Kv7.4-potassium channel expression plays a permissive role in skeletal muscle differentiation. The transcriptional repressor REST controls the changes in Kv7.4 levels during myogenesis by binding to regulatory regions in the Kv7.4 gene. This mechanism may be a target for intervention against abnormal repair and differentiation of skeletal muscle.
Abstract
Changes in the expression of potassium (K+) channels is a pivotal event during skeletal muscle differentiation. In mouse C2C12 cells, similarly to human skeletal muscle cells, myotube formation increased the expression of Kv7.1, Kv7.3, and Kv7.4, the last showing the highest degree of regulation. In C2C12 cells, Kv7.4 silencing by RNA interference reduced the expression levels of differentiation markers (myogenin, myosin heavy chain, troponinT-1, and Pax3) and impaired myotube formation and multinucleation. In Kv7.4-silenced cells, the differentiation-promoting effect of the Kv7 activator N-(2-amino-4-(4-fluorobenzylamino)-phenyl)-carbamic acid ethyl ester (retigabine) was abrogated. Expression levels for the repressor element-1 silencing transcription factor (REST) declined during myotube formation. Transcript levels for Kv7.4, as well as for myogenin, troponinT-1, and Pax3, were reduced by REST overexpression and enhanced upon REST suppression by RNA interference. Four regions containing potential REST-binding sites in the 5′ untranslated region and in the first intron of the Kv7.4 gene were identified by bioinformatic analysis. Chromatin immunoprecipitation assays showed that REST binds to these regions, exhibiting a higher efficiency in myoblasts than in myotubes. These data suggest that Kv7.4 plays a permissive role in skeletal muscle differentiation and highlight REST as a crucial transcriptional regulator for this K+ channel subunit.
INTRODUCTION
Highly coordinated changes in membrane potential of skeletal muscle cells regulate the early stages of myogenesis, allowing proliferating myoblasts to withdraw from the cell cycle and fuse in ordered arrays of large, multinucleated myotubes (Walsh and Perlman, 1997). In fact, membrane hyperpolarization is known to trigger myoblast commitment and fusion (Bijlenga et al., 2000). Potassium (K+) channels act as major controllers of membrane potential; thus, a specific pattern of sequential changes in the expression and function of distinct classes of voltage-sensitive and voltage-insensitive K+ channels is a critical step in myoblast to myotube transition both in vitro and in vivo (Lesage et al., 1992; Cooper, 2001). Indeed, Kv10.1 (Bijlenga et al., 1998), KIR2.1 (Wieland and Gong, 1995; Fischer-Lougheed et al., 2001), Kv1.5 (Villalonga et al., 2008), and IKCa channels (Fioretti et al., 2005) have been shown to play specific roles in various aspects of skeletal muscle cell proliferation, differentiation, and/or survival.
The Kv7 subclass of voltage-dependent K+ channels consists of five members (Kv7.1– Kv7.5), all implicated in membrane potential control in excitable and nonexcitable cells (Soldovieri et al., 2011). Kv7.1 subunits form the molecular basis of the cardiac repolarizing current IKs and are mutated in genetically determined human arrhythmias. Kv7.2, Kv7.3, Kv7.4, and Kv7.5 give rise to K+ currents widely distributed in neuronal and primary sensory cells. Heteromeric assembly of Kv7.2 and Kv7.3 subunits underlies the muscarinic-regulated current IKM, a primary controller of neuronal excitability (Wang et al., 1998); gene defects in Kv7.2 or Kv7.3 are responsible for benign familial neonatal seizures, an inherited epilepsy of the newborn (Maljevic et al., 2010). Kv7.4 is expressed in primary sensory cells of the inner ear, and its mutations cause a rare form of nonsyndromic autosomal-dominant hearing loss type 2 (DFNA2; Soldovieri et al., 2011). Expression and function of some Kv7 gene family members are not restricted to the nervous system but have been also detected in thyroid gland, pancreas, stomach, and intestine (Maljevic et al., 2010); moreover, Kv7.4 and Kv7.5 are expressed in smooth muscle cells from several vascular beds, where they contribute to vessel tone regulation (Mackie and Byron, 2008). Expression of Kv7 gene family members has also been detected in skeletal muscle cells (Lerche et al., 2000; Schroeder et al., 2000; Tsevi et al., 2005; Roura-Ferrer et al., 2008). In murine C2C12 cells, a widely used in vitro model for skeletal myogenesis, we previously showed that myotube formation was accompanied by an up-regulation of Kv7.1, Kv7.3, and Kv7.4. In these cells, N-(2-amino-4-(4-fluorobenzylamino)-phenyl)-carbamic acid ethyl ester (retigabine), a specific activator of neuronal Kv7.2–Kv7.5 subunits, reduced proliferation, accelerated myogenesis, and inhibited drug-induced myotoxic responses (Iannotti et al., 2010).
