Abstract
We assessed human mesenchymal stem cells (MSCs) harvested from breast and abdominal adipose tissues enriched in embryonic stage specific antigen (SSEA-4) expression for osteogenic and adipogenic differentiation in comparison to a mixed cell population. Human adipose was obtained from abdominal and breast tissues of females undergoing gastric bypass and breast reduction respectively. SSEA-4 expressing cells were enriched from the mixed cell population by magnetic cell sorting and expanded in culture. The results showed that freshly isolated cells from breast and abdominal tissues based on adipose from 3 patients comprised 12% and 10% SSEA-4+ cells respectively. At passage 0, 48 % of the cells from breast adipose tissue were positive for SSEA-4 while 12% of the cells from abdominal adipose tissue were positive for this antigen. The level of SSEA-4 expressing cells remained relatively constant with passaging; SSEA-4 expressing cells from breast tissue comprised 45 % of the total while 27% of the cells from abdominal adipose tissue expressed SSEA-4 at passage 5. Cells sorted for SSEA-4 expression exhibited a higher potential for differentiation toward osteogenic and adipogenc cell lineages in vitro when compared to a mixed population. Interestingly, SSEA-4 expression was lost upon differentiation suggesting that the antigen marks a subpopulation of MSCs. Taken together, the data demonstrate that breast adipose tissue is highly enriched in a subpopulation of MSCs expressing SSEA-4 and suggests that SSEA-4 may be a marker of a subpopulation of MSCs with high potential for osteogenic and adipogenic differentiation.
Keywords: Breast adipose tissue, mesenchymal stem cells, SSEA-4, stromal cells, differentiation
INTRODUCTION
Mesenchymal stem cells (MSCs) or multipotent mesenchymal stromal cells have generated interest and excitement because of the potential they hold in regenerative medicine. The cells are present in many tissues particularly bone marrow, muscle, fat and other tissues. (Pittenger et al. 1999; Gronthos et al. 2000; Lee et al. 2000; Zuk et al. 2001; De Bari et al. 2003; Bi et al. 2007). The most studied MSCs are those harvested from bone marrow; although adipose tissue has been shown to be an abundant source of these cells, it has received less attention. Adipose tissue is an attractive source of MSCs because it is plentiful and easy to obtain compared to most other types of mesenchymal tissues. Apart from their ease of isolation, adipose derived stem cells (ADSCs) and bone marrow derived mesenchymal stem cells (BMSCs) do not possess any major differences; however minor differences have been noted (Gimble et al. 2007; Yoshimura et al. 2007; Merceron et al. 2010; Niemeyer et al. 2010)
There are no established specific markers available for the identification of MSCs; thus MSCs are characterized in part by cell surface markers that are also expressed by other cell types with no stem cell characteristics (Chamberlain et al. 2007; D Docheva 2008; Alison and Islam 2009; Maddox et al. 2009). Some of the surface markers that identify MSCs include: CD49a, CD90, CD105, CD146, CD271, and STRO-1 among many others (Chamberlain et al. 2007; Crisan et al. 2008; D Docheva 2008; Alison and Islam 2009; Maddox et al. 2009; Halfon et al. 2010). These markers serve as a guide for characterizing MSCs harvested from various tissues.
Stage Specific Embryonic Antigen-4 (SSEA-4) is an early embryonic globoseries glycolipid antigen which is commonly used to identify human embryonic stem cells (Carpenter et al. 2003). This antigen has also been found to be expressed by MSCs, testicular germ cells, and aggressive cancer cell lines such as renal and breast cancer (Saito et al. 1991; Saito et al. 2003; Gang et al. 2007; Villadsen et al. 2007; Muller et al. 2008; Charafe-Jauffret et al. 2009; Gonzalez et al. 2009). The first report on SSEA-4 as a maker for MSC was by Perlingeiro and colleagues (Gang et. al., 2007). The authors reported that SSEA-4 positive cells were present in murine marrow and represented 1–2% of the cell population at day 0 of cell isolation. Similarly the authors reported that in human, SSEA-4+ cells comprised 2–4% of the cell population at day 0. The authors reported that the levels of cells expressing SSEA-4 increased with culturing reaching 78% of the cell population at day 21. The authors utilized SSEA-4 to isolate a homogenous population of multipotent bone marrow derived MSCs (BMSCs) and demonstrated that cells expressing this antigen exhibited multipotent differentiation (Gang et al. 2007). The authors concluded that SSEA-4 could prospectively be used to isolate MSCs from a mixed population of cells. The authors however did not examine uniqueness of the cells expressing this antigen in comparison to the mixed population. Since this report, few studies have followed expression of this antigen by cells harvested from other mesenchymal tissues.
A number of clinical trials are currently under way to assess effectiveness of ADSC in treating various pathological conditions. Some of these conditions include breast augmentation and reconstruction; immune related diseases, e.g. Crohn’s disease (Gimble et al. 2011). The outcomes of these clinical trials remain to be evaluated.
In the present investigation, we set out to determine the level of SSEA-4 expressing cells in preparations from adipose tissues and asked whether SSEA-4 could be considered a universal marker for the identification of MSCs. We also determined osteogenic and adipogenic differentiation of cells enriched in SSEA-4 expression and the mixed population. We report here that adipose tissue is enriched in cells expressing SSEA-4, especially cells in breast adipose tissue. The cells expressing this antigen exhibit a higher potential to differentiate into osteogenic and adipogenic cell lineages in vitro when compared to the mixed population. In contrast to the report by Gang et al (Gang et al. 2007) for cells in bone marrow and SSEA-4 expression; breast adipose tissue appeared to be enriched in a population of cells expressing this antigen at day 0 of isolation and its expression remained relatively constant.
MATERIALS AND METHODS
Isolation of the adipose derived mesenchymal stem cells (ADSCs)
Adipose derived stem cells (ADSCs) were isolated from female (ages 57–64) breast and abdominal adipose tissues. Breast adipose was obtained from patients undergoing breast reduction and abdominal fat was obtained from patients undergoing gastric bypass. In each case fat was harvested from 3 patients; however breast and abdominal adipose tissues were not harvested from the same patients but were of relatively close age ranges; 57–64 years. To isolate cells, adipose tissues were minced, washed extensively in sterile PBS (Invitrogen, Carisbad, CA) and digested with 0.075% Type II collagenase (Worthington, Lakewood, NJ) at 37°C for 30 minutes with gentle agitation as described previously (Zuk et al. 2001; Liao et al. 2008; Maddox et al. 2009). The digested fat was centrifuged at 2,000 rpm for 10 minutes. The pellet was washed repeatedly with PBS and then passed through 70 µm mesh filter. The filtrate was centrifuged as above and plated onto type I collagen coated tissue culture plates in DMEM (InVitrogen, Carisbad, CA) supplemented with 10% FBS, 1% penicillin/ streptomycin v/v (P/S) (All from Invitrogen) and incubated at 37°C in 5% CO2. Adipose tissue was obtained from 3 female patients in each case.
