The Ras-binding domain is conserved among fungal Ste11 MAPKKKs and is critical for mating in fungi. Its interaction with Ras1 is critical for Schizosaccharomyces pombe mating, whereas in Saccharomyces cerevisiae its interaction with the Ste5 PH domain plays the crucial role. The binding partner of RBD for fungal mating is shifted from Ras to a PH domain in fungi in which Ste5 exists.
Abstract
The Ste5 protein forms a scaffold that associates and regulates the components of the mitogen-activated protein (MAP) kinase cascade that controls mating-pheromone-mediated signaling in the yeast Saccharomyces cerevisiae. Although it is known that the MEK kinase of the pathway, Ste11, associates with Ste5, details of this interaction have not been established. We identified a Ras-binding-domain-like (RBL) region in the Ste11 protein that is required specifically for the kinase to function in the mating pathway. This module is structurally related to domains in other proteins that mediate Ras-MAP kinase kinase kinase associations; however, this RBL module does not interact with Ras, but instead binds the PH domain of the Ste5 scaffold. Structural and functional studies suggest that the key role of this PH domain is to mediate the Ste5–Ste11 interaction. Overall these two evolutionarily conserved modules interact with each other through a unique interface, and thus in the pheromone pathway the structural context of the RBL domain contribution to kinase activation has been shifted through a change of its interaction partner from Ras to a PH domain.
INTRODUCTION
The yeast Saccharomyces cerevisiae uses mitogen-activated protein (MAP) kinase pathways to respond to a variety of environmental cues to control cellular processes such as proliferation, differentiation, morphogenesis, and stress adaptation. The architecture of these signal transduction pathways is conserved from yeast to mammalian cells. The generic core of these pathways is the so-called MAP kinase cascade consisting of three protein kinases (designated MAP kinase kinase kinase [MAPKKK], MAP kinase kinase [MAPKK], and MAP kinase [MAPK]), which are sequentially activated through phosphorylation upon receiving an upstream activation signal (Herskowitz, 1995; Banuett, 1998; O’Rourke et al., 2002). Plasma membrane targeting of kinases is an important component of many signaling pathways, and a common strategy to accomplish this involves the association of the kinase with a small GTPase that provides a membrane-localizing hydrophobic C-terminus. Classic examples of this approach include the Ras–Raf association involved in proliferation control in higher eukaryotes and the p21–PAK linkages that regulate key signaling pathways throughout the eukaryotes (Leevers et al., 1994; Stokoe et al., 1994; Bartels et al., 1995; Lu and Mayer, 1999). In the pheromone response pathway involved in the mating process of fungi such as Schizosaccharomyces pombe, the fungal Ras homologue serves to link the MAPKKK of the mating pathway to the plasma membrane (Tu et al., 1997). However, in the branch of the ascomycetes leading to S. cerevisiae and Candida albicans, the MAPKKK membrane association function for mating has been shifted to the Ste5 scaffold protein, although the molecular changes that permitted this shift are unclear. In S. cerevisiae, there are multiple means through which Ste5 targets to plasma membrane (Whiteway et al., 1995; Winters et al., 2005; Garrenton et al., 2006).
The Ste11 MAPKKK of S. cerevisiae is involved in at least three MAP kinase pathways: those required for pheromone response, for regulation of osmotic stress, and for pseudohyphal growth. It thus serves as a good model for studying the mechanisms of pathway specificity and coordination/cross-talk among signal transduction pathways. Plasma membrane (PM) localization of Ste11 occurs through interactions with specific scaffold/adaptor proteins and, as in other eukaryotes, is critical for its differential activation/regulation of these pathways. Forced PM localization of Ste11 leads to simultaneous activation of all MAP kinase pathways that share the kinase (Winters et al., 2005; Wu et al., 2006). In the yeast pheromone response the Ste5 scaffold directs Ste11 to the PM and links the activation of a G protein–coupled receptor to the MAP kinase cascade (Elion, 1995, 2001; Whiteway et al., 1995; Wang and Dohlman, 2004). Ste5 binds all the components of the kinase cascade—Ste11 MAPKKK, Ste7 MAPKK, and Fus3 MAPK (Choi et al., 1994; Marcus et al., 1994; Printen and Sprague, 1994). The region of Ste5 required for Ste11 interaction largely overlaps with a cryptic PH domain that was found to bind specific phospholipids of plasma membrane, and the ability to bind the phospholipids has been proposed to be essential for Ste5 localization to the PM (Garrenton et al., 2006). The molecular basis of the Ste11–Ste5 interaction, essential for the pheromone response, remained unresolved. We previously identified a region of Ste11 that is critical for interaction with Ste5 and for kinase function in the pheromone response pathway (Wu et al., 1999). In this work, we use a structure-based bioinformatics approach, based on the Ste11 homologue in S. pombe, Byr2, to identify a Ras-binding-domain-like (RBL) structure within the regulatory region of Ste11. We narrow down the region of Ste5 that is essential for interaction with Ste11 and show that it is the PH domain that binds the RBLSte11 domain. This direct binding of the Ste5 scaffold PH domain to the Ste11 RBL domain is essential for the proper functioning of the pheromone-response MAP kinase pathway. This establishes the fact that although the binding partners have switched from the Ras protein to the scaffold PH domain, the ubiquitin fold–based Ras-binding module of the MAPKKK is commonly used to connect the kinase to a plasma membrane–targeting module of the signaling network.
RESULTS
Detection of RBLSte11 domains in fungal MAPKKKs by structural bioinformatics
The domain recognition methods based on primary sequence information have been only partially successful in detecting the ubiquitin fold of Ras-binding domains (RBDs) and Ras association (RA) domains. Although we were able to detect the RA domain in the S. cerevisiae Ste50 protein by application of simple homology tools (Ponting and Benjamin, 1996; Ekiel et al., 2009), initial database surveys (Ponting and Benjamin, 1996) suggested an absence of the RBD or RA motifs within the Ste11 protein, a kinase that serves as the MAPKKK for a variety of signaling pathways in yeast. To identify structural motifs that could be responsible for interaction of Ste11 with Ste5 scaffold, we reanalyzed the sequence of Ste11, applying structural bioinformatics approaches based on the state-of-the-art fold recognition methods assembled within the 3D-Jury meta-predictor (Ginalski and Rychlewski, 2003). Our analysis detected with statistical significance a ubiquitin fold–based RBD encompassing residues 117–240 within the previously identified Ste5-interacting region of the yeast Ste11 protein kinase. We termed this an RBD-like (RBL) motif, which shares all secondary structural elements characteristic of the archetypal ubiquitin β-grasp fold (Supplemental Figure S2(a)), with the highest 3D-JURY consensus fold recognition score to the RBD structure of the Ste11 homologue Byr2 of S. pombe (Scheffzek et al., 2001). This RBLSte11 motif is common to the Ste11MAPKKK homologues of a large number of fungi (Supplemental Figure S2(b)).
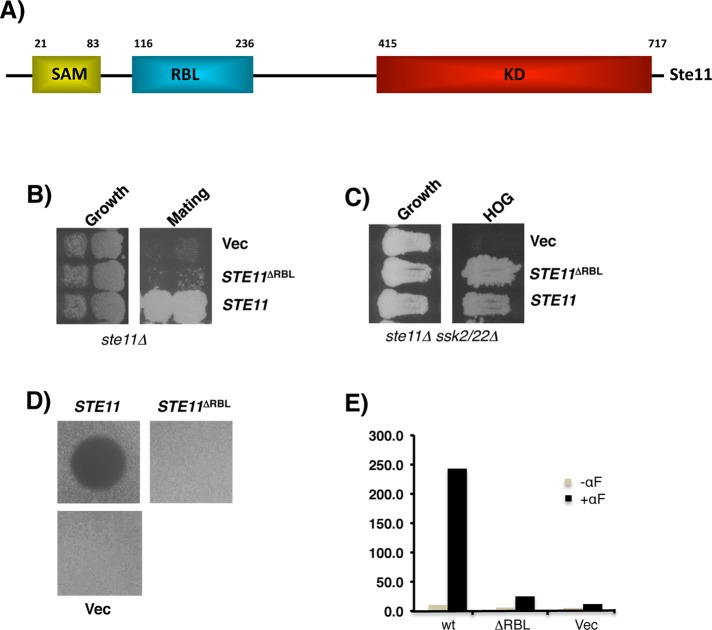
The RBL domain of Ste11 MAPKKK is essential for the pheromone response pathway
To directly assess the role of the RBL domain in signal transduction within the MAP kinase pathway required for pheromone response, we created a Ste11 mutant (Ste11ΔRBL) with an internal in-frame deletion of the region corresponding to the predicted RBL domain (residues 117–240). The resulting mutant was assayed for its ability to mate with wild-type cells of opposite mating type. Cells expressing Ste11ΔRBL are severely defective in mating and also in pheromone-induced cell cycle arrest, as well as in pheromone-induced transcriptional expression of a mating-specific reporter gene. Therefore cells containing Ste11 lacking the RBL have an essentially sterile phenotype (Figure 1). To ensure that Ste11ΔRBL is a functional kinase, we used the fact that Ste11 MAPKKK is shared among several MAP kinase pathways and essential also for high-osmolarity glycerol (HOG) synthesis. In the HOG pathway, the Ste11-SAM (for “sterile alpha mating”) domain, which is N-terminal and precedes the RBL domain, is necessary for the interaction of Ste11 with Ste50 adaptor for proper localization and activation of the Ste11 kinase. We therefore analyzed the function of the Ste11ΔRBL mutant in the activation of the HOG pathway and found that, in contrast to its behavior in the pheromone response pathway, it is fully capable of activating the HOG pathway, indicating that the Ste11ΔRBL retains kinase activity and suggesting that the RBL domain is uniquely crucial for Ste11 function in the activation of the MAP kinase pathway for pheromone response (Figure 1C).
