Morphology determination is critical for virulence of the human fungal pathogen Candida albicans. A genome-wide transcriptional analysis shows that genes associated with specifying the C. albicans pseudohyphal morphology represent a subset of hyphal genes and reveals fundamental differences between forward and reverse morphological transitions.
Abstract
Candida albicans, the most common cause of human fungal infections, undergoes a reversible morphological transition from yeast to pseudohyphal and hyphal filaments, which is required for virulence. For many years, the relationship among global gene expression patterns associated with determination of specific C. albicans morphologies has remained obscure. Using a strain that can be genetically manipulated to sequentially transition from yeast to pseudohyphae to hyphae in the absence of complex environmental cues and upstream signaling pathways, we demonstrate by whole-genome transcriptional profiling that genes associated with pseudohyphae represent a subset of those associated with hyphae and are generally expressed at lower levels. Our results also strongly suggest that in addition to dosage, extended duration of filament-specific gene expression is sufficient to drive the C. albicans yeast-pseudohyphal-hyphal transition. Finally, we describe the first transcriptional profile of the C. albicans reverse hyphal-pseudohyphal-yeast transition and demonstrate that this transition involves not only down-regulation of known hyphal-specific, genes but also differential expression of additional genes that have not previously been associated with the forward transition, including many involved in protein synthesis. These findings provide new insight into genome-wide expression patterns important for determining fungal morphology and suggest that in addition to similarities, there are also fundamental differences in global gene expression as pathogenic filamentous fungi undergo forward and reverse morphological transitions.
INTRODUCTION
Candida albicans, a fungus normally found as part of the human mucosal flora, is capable of causing a variety of diseases including oral and vaginal thrush, as well as systemic bloodstream infections (candidemia; Odds, 1988). Neonates, HIV/AIDS patients, cancer patients undergoing chemotherapy, and individuals with an otherwise compromised immune system are at higher risk for developing these infections (Cannon and Chaffin, 1999; Dongari-Bagtzoglou et al., 1999; Filler and Kullberg, 2002). Candida species are the fourth-leading cause of hospital-acquired bloodstream infections in the United States, with a mortality rate between 30 and 50% (Beck-Sague and Jarvis, 1993; Pfaller et al., 1998; Edmond et al., 1999; Wisplinghoff et al., 2004).
Several properties are known to contribute to the pathogenicity of C. albicans, including adherence to host epithelial and endothelial cells, secretion of enzymes that can degrade host tissues, and the ability to undergo a reversible morphological transition between yeast (single oval-shaped cells) and filaments (elongated cells, attached end to end; Odds, 1988; Yang, 2003; Filler and Sheppard, 2006; Zhu and Filler, 2010; Phan et al., 2007; Dalle et al., 2010). The C. albicans yeast–filament transition is important for invasion of epithelial and endothelial cell layers, lysis of macrophages and neutrophils, breaching of endothelial cells, and thigmotropism (Gow et al., 1994; Zink et al., 1996; Lo et al., 1997; Jong et al., 2001; Korting et al., 2003; Kumamoto and Vinces, 2005). This transition is known to occur in response to a variety of conditions present in the host, such as body temperature (37°C), serum, neutral pH, certain carbon sources (e.g., N-acetylglucosamine), and specific amino acids (e.g., proline). Several critical experiments have also indicated that the ability of C. albicans to undergo a reversible yeast–filament transition is required for virulence (Braun and Johnson, 1997; Braun et al., 2001; Lo et al., 1997; Mitchell, 1998; Brown and Gow, 1999; Brown, 2002; Ernst, 2000; Murad et al., 2001; Saville et al., 2003; Zheng et al., 2004; Carlisle et al., 2009).
C. albicans is known to form two distinct types of filaments: pseudohyphae and hyphae. Pseudohyphal filaments comprise ellipsoid-shaped cells attached end to end, have visible septal constrictions, and tend to be highly branched. Hyphal filaments, in contrast, have parallel-sided cells joined at true septal junctions (lacking constrictions; Sudbery et al., 2004). Unlike pseudohyphae, hyphae also possess specialized structures, such as a Spitzenkörper for tip growth, and undergo their first mitosis in the germ tube rather than the mother–bud neck (Sudbery et al., 2004; Crampin et al., 2005). Due to significant physical differences between pseudohyphae and hyphae, as well as studies demonstrating that very few mutants locked in the pseudohyphal form can be induced to form hyphae, it was believed for many years that these two morphologies are determined by genetically distinct mechanisms (Braun and Johnson, 1997; Bensen et al., 2002; Sudbery et al., 2004; Wightman et al., 2004). However, a study by our laboratory suggested that C. albicans yeast, pseudohyphal, and hyphal morphologies are actually determined by a common dosage-dependent transcriptional mechanism (Carlisle et al., 2009). This study involved placing a single allele of UME6, which encodes a key filament-specific transcriptional regulator, under control of a tetracycline-regulatable promoter. As UME6 levels increased, C. albicans cells were observed to sequentially transition from yeast to pseudohyphae to hyphae. Importantly, this experiment was carried out in the complete absence of filament-inducing conditions. Northern analysis indicated that as cells undergo the yeast-pseudohyphal-hyphal transition in response to UME6 expression levels there is an increase in the number of filament-specific genes expressed, as well as in their level of expression. Although these experiments suggested that similar genes are expressed in pseudohyphal and hyphal morphologies, they provided little information about global genome-wide expression patterns associated with the sequential yeast-pseudohyphal-hyphal transition or the relationship among gene sets associated with each morphology.
Several previous studies examined genome-wide transcriptional profiles as C. albicans undergoes the yeast–filament transition in response to filament-inducing conditions, such as the combination of serum at 37°C (Lane et al., 2001; Nantel et al., 2002; Kadosh and Johnson, 2005). These studies showed that genes involved in a wide variety of biological processes are induced. Whereas certain genes play direct roles in the mechanics of filament formation, others are associated with a number of virulence-related processes, including adhesion to host cells, biofilm formation, degradation of host cell membranes, and the ability to tolerate oxidative stress. A significant limitation of these studies, however, is that because filamentation was induced by an external environmental condition, it is not clear whether certain genes are specifically expressed in association with filaments per se or are induced in response to a variety of upstream regulators and signaling pathways that may only be associated with the environmental cue. In addition, all previous genome-wide transcriptional profiling experiments of C. albicans morphological transitions have only examined the forward transition from yeast to hyphae. Whereas the ability of C. albicans to undergo a reversible transition from yeast to filaments is required for virulence, very little, if any, information is available regarding gene expression patterns or mechanisms associated with the reverse, hyphal-to-yeast transition.
Because UME6 was previously shown to function as a major downstream target of numerous filamentous growth signaling and regulatory pathways, the tetracycline-regulatable UME6 expression system that we previously developed allows us to address many of the issues discussed above without added complications associated with external filament-inducing conditions (Banerjee et al., 2008; Zeidler et al., 2009; Martin et al., 2011; Shareck et al., 2011). Here, using this system in combination with genome-wide transcriptional profile analysis, we define sets of genes whose expression is specifically associated with C. albicans pseudohyphal and hyphal morphologies and determine the extent to which these gene sets overlap. A comparative analysis with a gene set induced in response to one of the strongest filament-inducing conditions, serum at 37°C, is also performed to identify a minimal set of genes specifically associated with hyphal growth. Finally, the UME6 expression system is used to provide the first genome-wide transcriptional profile of the C. albicans hyphal-pseudohyphal-yeast transition, allowing a direct comparison between gene expression patterns associated with forward and reverse morphological transitions.
RESULTS
Overlapping sets of genes are associated with specifying C. albicans pseudohyphal and hyphal morphologies in a dosage-dependent manner
We previously generated a strain (tetO-UME6) in which one allele of UME6 is under control of a tetracycline-regulatable promoter (tetO) driven by an Escherichia coli tetR DNA-binding domain–Saccharomyces cerevisiae HAP4 activation domain transactivator fusion protein expressed at high constitutive levels (Nakayama et al., 2000; Carlisle et al., 2009). In the presence of doxycycline (Dox), a tetracycline derivative, UME6 is not expressed, and C. albicans cells grow in the yeast form. As Dox levels decline, UME6 expression increases and cells undergo the transition from yeast to pseudohyphal to hyphal morphologies; hybrid yeast–pseudohyphal and pseudohyphal–hyphal filaments are also apparent (Carlisle et al., 2009). To better understand the relationship between gene sets associated with pseudohyphal and hyphal morphologies on a genome-wide scale, we performed a whole-genome DNA microarray analysis of C. albicans cells undergoing the yeast-pseudohyphal-hyphal transition in response to UME6 dosage in the tetO-UME6 strain. Gene expression values for hyphal cells were determined using the −Dox sample (nearly 100% of cells form hyphae under this condition), and values for pseudohyphal cells were determined using the 0.1 μg/ml Dox sample (Figure 1). At this Dox concentration, >80% of cells in the population form pseudohyphae; the large majority of the remaining cells are in yeast form (an example image is shown in Supplemental Figure S1A). The main criteria used to define pseudohyphae under this condition are as follows: 1) constrictions at septal junctions and the mother–bud neck, 2) unparallel cell sides, and 3) increased filament branching. All of these criteria were previously described by Sudbery et al. (2004). A control strain that expresses a tetR-HAP4 transactivator but lacks tet operator sequences was also grown overnight in the presence and absence of 20 μg/ml Dox.
FIGURE 1:
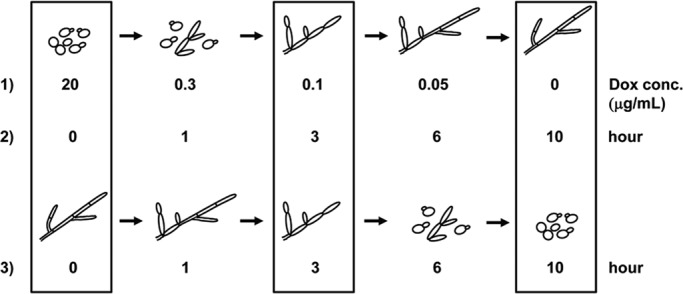
Schematic diagram of yeast-pseudohyphal-hyphal and hyphal-pseudohyphal-yeast transitions generated by induction or depletion, respectively, of UME6 in the absence of filament-inducing conditions. 1) Cultures of the tetO-UME6 strain were grown overnight in YEPD medium at 30°C in the presence of decreasing Dox concentrations (resulting in increasing UME6 levels). 2) The tetO-UME6 strain was grown overnight in YEPD medium at 30°C in the presence of Dox (UME6 off), washed 1× with YEPD, inoculated into YEPD medium prewarmed to 30°C in the absence of Dox (UME6 induced), and allowed to grow over a 10-h time course. 3) The tetO-UME6 strain was grown overnight in YEPD medium at 30°C in the absence of Dox (UME6 induced). After addition of Dox to the culture (UME6 shutoff), cells were grown over a 10-h time course. Cell morphologies were determined by differential interference contrast microscopy and were similar to those reported previously (Carlisle et al., 2009). Boxes indicate conditions used to represent yeast, pseudohyphal, and hyphal cultures.