Flow cytometry
The isolated cells were incubated in PE-labeled antibodies against CD45 and CD49a (BD Bioscience); and primary antibodies specific for SSEA-4 (Developmental Studies Hybridoma Bank, Iowa City, IA), CD105 (Endoglin) (eBioscience, San Diego CA), and CD146 (MelCAM) (Santa Cruz, Santa Cruz, CA) followed by incubation in secondary antibodies conjugated to PE-Cy5 (goat anti-mouse IgG PE-Cy5 conjugate; Santa Cruz, Santa Cruz, CA). Cells were incubated in the respective antibodies at a concentration of 20 µg/mL. The expression level of each antigen was assessed by flow cytometry. Three trials were completed for each marker; each trial was run in triplicate.
Sorting for SSEA-4 expressing cells with magnetic cell sorting system (MACS)
Since the focus was on cells isolated from breast adipose tissue, cells (ADSCs) isolated from breast adipose tissues were sorted for SSEA-4 by Magnetic Cell Sorting System (MACS, Miltenyi Biotec, Auburn, CA). Cells were incubated at 4°C for 30 minutes with SSEA-4 antibody at a concentration of 20µg/mL. The cells were washed, and then incubated at 4°C for 15 minutes with MACS (magnetic cell sorting) MicroBeads (20ml/107 cells). Cells were washed, resuspended in 500 µl of MACS buffer, and then added to the magnetized MACS MS magnetic column. The unlabeled cells passed through and were collected. The column was removed from the separation magnet; labeled cells passed through and were collected. The collected labeled cell fraction was resorted for further enrichment into cells expressing SSEA-4. The cells were then plated and expanded in culture.
Immunocytochemistry
Cells (1×103 cells) were plated in 8-well chamber slides and allowed to expand to 80% confluence. The wells were washed, and then fixed in ice-cold methanol/acetone (1:1) for 30 minutes at −20°C. Next, the cells were permeabilzed by washing with 0.1%Tween/PBS (Fisher, Pittsburgh, PA) 2 times. Then cells were blocked with 1% goat serum/PBS (Sigma Aldrich, St. Louis, MO) for 1hr at room temperature. The cells were washed 2–4 times with PBS. Primary SSEA-4 antibody in 1% goat serum/PBS was added overnight at 4°C. Cells were then washed 2–4 times with PBS. A secondary PE-Cy5 antibody (Santa Cruz) in 1% goat serum/PBS was added for 20 minutes. Cells were washed and then the slide was viewed under light microscope. In some experiments visualization was achieved by a secondary antibody conjugated to FITC and cells were also stained with DAPI for nuclei.
Differentiation assays
Alkaline phosphatase activity (ALP) assay
Cells cultured in DMEM supplemented with 10−7M dexamethasone (Sigma, St Louis MO), and 2×10−8 M- 2-Ascorbate (Sigma) were treated with 100 ng/ml of rhBMP-2 (Fitzgerald Industries, Acton, MA) for three days followed by analysis of ALP activity by densitometry using the Revelation Program (Dynex Tech, Virginia) and the methods described previously (Li et al. 2007; Liao et al. 2008). Cells (1×104) were plated in 24 well plates and then grown to 80% confluence before introduction of ALP induction media; each sample was run three independent times in triplicate.
Osteogenic differentiation
SSEA-4+ cells (1×104) were plated onto 24 well collagen coated plates, allowed to reach 80% confluence and then incubated in an osteogenic medium. Osteogenesis induction medium contained: α-minimum essential medium (α-MEM (Invitrogen), 5% FBS, 1% Penicillin /Streptomycin (P/S), 10mM Na-β-glycerophosphate (Sigma), 0.2mM Ascorbic Acid Phosphate, and 10−8 M Dexamethasone. Cells were stained with Alizarin Red S (Sigma) at 25 days after plating in osteogenic medium. Each sample was run three independent times in triplicate. To quantify the level of calcium deposit by the cells, deposited Alizarin Red S was extracted by 10% cetylpyridinium chloride in distilled water and assessed for absorbance at 560 nm. To further confirm the osteogenic potential of the ADSC, a different set of cells processed similarly except staining with Alizarin Red S were collected for osteogenic gene expression analysis.
Adipogenic differentiation
Breast adipose derived SSEA-4+ cells were stained with Oil Red O (Sigma) at 18 days after plating in adipogenic medium. Adipogenesis induction medium contained: DMEM, 10% FBS, 1% Penicillin /Streptomycin, 0.5mM 3-isobutyl-1-methylxanthine (IBMX), 1µM Indomethacin, 1µM Dexamethasone, and 1µM Insulin (all from Sigma). Cells (1×104) were plated in 24 well plates and cultured to 80% confluency before introduction of adipogenesis induction media. Each sample was run three independent times in triplicate. Oil Red O was extracted with 100% isopropanol by its absorbance at 490 nm to assess the level of fat deposit. To further confirm adipogenic potential of the ADSC, cells were collected for adipogenic gene expression analysis following culture in adipogenic differentiation medium.
Gene expression analysis
Gene expression analysis was performed as described previously (Li et al. 2007; Liao et al. 2008). Briefly, total RNA was extracted from 1×106 breast adipose derived SSEA-4+ sorted cells either cultured in osteogenic or maintained in regular maintenance medium using Trizol following manufacturer’s protocol. The mRNA was reverse transcribed to cDNA using SuperScript™ First-Strand Synthesis System for RT-PCR according to manufacturer’s instructions. cDNA was amplified using an Eppendorf PCR System at 94°C for 30 sec, 65°C for 30 sec, and 72°C for 1 min for 30 or 35 cycles, after initial denaturation at 94°C for 5 min. All primer sequences for the osteogenic or adipogenic genes that were used were determined using established GenBank sequences and are indicated in Table II. PCR fragments were analyzed by agarose gel electrophoresis. Gene expression analysis was performed a minimum of three independent times per sample. Triplicate PCR reactions were amplified using primers designed for β-actin as a control for assessing PCR efficiency.