FIGURE 1:
The RBLSte11 domain is essential for pheromone response signaling. (A) Schematic diagram of Ste11 MAPKKK with functional domains indicated: KD, kinase domain; RBL, RBD-like; SAM, sterile alpha mating. (B) The RBLSte11 domain is essential for mating. Yeast cells (ste11Δ) transformed with STE11 alleles were tested for their ability to mate with tester strain by diploid selection. (C) The RBL domain is not required for Ste11 signaling in the HOG pathway. Yeast cells (ste11Δ ssk2Δ ssk22Δ) transformed with STE11 alleles were tested for their ability to signal in the HOG pathway as indicated by growth on hyperosmotic medium. Halo assay (D) and β-galactosidase assay (E) of yeast cells (ste11Δ) transformed with STE11 alleles showing their ability to induce pheromone-dependent cell cycle arrest and transcriptional activation of mating-specific reporter gene.
Solution structure of the RBLSte11 domain
To confirm the structural bioinformatics prediction for the RBLSte11 domain, we determined its structure in solution by nuclear magnetic resonance (NMR) spectroscopy. The RBLSte11 domain (amino acids [aa] 116–236) was bacterially expressed in 15N- and 13C-enriched medium and purified. Using NMR experiments, we obtained chemical shift assignments for the protein backbone, as well as for aliphatic and aromatic side chains. Backbone assignments were complete, with exceptions for Cys-138 and Asp-142, for which signals were missing in 1H15N heteronuclear single quantum coherence (HSQC) spectra. An ensemble of 20 low-energy structures that satisfy NMR constraints was obtained using Cyana 2.1 (Güntert, 2004; Supplemental Figure S3).
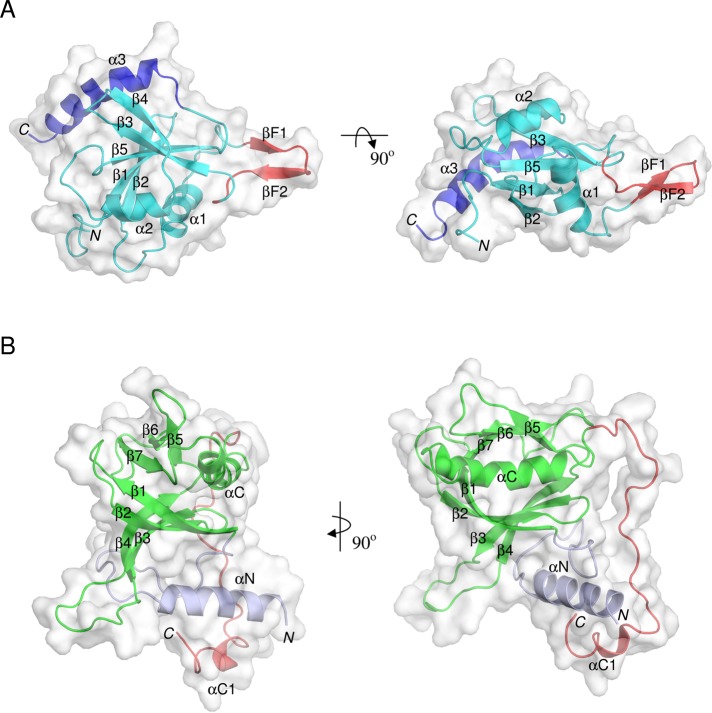
The NMR structure reveals that the predicted RBL domain indeed adopts a ubiquitin fold, comprising a mixed five-stranded β-sheet flanked by three α-helices (Figure 2A). The secondary structure assignments are as follows: strands β1 (aa 120–125) and β2 (aa 128–134), helix α1 (aa 142–151), strands β3 (aa 172–178) and β4 (aa 182–186), helix α2 (aa 189–196), and strand β5 (aa 206–211) are elements typical of the ubiquitin fold. The final, long C-terminal helix α3 (aa 218–233) is rarely present in this class of domains. In addition, we observed a short, unique insertion between helix α1 and strand β3 in RBLSte11 domain, which we call a β-finger, composed of two short strands βF1 (aa 158–160) and βF2 (aa 163–166). The ensemble of NMR structures shows significant protein backbone flexibility in the N-terminus preceding the ubiquitin fold, in the C-terminal end of helix α3, and also to a moderate degree in the β-finger insert (Supplemental Figure S3).
FIGURE 2:
Overall structures of the RBLSte11 and PHSte5 domains. (A) Solution NMR structure of the RBLSte11 domain. Canonical ubiquitin-like fold is in cyan, the inserted β-hairpin in red, and the helical C-terminal extension in blue. (B) Modeled PHSte5 domain. Canonical PH fold is in green and N- and C-terminal extensions are in light blue and red, respectively. Secondary structure elements are labeled. Two orthogonal views are presented in each case.
The structure of the RBLSte11 domain is most similar to the RBD domain of S. pombe Byr2 (Protein Data Bank [PDB] code 1K8R; Scheffzek et al., 2001), as indicated by various structural similarity metrics such as a Z-score of 8.0 and a Q-score of 0.38 (http://pdbe.org/fold). However, there are two major differences between these structures. First, the Byr2-RBD does not have the β-finger insertion, and its C-terminal helix α3 is much shorter than the corresponding helix in the RBLSte11 domain.
The RBLSte11 domain has no detectable association with small GTPases but interacts with the PH domain of Ste5 scaffold
To understand the role of this RBLSte11 domain in pheromone response signaling, we first searched for its interaction partner(s). Because of the similarity of the RBLSte11 domain to the Byr2-RBD of S. pombe, which has been shown to interact with Ras1 (Gronwald et al., 2001; Scheffzek et al., 2001), we tested the possibility of the RBLSte11 domain interacting with any of the S. cerevisiae small GTPases. To this end we performed in vitro resin-binding assays with small GTPases of the Ras and Rho family members (a total of 11, including Ras1, Ras2, Cdc42, Rho1, Rho2, Rho3, Rho4, Rho5, Rhb1, Rsr1, and Yhr022c) as glutathione S-transferase (GST) fusions from the yeast open reading frame library (Martzen et al., 1999) expressed in yeast and purified as previously described (Annan et al., 2008) and the RBLSte11 domain (aa 116–236) expressed as histidine (His)-tagged fusion protein in bacteria. For these assays the small GTPases were preloaded with the nonhydrolyzable GTP analogue GTPγS or with GTPβS (Truckses et al., 2006). No detectable binding of RBLSte11 to these GTPases was observed (unpublished data).
The presence of PH domains in some family members of Ste5-like and Far1-like proteins was predicted (Wiget et al., 2004; Garrenton et al., 2006; Cote et al., 2011). This prediction was derived from the application of fold-recognition methods (Supplemental Figure S1(a)), which detected the boundaries of the PH domain in a large number of Ste5 and Far1 fungal proteins (Supplemental Figure S1(b)). Simple application of homology tools was unable to detect this cryptic structural relationship due to the low sequence conservation characteristic of PH domains. Ste11 had been shown to interact with Ste5, and the Ste11-binding region on Ste5 was first mapped to residues 336–586 through deletion (Choi et al., 1994). This region was further delineated through mutagenesis to residues 463–514 (Inouye et al., 1997). It overlaps with the recently proposed lipid-binding PHSte5 domain (Garrenton et al., 2006), which maps to residues 400–512.