For the tetO-UME6 strain, we found that 238 genes were reproducibly induced >2-fold in hyphae when UME6 was expressed at its highest level in the absence of Dox. Based on data from the most highly expressed genes (>3-fold induction and higher), a subset of ∼30% was also found to be induced >2-fold in pseudohyphae (Table 1). The percentage of these genes expressed >4-fold and >10-fold in pseudohyphae was generally much lower. In addition, we observed that 86 genes are induced >2-fold in pseudohyphae, and that the large majority (>70%) of the most highly expressed pseudohyphal genes (>3-fold induction and higher) were also induced >2-fold in hyphae. Although the percentages of pseudohyphal genes expressed >4-fold and >10-fold in hyphae were somewhat lower, they were still significantly higher than the percentages of hyphal genes expressed at these levels in pseudohyphae (Table 1). None of these genes showed any reproducible change in expression when the tetR-HAP4 control strain was grown in the absence versus presence of Dox (expression of only two genes, orf19.6061 and orf19.7111.1, appeared to be affected by Dox). Overall these results suggest that genes associated with specifying the C. albicans pseudohyphal morphology represent a subset of those associated with hyphal growth and are generally expressed at lower levels.
TABLE 1:
Relative morphological distribution of genes induced by UME6 during the yeast-pseudohyphal-hyphal transition.
| Fold change relative to yeast expression | |||
|---|---|---|---|
| >3-fold | >4-fold | >10-fold | |
| Number of genes up in hyphal cellsa | 143 | 94 | 21 |
| Percentage of genes also up >2-fold in pseudohyphal cellsb | 28 | 29 | 38 |
| Percentage of genes also up >4-fold in pseudohyphal cellsc | 13 | 16 | 33 |
| Percentage of genes also up >10-fold in pseudohyphal cellsc | 3 | 4 | 19 |
| >2-fold | >3-fold | >4-fold | |
| Number of genes up in pseudohyphal cellsa | 86 | 46 | 22 |
| Percentage of genes also up >2-fold in hyphal cellsb | 55 | 72 | 86 |
| Percentage of genes also up >4-fold in hyphal cellsc | 31 | 52 | 68 |
| Percentage of genes also up >10-fold in hyphal cellsc | 9 | 15 | 32 |
Data exclude genes with expression values affected by Dox alone (as determined by the tetR-HAP4 control strain experiment). Cells of the tetO-UME6 strain were grown overnight in YEPD at 30°C to an OD600 of 1.0. Yeast cells, 20 μg/ml Dox; pseudohyphal cells, 0.1 μg/ml Dox; and hyphal cells, no Dox.
aFold changes are based on mean gene expression values from two independent experiments (n = 2). All genes were induced at least twofold in both experiments.
bPercentage of genes showing an induction of at least twofold in two independent experiments (n = 2) in the indicated cell morphology.
cPercentage of genes showing the indicated mean fold induction in the indicated morphology based on two independent experiments (n = 2). All genes were induced at least twofold in both experiments.
We also performed a hierarchical cluster analysis of genes induced in response to UME6 dosage during the yeast-pseudohyphal-hyphal transition. We observed that certain genes were induced at a low level during the yeast–pseudohyphal transition (20 to 0.1 μg/ml Dox) in response to low UME6 expression (Figure 2). As UME6 levels rise and cells transition from pseudohyphae to hyphae (0.1 to 0 μg/ml Dox), the expression level of these genes increases significantly. Additional genes also appear to be initially induced and show significantly increased expression as cells transition from pseudohyphae to hyphae. A larger cluster analysis, performed using all genes showing at least a twofold increase or decrease in expression in at least one data point, also shows certain genes that are expressed and reach a plateau level starting at low Dox concentrations (Supplemental Figure S2A). Of interest, very few, if any, genes appeared to be induced exclusively when cells grow as pseudohyphae (0.1 μg/ml Dox). In addition to providing further support for our conclusion that genes associated with specifying the pseudohyphal morphology represent a subset of those associated with hyphal growth, these results also corroborate our previous study (Carlisle et al., 2009) and strongly suggest that the expression level not only of UME6, but also of UME6-induced gene sets, plays an important role in determining C. albicans morphology in a dosage-dependent manner.
FIGURE 2:

Cluster diagram of genes induced in response to specific UME6 expression levels in a steady-state culture. Experimental data represents mean expression values based on two independent DNA microarray experiments (n = 2 biological replicates). Only a subset of genes showing fourfold or greater change in expression in at least one data point with >80% of data present are shown. Each data point represents fold change in gene expression relative to the 20 μg/ml Dox culture. Blue, increased expression; yellow, reduced expression; gray, no data. C = tetR-HAP4 control strain lacking a tet operator, −Dox vs. 20 μg/ml Dox.
Extended duration of UME6 expression is sufficient to drive the yeast-pseudohyphal-hyphal transition and gradually increases expression of filament-specific genes
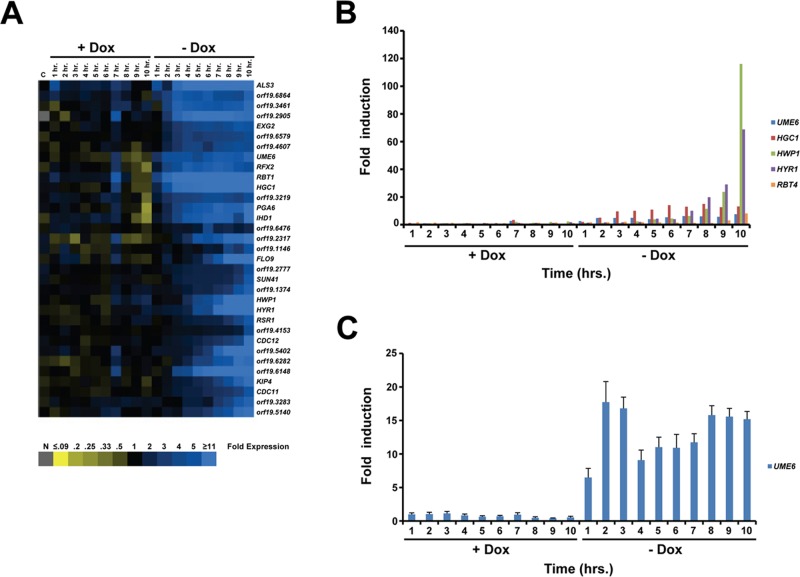
We also examined global changes in gene expression in response to increased UME6 expression over a time course. Cells of the tetO-UME6 strain were grown overnight in the presence of Dox, washed, and then inoculated into fresh yeast extract-peptone-dextrose (YEPD) medium in the presence or absence of Dox. In the absence of Dox, the tetO-UME6 strain transitions from yeast to pseudohyphae to hyphae over a 10-h time course: at the 0-h time point cells are in yeast form, at the 3-h time point >80% of cells are pseudohyphal (the large majority of the remaining cells are yeast; Supplemental Figure S1B), and by the 10-h time point nearly 100% of cells are hyphal (Figure 1; note that in our previous study, Carlisle et al., 2009, this transition was completed within 9 h, most likely due to differences in culture volume). The criteria used to define pseudohyphae are the same as those described for the UME6 dosage experiment. In the presence of Dox, UME6 is not expressed and cells remain in the yeast form. Over this time course cells were harvested at each hour from both +Dox and –Dox cultures for RNA extraction and DNA microarray analysis.
Overall we found that the expression pattern of genes in the –Dox time course was very similar to that observed in the steady-state culture UME6 dosage experiment: a subset of genes induced in hyphae are also induced in pseudohyphae, whereas the large majority of genes induced in pseudohyphae are also induced in hyphae; in addition, the percentage of pseudohyphal genes expressed at high levels (>4-fold and >10-fold) in hyphae was significantly greater than the percentage of hyphal genes expressed at these levels in pseudohyphae (Supplemental Table S1). As in the UME6 dosage experiment, none of these genes showed changes in expression when the tetR-HAP4 control strain was grown in the absence versus presence of Dox. Cluster analysis of the tetO-UME6 time-course experiment indicated that a large group of genes is induced at gradually increasing levels when UME6 is expressed in the absence of Dox (Figure 3A). Certain genes, such as HGC1, were induced early and reached a maximal expression level by the 3-h time point when C. albicans cells have completed the transition to pseudohyphae (Figure 3B). In contrast, other genes, such as HYR1 and HWP1, were induced at later time points and reached maximal levels only when cells had transitioned completely to the hyphal morphology at 10 h. As previously observed in the UME6 dosage experiment, a larger cluster analysis of the forward-transition time-course experiment, using all genes induced or reduced at least twofold in one data point, indicated that very few, if any, genes were expressed exclusively in pseudohyphae; as expected, however, a large number of genes showed changes in expression over the time course that were not dependent upon Dox (Supplemental Figure S2B). Overall these results provide further support for our conclusion that genes associated with specifying the C. albicans pseudohyphal morphology represent a subset of those associated with hyphal growth.
FIGURE 3:
Transcriptional analysis of the yeast-pseudohyphal-hyphal transition in response to UME6 expression over a time course. (A) Cluster diagram of genes induced. Experimental data represent mean expression values based on two independent DNA microarray experiments (n = 2 biological replicates). Only a subset of genes showing fourfold or greater change in expression in at least one data point with >80% of data present are shown. Each data point represents fold change in gene expression relative to the 0-h time point. Blue, increased expression; yellow, reduced expression; gray, no data. C = tetR-HAP4 control strain lacking a tet operator, −Dox vs. 20 μg/ml Dox. (B) Histogram showing mean fold induction (n = 2 biological replicates), relative to the 0-h time point, for several known hyphal-specific genes as determined by the DNA microarray analysis. (C) Fold induction of UME6 over the time course, as determined by real-time quantitative RT-PCR analysis. Mean fold induction for each time point, relative to the 0-h time point, is shown (n = 3). Bars, SE.