Table II.
Primer sequences used to assess expression of osteoblasts and adipocytes differentiation related genes.
| Runx2 | F5'-ACT GGG CCC TTT TTC AGA-3' R5'-CG GAA GCA TTC TGG AA-3' |
| Osterix | F5'-CCC AGG CAA CAC TCC TAC TC -3' R5'-GGC TGG ATT AAG GGG AGC AAA -3' |
| Osteopontin | F5'-CAC ATC GGA ATG CTC ATT GC-3' R5'-ATC ACC TGT GCC ATA CCA GT-3' |
| Osteocalcin | F5'-CTC ACA CTC CTC GCC CTA-3' R5'-CCT GAA AGC CGA TGT GGT-3' |
| Lipoprotein Lipase | F5'-GTC CGT GGC TAC CTG TCA T-3' R5'-AGC CCT TTC TCA AAG GCT TC-3' |
| Adipsin | F5'-GGT CAC CCA AGC AAC AAA GT-3' R5'-CCT CCT GCG TTC AAG TCA TC-3' |
| PPAR-γ | F5'-GCT GTT ATG GGT GAA ACT CT-3' R5'-ATA AGG TGG AGA TGC AGG CT-3' |
| Beta Actin | F5'-ACC ATG GAT GAT GAT ATC GCC-3' R5'-GTG CCA GAT TTT CTC CAT GTC-3' |
Statistical analysis
Data are expressed as mean±SD. Statistical analyses were performed using Student’s t test, and one-way or two-way analysis of variance followed by post hoc multiple comparison testing. A p-value of <0.05 was considered statistically significant. Experiments were done in triplicates and at least three separate times.
RESULTS
Isolation of adipose derived stem cells (ADSCs)
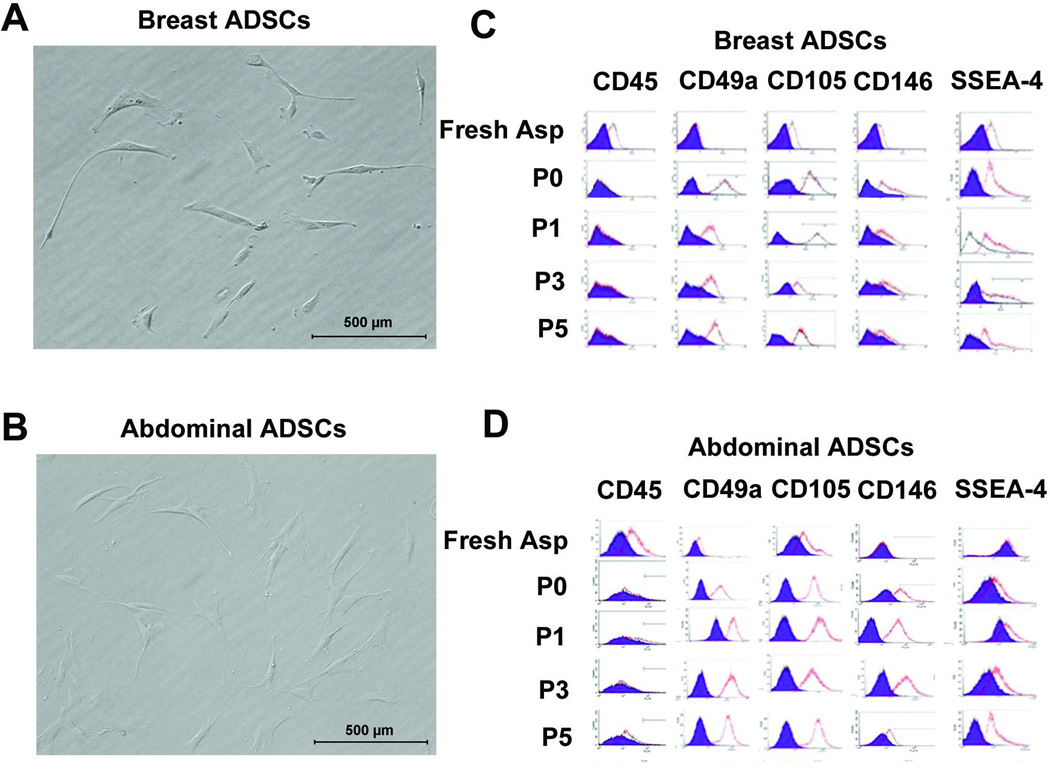
The cells isolated from breast and abdominal adipose tissues were assessed for selected marker expression either immediately after isolation prior to plating or after plating and passaging. The major focus was on SSEA-4 expression but other surface antigens examined were: CD45, CD49a, CD105 and CD146. Figures 1A and B show morphological appearance of cells isolated from breast and abdominal adipose tissues respectively. Expression profile of selected surface antigens by cells isolated from breast and abdominal adipose tissues is shown in Figure 1C and D respectively. Table I shows expression levels of each antigen at different passages in cells isolated from breast tissue. The results show that, prior to plating, the level of cells expressing SSEA-4 in cells isolated from breast tissue on average was 12%. Upon passaging, which removed most of the non-adherent cells, the level of SSEA-4+ cells in adipose derived cells from breast tissue averaged 48% of the adherent cells whereas in cells harvested from abdominal fat, SSEA-4 expressing cells averaged 12% (Table I) based on the adipose tissue harvested from 3 patients. With extended culturing, there was a slight increase in SSEA-4 expressing cells from abdominal tissues up to 27% at passage 5 while those from adipose tissue remained relatively constant at 45% on the average (Table I). These data suggest that a higher number of adherent cells isolated from breast adipose tissue expressed SSEA-4 while adherent cells harvested from abdominal fat tissue contained less cells expressing SSEA-4. This level of expression was consistent for the cells isolated from three different breast and abdominal adipose tissue donors. These data indicate that adipose tissues are enriched in cells expressing SSEA-4 but that breast adipose tissues is highly enriched in cells expressing this antigen.
Figure 1. Morphological appearance and surface marker profile expression of adipose derived mesenchymal stem cells (ADSCs) from breast and abdominal tissues.
(A) Morphological appearance of unsorted ADSCs from breast tissue and (B) abdominal tissue. (C) Selected marker profile as revealed by FACS for breast ADSCs and (D) abdominal ADSCs. Breast and abdominal derived cells display similar surface marker expression except SSEA-4 that is highly expressed by breast adipose derived cells after passage 1. The level of each marker expression is shown in table 1. Scale bar = .
