doi: 10.1016/j.cub.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 22.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70(2):1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebina H, Aoki J, Hatta S, Yoshida T, Koyanagi Y. Role of Nup98 in nuclear entry of human immunodeficiency virus type 1 cDNA. Microbes Infect. 2004;6(8):715–724. doi: 10.1016/j.micinf.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Ao Z, Danappa Jayappa K, Wang B, Zheng Y, Kung S, Rassart E, Depping R, Kohler M, Cohen EA, Yao X. Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J Virol. 2010;84(17):8650–8663. doi: 10.1128/JVI.00508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(a) Fassati A, Gorlich D, Harrison I, Zaytseva L, Mingot JM. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. Embo J. 2003;22(14):3675–3685. doi: 10.1093/emboj/cdg357. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zaitseva L, Cherepanov P, Leyens L, Wilson SJ, Rasaiyaah J, Fassati A. HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome. Retrovirology. 2009;6:11. doi: 10.1186/1742-4690-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaitseva L, Myers R, Fassati A. tRNAs Promote Nuclear Import of HIV-1 Intracellular Reverse Transcription Complexes. PLoS Biology. 2006;4(10):e332. doi: 10.1371/journal.pbio.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(a) Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266(5193):2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]; (b) Violot S, Hong SS, Rakotobe D, Petit C, Gay B, Moreau K, Billaud G, Priet S, Sire J, Schwartz O, Mouscadet JF, Boulanger P. The human polycomb group EED protein interacts with the integrase of human immunodeficiency virus type 1. J Virol. 2003;77(23):12507–12522. doi: 10.1128/JVI.77.23.12507-12522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.(a) Ao Z, Huang G, Yao H, Xu Z, Labine M, Cochrane AW, Yao X. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J Biol Chem. 2007;282(18):13456–13467. doi: 10.1074/jbc.M610546200. [DOI] [PubMed] [Google Scholar]; (b) Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M. Acetylation of HIV-1 integrase by p300 regulates viral integration. Embo J. 2005;24(17):3070–3081. doi: 10.1038/sj.emboj.7600770. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278(1):372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]; (d) Hamamoto S, Nishitsuji H, Amagasa T, Kannagi M, Masuda T. Identification of a novel human immunodeficiency virus type 1 integrase interactor, Gemin2, that facilitates efficient viral cDNA synthesis in vivo. J Virol. 2006;80(12):5670–5677. doi: 10.1128/JVI.02471-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.(a) Farnet CM, Bushman FD. HIV-1 cDNA Integration: Requirement of HMG I(Y) Protein for Function of Preintegration Complexes In Vitro. Cell. 1997;88(4):483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]; (b) Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. PNAS. 1998;95(26):15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(a) Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319(5865):921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]; (b) Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135(1):49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Rato S, Maia S, Brito PM, Resende L, Pereira CF, Moita C, Freitas RP, Moniz-Pereira J, Hacohen N, Moita LF, Goncalves J. Novel HIV-1 knockdown targets identified by an enriched kinases/phosphatases shRNA library using a long-term iterative screen in Jurkat T-cells. PLoS One. 2010;5(2):e9276. doi: 10.1371/journal.pone.0009276. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284(29):19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4(5):495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Goff SP. Knockdown screens to knockout HIV-1. Cell. 2008;135(3):417–420. doi: 10.1016/j.cell.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.