Of interest, we noted that the UME6 expression level appeared to peak very early at the 2-h time point, before completion of the yeast–pseudohyphal transition, and remained at a relatively constant high level for the remainder of the time course as cells transition to hyphae (Figure 3, A and B). This finding was verified using quantitative reverse transcription (RT)-PCR analysis (Figure 3C). Importantly, these results suggest that in addition to UME6 dosage, extended duration of UME6 expression is sufficient to induce overlapping sets of filament-specific genes and drive the C. albicans yeast-pseudohyphal-hyphal transition. Because certain UME6 target genes, such as HGC1, also reach peak levels early in the time course and then plateau, our results also suggest that expression of specific components of the C. albicans filamentation program for an extended duration is important for driving the yeast-pseudohyphal-hyphal transition.
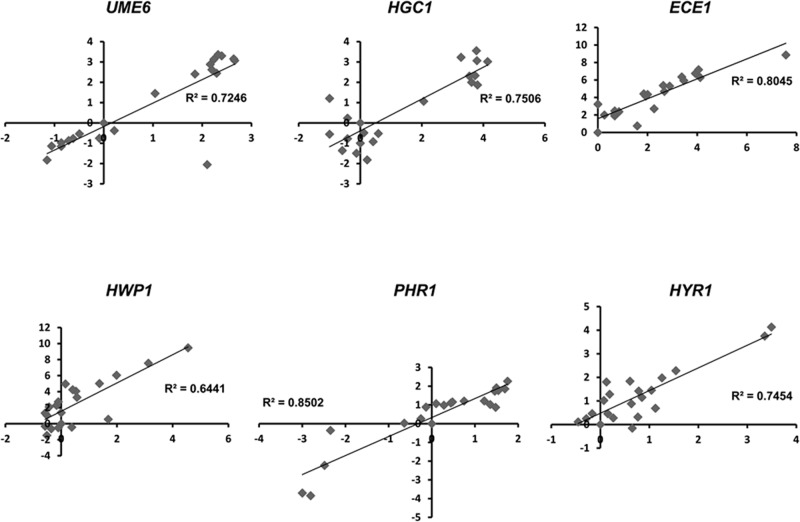
Overall validity of microarray data in the yeast-pseudohyphal-hyphal transition time-course experiment was confirmed by plotting microarray expression values versus quantitative RT-PCR values and generating a Pearson’s r (p ≤ 0.01) for six genes, including HGC1 (Figure 4). In addition, an independent DNA microarray analysis confirmed that several hyphal-associated genes, including ECE1, HYR1, SOD5, and HGC1, are induced upon ectopic expression of UME6 (Martin et al., 2011).
FIGURE 4:
Correlation of gene expression values obtained using DNA microarray vs. real-time quantitative RT-PCR data for the forward yeast-pseudohyphal-hyphal transition time course. For each gene, graphs represent mean change in gene expression (n = 2) as determined by DNA microarray (x-coordinate) plotted against mean gene expression changes (n = 3) determined using real-time quantitative RT-PCR (y-coordinate; values in log2). Pearson’s r was determined for each graph, and statistical significance was determined using the Student’s t test (p ≤ 0.01).
Genes induced in response to UME6 expression during the yeast-pseudohyphal-hyphal transition are involved in a variety of virulence-related processes and overlap with those induced in response to a strong natural filament-inducing condition
To gain a better understanding of biological processes important for UME6-driven hyphal formation and virulence, we classified genes that were reproducibly induced at least twofold in either the dosage or time-course UME6 expression experiments (Supplemental Data Sets S1 and S3) using the Candida Genome Database (CGD) Gene Ontology (GO) Slim Mapper tool available at www.candidagenome.org. GO Slim Mapper analysis revealed that genes induced during UME6-directed hyphal growth in both experiments are involved in a wide variety of biological processes, many of which have been shown to be important for virulence, including filamentous growth, adhesion, biofilm formation, carbohydrate metabolism, stress response, cell wall organization, signal transduction, cell cycle, and the ability to interact with the host (Supplemental Figures S3 and S4). When we performed this same analysis on the gene sets induced during pseudohyphal growth (Supplemental Data Sets S2 and S4), we found that many of the same biological processes were represented (Supplemental Figures S3 and S4). This finding is consistent with our previous observation that genes associated with pseudohyphal formation represent a subset of those associated with hyphal growth and suggests that these two morphologies share many of the same mechanisms important for C. albicans filament formation and virulence. We note, however, that additional mechanisms are also likely to play important roles in hyphal formation (see Discussion).
We next used the CGD GO Term Finder tool (also available at www.candidagenome.org) to identify gene classes induced in hyphae in response to UME6 expression over a time course that were overrepresented compared with their representation in the genome as a whole. Strikingly, nearly all overrepresented gene classes appear to be involved either in morphology or other virulence-related processes, including biofilm formation, adhesion, host interaction, microtubule-based movement, and iron assimilation (Figure 5B). Several previous studies showed that multiple virulence properties are coregulated with C. albicans morphogenesis (Staab et al., 1999; Braun et al., 2000; Calderone and Fonzi, 2001; Fu et al., 2002; Liu, 2002; Naglik et al., 2003; Zhao et al., 2004), and our results suggest that UME6 plays a key role in coordinating the expression of genes important for these properties.
FIGURE 5:
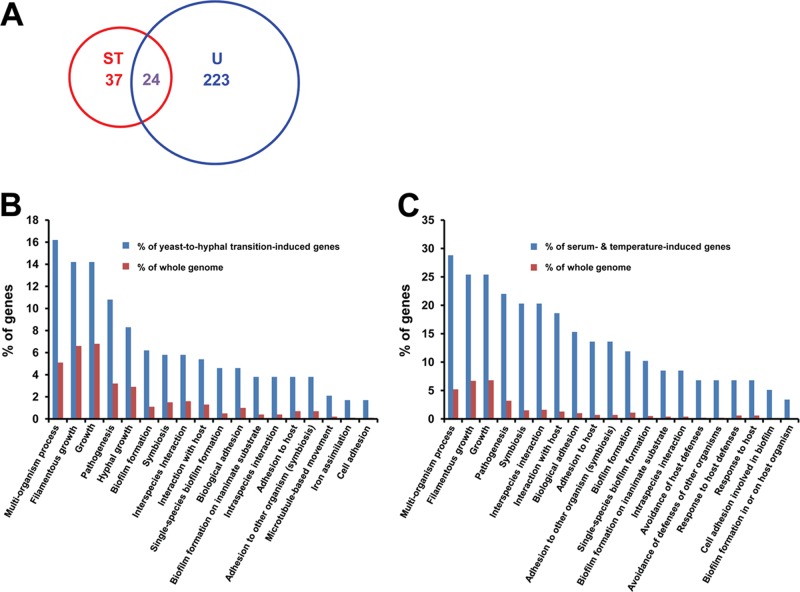
Gene expression profile of hyphal cells generated by UME6 expression over a time course strongly correlates with the profile of genes expressed upon filamentation of C. albicans in the presence of serum at 37°C. (A) Venn diagram indicating overlap between genes showing a mean induction greater than twofold at the 10-h time point in the absence of Dox relative to the 0-h time point (n = 2) in the tetO-UME6 strain, as defined in Supplemental Table S1 and listed in Supplemental Data Set S3 (U), and genes previously shown to be induced in response to growth of C. albicans in the presence of serum at 37°C (ST; Kadosh and Johnson, 2005). (B) Gene classes overrepresented in the set of genes showing a mean induction greater than twofold in hyphal cells of the tetO-UME6 strain (as defined in A) compared with their representation in the C. albicans genome as a whole. (C) Gene classes overrepresented in the set of 61 serum- and temperature-induced genes described by Kadosh and Johnson (2005) compared with their representation in the C. albicans genome as a whole. Data were generated using the GO Term Finder tool available at the Candida Genome Database website (default settings, p ≤ 0.1) and characterized by biological process gene ontology.
To determine the extent to which genes induced during UME6-driven hyphal formation overlapped with those induced in response to a strong filament-inducing condition, we compared genes in the UME6-induced gene set (time-course experiment, 10-h time point, –Dox) with a set of 61 genes previously shown to be induced in response to serum at 37°C (Kadosh and Johnson, 2005). We found that 24 of the serum- and temperature-induced genes were also induced at least twofold during hyphal growth in response to UME6 expression over a time course (Figure 5A). A comparison of gene classes overrepresented relative to the genome as a whole in the serum- and temperature-induced set versus the set of genes induced during UME6-driven hyphal growth in the time-course experiment indicated >75% correlation (Figure 5, B and C) (note that the previously reported [ Kadosh and Johnson, 2005] gene set was reclassified based on the most recent GO term assignments at CGD). Similarly, a strong correlation in overrepresented gene classes was also observed when this UME6-driven hyphal gene set was compared independently to the set of serum- and temperature-induced genes described by Nantel et al. (2002). These findings indicate that hyphal growth driven by UME6 bears many similarities, in terms of both genes expressed and gene classes overrepresented, to C. albicans hyphal growth under a strong natural filament-inducing condition.
By comparing the 24 genes induced by both UME6 expression over a time course and growth in serum at 37°C with those that are reproducibly induced in hyphae generated by high-level UME6 expression in a steady-state culture, we defined a minimal core set of 15 hyphal-associated genes (Table 2). Certain genes in this set are either known or likely to play important roles in the mechanics of hyphal formation: CDC10 and CDC12 encode septins (DiDomenico et al., 1994), RDI1 encodes a putative Rho GTPase inhibitor involved in organelle and cytoskeleton organization (Court and Sudbery, 2007), and KIP4 encodes a putative kinesin (Chua et al., 2007); although not identified as a serum- and temperature-induced gene in our previous DNA microarray analysis, HGC1 has also been shown to be induced under these conditions and is a UME6 target as well (Zheng et al., 2004; Carlisle et al., 2009). At least four genes in the minimal hyphal set encode cell wall/cell surface proteins: HWP1, PHR1, ALS3, and HYR1 (Bailey et al., 1996; Hoyer et al., 1998; Hoyer, 2001; Fonzi, 1999; Staab et al., 1999). Two genes in the set, HWP1 and ALS3, encode adhesins and play an important role in the ability of C. albicans to adhere to host cells and form biofilms (Hoyer et al., 1998; Hoyer, 2001; Staab et al., 1999; Nobile et al., 2006a, b). HYR1 has also been shown to be involved in protecting C. albicans from killing by macrophages and neutrophils (Luo et al., 2010). RBT4, which encodes a putative secreted protein related to plant pathogenesis-related (PR) proteins, is a member of the minimal hyphal gene set as well (Braun et al., 2000). Of note, the minimal hyphal gene set does not contain any transcriptional regulators aside from UME6. This observation is consistent with a previous finding that UME6 functions as a downstream target of multiple filamentous growth signaling and regulatory pathways (Zeidler et al., 2009).