Table I.
Expression profile of selected surface markers by Adipose derived stem cells.
| Breast ADSCs | |||||
|---|---|---|---|---|---|
| CD45 | CD49a | CD105 | CD146 | SSEA-4 | |
| Fresh Aspirate | 21% | 4% | 14% | 10% | 12% |
| P0 | 0% | 98% | 90% | 49% | 48% |
| P1 | 0% | 95% | 97% | 61% | 51% |
| P3 | 1% | 97% | 97% | 70% | 43% |
| P5 | 1% | 99% | 95% | 38% | 45% |
| Abdominal ADSCs | |||||
| CD45 | CD49a | CD105 | CD146 | SSEA-4 | |
| Fresh Aspirate | 26% | 1% | 17% | 4% | 10% |
| P0 | 0.5% | 95% | 92% | 55% | 12% |
| P1 | 0% | 96% | 99% | 69% | 14% |
| P3 | 1% | 96% | 97% | 78% | 21% |
| P5 | 0% | 97% | 98% | 45% | 27% |
Surface marker expression was assessed at different passages. Adipose derived stem cells from breast and abdominal tissues exhibited similar surface marker expression except breast derived cells were enriched in cells expressing SSEA-4.
Sorting of cells expressing SSEA-4
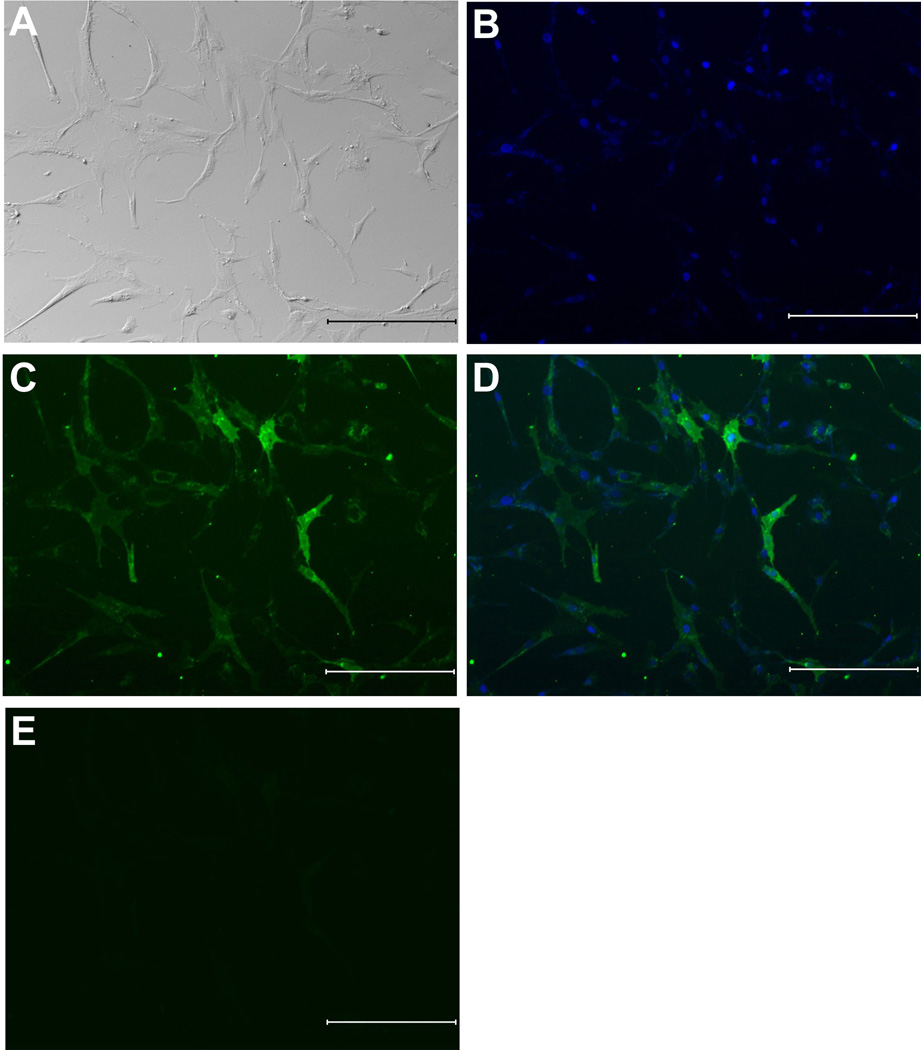
Cells isolated from breast and abdominal tissues were sorted using magnetic cell sorting system based on SSEA-4 expression. Because breast tissue contained a high level of these cells and abdominal and breast derived cells behaved similarly, we focused on cells isolated from this tissue in comparison to the mixed population. Figures 2A shows a bright field image of the morphological appearance of cells sorted for SSEA-4. The sorted cell population exhibited short and spindly morphological appearance (Fig. 2A). To demonstrate that the cells were positive for SSEA-4 they were immunostained for SSEA-4 expression. Following sorting for SSEA-4, 90 % of the cells expressed SSEA-4 as evidenced by the immunofluorescence staining (Fig. 2B, C and D). Sorted cells that were not treated with primary antibody but received secondary antibody to SSEA-4 did not show any immunofluorescence staining (Fig. 2E). These data demonstrate that cells sorted based on SSEA-4 expression were positive for SSEA-4.
Figure 2. Isolation by MACS and immunofluorescence staining of SSEA-4 expressing cells from breast adipose tissue.
(A) Morphological appearance of cells sorted based on SSEA-4 expression (B) SSEA-4+ cells stained with DAPI. (C) Immunofluorescence staining using an antibody specific for the SSEA-4 shows that nearly all cells were SSEA-4 positive following sorting. (D) Merged image of B and C showing that SSEA-4 antigen is expressed on the cell surface by the sorted cells. (E) Negative immunofluorescence staining by SSEA-4+ cells in which primary antibody to SSEA-4 was omitted. At passage 3, breast adipose derived cells were sorted and the resulting cells were assessed for SSEA-4 expression by immunofluorescence. Scale bar=500 µm
Differentiation of SSEA-4 expressing cells
Breast derived sorted cells expressing SSEA-4 were assessed for differentiation toward osteogenic and adipogenic cell lineages. First, cells were analyzed for alkaline phosphatase activity (ALP) expression in vitro following BMP-2 treatment. Results showed that SSEA-4 sorted cells from abdominal and breast adipose tissue expressed higher levels of alkaline phosphatase activity upon treatment with BMP-2 (Fig. 3A, B). Interestingly, SSEA-4+ cells expressed higher levels of ALP activity (1.75× higher) than the mixed population (Fig. 3A, B). These data suggested that SSEA-4+ cells respond to BMP-2 efficiently to differentiate toward osteoblast lineage.