(a) Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80(18):9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Denard J, Rundwasser S, Laroudie N, Gonnet F, Naldini L, Radrizzani M, Galy A, Merten OW, Danos O, Svinartchouk F. Quantitative proteomic analysis of lentiviral vectors using 2-DE. Proteomics. 2009;9(14):3666–3676. doi: 10.1002/pmic.200800747. [DOI] [PubMed] [Google Scholar]; (c) Saphire AC, Gallay PA, Bark SJ. Proteomic analysis of human immunodeficiency virus using liquid chromatography/tandem mass spectrometry effectively distinguishes specific incorporated host proteins. J Proteome Res. 2006;5(3):530–538. doi: 10.1021/pr050276b. [DOI] [PubMed] [Google Scholar]
- 33.(a) Chan EY, Qian W-J, Diamond DL, Liu T, Gritsenko MA, Monroe ME, Camp DG, II, Smith RD, Katze MG. Quantitative Analysis of Human Immunodeficiency Virus Type 1-Infected CD4+ Cell Proteome: Dysregulated Cell Cycle Progression and Nuclear Transport Coincide with Robust Virus Production. J. Virol. 2007;81(14):7571–7583. doi: 10.1128/JVI.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chan EY, Sutton JN, Jacobs JM, Bondarenko A, Smith RD, Katze MG. Dynamic host energetics and cytoskeletal proteomes in human immunodeficiency virus type 1-infected human primary CD4 cells: analysis by multiplexed label-free mass spectrometry. J Virol. 2009;83(18):9283–9295. doi: 10.1128/JVI.00814-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Melendez LM, Colon K, Rivera L, Rodriguez-Franco E, Toro-Nieves D. Proteomic analysis of HIV-infected macrophages. J Neuroimmune Pharmacol. 2011;6(1):89–106. doi: 10.1007/s11481-010-9253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Rasheed S, Yan JS, Lau A, Chan AS. HIV replication enhances production of free fatty acids, low density lipoproteins and many key proteins involved in lipid metabolism: a proteomics study. PLoS One. 2008;3(8):e3003. doi: 10.1371/journal.pone.0003003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Ringrose JH, Jeeninga RE, Berkhout B, Speijer D. Proteomic studies reveal coordinated changes in T-cell expression patterns upon infection with human immunodeficiency virus type 1. J Virol. 2008;82(9):4320–4330. doi: 10.1128/JVI.01819-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coiras M, Camafeita E, Urena T, Lopez JA, Caballero F, Fernandez B, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. Modifications in the human T cell proteome induced by intracellular HIV-1 Tat protein expression. Proteomics. 2006;6(Suppl 1):S63–S73. doi: 10.1002/pmic.200500437. [DOI] [PubMed] [Google Scholar]
- 35.(a) Ciborowski P, Kadiu I, Rozek W, Smith L, Bernhardt K, Fladseth M, Ricardo-Dukelow M, Gendelman HE. Investigating the human immunodeficiency virus type 1-infected monocyte-derived macrophage secretome. Virology. 2007;363(1):198–209. doi: 10.1016/j.virol.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rozek W, Ricardo-Dukelow M, Holloway S, Gendelman HE, Wojna V, Melendez LM, Ciborowski P. Cerebrospinal fluid proteomic profiling of HIV-1-infected patients with cognitive impairment. J Proteome Res. 2007;6(11):4189–4199. doi: 10.1021/pr070220c. [DOI] [PubMed] [Google Scholar]; (c) Wiederin J, Rozek W, Duan F, Ciborowski P. Biomarkers of HIV-1 associated dementia: proteomic investigation of sera. Proteome Sci. 2009;7:8. doi: 10.1186/1477-5956-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raghavendra NK, Shkriabai N, Graham R, Hess S, Kvaratskhelia M, Wu L. Identification of host proteins associated with HIV-1 preintegration complexes isolated from infected CD4+ cells. Retrovirology. 2010;7:66. doi: 10.1186/1742-4690-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D'Orso I, Fernandes J, Fahey M, Mahon C, O'Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. Global landscape of HIV-human protein complexes. Nature. 2011;481(7381):365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.(a) Belshan M, Schweitzer CJ, Donnellan MD, Lu R, Engelman A. In vivo biotinylation and capture of HIV-1 matrix and integrase proteins. J Virol Methods. 2009;159:178–184. doi: 10.1016/j.jviromet.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Engelman A. Isolation and analysis of HIV-1 preintegration complexes. In: Prasad VR, Kalpana GV, editors. HIV Protocols. 2nd Edition. Totowa, NJ: The Humana Press; 2009. pp. 135–149. [DOI] [PubMed] [Google Scholar]