TABLE 2:
Minimal set of genes associated with C. albicans hyphal growth.
| Gene namea | Reference number | Descriptiona | Fold induction in response to serum at 37°Cb | Fold induction in response to UME6 expressionc |
|---|---|---|---|---|
| ALS3 | orf19.1816 | Agglutinin-like protein, adhesin | 97.8 | 397.5 |
| ECE1 | orf19.3374 | Protein associated with extent of cell elongation | 555.3 | 281.6 |
| HWP1 | orf19.1321 | Hyphal wall protein, adhesin, host transglutaminase, substrate mimic | 72.1 | 116.1 |
| HYR1 | orf19.4975 | Hyphal-induced, GPI-anchored cell wall protein | 187.2 | 68.9 |
| IHD1 | orf19.5760 | Putative GPI-anchored protein | 34.8 | 22.5 |
| CDC12 | orf19.3013 | Septin | 5.0 | 11.0 |
| KIP4 | orf19.5265 | Kinesin heavy-chain homologue | 6.6 | 9.9 |
| RBT4 | orf19.6202 | Pathogenesis-related (PR) protein, repressed by Tup1 | 17.2 | 8.1 |
| UME6 | orf19.1822 | Transcriptional regulator of filamentous growth | 5.0 | 7.6 |
| CBP1 | orf19.7323 | Corticosteroid-binding protein | 2.7 | 6.4 |
| FAV2 | orf19.1120 | Putative adhesin-like protein | 13.5 | 4.9 |
| CDC10 | orf19.548 | Septin | 5.8 | 3.8 |
| PHR1 | orf19.3829 | pH-regulated, putative cell surface glycosidase | 25.1 | 3.4 |
| orf19.6705 | orf19.6705 | Putative guanyl nucleotide exchange factor with Sec7 domain | 19.2 | 3.2 |
| RDI1 | orf19.5968 | Putative Rho GDP dissociation inhibitor | 2.8 | 2.8 |
GPI, glycosylphosphatidylinositol.
aGene names and descriptions based on Candida Genome Database annotation (www.candidagenome.org) and BLAST (blast.ncbi.nlm.nih.gov/Blast.cgi) analysis.
bIndicates mean fold induction (n = 2) in response to growth in YEPD + 10% serum at 37°C at the 1-h time point as described previously (Kadosh and Johnson, 2005).
cIndicates mean fold induction (n = 2) in the tetO-UME6 strain at the 10-h time point in the absence of Dox as described in Supplemental Table S1.
Transcriptional profile of the C. albicans reverse hyphal-pseudohyphal-yeast morphological transition
Our ability to modulate expression levels of UME6 in the absence of filament-inducing conditions allows us to study gene expression patterns specifically associated with the reverse hyphal-pseudohyphal-yeast transition. We previously showed that when the tetO-UME6 strain is growing in hyphal form (in the absence of Dox), reducing UME6 expression (by the addition of Dox) results in a transition from hyphae to pseudohyphae to yeast over a time course (Carlisle et al., 2009). A similar experiment was performed and cells were harvested at hourly time points for RNA extraction. By the 3-h time point the large majority of C. albicans cells are in the pseudohyphal form, and by 10 h the cells have undergone a nearly complete transition to yeast form (Figure 1; note that due to the larger culture volume size, we observed that the reverse transition is complete by 10 h rather than 9 h as reported in our previous study; Carlisle et al., 2009). Cells from a control tetO-UME6 culture, which did not receive Dox treatment and remained in the hyphal form over the time course, were harvested as well. RNA from each sample was used to generate cDNA for C. albicans whole-genome DNA microarray analysis.
We identified 203 genes that showed at least twofold reduction in expression as the tetO-UME6 strain transitions from hyphae to yeast. We also observed that a significant fraction of genes showing the most highly reduced expression (>4-fold) in yeast also showed >2-fold reduced expression in pseudohyphae (Table 3). About two-thirds of genes showing the greatest reduction in expression in pseudohyphae (>3-fold) were also found to have >2-fold reduced expression in yeast cells. In general, a higher proportion of genes reduced in pseudohyphae showed >4-fold and >10-fold reduction in yeast than the proportion of genes down-regulated in yeast that were also reduced to an equivalent level in pseudohyphae. These results suggest that there is a significant degree of overlap between genes whose expression is reduced in pseudohyphae and yeast during the reverse hyphal-pseudohyphal-yeast transition.
TABLE 3:
Relative morphological distribution of genes showing reduced expression upon depletion of UME6 during the hyphal-pseudohyphal-yeast transition.
| Fold change relative to hyphal expression | |||
|---|---|---|---|
| >3-fold | >4-fold | >10-fold | |
| Number of genes down in yeast cellsa | 87 | 37 | 7 |
| Percentage of genes also down >2-fold in pseudohyphal cellsb | 23 | 41 | 86 |
| Percentage of genes also down >4-fold in pseudohyphal cellsc | 11 | 22 | 57 |
| Percentage of genes also down >10-fold in pseudohyphal cellsc | 1 | 3 | 14 |
| >2-fold | >3-fold | >4-fold | |
| Number of genes down in pseudohyphal cellsa | 66 | 32 | 17 |
| Percentage of genes also down >2-fold in yeast cellsb | 53 | 69 | 76 |
| Percentage of genes also down >4-fold in yeast cellsc | 27 | 47 | 59 |
| Percentage of genes also down >10-fold in yeast cellsc | 11 | 22 | 29 |
Data exclude genes with expression values affected in a control strain by Dox alone (as determined by the tetR-HAP4 control experiment) and genes reduced at least twofold in two independent experiments (n = 2) in the tetO-UME6 strain time-course control culture (for genes down in pseudohyphal cells, control sample is 3-h time point, −Dox; for genes down in yeast cells, control sample is 10-h time point, −Dox). The tetO-UME6 strain was grown overnight in YEPD −Dox at 30°C to an OD600 of ∼0.01. Dox, 20 μg/ml, was added to the culture, and cells were grown in YEPD at 30°C over a 10-h time course. Yeast cells, 10-h time point; pseudohyphal cells, 3-h time point; and hyphal cells, 0-h time point, just before the addition of Dox.
aFold changes are based on mean gene expression values from two independent experiments (n = 2). All genes showed at least a twofold reduction in expression in both experiments.
bPercentage of genes showing reduced expression of at least twofold in two independent experiments (n = 2) in the indicated cell morphology.
cPercentage of genes showing the indicated mean fold reduction in the indicated morphology based on two independent experiments (n = 2). All genes were reduced at least twofold in both experiments.
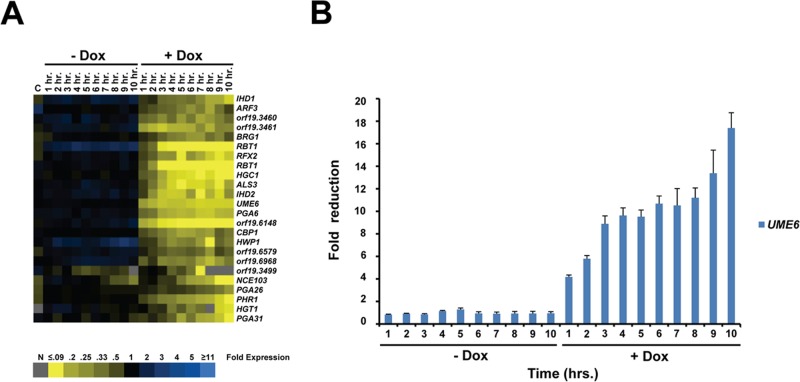
A hierarchical cluster analysis of data obtained from the reverse hyphal-pseudohyphal-yeast transition experiment indicated that many genes generally showed increasingly reduced expression over the time course (Figure 6A). Certain genes (e.g., RBT1) reached their lowest expression level early, by the 3- to 4-h time point, soon after the hyphal–pseudohyphal transition, and maintained this expression level throughout the time course, whereas other genes (e.g., PHR1) did not show maximal reduction in expression until the 10-h time point, when cells had transitioned to yeast.
FIGURE 6:
Transcriptional profile of genes down-regulated during the reverse hyphal-pseudohyphal-yeast transition in response to depletion of UME6 over a time course. (A) Cluster diagram of down-regulated genes. Experimental data represent mean expression values based on two independent DNA microarray experiments (n = 2 biological replicates). Only a subset of genes showing fourfold or greater change in expression in at least one data point with >80% of data present are shown. Each data point represents fold change in gene expression relative to the 0-h time point. Blue, increased expression; yellow, reduced expression; gray, no data. C = tetR-HAP4 control strain lacking a tet operator, −Dox vs. 20 μg/ml Dox. (B) Fold reduction in UME6 gene expression over the time course, as determined by real-time quantitative RT-PCR analysis. Mean fold reduction for each time point relative to the respective 0-h time point for +Dox and –Dox cultures is shown (n = 3). Bars, SE.
To specifically monitor UME6 expression levels during the reverse hyphal-pseudohyphal-yeast transition, we performed a quantitative RT-PCR analysis (Figure 6B). Of interest, we observed that the largest reduction in UME6 transcript (about ninefold) occurred between the 0- and 3-h time points as cells transition from hyphae to pseudohyphae. UME6 levels further decreased about twofold during the pseudohyphal–yeast transition between the 3- and 10-h time points. These results suggest that an initial large reduction in the expression of UME6, accompanied by an initial reduction in the expression of several filament-specific genes, is sufficient to trigger the reverse hyphal–pseudohyphal transition. In contrast to the forward yeast-pseudohyphal-hyphal transition, the majority of change in UME6 gene expression is observed in the transition between hyphae and pseudohyphae, and the early decrease in UME6 levels appears to set in motion a series of events that, over time, lead to the final transition to yeast form. In addition to UME6, quantitative RT-PCR analysis of four additional genes verified the overall validity of the reverse hyphal-pseudohyphal-yeast transition microarray data (Supplemental Figure S5).