Figure 3. Alkaline phosphatase activity of ADSCs harvested from breast and abdominal adipose tissue.
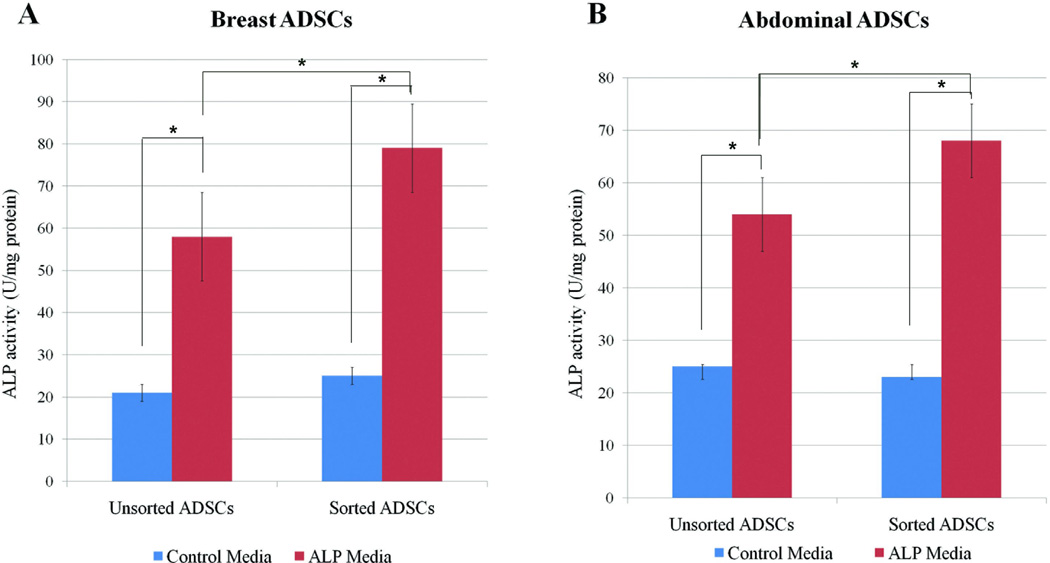
ADSCs sorted for SSEA-4 were treated with or without BMP-2 and ALP activity was assessed at 3 days following treatment. In all cases, ADSCs treated with BMP-2 showed significant increases in ALP activity than non BMP-2 treated. Cells enriched in SSEA-4 expressed higher levels of ALP activity than the mixed cell population for both the (A) breast derived ADSCs and (B) abdominal derived ADSCs. The data is from 3 patients and experiment was performed 3 times. Data is expressed as means of ±SD. P < 0.05. * indicates statistically significant differences.
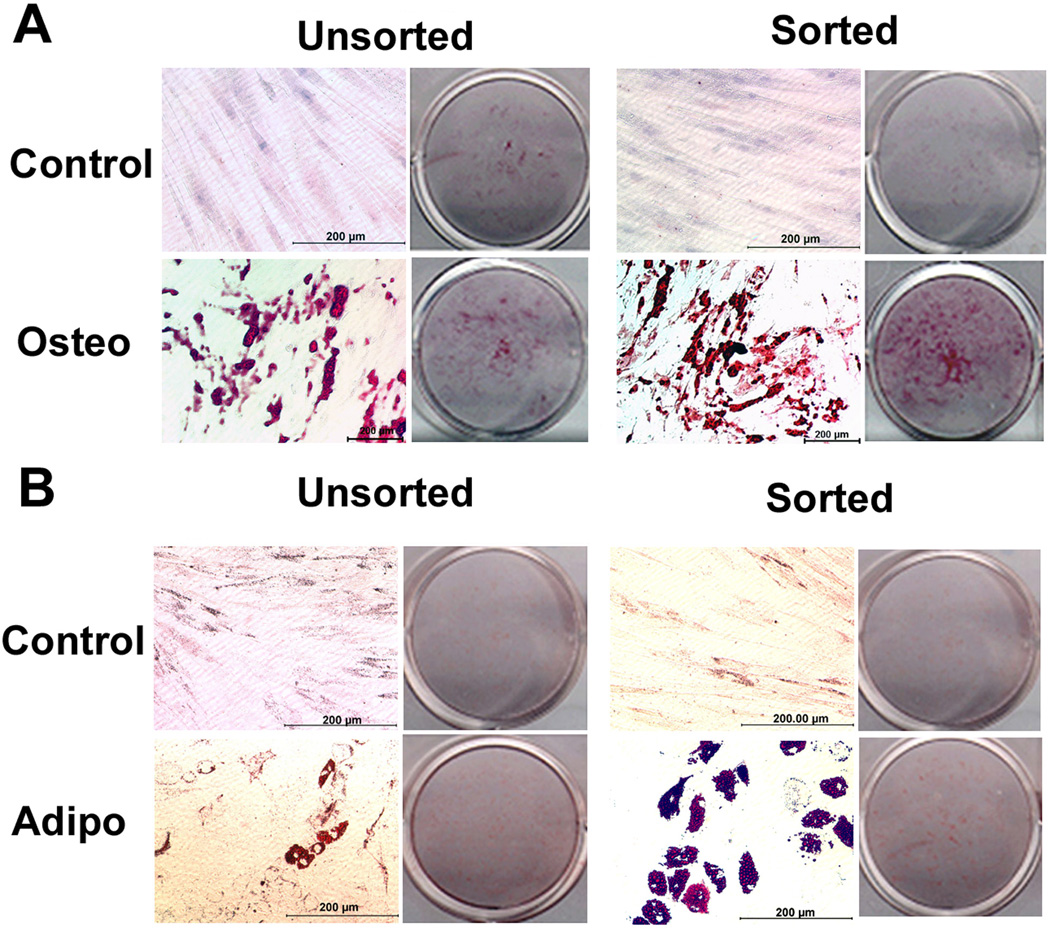
Next, SSEA-4+ sorted and unsorted cells from breast tissues were assessed for their ability to differentiate into osteoblasts and adipocytes when cultured in respective media for 21 days. The results showed that SSEA-4+ cells deposited higher levels of calcium (6× higher) than unsorted population consistent with ALP activity results (Fig. 4A). Similarly, breast adipose derived cells sorted for SSEA-4 expression were more efficient in differentiating toward adipogenic lineage than the mixed population (Fig. 4B). Abdominal derived SSEA-4 sorted cells showed similar findings as breast derived cells and therefore not reported here.