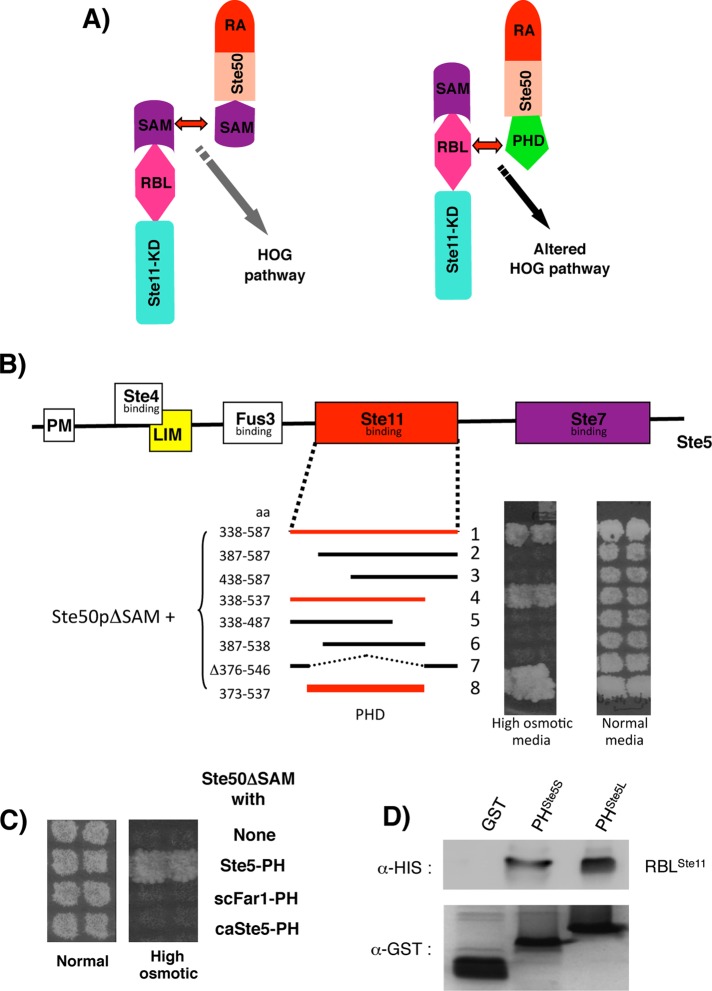
To further delineate the domain boundary for the region of Ste5 that is specifically required for the interaction with the RBLSte11 domain, we used a modified cytoplasmic yeast, two-hybrid system (Wu, Jansen, and Yerko, unpublished data) developed based on our finding that the interaction of Ste50 and Ste11 through their SAM domains, in the HOG pathway activation, can be replaced with other protein–protein interacting modules (Wu et al., 2006).
We replaced the Ste50-SAM domain with the Ste11-interacting region of Ste5, so that the activity of the modified HOG pathway in cells (ste50Δ ssk2Δ ssk22Δ) depended on the ability of the Ste5 fragment in the Ste50 chimera to interact with Ste11 (Figure 3A). Deletions from both the N- and C-termini of the Ste5 fragment delineated a Ste5 fragment composed of residues 373–537 that was able to activate the HOG pathway when fused to Ste50ΔSAM (Figure 3B). Further deletion analysis indicated that a Ste5 fragment of aa 373–523 was still functional, albeit with somewhat reduced activity compared with the larger fragment. However, the fragment consisting of aa 373–515 of Ste5 was unable to activate HOG pathway (unpublished data). These results show that the Ste5 region interacting with Ste11 largely overlaps with the PH domain of Ste5. Because we showed previously that the Ste11 regulatory region encompassing the RBL domain is required for interactions with Ste5 (Wu et al., 1999), we concluded that the Ste5–Ste11 association is mediated through PH–RBL domain interactions. This interaction of the PHSte5 domain with the RBLSte11 domain is specific, as other versions of PH domains show no interaction with the RBL domain (Figure 3C).
FIGURE 3:
Ste11 interacts with the PHSte5 domain. (A) Schematic representation of the interaction of Ste11 with Ste50 through their respective SAM domains in the natural HOG pathway (left) and through the RBL and grafted PHSte5 domain in the altered HOG pathway. (B) The altered HOG pathway format in A was used to delineate the boundary of the PHSte5 domain in yeast strain YCW1476 (ste50Δ ssk2Δ ssk22Δ) by monitoring its ability to grow on hyperosmotic media (with 0.75 M NaCl). (C) The RBLSte11 domain specifically interacts with the PHSte5 domain. Other PH domains replacing the Ste50-SAM domain were unable to activate the altered HOG pathway, indicating that they do not interact with the RBLSte11 domain. (D) The RBLSte11 domain interacts with the PHSte5 domain in vitro. Bacterially expressed, His-tagged RBLSte11 was incubated with glutathione–Sepharose bead–immobilized Ste5 fragments of either aa 373–537 (PHSte5L) or 373–523 (PHSte5S) as GST fusion or GST alone as control. His-tagged BRLSte11 copurified with the glutathione–Sepharose beads was revealed by Western blotting analysis with anti-His antibody (top) and the GST fusion with anti-GST antibody (bottom).
To test whether the RBLSte11 domain and PHSte5 domain can interact directly in vitro, we performed pull-down resin-binding assays with independently expressed and differentially tagged fragments: a Ste11 fragment consisting of aa 116–236 with two Ste5 constructs encompassing aa 373–537 and aa 373–523, respectively. When bacterially expressed GST fusions of the Ste5 fragments were mixed with a bacterial extract containing the His-tagged RBLSte11, both Ste5 fragments were able to pull down the RBLSte11 domain, although the smaller Ste5 (373–523) fragment showed somewhat lower efficiency. The GST protein alone was used as a negative control, and no Ste11 fragment was retained on the column. These results established a direct interaction between Ste5 and Ste11 through a PH domain and an RBD-like domain. Further analysis indicated that the longer form of the PHSte5 domain behaved better in solution, and it was chosen along with the RBLSte11 domain for subsequent studies.
The bacterially expressed and purified RBLSte11 domain and the PHSte5 domain appeared to be predominantly monomers and were able to form a complex at a 1:1 ratio as judged by size exclusion chromatography. The complex appeared to be more stable in solution than either partner separately. The apparent affinity of the interaction of the complex was determined to be ∼200 nM using surface plasmon resonance with the immobilized PHSte5 domain on the surface and the RBLSte11 domain in the flowing phase (Supplemental Figure S4(a)). The physical interaction of these two domains was also demonstrated by NMR analysis using an 15N‑labeled RBLSte11 domain titrated with an unlabeled PHSte5 domain. A set of amino acid residues showed specific chemical shifts upon the addition of PHSte5 domain, indicating their involvement in the interaction (Supplemental Figure S4(b)).
Mutational analysis of the PHSte5 domain and structural mapping of the Ste11-interacting site
The RBL domain–PH domain interaction represents a new type of RBD interaction complex, as Ras-binding domain modules typically associate with small GTPases. To probe the structural basis of this interaction and the role of this interaction in signal transduction in the pheromone response MAP kinase pathway, we identified functionally important residues of the PHSte5 domain by both random and site-directed mutagenesis. We first screened for mutations that disrupted the function of the PH domain in the context of the PHSte5-Ste50 chimera shown in Figure 3A. Approximately 150 clones that satisfied this criterion were selected after sequencing and classification according to the nature of the substitutions, and 10 representative mutants from random mutagenesis were chosen along with 6 mutants of site-directed mutagenesis, including I504T mutant based on previous work (Inouye et al., 1997), for further functional analysis (Table 1).
TABLE 1:
PHSte5-domain mutants.
| Mutation | Interaction with Ste11 | Mating (% of wild type) |
| R379G | − | — (<<0.1) |
| R407K | Decreased | 9.7 |
| R462E | nd | 92 |
| T465A | Severely decreased | 0.8 |
| K472E | − | — (<<0.1) |
| S484P | Severely decreased | 0.9 |
| Y487H | Decreased | 10.3 |
| N491I | Severely decreased | 0.7 |
| S494P | Severely decreased | 0.7 |
| T498A | + | 100 |
| T499A | nd | 91 |
| Q501R | − | — (<<0.1) |
| K502R | Severely decreased | 0.8 |
| I504T | − | — (<<0.1) |
| L508S | Severely decreased | 0.5% |
| F514L | − | — (<<0.1) |
| WT | + | 100 |
Protein–protein interaction was assayed using the tailed two-hybrid system as described in the text. The relative amount of cell growth reflects the extent of PHSte5-domain interaction with Ste11. nd, not determined.
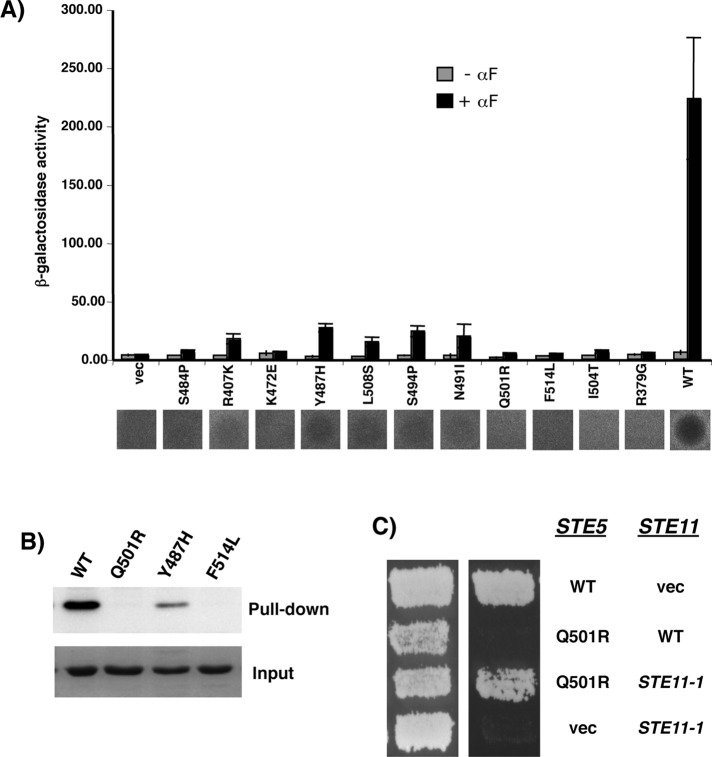
These mutants were transferred into STE5 under the control of its own promoter using in vivo recombination in yeast cells deleted for the endogenous STE5. The resulting yeast strains bearing different single point mutations in the PHSte5 domain were assayed for their ability to direct pheromone signaling. All the mutants showed severe defects in pheromone response, with some exhibiting a totally sterile phenotype (Figure 4 and Table 1).