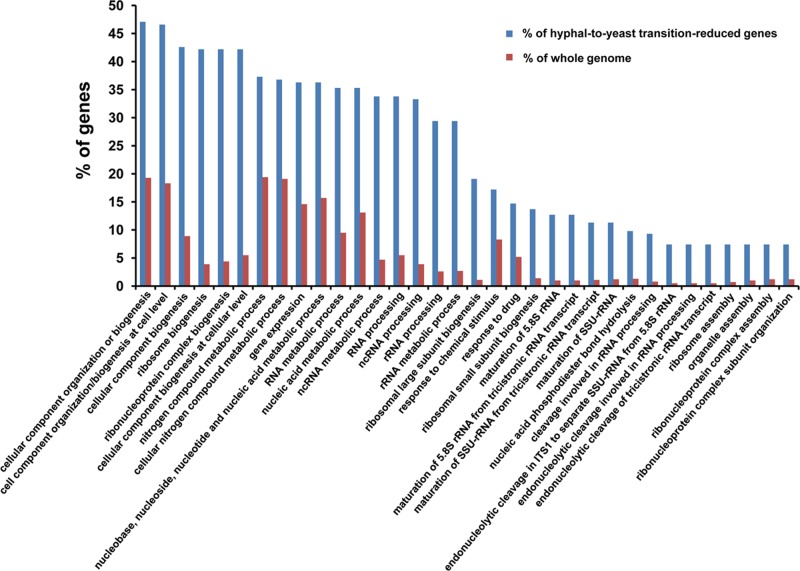
We next sought to identify specific gene classes that showed down-regulation during the hyphal-pseudohyphal-yeast transition. A CGD GO Slim Mapper analysis of genes showing a greater than twofold reduction in expression upon UME6 depletion indicated, not surprisingly, that several of the same gene classes induced during the forward yeast-pseudohyphal-hyphal transition (e.g., filamentous growth, cell wall organization, response to stress, cell cycle, and interaction with host) were also down-regulated in both pseudohyphae and yeast during the reverse transition (compare Supplemental Figures S3 and S4 with Supplemental Figure S6). However, several gene classes were highly represented in the set of genes down-regulated in yeast and either poorly represented or not represented at all in the set of genes down-regulated in pseudohyphae. These gene classes include ribosome biogenesis, RNA metabolic process, response to chemical stimulus, and response to drug (Supplemental Figure S6). A CGD GO Term Finder analysis indicated that most gene classes overrepresented in the +Dox 10-h time-point set compared with their representation in the genome as a whole were involved in protein synthesis (Figure 7 and Supplemental Figure S7). Because these genes appear to be specifically down-regulated during the pseudohyphal–yeast transition, our results suggest that this last phase of the C. albicans reverse morphological transition is associated with significantly reduced protein production.
FIGURE 7:
Gene classes involved in protein synthesis are overrepresented, compared with their representation in the genome as a whole, in the set of genes showing reduced expression as C. albicans undergoes the reverse hyphal-pseudohyphal-yeast transition in response to depletion of UME6. Representation of gene classes in the set of genes showing a mean reduction greater than twofold at the 10-h time point in the presence of Dox relative to the 0-h time point (n = 2) in the tetO-UME6 strain, as defined in Table 3 and listed in Supplemental Data Set S5, was compared with their representation in the C. albicans genome as a whole. Data were generated using the GO Term Finder tool available at the Candida Genome Database website (default settings, p ≤ 0.1) and characterized by process gene ontology. Note that this histogram shows data for gene classes that represent ≥7% of the gene set. Data for genes that represent <7% of the gene set are displayed in Supplemental Figure S7.
Interestingly, we also observed that the reverse transition was associated with strong induction of a significant number of genes (66 genes were induced greater than fourfold). In this set, genes encoding transporters, as well as those involved in carbohydrate and amino acid (particularly methionine) metabolism, were highly represented (Supplemental Data Set S13). Consistent with previous findings, NRG1, which encodes a strong transcriptional repressor of filament-specific genes (Braun et al., 2001; Murad et al., 2001), also showed increased expression during the reverse transition. Overall our results suggest that in addition to similarities, there are also several fundamental differences in the major genes and gene classes whose expression is significantly affected during the forward versus reverse C. albicans morphological transition.
DISCUSSION
Genes associated with determination of the C. albicans pseudohyphal morphology are a subset of those associated with hyphal formation
For many years it was believed that pseudohyphal and hyphal morphologies of C. albicans and other fungal species were determined by genetically distinct mechanisms. In addition to observed differences in phenotype and cell cycle, this assumption was also based on the finding that, with one important exception (Murad et al., 2001), most C. albicans mutants locked in the pseudohyphal form are not able to transition to hyphae in the presence of strong filament-inducing conditions (Braun and Johnson, 1997; Bensen et al., 2002; Sudbery et al., 2004; Wightman et al., 2004). However, the paradigm of genetically distinct morphology determination was challenged by our more recent observation that expression levels of UME6 are sufficient to specify C. albicans pseudohyphal and hyphal morphologies in a dosage-dependent manner (Carlisle et al., 2009). Because UME6 levels determine C. albicans morphology in the absence of filament-inducing conditions, our tetO-UME6 strain provides a powerful strategy to identify gene expression patterns specifically associated with pseudohyphae and hyphae and not associated with external environmental cues that promote filamentation. We previously suggested a model in which genes important for C. albicans pseudohyphal formation represent a subset of those involved in the generation of hyphae (Carlisle et al., 2009). This model was based on our initial observation by Northern analysis that there is an increase in both the number of filament-specific genes expressed and their level of expression as increased UME6 levels drive the yeast-pseudohyphal-hyphal transition. In large part, this model appears to be confirmed on a genome-wide scale based on results of our DNA microarray analysis, as well as an independent DNA microarray analysis of nearly pure populations of C. albicans morphologies generated using clearly defined environmental conditions (P. Sudbery and J. Berman, personal communication). In addition, our DNA microarray analysis allows us to make several new and important observations regarding the relationship between pseudohyphal and hyphal gene sets. First, very few, if any, genes appear to be expressed exclusively in pseudohyphae; Sudbery and Berman independently made a similar observation (personal communication). Second, based on data obtained in both of our yeast-pseudohyphal-hyphal transition experiments (UME6 dosage and expression time course), very few, if any, genes appear to be expressed exclusively at the yeast–pseudohyphal and pseudohyphal–hyphal transition points. Both of these observations strongly suggest that, on a genome-wide scale, genes associated with C. albicans pseudohyphal formation represent a subset of those associated with hyphal growth and that the yeast-pseudohyphal-hyphal transition involves an increase in the number of filament-specific genes expressed, as well as in their expression level. These conclusions are supported by the observation that, unlike pseudohyphae, hyphae possess specialized structures, such as true septa and the Spitzenkörper, that might require additional genes to be expressed (Sudbery et al., 2004; Crampin et al., 2005) and are also consistent with a recent hypothesis that morphology of Candida species evolved in a stepwise manner from yeast to pseudohyphae to hyphae (Bastidas and Heitman, 2009; Thompson et al., 2011).
Duration of filament-specific gene expression plays an important role in C. albicans morphology determination
Whereas previous studies indicated that UME6 expression level clearly plays an important role in determination of C. albicans morphology (Carlisle et al., 2009; Zeidler et al., 2009), they did not address the role of duration of UME6 expression. Of interest, our forward-transition time-course experiment shows that the UME6 expression level peaks at a very early time point, well before completion of the morphological transition, and remains high through the end of the time course. A significant number of UME6 target genes show a similar transcriptional profile, whereas others are initially expressed at later time points and gradually increase through the time course. These findings strongly suggest that extended duration of UME6 expression, as well as expression of filament-specific target genes, plays an important role in driving the yeast-pseudohyphal-hyphal transition. This conclusion is supported by our previous finding that the ume6Δ/Δ mutant not only is defective for hyphal extension but also shows a clear reduction in both the level and duration of filament-specific gene expression upon induction by serum at 37°C (Banerjee et al., 2008). Our conclusion is also supported by a recent study indicating that C. albicans hyphal development requires temporally linked initiation and maintenance phases (Lu et al., 2011). The initiation phase involves transient removal of the Nrg1 repressor from hyphal-specific promoters by activation of the cAMP-PKA pathway, whereas maintenance involves recruitment of the Hda1 histone deacetylase. We previously showed that Nrg1 is a repressor of Ume6 and that the two genes function together in a feedback loop that, under strong filament-inducing conditions, results in an amplification of filament-specific gene expression (Banerjee et al., 2008); a recent independent study also suggested that Ume6 functions as a positive feedback regulator of hyphal genes (Lu et al., 2012). Ume6 is therefore likely to play an important role in the maintenance phase of hyphal development by promoting a significant increase in both the level and duration of filament-specific gene expression.
Why is extended duration of filament-specific gene expression important for determination of C. albicans morphology? Although the answer to this question is not entirely clear, at least one example of the critical importance of long-lasting expression of a filament-specific gene has been reported. We showed that Ume6 drives hyphal growth via a mechanism involving the Hgc1 cyclin-related protein and that maintenance of high-level HGC1 expression at the later time points of a serum- and temperature-induction time course is associated with extended hyphal development (Carlisle and Kadosh, 2010). Hgc1 is known to form a complex with Cdc28 kinase that is involved in promoting hyphal growth by septin phosphorylation, Cdc42 activation, and inhibition of cell separation (Zheng et al., 2004, 2007; Sinha et al., 2007; Gonzalez-Novo et al., 2008; Wang et al., 2009). Maintaining sufficiently high levels of the Hgc1–Cdc28 complex for an extended time period therefore appears to be critical for proper hyphal development.