Figure 4. Osteogenic and adipogenic differentiation of cells enriched in SSEA-4+ and a mixed population from breast adipose tissue.
(A) Osteogenic differentiation as indicated by Alizarin Red staining for mineral deposition in osteogenic culture of unsorted and sorted cell populations. A higher level mineral deposition is evident in sorted cells. (B) Adipogenic differentiation of sorted and unsorted cells populations. A higher level of cells positive for oil Red O staining is evident in sorted cells. Equal numbers of cells sorted or unsorted were cultured either in osteogenic or adipogenic medium for 21 days. Control cells were maintained in regular maintenance medium. Osteo= osteogenic medium, Adipo=adipogenic medium. Scale bar = .
Calcium deposition was quantified indirectly by determining the level of Alizarin Red S deposition. Similarly, adipogenic level was assessed by determining the level of oil Red O deposited by the cells. The results demonstrated that sorted breast cells deposited more Alizarin Red S (6×) and oil Red O (1.3×) than the mixed cell population, confirming results shown in figure 4 (Fig. 5A, C). Osteogenic and adipogenic gene expression was assessed to confirm osteogenic and adipogenic differentiation. Semi quantitative PCR was used to assess osteoblast and adipogenic related genes; quantitative PCR was not performed (Fig. 5B, D) because the focus was not to quantitate gene expression but to confirm differentiation of the cells to respective lineages. The results showed expression of genes associated with osteoblasts and adipocytes differentiation (Fig 5B, D).
Figure 5. Quantitation of the level of mineral and oil red staining by sorted and unsorted breast adipose cells.
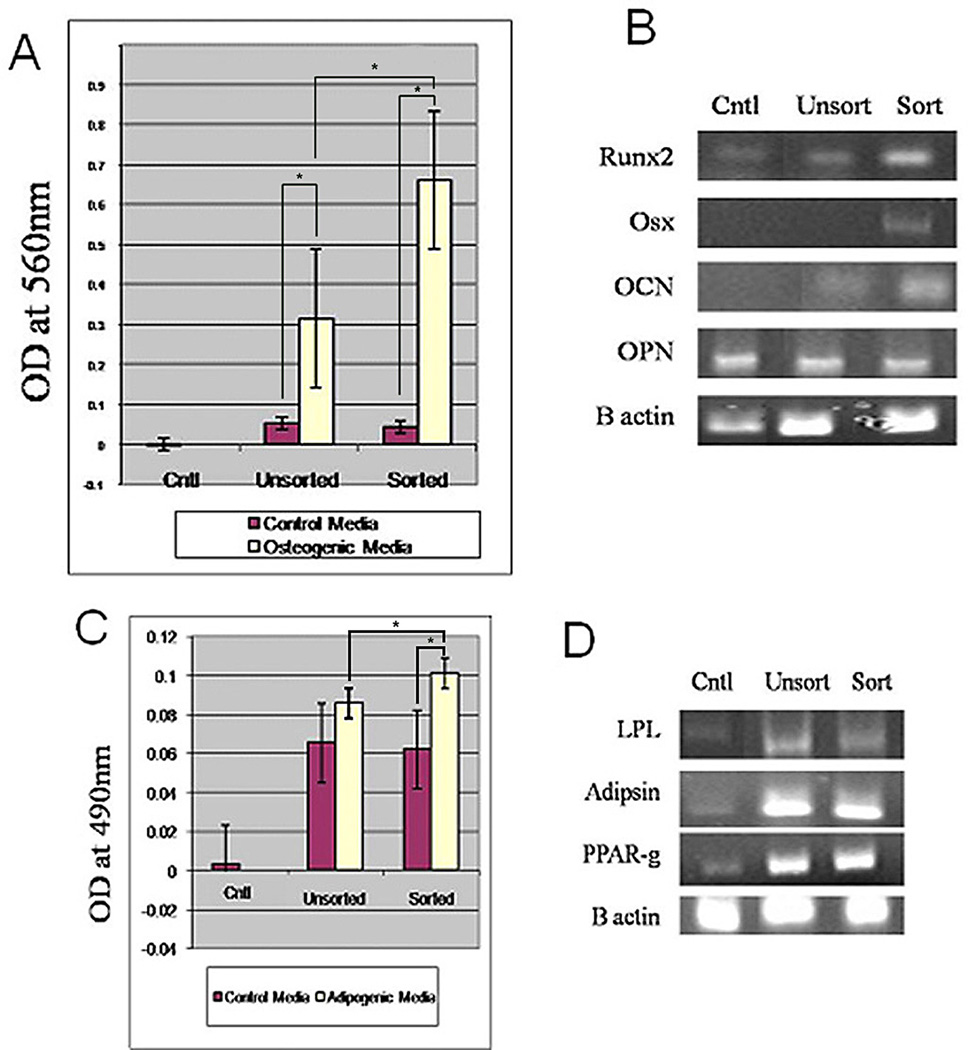
A) Relative level of mineral deposition by sorted and unsorted breast adipose cells. Sorted cells deposit higher levels of mineral deposition (6×) than unsorted cells. B) Expression of osteogenic genes by sorted and unsorted cell populations demonstrated genes associated with osteoblasts differentiation. C) Relative level of oil red deposition by unsorted and sorted cells. Sorted cells show a slight increase (1.3×) in oil red deposition than unsorted cells. D) Expression of genes associated with adipogenic differentiation. Osx= osterix, OCN= osteoclacin, OPN= osteopontin. Cntl= control cells maintained in regular medium, sort= sorted, unsort= unsorted. Data is expressed as means±SD. P≤=0.05.
DISCUSSION
Mesenchymal stem cells have great potential in regenerative medicine. Clinical trials have demonstrated promise of adult derived stem cells in ameliorating effects of Parkinson’s disease, Crohn’s disease, and congestive heart failure (Segers and Lee 2008; Garcia-Olmo et al. 2009; Ko et al. 2010; Lindvall and Kokaia 2010). The therapeutic effects of MSCs are presently attributed in part to trophic factors generated by the cells that modulate microenvironment for tissue repair (Gnecchi et al. 2005; Caplan 2007). The data presented here have demonstrated that the SSEA-4+ cell population has a higher potential for differentiation toward osteoblasts and adipocytes at least in vitro. Present MSCs therapies have focused on using bone marrow derived stem cells (BMSCs); however MSCs may be derived from alternate sources. Adipose tissue has become an appealing source of stem cells as it is abundant, and can be quickly and easily harvested. Adipose derived stem cells (ADSCs) have been shown to exhibit characteristics similar to BMSCs; the present data however suggest that adipose derived MSCs are more enriched in SSEA-4 expressing cells than marrow derived MSCs (Gang et al. 2007).