- 39.Brown HE, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J Virol. 1999;73(11):9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74(21):10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler S, Hansen M, Bushman F. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 42.(a) Kraft-Terry S, Gerena Y, Wojna V, Plaud-Valentin M, Rodriguez Y, Ciborowski P, Mayo R, Skolasky R, Gendelman HE, Melendez LM. Proteomic analyses of monocytes obtained from Hispanic women with HIV-associated dementia show depressed antioxidants. Proteomics Clin Appl. 2010;4(8–9):706–714. doi: 10.1002/prca.201000010. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wiederin JL, Donahoe RM, Anderson JR, Yu F, Fox HS, Gendelman HE, Ciborowski PS. Plasma Proteomic Analysis of Simian Immunodeficiency Virus Infection of Rhesus Macaques. Journal of Proteome Research. 2010;9(9):4721–4731. doi: 10.1021/pr1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu R, Vandegraaff N, Cherepanov P, Engelman A. Lys-34, Dispensable for Integrase Catalysis, Is Required for Preintegration Complex Function and Human Immunodeficiency Virus Type 1 Replication. J. Virol. 2005;79(19):12584–12591. doi: 10.1128/JVI.79.19.12584-12591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.(a) Iordanskiy S, Berro R, Altieri M, Kashanchi F, Bukrinsky M. Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin. Retrovirology. 2006;3(1):4. doi: 10.1186/1742-4690-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hulme AE, Perez O, Hope TJ. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci U S A. 2011;108(24):9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.(a) Chen P, Hubner W, Spinelli MA, Chen BK. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007;81(22):12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Martin N, Welsch S, Jolly C, Briggs JA, Vaux D, Sattentau QJ. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J Virol. 2010;84(7):3516–3527. doi: 10.1128/JVI.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sato H, Orenstein J, Dimitrov D, Martin M. Cell-to-cell spread of HIV-1 occurs within minutes and may not involve the participation of virus particles. Virology. 1992;186(2):712–724. doi: 10.1016/0042-6822(92)90038-q. [DOI] [PubMed] [Google Scholar]
- 46.(a) Kewalramani VN, Park CS, Gallombardo PA, Emerman M. Protein Stability Influences Human Immunodeficiency Virus Type 2 Vpr Virion Incorporation and Cell Cycle Effect. Virology. 1996;218(2):326. doi: 10.1006/viro.1996.0201. [DOI] [PubMed] [Google Scholar]; (b) Liska V, Spehner D, Mehtali M, Schmitt D, Kirn A, Aubertin AM. Localization of viral protein X in simian immunodeficiency virus macaque strain and analysis of its packaging requirements. J Gen Virol. 1994;75(Pt 11):2955–2962. doi: 10.1099/0022-1317-75-11-2955. [DOI] [PubMed] [Google Scholar]; (c) Wyma DJ, Kotov A, Aiken C. Evidence for a stable interaction of gp41 with Pr55(Gag) in immature human immunodeficiency virus type 1 particles. J Virol. 2000;74(20):9381–9387. doi: 10.1128/jvi.74.20.9381-9387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Yu X, Matsuda Z, Yu QC, Lee TH, Essex M. Vpx of simian immunodeficiency virus is localized primarily outside the virus core in mature virions. J Virol. 1993;67(7):4386–4390. doi: 10.1128/jvi.67.7.4386-4390.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Hum Retroviruses. 1993;9(9):817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, Martin SL, Bushman FD. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. Embo J. 2001;20(12):3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Y, Ao Z, Wang B, Danappa Jayappa K, Yao X. Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J Biol Chem. 2011;286:17722–17735. doi: 10.1074/jbc.M110.184739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.(a) Agostini I, Popov S, Li J, Dubrovsky L, Hao T, Bukrinsky M. Heat-shock protein 70 can replace viral protein R of HIV-1 during nuclear import of the viral preintegration complex. Exp Cell Res. 2000;259(2):398–403. doi: 10.1006/excr.2000.4992. [DOI] [PubMed] [Google Scholar]; (b) Gurer C, Cimarelli A, Luban J. Specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J Virol. 2002;76(9):4666–4670. doi: 10.1128/JVI.76.9.4666-4670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Iordanskiy S, Zhao Y, DiMarzio P, Agostini I, Dubrovsky L, Bukrinsky M. Heat-shock protein 70 exerts opposing effects on Vpr-dependent and Vpr-independent HIV-1 replication in macrophages. Blood. 2004;104(6):1867–1872. doi: 10.1182/blood-2004-01-0081. [DOI] [PubMed] [Google Scholar]
- 51.Madrid R, Janvier K, Hitchin D, Day J, Coleman S, Noviello C, Bouchet J, Benmerah A, Guatelli J, Benichou S. Nef-induced alteration of the early/recycling endosomal compartment correlates with enhancement of HIV-1 infectivity. J Biol Chem. 2005;280(6):5032–5044. doi: 10.1074/jbc.M401202200. [DOI] [PubMed] [Google Scholar]
- 52.(a) Ou SH, Garcia-Martinez LF, Paulssen EJ, Gaynor RB. Role of flanking E box motifs in human immunodeficiency virus type 1 TATA element function. J Virol. 1994;68(11):7188–7199. doi: 10.1128/jvi.68.11.7188-7199.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhou Q, Sharp PA. Novel mechanism and factor for regulation by HIV-1 Tat. Embo J. 1995;14(2):321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.(a) Brauksiepe B, Mujica A, Herrmann H, Schmidt E. The Serine/threonine kinase Stk33 exhibits autophosphorylation and phosphorylates the intermediate filament protein Vimentin. BMC Biochemistry. 2008;9(1):25. doi: 10.1186/1471-2091-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mujica AO, Hankeln T, Schmidt ER. A novel serine/threonine kinase gene, STK33, on human chromosome 11p15.3. Gene. 2001;280(1–2):175–181. doi: 10.1016/s0378-1119(01)00780-6. [DOI] [PubMed] [Google Scholar]
- 54.Karczewski MK, Strebel K. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. Journal of Virology. 1996;70(1):494–507. doi: 10.1128/jvi.70.1.494-507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.(a) Shoeman RL, Höner B, Stoller TJ, Kesselmeier C, Miedel MC, Traub P, Graves MC. Human immunodeficiency virus type 1 protease cleaves the intermediate filament proteins vimentin, desmin, and glial fibrillary acidic protein. Proceedings of the National Academy of Sciences. 1990;87(16):6336–6340. doi: 10.1073/pnas.87.16.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Snášel J, Shoeman R, Hořejšı; amp x, Magda, Hrušková-Heidingsfeldová O, Sedláček J, Ruml T, Pichová I. Cleavage of Vimentin by Different Retroviral Proteases. Archives of Biochemistry and Biophysics. 2000;377(2):241–245. doi: 10.1006/abbi.2000.1776. [DOI] [PubMed] [Google Scholar]
- 56.Levi G, Patrizio M, Bernardo A, Petrucci TC, Agresti C. Human immunodeficiency virus coat protein gp120 inhibits the beta-adrenergic regulation of astroglial and microglial functions. Proceedings of the National Academy of Sciences. 1993;90(4):1541–1545. doi: 10.1073/pnas.90.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas EK, Connelly RJ, Pennathur S, Dubrovsky L, Haffar OK, Bukrinsky MI. Anti-idiotypic antibody to the V3 domain of gp120 binds to vimentin: a possible role of intermediate filaments in the early steps of HIV-1 infection cycle. Viral Immunol. 1996;9(2):73–87. doi: 10.1089/vim.1996.9.73. [DOI] [PubMed] [Google Scholar]