Identification of a minimal set of genes associated with C. albicans hyphal formation
Previous whole-genome transcriptional analyses identified sets of genes induced during C. albicans hyphal formation (Nantel et al., 2002; Kadosh and Johnson, 2005). However, a major limitation of these studies is that because cells were grown in the presence of strong filament-inducing conditions, serum at 37°C, it was unclear whether the induced genes were expressed specifically in association with hyphal formation or the environmental cue per se. Because the tetO-UME6 strain is capable of generating hyphae in the complete absence of filament-inducing conditions, a comparison of genes induced by UME6 with those induced in response to serum at 37°C has allowed us to identify a minimal set of 15 genes that are specifically associated with C. albicans hyphal growth. Genes in this set are involved in a wide variety of virulence-related processes, and several have previously been shown to be required for virulence (Braun et al., 2000; Zheng et al., 2004). This observation provides an additional example of the coregulation of C. albicans hyphal growth with pathogenesis and highlights the close relationship between these processes. Interestingly, given the complexity of hyphal development, the minimal set of hyphal-associated genes as a whole is rather small. Only about one-half of these genes (CDC10, CDC12, UME6, KIP4, PHR1, orf19.6705, RDI1) are known to affect filamentation, based on direct experimental evidence (Yesland and Fonzi, 2000; Warenda and Konopka, 2002; Uhl et al., 2003; Banerjee et al., 2008; Elson et al., 2009; Zeidler et al., 2009; Li et al., 2012); similarly, only about one-fourth of all serum- and temperature-induced genes play a role in filamentous growth (for all genes assigned the GO term “filamentous growth” this assignment is based on experimental evidence rather than solely the expression pattern; www.candidagenome.org). An even smaller number of genes in both the minimal hyphal set and the serum- and temperature-induced set appear to be directly involved in the physical process of polarized growth. These observations, in combination with our previous findings, suggest that prolonged high-level expression of genes with direct roles in the mechanics of hyphal growth sets in motion an extensive series of posttranslational events to drive hyphal development. This notion is supported by the example of HGC1 and the multiple roles that the Hgc1–Cdc28 complex plays in promoting hyphal extension (Zheng et al., 2004, 2007; Sinha et al., 2007; Gonzalez-Novo et al., 2008; Wang et al., 2009).
Interestingly, all genes in the minimal hyphal gene set are expressed at a lower level in pseudohyphae. We also showed that genes in the larger gene set associated with pseudohyphal growth are involved in many of the same virulence-related processes as genes induced in hyphae. Although difficult to prove, these observations might suggest that the C. albicans pseudohyphal morphology is associated with low-level virulence in the host.
An important limitation of the approach we have taken to define the minimal set of hyphal genes, as well as all other gene sets, is that because UME6 encodes a transcriptional regulator, there is always the possibility that additional genes, not specifically associated with pseudohyphal/hyphal morphology, may be induced in response to UME6. Although we cannot exclude this possibility, for several reasons we believe that it is significantly minimized when compared with other approaches that have been used to examine gene expression patterns associated with C. albicans morphologies. First, unlike most other transcriptional regulators of C. albicans filamentous growth, UME6 is only expressed in the presence of filament-inducing conditions and serves as a major downstream target of multiple filamentous growth signaling pathways and regulators (Banerjee et al., 2008; Zeidler et al., 2009); UME6 targets therefore provide a more accurate “readout” of genes specifically associated with filamentation. Second, the strength of UME6 induction correlates very well with the strength of specific filament-inducing conditions (Banerjee et al., 2008). Finally, many of the same gene classes involved in pathogenicity, filamentous growth, adhesion, and other virulence-related properties are induced in response to both strong filament-inducing conditions and UME6 expression.
Similarities and differences between the gene expression profiles of forward and reverse morphological transitions in C. albicans
Our ability to drive both the C. albicans forward, yeast-pseudohyphal-hyphal and reverse, hyphal-pseudohyphal-yeast transition upon induction or depletion, respectively, of UME6 in the same strain places us in a unique position to directly compare gene expression patterns associated with each transition. Similar to the forward transition, initial changes in gene expression occur at very early time points of the reverse transition. While the large majority of change in UME6 transcript level occurs early (by the 3-h time point) in both the forward and reverse transition time-course experiments, certain genes plateau at their peak level of induction or down-regulation early, whereas others gradually reach their peak levels toward the end of each time course. This observation suggests that both induction and depletion of UME6 sets in motion a complex series of temporal events that are involved in morphological transition; in addition to transcriptional changes, the cells are also most likely undergoing significant posttranslational changes as well.
Interestingly, while the forward-transition time course indicates that genes induced in pseudohyphae are a subset of those induced in hyphae, the reverse-transition experiment suggests that in addition to many similarities, there are also several differences between genes that are down-regulated in pseudohyphae and in yeast. Several genes and gene classes involved in morphogenesis and virulence that have reduced expression in both pseudohyphae and yeast during the reverse transition were also induced during the forward transition. For example, genes encoding Rbt1 and Phr1—cell wall proteins that are strongly induced during filamentation—are among the most highly repressed genes during reverse morphogenesis. Other filament-specific genes, such as Hgc1, show a similar pattern. These observations suggest that, to some extent, there is an inverse relationship between genes expressed during the forward and reverse C. albicans morphological transitions. Many of the gene classes involved in filamentation and virulence appear to be initially down-regulated during the reverse hyphal–pseudohyphal transition and remain down-regulated through the pseudohyphal–yeast transition. Interestingly, however, additional major gene classes, particularly those involved in protein synthesis, also appear to show significantly reduced expression specifically during the reverse pseudohyphal–yeast transition. This observation not only highlights an important difference between forward and reverse C. albicans morphological transitions, but it also suggests that the final transition from filaments to yeast involves a significant “down-scaling” in protein production that is required to generate and maintain filamentous morphologies.
Another important difference between the C. albicans forward and reverse transitions is related to the number of genes that show reduced expression during filament induction versus increased expression during filament reduction. Similar to previous transcriptional profiling studies that examined the transition from yeast to hyphae in response to serum at 37°C (Nantel et al., 2002; Kadosh and Johnson, 2005), fewer genes showed strongly reduced, rather than increased, expression during both the forward-transition steady-state culture and time-course experiments. Genes consistently reduced in pseudohyphae in both experiments included MNN22, a putative Golgi α-1,2 mannosyltransferase, and RBE1, a cell wall protein previously shown to be down-regulated by several filamentous growth transcription factors (Supplemental Data Sets S14 and S15). Both of these genes, as well as CHT3, encoding a major secreted chitinase involved in cell separation, were also observed to be down-regulated in hyphae; in the steady-state culture experiment, genes involved in glycolysis also showed reduced expression in hyphae (Supplemental Data Sets S11 and S12). In contrast, the reverse transition was associated with strong induction of a significant number of genes. Several are involved in transport, as well as in carbohydrate and amino acid metabolism. We hypothesize that these genes may play roles in directing major changes in cell wall organization and cellular metabolism during the filament-to-yeast transition.
Our comparison of both forward and reverse morphological transitions in C. albicans also provides new information to address the question of whether the yeast form may be considered a “default” state. Multiple studies, including this one, show that hyphal formation involves strong induction of a significant number of new genes that play roles in filament- and virulence-related processes (Nantel et al., 2002; Kadosh and Johnson, 2005; Kumamoto and Vinces, 2005), and here we show that several of these genes are also down-regulated during the reverse hyphal-pseudohyphal-yeast transition. These observations alone strongly support the notion that C. albicans yeast cells are a “default” morphology. However, our finding that a significant number of new genes involved in different major processes are both down-regulated and induced as cells transition from hyphae to yeast suggests a more complex scenario in which specific patterns of gene expression may be required to maintain the yeast state. Although results of our analysis do not completely resolve this question, they do highlight several important similarities and differences between forward and reverse morphological transitions in C. albicans. While a wide variety of fungal pathogens are known to undergo morphological transitions that are associated with virulence, very little is known about how and why these transitions are reversed. Our study, in addition to future work, should help to address this important question and shed more light on the relationship between forward and reverse morphological transitions in pathogenic fungi.
MATERIALS AND METHODS
Strains, media, and growth conditions
The tetO-UME6 strain (MBY38) and respective tetR-HAP4 control strain (PCY87) were described previously (Carlisle et al., 2009; Carlisle and Kadosh, 2010). In all experiments cells were grown under non–filament-inducing conditions: YEPD medium at 30°C. For the experiment in which C. albicans morphologies were examined in static culture in response to UME6 dosage, a saturated overnight culture of the tetO-UME6 (MBY38) strain was diluted into 50 ml of YEPD medium plus various concentrations of Dox (20, 0.4, 0.35, 0.3, 0.25, 0.2, 0.15, 0.1, 0.05, 0.02, and 0 μg/ml) and grown overnight at 30°C to an OD600 of 1.0. Cells were then harvested for RNA extraction. For the experiment in which C. albicans undergoes the yeast-pseudohyphal-hyphal transition in response to increased UME6 levels over a time course, the tetO-UME6 (MBY38) strain was grown overnight at 30°C in YEPD medium plus 1.0 μg/ml Dox to an OD600 of ∼0.5. Then 50-ml aliquots of cells were washed 1× in prewarmed YEPD medium at 30°C and used to inoculate 1.5 l of YEPD medium in the presence or absence of 1.0 μg/ml Dox. Cultures were grown at 30°C, and cells were harvested for RNA extraction at each hour for 10 h. Cells for the 0-h time point were collected from the tetO-UME6 overnight culture just before washing.
To carry out the reverse hyphal-pseudohyphal-yeast transition experiment, the tetO-UME6 strain was grown overnight in 1.5 l of YEPD medium at 30°C in the absence of Dox. When the culture reached an OD600 of ∼0.01, Dox was added to a concentration of 20 μg/ml and cells were harvested for RNA extraction at each hour for 10 h. Cells for the 0-h time point were collected immediately before the addition of Dox. Over the time course cells were also harvested from an additional control culture in which the tetO-UME6 strain was grown in the same manner but in the absence of Dox.
To determine the effect of Dox alone on C. albicans global gene expression, the tetR-HAP4 control strain was grown to saturation and diluted into 50 ml of YEPD medium in the presence or absence of 20 μg/ml Dox. These cultures were grown overnight at 30°C, and cells were harvested for RNA preparation at an OD600 of 1.0.
DNA microarray experiments and analysis
RNA preparation.
RNA was isolated from C. albicans cells using the hot acid phenol method (Ausubel et al., 1992). Total RNA quality was assessed using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA) according to manufacturer’s recommendations.
Labeling of RNA transcripts, cDNA synthesis, and hybridization.