Initial studies on SSEA-4 as a marker of BMSCs suggested this antigen was a potential marker for MSCs (Gang et al). The level of cells expressing this antigen was however, very low and was estimated at about 2% upon initial isolation. Present findings however have demonstrated that cells expressing this antigen are abundant upon initial isolation from adipose tissue. After the first passage there was a slight increase in cells expressing this antigen from abdominal tissue but those from breast tissue expressing this antigen remained relatively constant. These data are however contrary to those of Gang et al (Gang et al.2007) who showed increased expression of SSEA-4 expressing cells with extended culturing of marrow derived cells. The present findings demonstrated that breast adipose tissue is enriched in cells expressing SSEA-4 and that the level of cells expressing this antigen remained relatively constant with culturing. Interestingly though, upon differentiation, expression of this antigen was lost suggesting that SSEA-4 marks a subset of cell population within adipose tissue that exhibit MSCs characteristics with a higher potential for differentiation toward osteogenic and adipogenic cell lineages.
Stage specific embryonic antigen-4 (SSEA-4) is an early embryonic cell surface antigen commonly used to identify human embryonic stem cells (ESCs). Other reports have shown expression of SSEA-4 by cells in marmoset testis at 9 %, fetal marrow at 2.5 % and by cells in marrow, skin, and adipose at 7, 8 and 4 % respectively (Gang et al. 2007; Riekstina et al. 2009) These levels are much lower than what we have shown in breast adipose tissue in which about 50% adherent cells express this antigen. It is tempting to speculate that cells expressing SSEA-4 would be more primitive than cells that do not express SSEA-4. Indeed some studies have shown that adult derived stem cells which express SSEA-4 also express embryonic stem cell markers like Oct-4 or nanog.(Villadsen et al. 2007; Trubiani et al. 2010)
Breast tissue is comprised of many cell types including cells that express SSEA-4 which are present within the ducts. The SSEA-4 positive cells in breasts ducts have been described as stem cells that give rise to epithelial cells (Wojakowski et al. 2009). It is not clear if the population of cells expressing SSEA-4 isolated from breast adipose tissue is contaminated with this cell population. This is unlikely, because SSEA-4 expressing cells present in breast ducts represent a small population. In addition, following sorting, cells expressing SSEA-4 were spindle shaped which is a characteristic of MSCs; SSEA-4+ cells also expressed high levels of MSC markers CD44, CD49a, and CD105. It is possible that epithelial mesenchymal trasnsition (EMT)could account for presence of cells with MSCs characteristics expressing SSEA-4 in breast tissue. This is however unlikely because, this phenomenon occurs during pathological conditions (Kalluri and Weinberg 2009);the adipose tissues used for harvesting MSCs were from normal individuals. Cells expressing SSEA-4 increased in culture during expansion; this observation suggests that culture conditions could be responsible for the increase in cells expressing this antigen. This is a possibility; however after passage 1, expression of this antigen was relatively constant suggesting culture conditions were not responsible for the expression of this antigen by the cells. Hematopoietic stem cells express SSEA-4, these cells are removed during media changes; FACS analysis data showed that, these cells were eliminated by passage 1 thus indicating that adherent cells are free of CD45 expressing cells thus SSEA-4 is expressed by a subpopulation of MSCs.
Taken together, the SSEA-4 antigen appears to mark a subset of cells of MSCs with a higher potential for differentiation toward osteogenic and adipogenic cell lineages at least in vitro.
Acknowledgements
The SSEA-4 antibody (MC-813-70) developed by D. Solter and B.B. Knowles was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. This work was funded in part by NIH 1R21AR059383-01 and Penn State College of Medicine Department of Orthopaedics.
Literature Cited
- Alison MR, Islam S. Attributes of adult stem cells. J Pathol. 2009;217:144–160. doi: 10.1002/path.2498. [DOI] [PubMed] [Google Scholar]
- Bi Y, Ehirchiou D, Kilts TM, Inkson CA, Embree MC, Sonoyama W, Li L, Leet AI, Seo BM, Zhang L, Shi S, Young MF. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13:1219–1227. doi: 10.1038/nm1630. [DOI] [PubMed] [Google Scholar]
- Caplan AI. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol. 2007;213:341–347. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
- Carpenter MK, Rosler E, Rao MS. Characterization and differentiation of human embryonic stem cells. Cloning Stem Cells. 2003;5:79–88. doi: 10.1089/153623003321512193. [DOI] [PubMed] [Google Scholar]
- Chamberlain G, Fox J, Ashton B, Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25:2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, Norotte C, Teng PN, Traas J, Schugar R, Deasy BM, Badylak S, Buhring HJ, Giacobino JP, Lazzari L, Huard J, Peault B. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Docheva FH, Schieker M. Mesenchymal Stem Cells and Their Cell Surface Receptor. Cur Rheumatology Rev. 2008;4:1–5. [Google Scholar]
- De Bari C, Dell'Accio F, Vandenabeele F, Vermeesch JR, Raymackers JM, Luyten FP. Skeletal muscle repair by adult human mesenchymal stem cells from synovial membrane. J Cell Biol. 2003;160:909–918. doi: 10.1083/jcb.200212064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang EJ, Bosnakovski D, Figueiredo CA, Visser JW, Perlingeiro RC. SSEA-4 identifies mesenchymal stem cells from bone marrow. Blood. 2007;109:1743–1751. doi: 10.1182/blood-2005-11-010504. [DOI] [PubMed] [Google Scholar]