FIGURE 4:
Mutational analysis of the essential role of the PHSte5 domain in pheromone response. (A) β-Galactosidase assay (top) and Halo assay (bottom) of yeast cells (ste5Δ) transformed with STE5 alleles carrying mutations in the PH domain for their ability to induce pheromone-dependent transcriptional activation of mating-specific reporter gene and cell cycle arrest. (B) Pull-down assay with bacterially expressed GST-RBLSte11- and His-tagged PHSte5-domain mutants. Western blot analysis of pull-down and input of PHSte5-domain mutants was carried out with anti-His antibody. (C) Activated STE11 allele bypasses the mating defect of the PHSte5-domain mutant. Yeast cells (ste5Δ) cotransformed with either STE5 or STE11 allele or vectors in combination as indicated were assayed for their ability to mate, and the mating products were revealed on selective medium (right).
To confirm that these PHSte5-domain mutants have altered interactions with the RBLSte11, were cloned Ste5 mutants corresponding to residues 373–537, expressed them in bacteria, and used them for the in vitro binding assay with a bacterially expressed RBLSte11 domain (116–236). PHSte5-domain mutants showed severely decreased or no binding to the RBLSte11 domain, and the extent of the decrease in the interaction correlated well with that of the decease in the pheromone response (Figure 4, A and B). These results indicate that the interaction of the PHSte5 domain with the RBLSte11 domain is essential for the pheromone response signal transduction pathway. Further analysis indicated that this interaction played a critical role in Ste11 MAPKKK activation, as an activated allele of STE11 largely bypassed the signaling defects of the Ste5 mutants (Figure 4C).
Modeled structure of the PHSte5 domain
To analyze the spatial relationship between the loss-of-binding mutants and to gain insight into the residues forming the interface between the RBL domain and the PH domain, we applied structural bioinformatics and homology modeling to construct a three-dimensional (3D) model of the PHSte5 domain encompassing residues 374–537. This model was refined by a 20-ns molecular dynamics (MD) simulation, at which point it attained structural convergence, with only a solvent-exposed loop region in the C-terminal end showing significant fluctuations at room temperature over the last 5 ns of MD simulation (Supplemental Figure S5). The average minimized structured over the last 1 ns of MD simulation is of good quality as validated by several methods (Supplemental Figure S6).
The modeled structure of the PHSte5 domain (Figure 2B) was based on the PH domain of the guanine nucleotide exchange factor collybistin (PDB code 2DFK; Xiang et al., 2006). It comprises the canonical PH fold (residues Leu-406–Asp-511) consisting of an antiparallel seven-stranded β-barrel (β4-β3-β2-β1-β7-β6-β5) capped by a C-terminal α-helix (αC). The axis of αC has a noticeable curvature. The canonical PH fold is flanked by N- and C-terminal extensions (Thr-374–Leu-405 and Phe-512–Gly-537) that interact with each other via helical regions present within these extensions and contact the outside of the β-barrel on strands β1-β2-β3. The N-terminal helix αN (Leu-375–Asn-389) is well formed and sandwiched between the β-barrel and the short C-terminal helix αC1 (residues Ile-529–Phe-532). Terminal helical extensions are predicted also in other PH domains from fungal Ste5 and Far1 homologues, and these are linked to the canonical PH fold by structurally varying linkers (Supplemental Figure S1(a)). The longer linker connecting the αC1 helix appears particularly flexible (i.e., unstructured) in our 3D model of the yeast PHSte5 domain (Supplemental Figure S5).
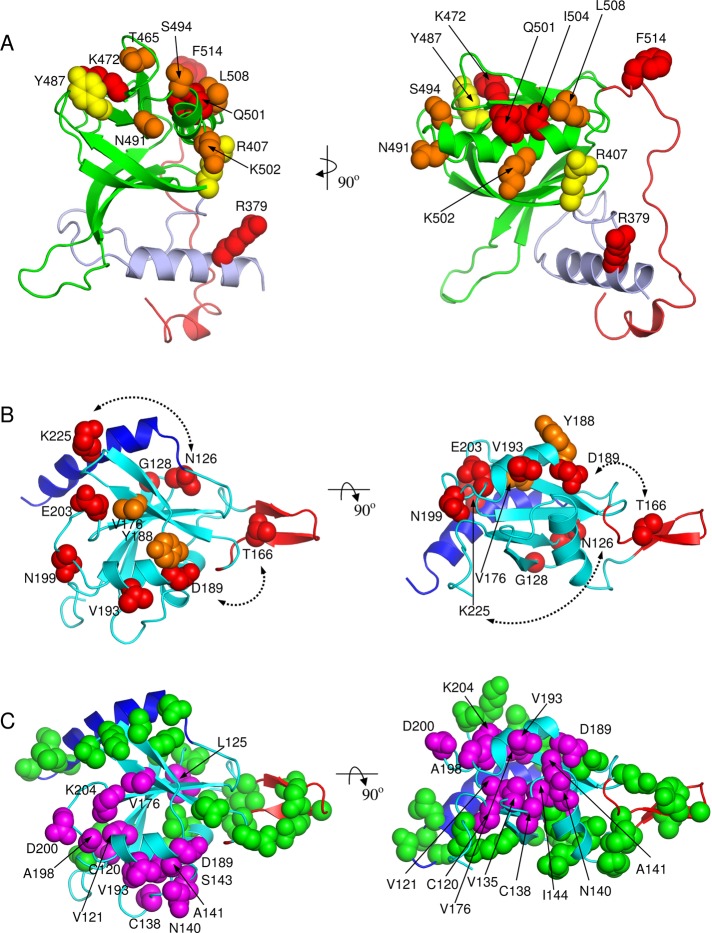
The mutagenized residues of the PHSte5 domain that affected mating and that modified Ste11 binding ability were projected on this 3D model (Figure 5A and Table 1). Mutations that significantly affected the mating activity (<1%) and had undetectable or severely decreased Ste11 binding define a contiguous surface patch on one face of the PHSte5-domain structure. They are located in the β5-β6-β7-αC region: T465A (β5–β6 loop), K472E (β6), S484P (β6–β7 loop), N491I (β7-–αC loop), and S494P, Q501R, K502R, and I504, in the αC helix. The exceptions are R379G in the αN helix in the N-terminal extension and F514L at the beginning of the loop following the αC helix. Two mutations affecting mating to a lesser extent (∼10%) and having decreased Ste11 binding relative to wild-type Ste5 are R407K (at the beginning of the β1 strand) and Y487H (β7) map also to the same surface area. Overall the Ste11-interacting surface of the PHSte5 domain is centered on the αC helix.
FIGURE 5:
Structural mapping of functional data. (A) Mutagenesis data (Table 1) mapped on the modeled PHSte5 domain. Mutated residues are shown as CPK models color coded by activity change upon mutation relative to wild-type Ste5: red, <<0.1% mating activity and no Ste11 binding; orange, <1% mating activity and severely decreased Ste11 interaction; yellow, ∼10% mating activity and decreased Ste11 interactions. (B) Mutagenesis data (Table 2) mapped on the solution NMR structure of the RBLSte11 domain. Mutated residues are shown as CPK models color coded by activity change upon mutation relative to wild-type Ste11: red, <<0.1% mating activity; orange, <1% mating activity. Dashed lines indicate double mutants. (C) NMR interaction data mapped on the solution NMR structure of the RBLSte11 domain. CPK models indicate residues affected (in magenta, labeled) and not affected (in green, not labeled) upon binding to PHSte5 domain. Two orthogonal views are presented in each case corresponding to those in Figure 2.
PHSte5-domain mutants are defective only in the interaction with Ste11 and show normal cellular localization
Some known PH domains are capable of binding inositol phosphates and phosphatidylinositides (PIs) and may be functionally involved in plasma membrane targeting and association or subcellular localization (Lemmon, 2004, 2008). The PHSte5 domain has recently been shown to bind phospholipids and to be required for plasma membrane localization of the protein. This property has been proposed to be required for the function of Ste5 in the activation of the pheromone response pathway (Garrenton et al., 2006). A detailed analysis of the putative PI-binding sites of the PHSte5 domain is given in the Supplemental Figure S7. Briefly, the Ste11-interacting surface of the PHSte5 domain mapped by mutagenesis is distant from the canonical PI-binding site but partially overlaps with the general location of the noncanonical PI-binding site.