RNA transcript labeling, cDNA synthesis, and hybridization to microarrays were performed at the Washington University Genome Center using the protocols described in this section. First-strand cDNA was generated by oligo-dT–primed reverse transcription (Superscript II; Invitrogen, Carlsbad, CA) using the 3DNA Array 350 kit (Genisphere, Hatfield, PA). Modified oligo-dT primers were used in which a fluorophore/dendrimer-specific oligonucleotide sequence is attached to the 5′ end of the dT primer. For cDNA synthesis, 1 μl of fluorophore-specific oligo-dT primer was added to 8 μg of total RNA, and the solution was incubated at 80˚C for 10 min and then cooled on ice for 2 min. To each sample was added RNase inhibitor (Superase-In; Ambion, Austin, TX) (1 μl), 5× first strand buffer (4 μl), dNTP mix (10 mM each dATP, dCTP, dGTP, and dTTP; 1 μl), 0.1 M dithiothreitol (2 μl), and Superscript II RNase H-Reverse Transcriptase (1 μl). Reverse transcription was carried out at 42°C for 2 h. The reaction was terminated by adding 0.5 M NaOH/50 mM EDTA (3.5 μl) and incubating at 65°C for 15 min, then neutralized with 1 M Tris-HCl, pH 7.5 (5 μl). For RNA expression-level comparison, samples were paired and concentrated using Microcon YM30 microconcentrators (Millipore, Billerica, MA) according to the manufacturer’s protocol. All individual experimental samples were hybridized along with a reference sample comprised of a mixture containing equal aliquots of all samples in a given experiment (for the experiment to determine the effect of Dox alone on C. albicans gene expression, the reference consisted of equal aliquots of samples from the UME6 dosage experiment). Each sample pair (∼20 μl) was suspended in formamide-based hybridization buffer (26 μl, vial 7; Genisphere), Array 50dT blocker (2 μl; Genisphere), and RNase/DNase-free water (4 μl). Two hybridizations were carried out in a sequential manner. The primary hybridization was performed by adding 48 μl of sample to the microarray under a supported glass coverslip (Erie Scientific, Portsmouth, NH) at 43°C for 16–20 h at high humidity. Before the secondary hybridization, slides were gently submerged into 2× SSC (saline sodium citrate), 0.2% SDS (at 43°C) for 11 min, transferred to 2× SSC (at room temperature) for 11 min, transferred to 0.2× SSC (at room temperature) for 11 min, and then spun dry by centrifugation. Secondary hybridization was carried out using the complimentary capture reagents provided in the 3DNA Array 350 kit (Genisphere). For each reaction, the following were added: 3DNA capture reagent with Cy3 (2.5 μl), 3DNA capture reagent with Cy5 (2.5 μl), SDS-based hybridization buffer (26 μl, vial 6; Genisphere), and RNase/DNnase-free water (21 μl). The secondary hybridization solution was incubated in the dark at 80°C for 10 min and then at 50°C for 15 min. Hybridization was performed by adding 48 μl of secondary hybridization solution to the slide under a supported glass coverslip and carried out at 65°C for 3 h at high humidity in the dark. At hybridization termination, arrays were gently submerged into 2× SSC, 0.2% SDS (at 65°C) for 11 min, transferred to 2× SSC (at room temperature) for 11 min, transferred to 0.2× SSC (at room temperature) for 11 min, and then spun dry by centrifugation. To prevent fluorophore degradation, the arrays were treated with Dyesaver (Genisphere). Spotted C. albicans 70-mer oligonucleotide DNA microarrays used for hybridizations were described previously (Brown et al., 2006).
Data analysis
Slides were scanned using an Axon 4000B scanner (Molecular Devices, Sunnyvale, CA) to detect Cy3 and Cy5 fluorescence. Laser power was kept constant for Cy3/Cy5 scans, and PMT (photomultiplier tube) values were varied for each experiment based on optimal signal intensity with lowest possible background fluorescence. Gridding and analysis of images was performed using ScanArray, version 3 (PerkinElmer, Waltham, MA). Microarray data were normalized using the Lowess method (Yang et al. 2001) and filtered using Partek software to only include spots showing a median signal intensity of >200 and signal-to-noise ratio of >2. The UME6 dosage microarray experiment was performed in biological duplicate. For this experiment, the signal ratios (ratio of the median signal intensity of each spot) for each Dox concentration sample versus reference were divided by the median signal ratio of the 20 μg/ml Dox sample versus reference. For each biological replicate of the UME6 dosage experiment, the median signal ratio of the 20 μg/ml Dox sample versus reference was determined based on three technical replicates (one of which was performed using reverse fluors). The forward yeast-pseudohyphal-hyphal transition time-course experiment was performed in biological duplicate. In this experiment, signal ratios for each time-point sample versus reference were divided by the median signal ratio for the zero time point versus reference. For each biological replicate of the forward transition experiment, the median signal ratio for the zero time point versus reference was determined based on three technical replicates, one of which was performed using reverse fluors. The reverse hyphal-pseudohyphal-yeast transition experiment was performed in biological duplicate, and data transformations were carried out in a nearly identical manner to those of the forward-transition experiment except that separate median 0-h time point versus reference samples were used for +Dox and –Dox time points. For the experiment to determine the effect of Dox alone on C. albicans global gene expression, signal ratios for the –Dox sample versus reference were divided by signal ratios for the 20 μg/ml Dox sample versus reference. This experiment was performed using five independent biological replicates. Three of the biological replicates were labeled with normal fluors, one of which was repeated as a technical replicate using reverse fluors. The remaining two biological replicates were performed using reverse fluors. Genes affected by Dox were defined as those showing a greater than twofold increase or decrease in at least three of the five biological replicates. Cluster analysis was performed using Cluster, version 3.0 (Eisen et al., 1998), and data were visualized using Treeview, version 1.60 (http://rana.lbl.gov/EisenSoftware.htm). Gene annotation for the spotted C. albicans oligonucleotide microarrays was described previously and was based on assembly 19 (Braun et al., 2005; Brown et al., 2006). Gene classification was based on C. albicans GO terms and performed using the GO Term Mapper and GO Term Finder tools available at the Candida Genome Database (www.candidagenome.org).
Complete transformed data for all experiments is provided in the supplemental materials (Supplemental Data Sets S7–S10). Primary RNA expression microarray data associated with this study have also been deposited in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE39677.
Real-time quantitative RT-PCR analysis
Total RNA isolated for both forward- and reverse-transition DNA microarray time-course experiments (two independent biological replicates each), along with RNA from a third independent biological replicate of each time-course condition, was used for real-time quantitative RT-PCR analysis. We treated 2 μg of total RNA with DNase I (Invitrogen) and prepared cDNA (High Capacity cDNA Reverse Transcription Kit; Applied Biosystems, Foster City, CA) according to the manufacturer’s directions. Real-time PCR was performed with four technical replicates for each sample, using a Chromo4 Four-Color Real-Time PCR Detection System (Bio-Rad, Hercules, CA). PCRs were carried out in 25-μl volumes containing 5 μl of 1:50 diluted cDNA, 12.5 μl GoTaq qPCR Master Mix (Promega, Madison, WI), 1.6 μl each of forward and reverse primers, and 4.3 μl of distilled H2O. Primer sequences are listed in Supplemental Table S2. The following parameters were used for each quantitative PCR: step 1, 95°C for 2 min; step 2, 95°C for 30 s; step 3, annealing temperature (determined for each primer pair) for 1 min; step 4, read plate; step 5, repeat steps 2–4 for 39 times, step 6, 72°C for 5 min; and step 7, melting curve 50–95°C every 0.4°C, hold 1 s, and read plate. Standard curves were generated using seven serial dilutions of a pool of all cDNA samples prepared from the first forward yeast-pseudohyphal-hyphal transition time-course experiment. Raw expression levels for each gene were determined using the Pfaffl method (Pfaffl, 2001) and normalized to levels of an ACT1 internal control gene. Normalized mean expression values for each time point were used to calculate fold induction or fold reduction relative to the normalized mean expression value for the respective 0-h time point.
Supplementary Material
Acknowledgments
We thank Mohua Banerjee and Delma Childers for technical assistance during the course of the experiments, John Bennett (National Institutes of Health/National Institute of Allergy and Infectious Diseases) for the use of facilities, Kathleen Meyer (National Institutes of Health/Center for Information Technology) for bioinformatics assistance, and Mike Heinz and Seth Crosby (Washington University Genome Technology Access Center) for performing DNA microarray hybridizations and assisting with data analysis. We also thank Martha Arnaud (Candida Genome Database, www.candidagenome.org/) for assistance with gene classifications and Hervé Tettelin (University of Maryland, Baltimore, MD) for assistance with generating r graphs. Candida albicans sequence data were provided by the Stanford Genome Technology Center (www-sequence.stanford.edu/group/candida). We are grateful to Pete Sudbery and Judy Berman for sharing unpublished results. We also thank Michael Lorenz, Brian Wickes, José López-Ribot, David Kolodrubetz, and Cristina Villar for comments on the manuscript, as well as for useful advice and suggestions. P.L.C. was supported by a Ruth L. Kirschstein National Research Service Award for Individual Predoctoral Fellows (5F31DE020214–03) and a COSTAR fellowship (National Institute of Dental and Craniofacial Research Grant T32DE14318–07). This work was also supported by a Research Scholar Grant from the American Cancer Society (MPC-117450) and by Grant 5RO1AI083344–03 from the National Institute of Allergy and Infectious Diseases to D.K.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institute of Dental and Craniofacial Research, or the National Institutes of Health.
Abbreviations used:
- CGD
Candida genome database
- Dox
doxycycline
- GO
gene ontology
- RT-PCR
reverse transcription PCR
- YEPD
yeast extract–peptone–dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-01-0065) on December 14, 2012.