- Garcia-Olmo D, Herreros D, Pascual I, Pascual JA, Del-Valle E, Zorrilla J, De-La-Quintana P, Garcia-Arranz M, Pascual M. Expanded adipose-derived stem cells for the treatment of complex perianal fistula: a phase II clinical trial. Dis Colon Rectum. 2009;52:79–86. doi: 10.1007/DCR.0b013e3181973487. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Bunnell BA, Chiu ES, Guilak F. Adipose-derived Stromal Vascular Fraction Cells and Stem Cells: Let's Not Get Lost in Translation. Stem Cells. 2011 doi: 10.1002/stem.629. 2011. [DOI] [PubMed] [Google Scholar]
- Gimble JM, Katz AJ, Bunnell BA. Adipose-derived stem cells for regenerative medicine. Circ Res. 2007;100:1249–1260. doi: 10.1161/01.RES.0000265074.83288.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnecchi M, He H, Liang OD, Melo LG, Morello F, Mu H, Noiseux N, Zhang L, Pratt RE, Ingwall JS, Dzau VJ. Paracrine action accounts for marked protection of ischemic heart by Akt-modified mesenchymal stem cells. Nat Med. 2005;11:367–368. doi: 10.1038/nm0405-367. [DOI] [PubMed] [Google Scholar]
- Gonzalez R, Griparic L, Vargas V, Burgee K, Santacruz P, Anderson R, Schiewe M, Silva F, Patel A. A putative mesenchymal stem cells population isolated from adult human testes. Biochem Biophys Res Commun. 2009;385:570–575. doi: 10.1016/j.bbrc.2009.05.103. [DOI] [PubMed] [Google Scholar]
- Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon S, Abramov N, Grinblat B, Ginis I. Markers Distinguishing Mesenchymal Stem Cells from Fibroblasts Are Downregulated with Passaging. Stem Cells Dev. 2010 doi: 10.1089/scd.2010.0040. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko IK, Kim BG, Awadallah A, Mikulan J, Lin P, Letterio JJ, Dennis JE. Targeting improves MSC treatment of inflammatory bowel disease. Mol Ther. 2010;18:1365–1372. doi: 10.1038/mt.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Qu-Petersen Z, Cao B, Kimura S, Jankowski R, Cummins J, Usas A, Gates C, Robbins P, Wernig A, Huard J. Clonal isolation of muscle-derived cells capable of enhancing muscle regeneration and bone healing. J Cell Biol. 2000;150:1085–1100. doi: 10.1083/jcb.150.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang X, Niyibizi C. Distribution of single-cell expanded marrow derived progenitors in a developing mouse model of osteogenesis imperfecta following systemic transplantation. Stem Cells. 2007;25:3183–3193. doi: 10.1634/stemcells.2007-0466. [DOI] [PubMed] [Google Scholar]
- Liao X, Li F, Wang X, Yanoso J, Niyibizi C. Distribution of murine adipose-derived mesenchymal stem cells in vivo following transplantation in developing mice. Stem Cells Dev. 2008;17:303–314. doi: 10.1089/scd.2007.0086. [DOI] [PubMed] [Google Scholar]
- Lindvall O, Kokaia Z. Stem cells in human neurodegenerative disorders--time for clinical translation? J Clin Invest. 2010;120:29–40. doi: 10.1172/JCI40543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox JR, Liao X, Li F, Niyibizi C. Effects of Culturing on the Stability of the Putative Murine Adipose Derived Stem Cells Markers. Open Stem Cell J. 2009;1:54–61. doi: 10.2174/1876893800901010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merceron C, Vinatier C, Portron S, Masson M, Amiaud J, Guigand L, Cherel Y, Weiss P, Guicheux J. Differential effects of hypoxia on osteochondrogenic potential of human adipose-derived stem cells. Am J Physiol Cell Physiol. 2010;298:C355–C364. doi: 10.1152/ajpcell.00398.2009. [DOI] [PubMed] [Google Scholar]
- Muller T, Eildermann K, Dhir R, Schlatt S, Behr R. Glycan stem-cell markers are specifically expressed by spermatogonia in the adult non-human primate testis. Hum Reprod. 2008;23:2292–2298. doi: 10.1093/humrep/den253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer P, Fechner K, Milz S, Richter W, Suedkamp NP, Mehlhorn AT, Pearce S, Kasten P. Comparison of mesenchymal stem cells from bone marrow and adipose tissue for bone regeneration in a critical size defect of the sheep tibia and the influence of platelet-rich plasma. Biomaterials. 2010;31:3572–3579. doi: 10.1016/j.biomaterials.2010.01.085. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Riekstina U, Cakstina I, Parfejevs V, Hoogduijn M, Jankovskis G, Muiznieks I, Muceniece R, Ancans J. Embryonic stem cell marker expression pattern in human mesenchymal stem cells derived from bone marrow, adipose tissue, heart and dermis. Stem Cell Rev. 2009;5:378–386. doi: 10.1007/s12015-009-9094-9. [DOI] [PubMed] [Google Scholar]
- Saito S, Aoki H, Ito A, Ueno S, Wada T, Mitsuzuka K, Satoh M, Arai Y, Miyagi T. Human alpha2,3-sialyltransferase (ST3Gal II) is a stage-specific embryonic antigen-4 synthase. J Biol Chem. 2003;278:26474–26479. doi: 10.1074/jbc.M213223200. [DOI] [PubMed] [Google Scholar]
- Saito S, Orikasa S, Ohyama C, Satoh M, Fukushi Y. Changes in glycolipids in human renal-cell carcinoma and their clinical significance. Int J Cancer. 1991;49:329–334. doi: 10.1002/ijc.2910490303. [DOI] [PubMed] [Google Scholar]
- Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- Trubiani O, Zalzal SF, Paganelli R, Marchisio M, Giancola R, Pizzicannella J, Buhring HJ, Piattelli M, Caputi S, Nanci A. Expression profile of the embryonic markers nanog, OCT-4, SSEA-1, SSEA-4, and frizzled-9 receptor in human periodontal ligament mesenchymal stem cells. J Cell Physiol. 2010;225:123–131. doi: 10.1002/jcp.22203. [DOI] [PubMed] [Google Scholar]
- Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, Bissell MJ, Petersen OW. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojakowski W, Tendera M, Kucia M, Zuba-Surma E, Paczkowska E, Ciosek J, Halasa M, Krol M, Kazmierski M, Buszman P, Ochala A, Ratajczak J, Machalinski B, Ratajczak MZ. Mobilization of bone marrow-derived Oct-4+ SSEA-4+ very small embryonic-like stem cells in patients with acute myocardial infarction. J Am Coll Cardiol. 2009;53:1–9. doi: 10.1016/j.jacc.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura H, Muneta T, Nimura A, Yokoyama A, Koga H, Sekiya I. Comparison of rat mesenchymal stem cells derived from bone marrow, synovium, periosteum, adipose tissue, and muscle. Cell Tissue Res. 2007;327:449–462. doi: 10.1007/s00441-006-0308-z. [DOI] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]