- 58.Bukrinskaya A, Brichacek B, Mann A, Stevenson M. Establishment of a Functional Human Immunodeficiency Virus Type 1 (HIV-1) Reverse Transcription Complex Involves the Cytoskeleton. J. Exp. Med. 1998;188(11):2113–2125. doi: 10.1084/jem.188.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moss SE, Morgan RO. The annexins. Genome Biol. 2004;5(4):219. doi: 10.1186/gb-2004-5-4-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Enrich C, Rentero C, de Muga SV, Reverter M, Mulay V, Wood P, Koese M, Grewal T. Annexin A6-Linking Ca(2+) signaling with cholesterol transport. Biochim Biophys Acta. 2011;1813(5):935–947. doi: 10.1016/j.bbamcr.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 61.Ryzhova EV, Vos RM, Albright AV, Harrist AV, Harvey T, Gonzalez-Scarano F. Annexin 2: a novel human immunodeficiency virus type 1 Gag binding protein involved in replication in monocyte-derived macrophages. J Virol. 2006;80(6):2694–2704. doi: 10.1128/JVI.80.6.2694-2704.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rai T, Mosoian A, Resh MD. Annexin 2 is not required for human immunodeficiency virus type 1 particle production but plays a cell type-dependent role in regulating infectivity. J Virol. 2010;84(19):9783–9792. doi: 10.1128/JVI.01584-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cubells L, Vila de Muga S, Tebar F, Wood P, Evans R, Ingelmo-Torres M, Calvo M, Gaus K, Pol A, Grewal T, Enrich C. Annexin A6-induced alterations in cholesterol transport and caveolin export from the Golgi complex. Traffic. 2007;8(11):1568–1589. doi: 10.1111/j.1600-0854.2007.00640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang XM, Nadeau PE, Lin S, Abbott JR, Mergia A. Caveolin 1 inhibits HIV replication by transcriptional repression mediated through NF-kappaB. J Virol. 2011;85(11):5483–5493. doi: 10.1128/JVI.00254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campbell LH, Borg KT, Haines JK, Moon RT, Schoenberg DR, Arrigo SJ. Human immunodeficiency virus type 1 Rev is required in vivo for binding of poly(A)-binding protein to Rev-dependent RNAs. Journal of Virology. 1994;68(9):5433–5438. doi: 10.1128/jvi.68.9.5433-5438.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Álvarez E, Castelló A, Menéndez-arias L, Carrasco L. HIV protease cleaves poly(A)-binding protein. Biochem J. 2006;396(2):219–226. doi: 10.1042/BJ20060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17(15):4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daher A, Laraki G, Singh M, Melendez-Peña CE, Bannwarth S, Peters AHFM, Meurs EF, Braun RE, Patel RC, Gatignol A. TRBP Control of PACT-Induced Phosphorylation of Protein Kinase R Is Reversed by Stress. Molecular and Cellular Biology. 2009;29(1):254–265. doi: 10.1128/MCB.01030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith AJ, Li Q, Wietgrefe SW, Schacker TW, Reilly CS, Haase AT. Host Genes Associated with HIV-1 Replication in Lymphatic Tissue. The Journal of Immunology. 2010;185(9):5417–5424. doi: 10.4049/jimmunol.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.(a) Kiesler P, Haynes PA, Shi L, Kao PN, Wysocki VH, Vercelli D. NF45 and NF90 regulate HS4-dependent interleukin-13 transcription in T cells. J Biol Chem. 2010;285(11):8256–8267. doi: 10.1074/jbc.M109.041004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pfeifer I, Elsby R, Fernandez M, Faria PA, Nussenzveig DR, Lossos IS, Fontoura BM, Martin WD, Barber GN. NFAR-1 and-2 modulate translation and are required for efficient host defense. Proc Natl Acad Sci U S A. 2008;105(11):4173–4178. doi: 10.1073/pnas.0711222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.(a) Urcuqui-Inchima S, Castano ME, Hernandez-Verdun D, St-Laurent G, 3rd, Kumar A. Nuclear Factor 90, a cellular dsRNA binding protein inhibits the HIV Rev-export function. Retrovirology. 2006;3:83. doi: 10.1186/1742-4690-3-83. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Urcuqui-Inchima S, Patino C, Zapata X, Garcia MP, Arteaga J, Chamot C, Kumar A, Hernandez-Verdun D. Production of HIV particles is regulated by altering sub-cellular localization and dynamics of Rev induced by double-strand RNA binding protein. PLoS One. 2011;6(2):e16686. doi: 10.1371/journal.pone.0016686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.