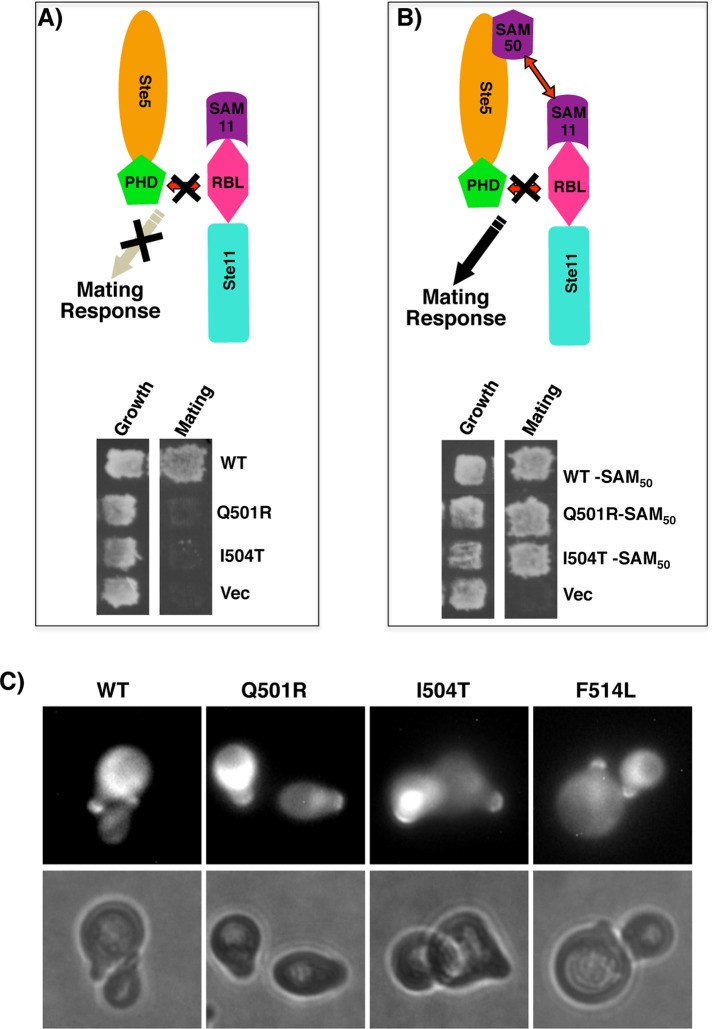
To demonstrate that the defect of our PHSte5-domain mutants for pheromone response is due only to their inability to interact with Ste11 and not in membrane association, we constructed Ste5 mutants with a protein–protein interaction module that restores interaction with Ste11 and tested their ability to mate. To this end, we made an in-frame fusion of the SAM domain of Ste50 to the C-terminal region of Ste5. The fusion proteins are expected to restore the ability to interact with the Ste11 by taking advantage of the fact that Ste50 and Ste11 interact through their respective SAM domains (Wu et al. 1999, 2006). Two Ste5 mutants chosen to be modified and assayed were Q501R and I504T, as these mutants showed a nearly sterile phenotype. These sterile Ste5 mutants became mating competent when they were fused with the SAM domain of Ste50 (Figure 6, A and B), demonstrating that reestablishing the association between Ste5 mutants and Ste11 is critical for the signal transduction of the pheromone response pathway and that the mating incompetence of the Ste5 mutants is due to their inability to interact with Ste11.
FIGURE 6:
PHSte5-domain mutants defective primarily in the binding of Ste11 MAPKKK. (A) Yeast cells (ste5Δ) carrying PHSte5-domain mutants that are defective in binding to RBLSte11 are defective in mating. (B) Reestablishing the interaction of Ste5 mutants in A with Ste11 through another protein–protein interaction alleviates the mating defect. Yeast cells in A were transformed with Ste5 alleles as indicated and carrying the SAM domain of Ste50 (SAM50), which interacts with the SAM domain of Ste11, were assayed for the ability to mate. (C) The PHSte5-domain mutants have normal cellular localization. Yeast strain YCW338 (MATa sst1) carrying GFP-tagged Ste5 alleles as indicated were induced in galactose media, treated with α-factor mating pheromone (αF) (1 μM) for 1 h, and photographed using a Leitz photomicroscope equipped with a 100× objective and a MicroMax camera (bottom); GFP fluorescence photographs were acquired and processed as described in Materials and Methods.
We also constructed N-terminally green fluorescent protein (GFP)–tagged PHSte5 domain mutants to examine the subcellular localization of PHSte5-domain mutants using fluorescence microscopy. We chose three mutants with the most severe mating defects (Q501R, I504T, and F514L) to make the GFP-tagged constructs. These constructs, along with GFP-tagged wild-type Ste5, were transformed into wild-type yeast cells that are capable of forming pheromone-induced shmoos. All the mutants showed a subcellular localization similar to the wild-type Ste5: general cytoplasmic and nuclear distribution both in the absence and presence of pheromone and a sharp crescent-shaped localization to the shmoo tip in the presence of pheromone (Figure 6C). These results demonstrated that the Ste5 mutants are competent in plasma membrane recruitment, suggesting that the observed mating defect is likely due to the defect in the ability of these PH-domain mutants to interact with the RBL domain of Ste11.
Mutational analysis of the RBLSte11 domain and structural mapping of residues essential for pheromone response
We were interested in defining the face of the RBL module that interacts with this PHSte5 domain. To identify those residues in the RBLSte11 domain critical for the binding of the PHSte5 region, we carried out a random mutagenesis analysis. We showed previously that deleting a region encompassing the RBL domain of Ste11 permits a reduced response to mating pheromone in an otherwise wild-type background but causes sterility in a ste50Δ yeast strain (Wu et al., 1999). We used this observation and performed the mutagenesis analysis in a ste50Δ strain so that the interaction of the RBLSte11 and PHSte5 domains is the only driver of the pheromone response. Mutants that were pheromone response negative were selected and then further analyzed for their ability to activate the HOG pathway in the presence of Ste50 to eliminate nonfunctional nonsense mutations, as Ste11 lacking the RBL motif is fully capable of activating the HOG pathway (Wu et al., 1999). Approximately 100 clones were selected for sequencing analysis and, after classification, eight representative clones with either single or double mutation were chosen for further studies. Two additional mutants generated by site-directed mutagenesis (D178R and D173R/F175A) were used as controls to show that RBLSte11 can tolerate radical mutations, as Ste11 with these mutations has a nearly normal signaling function (Table 2).
TABLE 2:
RBLSte11-domain mutants.
| Mutation | Mating (% of wild type) | HOG pathway |
|---|---|---|
| G128D | — (<<0.1) | + |
| V176R | 0.4 | + |
| D178R | 91 | + |
| V193A | — (<<0.1) | + |
| N199S | — (<<0.1) | + |
| E203G | — (<<0.1) | + |
| N126T, K225I | — (<<0.1) | + |
| T166A, D189G | — (<<0.1) | + |
| D173R, F175A | 97 | + |
| Y188C, S241R | 0.7 | + |
| WT | 100 | + |
Semiquantitative mating assay was performed with yeast cells (ste11Δ) carrying the Ste11 alleles indicated. Mating efficiency is expressed as the percentage of the wild type. The HOG pathway activity was assayed on hyperosmotic medium as described (Wu et al. 1999) for growth of yeast cells (ste11Δ ssk2Δ ssk22Δ) carrying the Ste11 alleles.
These Ste11 mutants were assayed for their function in the pheromone response pathway through their ability to allow formation of diploids, to permit pheromone-dependent cell cycle arrest, and to facilitate mating-specific gene transcription; all the mutants selected from the screening showed severe defects up to an essentially sterile phenotype. All these mutants were, however, fully able to activate the HOG pathway. This indicates that the mutational effects are specific for the function that requires the RBLSte11 domain and not due to a general loss of function of Ste11. The residues defined by the defective mutants were mapped onto the RBLSte11 structure (Figure 5B).
NMR-based mapping of the PHSte5 domain-binding interface on the RBLSte11-domain surface
The residues of the RBLSte11 domain involved in binding to the PHSte5 domain were also interrogated by NMR spectroscopy. To this end, we used a chemical shift perturbation approach through analysis of 1H15N HSQC spectra of the RBLSte11 domain upon addition of increasing amounts of an unlabeled PHSte5 domain. The majority of NMR signals experience some line broadening due to the large size of the complex. This indicates that the RBL domain does not undergo substantial conformational changes upon complex formation. However, a subset of amide signals undergoes a chemical shift change or extensive line broadening (Supplemental Figure S3B), allowing identification of the interface. The results are summarized in Figure 5C, where the affected residues are highlighted in magenta. The regions most affected by the binding of the PH domain include the N-terminal end of the strand β1, the β1/β2 loop, one residue from strand β3, the helix α2, and the following loop leading to strand β5 (Figure 5C). All of these regions are located close to each other in space, forming a continuous patch on the surface of the RBL domain and identifying the interface. This interface is in excellent agreement with the functional results from mutagenesis (Figure 5B); of eight mutants with drastically reduced mating (Table 2), seven were also picked up by NMR experiments, either identifying the same amino acid or its direct neighbor in the sequence, suggesting that the mutational analysis picked up critical residues involved in RBLSte11- and PHSte5-domain interaction. The number of identified residues that functionally impair mating is lower than the number of amino acids identified on the interface by NMR, indicating incomplete coverage by mutagenesis. However, the NMR data did not identify participation of the loop βF1/βF2 in binding the PH domain (Figures 2A and 5C). Hence, this β-finger insert does not appear to be involved in the interaction with the Ste5 scaffold, although it may play a different functional role. The NMR data also help to interpret the three cases of impaired double mutants identified by random mutagenesis (Table 2), suggesting that most likely Asp-189 (rather than Thr-166), Asn-126 (rather than Lys-225), and Tyr-188 (rather than Ser-241) are responsible for the reduced mating. The only region located outside of this contiguous interface is the loop β1/β2, which was picked up by both mutagenesis and NMR (Asn-126 and Gly-128; Table 2) or only by NMR experiments (the neighboring amino acid Leu-125). Most likely, some small conformational changes in this region are responsible for the observed effect.