REFERENCES
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Greene Publishing Associates and Wiley-Interscience; 1992. [Google Scholar]
- Bailey DA, Feldmann PJ, Bovey M, Gow NA, Brown AJ. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, Lopez-Ribot JL, Kadosh D. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell. 2008;19:1354–1365. doi: 10.1091/mbc.E07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas RJ, Heitman J. Trimorphic stepping stones pave the way to fungal virulence. Proc Natl Acad Sci USA. 2009;106:351–352. doi: 10.1073/pnas.0811994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck-Sague C, Jarvis WR. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. National Nosocomial Infections Surveillance System. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- Bensen ES, Filler SG, Berman J. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot Cell. 2002;1:787–798. doi: 10.1128/EC.1.5.787-798.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Head WS, Wang MX, Johnson AD. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics. 2000;156:31–44. doi: 10.1093/genetics/156.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Braun BR, Kadosh D, Johnson AD. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001;20:4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, et al. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005;1:36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ. Morphogenetic signaling pathways in Candida albicans. In: Calderone RA, editor. Candida and Candidiasis. Washington, DC: ASM Press; 2002. pp. 95–106. [Google Scholar]
- Brown AJ, Gow NA. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 1999;7:333–338. doi: 10.1016/s0966-842x(99)01556-5. [DOI] [PubMed] [Google Scholar]
- Brown V, Sexton JA, Johnston M. A glucose sensor in Candida albicans. Eukaryot Cell. 2006;5:1726–1737. doi: 10.1128/EC.00186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends Microbiol. 2001;9:327–335. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- Cannon RD, Chaffin WL. Oral colonization by Candida albicans. Crit Rev Oral Biol Med. 1999;10:359–383. doi: 10.1177/10454411990100030701. [DOI] [PubMed] [Google Scholar]
- Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, Lopez-Ribot JL, Kadosh D. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc Natl Acad Sci USA. 2009;106:599–604. doi: 10.1073/pnas.0804061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle PL, Kadosh D. Candida albicans Ume6, a filament-specific transcriptional regulator, directs hyphal growth via a pathway involving Hgc1 cyclin-related protein. Eukaryot Cell. 2010;9:1320–1328. doi: 10.1128/EC.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PR, et al. Effective killing of the human pathogen Candida albicans by a specific inhibitor of non-essential mitotic kinesin Kip1p. Mol Microbiol. 2007;65:347–362. doi: 10.1111/j.1365-2958.2007.05787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court H, Sudbery P. Regulation of Cdc42 GTPase activity in the formation of hyphae in Candida albicans. Mol Biol Cell. 2007;18:265–281. doi: 10.1091/mbc.E06-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampin H, Finley K, Gerami-Nejad M, Court H, Gale C, Berman J, Sudbery P. Candida albicans hyphae have a Spitzenkorper that is distinct from the polarisome found in yeast and pseudohyphae. J Cell Sci. 2005;118:2935–2947. doi: 10.1242/jcs.02414. [DOI] [PubMed] [Google Scholar]
- Dalle F, Wachtler B, L’Ollivier C, Holland G, Bannert N, Wilson D, Labruere C, Bonnin A, Hube B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol. 2010;12:248–271. doi: 10.1111/j.1462-5822.2009.01394.x. [DOI] [PubMed] [Google Scholar]
- DiDomenico BJ, Brown NH, Lupisella J, Greene JR, Yanko M, Koltin Y. Homologs of the yeast neck filament associated genes: isolation and sequence analysis of Candida albicans CDC3 and CDC10. Mol Gen Genet. 1994;242:689–698. doi: 10.1007/BF00283424. [DOI] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A, Wen K, Lamster IB. Candida albicans triggers interleukin-6 and interleukin-8 responses by oral fibroblasts in vitro. Oral Microbiol Immunol. 1999;14:364–370. doi: 10.1034/j.1399-302x.1999.140606.x. [DOI] [PubMed] [Google Scholar]
- Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson SL, Noble SM, Solis NV, Filler SG, Johnson AD. An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS Genet. 2009;5:e1000664. doi: 10.1371/journal.pgen.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JF. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology. 2000;146:1763–1774. doi: 10.1099/00221287-146-8-1763. [DOI] [PubMed] [Google Scholar]
- Filler SG, Kullberg BJ, Calderone RA. Candida and Candidiasis. Washington DC: ASM Press; 2002. Deep-seated candidal infections; pp. 341–348. [Google Scholar]
- Filler SG, Sheppard DC. Fungal invasion of normally non-phagocytic host cells. PLoS Pathog. 2006;2:e129. doi: 10.1371/journal.ppat.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans. J Bacteriol. 1999;181:7070–7079. doi: 10.1128/jb.181.22.7070-7079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, Cutler JE, Filler SG, Edwards JE., Jr Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol Microbiol. 2002;44:61–72. doi: 10.1046/j.1365-2958.2002.02873.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Novo A, Correa-Bordes J, Labrador L, Sanchez M, Vazquez de Aldana CR, Jimenez J. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol Biol Cell. 2008;19:1509–1518. doi: 10.1091/mbc.E07-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NA, Perera TH, Sherwood-Higham J, Gooday GW, Gregory DW, Marshall D. Investigation of touch-sensitive responses by hyphae of the human pathogenic fungus Candida albicans. Scanning Microsc. 1994;8:705–710. [PubMed] [Google Scholar]
- Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9:176–180. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Payne TL, Bell M, Myers AM, Scherer S. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr Genet. 1998;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- Jong AY, Stins MF, Huang SH, Chen SH, Kim KS. Traversal of Candida albicans across human blood-brain barrier in vitro. Infect Immun. 2001;69:4536–4544. doi: 10.1128/IAI.69.7.4536-4544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korting HC, Hube B, Oberbauer S, Januschke E, Hamm G, Albrecht A, Borelli C, Schaller M. Reduced expression of the hyphal-independent Candida albicans proteinase genes SAP1 and SAP3 in the efg1 mutant is associated with attenuated virulence during infection of oral epithelium. J Med Microbiol. 2003;52:623–632. doi: 10.1099/jmm.0.05125-0. [DOI] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Lane S, Birse C, Zhou S, Matson R, Liu H. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J Biol Chem. 2001;276:48988–48996. doi: 10.1074/jbc.M104484200. [DOI] [PubMed] [Google Scholar]
- Li L, Zhang C, Konopka JB. A Candida albicans temperature-sensitive cdc12–6 mutant identifies roles for septins in selection of sites of germ tube formation and hyphal morphogenesis. Eukaryot Cell. 2012;11:1210–1218. doi: 10.1128/EC.00216-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int J Med Microbiol. 2002;292:299–311. doi: 10.1078/1438-4221-00215. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lu Y, Su C, Liu H. A GATA transcription factor recruits Hda1 in response to reduced Tor signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog. 2012;8:e1002663. doi: 10.1371/journal.ppat.1002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Su C, Wang A, Liu H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 2011;9:e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Ibrahim AS, Spellberg B, Nobile CJ, Mitchell AP, Fu Y. Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J Infect Dis. 2010;201:1718–1728. doi: 10.1086/652407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Moran GP, Jacobsen ID, Heyken A, Domey J, Sullivan DJ, Kurzai O, Hube B. The Candida albicans-specific gene EED1 encodes a key regulator of hyphal extension. PLoS One. 2011;6:e18394. doi: 10.1371/journal.pone.0018394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AP. Dimorphism and virulence in Candida albicans. Curr Opin Microbiol. 1998;1:687–692. doi: 10.1016/s1369-5274(98)80116-1. [DOI] [PubMed] [Google Scholar]
- Murad AMA, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Mio T, Nagahashi S, Kokado M, Arisawa M, Aoki Y. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect Immun. 2000;68:6712–6719. doi: 10.1128/iai.68.12.6712-6719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006a;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Andes DR, Mitchell AP. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell. 2006b;5:1604–1610. doi: 10.1128/EC.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis. London: Baillière Tindall; 1988. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Lockhart SR, Pujol C, Swails-Wenger JA, Messer SA, Edmond MB, Jones RN, Wenzel RP, Soll DR. Hospital specificity, region specificity, and fluconazole resistance of Candida albicans bloodstream isolates. J Clin Microbiol. 1998;36:1518–1529. doi: 10.1128/jcm.36.6.1518-1529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE, Jr, Filler SG. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007;5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareck J, Nantel A, Belhumeur P. Conjugated linoleic acid inhibits hyphal growth in Candida albicans by modulating Ras1p cellular levels and downregulating TEC1 expression. Eukaryot Cell. 2011;10:565–577. doi: 10.1128/EC.00305-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha I, Wang YM, Philp R, Li CR, Yap WH, Wang Y. Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev Cell. 2007;13:421–432. doi: 10.1016/j.devcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Thompson DS, Carlisle PL, Kadosh D. Coevolution of morphology and virulence in Candida species. Eukaryot Cell. 2011;10:1173–1182. doi: 10.1128/EC.05085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl MA, Biery M, Craig N, Johnson AD. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 2003;22:2668–2678. doi: 10.1093/emboj/cdg256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Raniga PP, Lane S, Lu Y, Liu H. Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol Cell Biol. 2009;29:4406–4416. doi: 10.1128/MCB.01502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warenda AJ, Konopka JB. Septin function in Candida albicans morphogenesis. Mol Biol Cell. 2002;13:2732–2746. doi: 10.1091/mbc.E02-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R, Bates S, Amornrrattanapan P, Sudbery P. In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J Cell Biol. 2004;164:581–591. doi: 10.1083/jcb.200307176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Speed TP. Normalization for cDNA microarray data. In: Bittner ML, Chen Y, Dorsel AN, Dougherty ER, editors. Microarrays: Optical Technologies and Informatics. San Jose, CA: SPIE, Society for Optical Engineering; 2001. pp. 141–152. [Google Scholar]
- Yang YL. Virulence factors of Candida species. J Microbiol Immunol Infect. 2003;36:223–228. [PubMed] [Google Scholar]
- Yesland K, Fonzi WA. Allele-specific gene targeting in Candida albicans results from heterology between alleles. Microbiology. 2000;146:2097–2104. doi: 10.1099/00221287-146-9-2097. [DOI] [PubMed] [Google Scholar]
- Zeidler U, Lettner T, Lassnig C, Muller M, Lajko R, Hintner H, Breitenbach M, Bito A. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 2009;9:126–142. doi: 10.1111/j.1567-1364.2008.00459.x. [DOI] [PubMed] [Google Scholar]
- Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, Leng RP, Brown AJ, Hoyer LL. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology. 2004;150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wang Y, Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004;23:1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XD, Lee RT, Wang YM, Lin QS, Wang Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007;26:3760–3769. doi: 10.1038/sj.emboj.7601814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Filler SG. Interactions of Candida albicans with epithelial cells. Cell Microbiol. 2010;12:273–282. doi: 10.1111/j.1462-5822.2009.01412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink S, Nass T, Rosen P, Ernst JF. Migration of the fungal pathogen Candida albicans across endothelial monolayers. Infect Immun. 1996;64:5085–5091. doi: 10.1128/iai.64.12.5085-5091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.