DISCUSSION
Modular domains of signaling proteins
Eukaryotic cells use a wide range of protein modules to wire together signaling networks. These modules include catalytic elements such as kinases, phosphatases, and other enzymatic elements, as well as interaction modules such as SAM, SH2, and SH3 domains. The RBDs or RA domains and the PH domains are other very common modules implicated primarily in protein–protein interactions. The RBDs (or RA domains) belong to the ubiquitin superfold, and one characteristic feature of ubiquitin is the diversity of its binding partners and their modes of interaction (Schnell and Hicke, 2003; Hurley et al., 2006; Kiel and Serrano, 2006). However, although the RBD/RA domains are ubiquitous within signaling networks, their function has been found to be primarily focused on transmitting signals by binding specifically to GTPases and thus activating effector functions.
PH domains are also among the most common domains in signaling proteins; however, up to now there has been no example of their interaction with RBD or RBD-like domains. Although ubiquitin has shown interactions with PH domains, these associations have been reported to function in protein ubiquitination and degradation but not signaling (Alam et al., 2006; Hirano et al., 2006; Schreiner et al., 2008). In this work, our structure–function data demonstrate that RBL–PH domain interactions can occur in signaling networks and that this interaction is critical for the proper functioning of a MAPK signaling pathway. In fact, this complex has a unique interface, different from all known ubiquitin–receptor complexes, and has higher affinity than that found for any of these ubiquitin-based complexes. It is intriguing that this interaction between the RBL domain and the scaffold PH domain in S. cerevisiae serves the same molecular function as the Byr2–Ras1 interaction in S. pombe—to connect a MAPKKK to a potential membrane-tethering molecule.
The interaction of RBL and PH domains
The interaction between the PH domain and the RBL module exploits a new surface of the ubiquitin-fold structure that is different from the canonical surface involved in binding Ras-like proteins. The latter surface is centered on the β2 strand (for antiparallel interactions with the β-sheet of the small GTPase) and the C-terminal end of the subsequent α-helix (positively charged for complementarity with the negatively charged switch I region of the small GTPase; Nassar et al., 1995; Scheffzek et al., 2001). In contrast, the RBLSte11-domain surface that interacts with the PHSte5 domain is centered on loop β2–α1, helix α2, and loop α2–β5, a surface adjacent to, but distinct from, the canonical Ras-binding face (Figures 2A and 5, B and C).
In addition, the surface of the PHSte5 domain that interacts with the RBLSte11, centered on the αC helix, appears to be marginally overlapping with, yet clearly distinct from, the ubiquitin-contacting surfaces in other PH domains (Figure 7). For example, in the case of the GLUE–ubiquitin complex (Alam et al., 2006; Hirano et al., 2006), the interaction surface on the PH domain (GLUE) is centered on the β5 strand, with less contribution from the αC helix, greater contribution from the β6–β7, and no contribution from the (absent) N- and C-terminal elements. In the Rpn13–ubiquitin complex (Schreiner et al., 2008), the interaction is located at the β-barrel side corresponding to the C-terminal end of the αC helix. The two complexes with ubiquitin use the “Ile44 face” of ubiquitin (Figure 7), whereas in RBLSte11 the “Ile44 face” of the ubiquitin fold is blocked by the C-terminal helical extension (α3) and cannot be used for interactions with the PHSte5 domain. These comparisons underscore the structural variability and versatility for interactions not only of the participating folds, but also for the pair of interacting folds, which expands their ability to introduce specificity of interactions.
FIGURE 7:
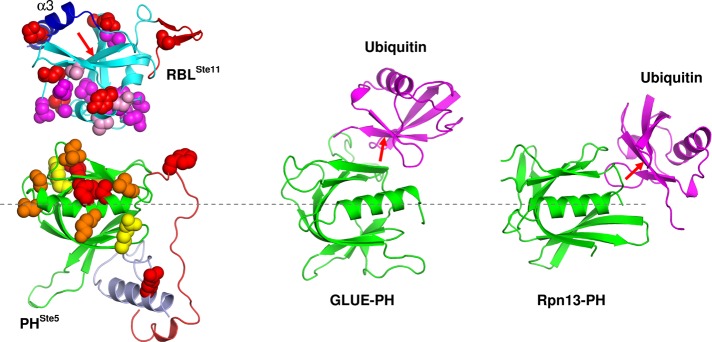
Comparison of the inferred interface between the RBLSte11 and PHSte5 domains with other known ubiquitin–PH domain complexes. Here PH domains are structurally aligned between these complexes (dashed line through the axis of the αC helix). RBLSte11-domain and PHSte5-domain interacting surfaces mapped in this study are indicated by CPK models. Functional residue color coding is as in Figure 5 for the PHSte5 domain, whereas in the case of the RBLSte11 domain all residues mapped by mutagenesis are in red, those mapped by NMR-based interaction are in magenta, and those mapped by both mutagenesis and NMR are in purple. The orientation of the RBLSte11 domain relative to the PHSte5 domain was generated manually and is only suggestive of a putative docking approach between these domains based on functional data. The PH domains (shown in green) of both GLUE and Rpn13 appear to interact with the ubiquitin fold (shown in magenta) at partially overlapping but distinct surfaces relative to the PHSte5-domain interaction with the RBLSte11 domain. The red arrow indicates the “Ile44 face” of the fold, which is engaged in interactions of ubiquitin with GLUE-PH and Rpn13-PH domains but is blocked by a C-terminal helix (α3) in the RBLSte11 domain.
Phosphoinositide-binding sites on the PHSte5 domain
Approximately one-third of the known PH domains are capable of binding inositol phosphates/PIs, and those may be functionally involved in plasma membrane targeting and association or subcellular localization (Lemmon, 2004, 2008). Only a small fraction of PH domains bind reasonably strongly and with some specificity to phosphoinositides in vitro, whereas the majority of PH domains appear to bind very weakly to phosphoinositides according to lipid overlay assays (Lemmon, 2007). Perhaps one of the biggest challenges for understanding the general properties of this large class of domains is to determine whether the frequently observed low-affinity and promiscuous phosphoinositide binding has functional importance. A version of the yeast Ste5 PH domain (131 residues, 388–518) shorter than the one studied here (165 residues, 373–537) was shown to bind in vitro to phosphatidylinositol 4,5-bisphosphate directly and reasonably specifically but with relatively modest affinity (Garrenton et al., 2006).
PH domains can bind PI ligands at two distinct sites: a canonical site located between the β1–β2 and β3–β4 loops and a noncanonical site located on the other side of the β1–β2 loop, in a pocket between the β1–β2 and β6–β7 loops (Alam et al., 2006). Positioning of these PI-binding sites onto the modeled structure of the yeast PHSte5 domain, based on overlays onto PH–PI complexes binding PI at either site, is indicated in Supplemental Figure S7. It is clear that despite the presence of the basic Lys-416 and Arg-420 residues in the β1–β2 loop, the canonical PI-binding site is quite acidic (Asp-417, Glu-437, Glu-453, Asp-482) and hydrophobic (Ile-414, Ile-422, Cys-424, Ile-435, Val-454, Leu-486, Leu-483). Binding at the canonical site of PIs with negative formal charges of at least −6e requires the presence of side chains that are both positively charged and H-bond-donor capable at most of these positions (e.g., PH–PI interactions in the structures with PDB code 1W1D or 1FAO). It is hence unlikely that PI binding occurs at the canonical site on the PHSte5 structure. This agrees with the in vitro data for the double mutation of the two positively charged residues in the canonical PI-binding site of PHSte5 (K416S, R420S), which does not change the apparent weak PI-binding capacity of this domain (Garrenton et al., 2006). At the noncanonical site on the PHSte5-domain model, there is only one H-bond donor group (Tyr-421). The modeled conformation of the β1–β2 hairpin loop will also require a change in order to alleviate a steric collision in accommodating the PI at this site (Supplemental Figure S7(b)). Such electrostatic and steric properties do not support PI binding at the noncanonical site of PHSte5 either. For example, at least four positively charged side chains and two H-bond-donating groups are present for PI binding at the noncanonical site of β-spectrin PH domain (PDB code 1BTN).
Of interest, a double mutant of two positively charged residues (R407S, K411S) appeared to abolish the apparent moderate PI binding of the PHSte5 construct of aa 388–511 in vitro and to impair membrane targeting of Ste5 in vivo, suggesting that they might constitute a PI-binding site (Garrenton et al., 2006). However, our model, as well as the secondary structure prediction of that previous report, indicate that these two basic residues are located not in the β1–β2 loop, but instead upstream in the β1 strand, which positions them far from each other and also far from both the canonical and noncanonical PI-binding sites of a PH domain, particularly in the case of Arg-407 (Supplemental Figure S7(a)). Whether one or both these sites indeed represent PI-interacting residues remains to be clarified by experimental structural analysis. Residues R407 and K411 are partially solvent exposed in our model of PHSte5 and have relatively minor structural roles. We note that in our hands, even the conservative single mutation R407K, which preserves the positive charge at this position, exhibited only ∼10% of mating activity relative to wild-type Ste5 and also showed reduced Ste11 binding, in contrast with the earlier report that the (R407S, K411S) double mutant retains Ste11 binding as judged by coimmunoprecipitation from cell extracts (Garrenton et al., 2006). The discrepancy of the mutational effect on residue R407 between our study and the previous report cannot be easily explained, because the mutation in our study is more conserved (R to K) than those in the previous study (R to S). It might be due to the fact that our analyses used an isolated Ste5 PH domain expressed in yeast as a fusion in a heterologous context, which could minimize the apparent association due to bridging effects that can occur in coimmunoprecipitation assays. Another possibility, although unlikely, could be that the second mutation K411S in the previous study has compensatory effects to mutations at residue R407 that render the double mutant apparently normal for Ste11 binding.
The mapped Ste11-interacting surface as defined by mutagenesis is distant from the canonical PI-binding site but partially overlaps with the general location of the noncanonical PI-binding site. However, as discussed earlier, our analysis indicates that stable and specific PI binding appears to be incompatible with the nature of these sites as predicted by the modeled structure of the PHSte5 domain. Additional detailed structure studies will be required to solve the issue.
Role of the RBL–PH domain interaction in yeast mating
The RBL–PH domain interaction that connects Ste5 and Ste11 has a critical function in yeast mating. The Ste11 MAPKKK plays essential roles in multiple MAPK signaling pathways in yeast. It interacts through its N-terminal regulatory region with different scaffold/adaptor proteins, which in turn direct plasma membrane targeting of Ste11, and this adaptor-specific targeting serves to ensure that signaling is properly linked to different environmental cues. The SAM domain of the kinase is essential for Ste11 function in the HOG pathway, whereas the RBL domain that lies C-terminal to the SAM domain is required for pheromone response through interaction with the Ste5 scaffold (Choi et al., 1994; Marcus et al., 1994; Printen and Sprague, 1994; Wu et al., 1999; Wang and Elion, 2003). This RBD-like domain is conserved in Ste11 MAPKKK homologues among fungal species.
The RBL domain is structurally similar to the RBD domain of S. pombe, yet the two domains have very different modes of interaction. We showed that the RBL domain of Ste11 interacts with the PH domain of Ste5, whereas in S. pombe the Byr2–RBD forms a complex with the small GTPase Ras1 (Tu et al., 1997; Scheffzek et al., 2001). We were unable to demonstrate detectable interaction of the RBLSte11 domain with any known small GTPase of Ras and Rho family in S. cerevisiae using a resin-binding assay. We tested the possibility that the β-finger “insert” sequence, which is present in the RBLSte11 domain and lacking in the RBDByr2 (Supplemental Figure S2(a)), may contribute to interaction partner selection. However, deleting the sequence (aa 158–173) encompassing the “insert” did not affect Ste11 function in either the pheromone response or the HOG pathways (unpublished data).
The RBL–PH domain interaction is highly specific, as RBLSte11 does not bind the PH domains of Ste5 homologues from other species (i.e., Ste5 of C. albicans) or the PH domains of the scaffold Far1 proteins of S. cerevisiae (Figure 3C). Similar specificity has also been observed in C. albicans, in which caSte11 interacts with the PHcaSte5 domain but not the PHcaFar1 domain (Cote et al., 2011). There appears to be limited residue conservation of the mapped Ste11-interacting site of the yeast PHSte5 domain across all Ste5 species homologues that possess PH domains (Supplemental Figure S1(b)), possibly relating to correlated substitutions in the corresponding RBLSte11 domains (Supplemental Figure S2(b)). Residue variations at these positions are also evident relative to the PH domains across the Far1-like proteins. The specificity of the interaction of two structurally conserved protein domains suggests that these functional partners have coevolved and that it is the determinant residues, not the general fold, that select the interaction partner.
Our structural and functional analyses clearly demonstrate that the RBLSte11–PHSte5 domain interaction is essential for Ste11 activation during pheromone response. The PHSte5-domain mutants are defective only in Ste11 binding and do not seem to be affected in their subcellular localization. Of interest, the function of these Ste5 mutants could be restored when the Ste11–Ste5 interaction was reestablished through another protein–protein interaction module (Figure 6), suggesting that the interaction between RBLSte11 and PHSte5 domains does not involve a conformational change that is required for Ste11 activation. Consistent with this, a constitutively active Ste11 bypasses the pheromone-signaling defect of the PHSte5-domain mutants that are impaired for Ste11 binding.
It is interesting to note that Ste11 has a more general presence among fungal species than Ste5. Some species contain both Ste5 and Far1 scaffold proteins, each with a PH domain, such as S. cerevisiae and C. albicans; some have only the Far1 scaffold, and yet other species, such as S. pombe and Schizosaccharomyces japonicus, have neither (Cote et al., 2011). Whereas all Ste11 MAPKKK homologues contain an RBL domain, only the RBD of Byr2 in S. pombe has been shown to bind the Ras1 small GTPase, and this binding contributes to mating signaling (Tu et al., 1997; Scheffzek et al., 2001). In contrast, the RBLSte11 domains in S. cerevisiae and C. albicans bind to their respective PHSte5 domains (Figure 3C; Cote et al., 2011). In agreement with this protein interaction profile, no significant role for Ras in mating pheromone response has been uncovered in S. cerevisiae (Mosch et al., 1996). It follows that the RBL domains are capable of binding to either small GTPases or PH domains. Understanding how the Ste11 MAPKKK RBL domain evolved from Ras-binding and differentiated toward PHSte5-domainbinding requires further detailed structural information of the complex of these two conserved but functionally versatile domains.
MATERIALS AND METHODS
Yeast strains and assays
Yeast media, culture conditions, and manipulations of yeast strains were as described (Rose et al., 1990). Yeast transformations were carried out with the lithium acetate method (Rose et al., 1990; Gietz et al., 1992). The yeast strains used in this study are listed in the Supplemental Experimental Procedures. Halo assays, quantitative β-galactosidase reporter assays for the pheromone response and HOG pathways, and mating assays were performed as described (Wu et al., 1999; Tatebayashi et al., 2006). GFP fluorescence photomicroscopy was performed as previously described (Wu et al., 1999).
Plasmid construction and mutagenesis
The protein-interaction boundary of the Ste5-PH domain with Ste11 was defined using the yeast two-hybrid system developed recently in our lab for the detection of protein–protein interactions in cytoplasm (Cote et al., 2011; see the Supplemental Experimental Procedures for details). Random mutageneses of the RBLSte11 and PHSte5 domains were performed using error-prone PCR. Site-directed mutagenesis of STE5 and STE11 were performed with either mutagenic sewing PCR (Ho et al., 1989) or a QuikChange Mutagenesis Kit (Agilent Technology, Santa Clara, CA) according to manufacturer’s protocol. All desired mutations were confirmed by sequencing. Plasmids and oligos used in this study are listed in the Supplemental Experimental Procedures.
Protein expression and purification
All His-tagged fusion proteins were expressed and purified by affinity chromatography using nickel-nitriloacetic acid resin (GE Healthcare Bio-Sciences, Piscataway, NJ) using standard protocols. The GST fusion proteins were purified by affinity chromatography on glutathione–Sepharose according to a modification of the manufacturer’s recommendation.
Surface plasmon resonance analysis of protein–protein interaction
Surface plasmon resonance analysis of the RBLSte11–PHSte5 domain interaction was performed using a Biacore 3000 (GE Healthcare Biosciences). Sensograms were aligned and double referenced to the control surface using buffer injections and analyzed by both global fitting to a 1:1 interaction and steady-state analysis with BiaEvaluation (version 3.2) software.
Structural bioinformatics and PHSte5 model building
Homologous protein sequences were retrieved by either BLASTP or TBLASTN search (Altschul and Lipman, 1990) in the Fungal Genome Database (www.yeastgenome.org/) and by protein domain architecture search in the SMART database (http://smart.embl.de/; Letunic et al., 2006). Structural fold detection for regions of representative Ste5, Far1, and Ste11 fungal proteins was carried out at the Structure Prediction Meta Server (http://meta.bioinfo.pl/), which provides a consensus sequence–to–structure fold recognition score using the 3D-Jury meta-predictor (Ginalski and Rychlewski, 2003). Homology modeling of the PH domain of S. cerevisiae Ste5 was done in MODELLER 9.1 (Fiser and Sali, 2003a, b; Marti-Renom et al., 2000) starting from a multiple-queries/multiple-templates sequence alignment (Supplemental Figure S1). The resulting structure was further refined in AMBER 9 (Case et al., 2005) by 20 ns of classical molecular dynamics simulation in explicit solvent using the FF03 force field parameters (Duan et al., 2003; Lee and Duan, 2004). Model validation was carried out with PROCHECK (Laskowski et al., 1993), ProSA (Wiederstein and Sippl, 2007), and Verify3D (Luthy et al., 1992).
NMR spectroscopy and structure calculation
The RBLSte11 domain was produced in minimal medium (M9) enriched with 15N-ammonium chloride or 15N-ammonium chloride/13C-glucose and purified using Ni2+-nitriloacetic acid resin (Qiagen, Valencia, CA). Samples for NMR measurements were prepared in 50 mM sodium phosphate buffer, pH 6.8, 200 mM NaCl, 5 mM dithiothreitol, and 0.02% (wt/vol) NaN3 at protein concentrations of 1.2–1.5 mM. All NMR data were collected at 300 K on a Bruker Avance 500 spectrometer equipped with a triple-resonance cryoprobe and with z-gradient pulse field gradient accessories. The 3D 1H15N nuclear Overhauser effect spectroscopy (NOESY)-HSQC, 3D 1H13C NOESY-HSQC (in 2H2O) and 2D homonuclear NOESY experiments were used to collect NOE-restraints for structure calculation using CYANA 2.1 (Güntert, 2004). A summary of the results is given in Table S1. See the Supplemental Experimental Procedures for details.
Supplementary Material
Acknowledgments
We thank Gregor Jansen and Peter Pryciak for plasmids and strains and Maria Kowalik for preparing proteins for NMR experiments. This work was partially supported by the Genomic Health Initiative of the National Research Council Canada and by Grant GSP-48370 from the Canadian Institute of Health Research (to M.C. and I.E.). This is National Research Council Canada Publication Number 50653.
Abbreviations used:
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HOG
high-osmolarity glycerol
- MAP
mitogen-activated protein
- NMR
nuclear magetic resonance
- PDB
Protein Data Bank
- PH
pleckstrin homology
- RA
Ras association
- RBD
Ras-binding domain
- RBL
Ras-binding domain–like
- SAM
sterile alpha mating
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-07-0516) on December 14, 2012.
Present addresses: *Douglas Institute Research Center, LaSalle, Montreal, QC H4H 1R3, Canada;
†Department of Biochemistry, University of Saskatchewan, Saskatoon, SK S7N 5E5, Canada.
None of the authors has a financial interest related to this work.
REFERENCES
- Alam SL, Langelier C, Whitby FG, Koirala S, Robinson H, Hill CP, Sundquist WI. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat Struct Mol Biol. 2006;13:1029–1030. doi: 10.1038/nsmb1160. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci USA. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annan RB, Wu C, Waller DD, Whiteway M, Thomas DY. Rho5p is involved in mediating the osmotic stress response in Saccharomyces cerevisiae, and its activity is regulated via Msi1p and Npr1p by phosphorylation and ubiquitination. Eukaryot Cell. 2008;7:1441–1449. doi: 10.1128/EC.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- Cote P, Sulea T, Dignard D, Wu C, Whiteway M. Evolutionary reshaping of fungal mating pathway scaffold proteins. MBio. 2011;2:e00230–e00210. doi: 10.1128/mBio.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Ekiel I, Sulea T, Jansen G, Kowalik M, Minailiuc O, Cheng J, Harcus D, Cygler M, Whiteway M, Wu C. Binding the atypical RA domain of Ste50p to the unfolded Opy2p cytoplasmic tail is essential for the high-osmolarity glycerol pathway. Mol Biol Cell. 2009;20:5117–5126. doi: 10.1091/mbc.E09-07-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA. Ste5: a meeting place for MAP kinases and their associates. Trends in Cell Biol. 1995;5:322–327. doi: 10.1016/s0962-8924(00)89055-8. [DOI] [PubMed] [Google Scholar]
- Elion EA. The Ste5p scaffold. J Cell Sci. 2001;114:3967–3978. doi: 10.1242/jcs.114.22.3967. [DOI] [PubMed] [Google Scholar]
- Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003a;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003b;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- Garrenton LS, Young SL, Thorner J. Function of the MAPK scaffold protein, Ste5, requires a cryptic PH domain. Genes Dev. 2006;20:1946–1958. doi: 10.1101/gad.1413706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K, Rychlewski L. Detection of reliable and unexpected protein fold predictions using 3D-Jury. Nucleic Acids Res. 2003;31:3291–3292. doi: 10.1093/nar/gkg503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronwald W, Huber F, Grunewald P, Sporner M, Wohlgemuth S, Herrmann C, Kalbitzer HR. Solution structure of the Ras binding domain of the protein kinase Byr2 from Schizosaccharomyces pombe. Structure. 2001;9:1029–1041. doi: 10.1016/s0969-2126(01)00671-2. [DOI] [PubMed] [Google Scholar]
- Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hirano S, Suzuki N, Slagsvold T, Kawasaki M, Trambaiolo D, Kato R, Stenmark H, Wakatsuki S. Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat Struct Mol Biol. 2006;13:1031–1032. doi: 10.1038/nsmb1163. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye C, Dhillon N, Durfee T, Zambryski PC, Thorner J. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: application of a differential interaction trap assay for examining protein–protein interactions. Genetics. 1997;147:479–492. doi: 10.1093/genetics/147.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C, Serrano L. The ubiquitin domain superfold: structure-based sequence alignments and characterization of binding epitopes. J Mol Biol. 2006;355:821–844. doi: 10.1016/j.jmb.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Lee MC, Duan Y. Distinguish protein decoys by using a scoring function based on a new AMBER force field, short molecular dynamics simulations, and the generalized born solvent model. Proteins. 2004;55:620–634. doi: 10.1002/prot.10470. [DOI] [PubMed] [Google Scholar]
- Leevers SJ, Paterson HF, Marshall CJ. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Pleckstrin homology domains: not just for phosphoinositides. Biochem Soc Trans. 2004;32:707–711. doi: 10.1042/BST0320707. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp. 2007:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucl Acid Res. 2006;34:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Mayer BJ. Mechanism of activation of Pak1 kinase by membrane localization. Oncogene. 1999;18:797–806. doi: 10.1038/sj.onc.1202361. [DOI] [PubMed] [Google Scholar]
- Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Martzen MR, McCraith SM, Spinelli SL, Torres FM, Fields S, Grayhack EJ, Phizicky EM. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- Mosch HU, Roberts RL, Fink GR. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375:554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- O’Rourke SM, Herskowitz I, O’Shea EK. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 2002;18:405–412. doi: 10.1016/s0168-9525(02)02723-3. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Benjamin DR. A novel family of Ras-binding domains. Trends Biochem Sci. 1996;21:422–425. doi: 10.1016/s0968-0004(96)30038-8. [DOI] [PubMed] [Google Scholar]
- Printen JA, Sprague GF., Jr Protein–protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Scheffzek K, Grunewald P, Wohlgemuth S, Kabsch W, Tu H, Wigler M, Wittinghofer A, Herrmann C. The Ras-Byr2RBD complex: structural basis for Ras effector recognition in yeast. Structure. 2001;9:1043–1050. doi: 10.1016/s0969-2126(01)00674-8. [DOI] [PubMed] [Google Scholar]
- Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem. 2003;278:35857–35860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokoe D, Macdonald SG, Cadwallader K, Symons M, Hancock JF. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- Tatebayashi K, Yamamoto K, Tanaka K, Tomida T, Maruoka T, Kasukawa E, Saito H. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 2006;25:3033–3044. doi: 10.1038/sj.emboj.7601192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truckses DM, Bloomekatz JE, Thorner J. The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:912–928. doi: 10.1128/MCB.26.3.912-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H, Barr M, Dong DL, Wigler M. Multiple regulatory domains on the Byr2 protein kinase. Mol Cell Biol. 1997;17:5876–5887. doi: 10.1128/mcb.17.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dohlman HG. Pheromone signaling mechanisms in yeast: a prototypical sex machine. Science. 2004;306:1508–1509. doi: 10.1126/science.1104568. [DOI] [PubMed] [Google Scholar]
- Wang Y, Elion EA. Nuclear export and plasma membrane recruitment of the Ste5 scaffold are coordinated with oligomerization and association with signal transduction components. Mol Biol Cell. 2003;14:2543–2558. doi: 10.1091/mbc.E02-10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway MS, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas DY, Leberer E. Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiget P, Shimada Y, Butty AC, Bi E, Peter M. Site-specific regulation of the GEF Cdc24p by the scaffold protein Far1p during yeast mating. EMBO J. 2004;23:1063–1074. doi: 10.1038/sj.emboj.7600123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters MJ, Lamson RE, Nakanishi H, Neiman AM, Pryciak PM. A membrane binding domain in the ste5 scaffold synergizes with gbetagamma binding to control localization and signaling in pheromone response. Mol Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Wu C, Jansen G, Zhang J, Thomas DY, Whiteway M. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev. 2006;20:734–746. doi: 10.1101/gad.1375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Leberer E, Thomas DY, Whiteway M. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2425–2440. doi: 10.1091/mbc.10.7.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Kim EY, Connelly JJ, Nassar N, Kirsch J, Winking J, Schwarz G, Schindelin H. The crystal structure of Cdc42 in complex with collybistin II, a gephyrin-interacting guanine nucleotide exchange factor. J Mol Biol. 2006;359:35–46. doi: 10.1016/j.jmb.2006.03.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.