In the present study, we explore the functional role and transcriptional regulation in myogenic development of Kv7.4, the Kv7 gene member showing the highest degree of up-regulation. To this aim, we modified expression levels of Kv7.4 by gene silencing or overexpression techniques in C2C12 cells; we evaluated skeletal myogenesis by means of quantitative PCR (qPCR), Western blot, and immunocytochemical analysis of skeletal muscle–specific markers such as troponinT-1 (TnT-1), myogenin (Myog), Pax3, and myosin heavy chain (MyHC). In Kv7.4-silenced and Kv7.4-overexpressing cells, we assessed the effect of the Kv7 modulator retigabine on skeletal myogenesis and Kv7.4 currents by biochemical and electrophysiological experiments, respectively. Because in neuronal cells, the repressor element-1 silencing transcription factor (REST; Chong et al., 1995; Schoenherr and Anderson, 1995) controls the expression of genes involved in the acquisition of the differentiated phenotype (D’Alessandro and Meldolesi, 2010) and members of the Kv7 gene family are directly regulated by REST (Mucha et al., 2010), we investigated the REST-dependent transcriptional regulation of Kv7.4, as well as sites potentially involved, by bioinformatic analysis and chromatin immunoprecipitation (ChIP) assays.
RESULTS
Changes in Kv7 expression during in vitro myogenesis
Murine C2C12 myoblasts exposed for 48 h to differentiation medium (DM) showed a large increase in the expression levels of skeletal muscle–specific proteins such as TnT-1 and Myog and of myosin heavy chain (MyHC), a marker of late skeletal muscle cell maturation (Bennett and Tonks, 1997; Iannotti et al., 2010). Proliferating human primary myoblasts (hSkM-S), similarly to C2C12 myoblasts, expressed transcripts encoding for all members of the Kv7 subfamily; when these cells were cultured in DM for 48 h, the expression levels for Kv7.1, Kv7.3, and Kv7.4 were increased, with Kv7.4 showing the highest degree of modulation; by contrast, transcript levels for Kv7.2 and Kv7.5 were not modified (Supplemental Figure S1). Given the similarities between murine C2C12 cells and human primary myoblasts with regard to the expression changes of the Kv7 subfamily members during in vitro myogenesis, all of the following experiments were performed using the clonal murine cellular model.
Regulation of in vitro skeletal muscle differentiation by Kv7.4
To evaluate the role played by Kv7.4 in skeletal muscle differentiation, C2C12 cells were transiently transfected with a plasmid encoding for a short hairpin RNA (shRNA) directed against Kv7.4 mRNA (pLKO.1-shKv7.4) 1 d before DM exposure for 48 h; in these experiments, C2C12 cells transfected with a plasmid carrying a nonsense sequence (pLKO.1-scramble) served as controls. In C2C12 myoblasts, pLKO.1-shKv7.4 specifically suppressed Kv7.4 mRNA expression, with no effect on the transcripts encoding for other Kv7 members (Supplemental Figure S2). In shKv7.4-transfected myotubes, Kv7.4 mRNA (Figure 1A) and protein (Figure 1B) expression levels were markedly reduced (by >90% and ∼50%, respectively); in these cells, the mRNA levels for the closely related Kv7.2 gene were unaffected by pLKO.1-shKv7.4 transfection (Figure 1A). In shKv7.4-transfeced C2C12 myotubes, a marked reduction of the mRNA levels for Myog, Pax3, and TnT-1 (Figure 1A), as well as of MyHC protein expression (Figure 1B), was observed.
FIGURE 1:
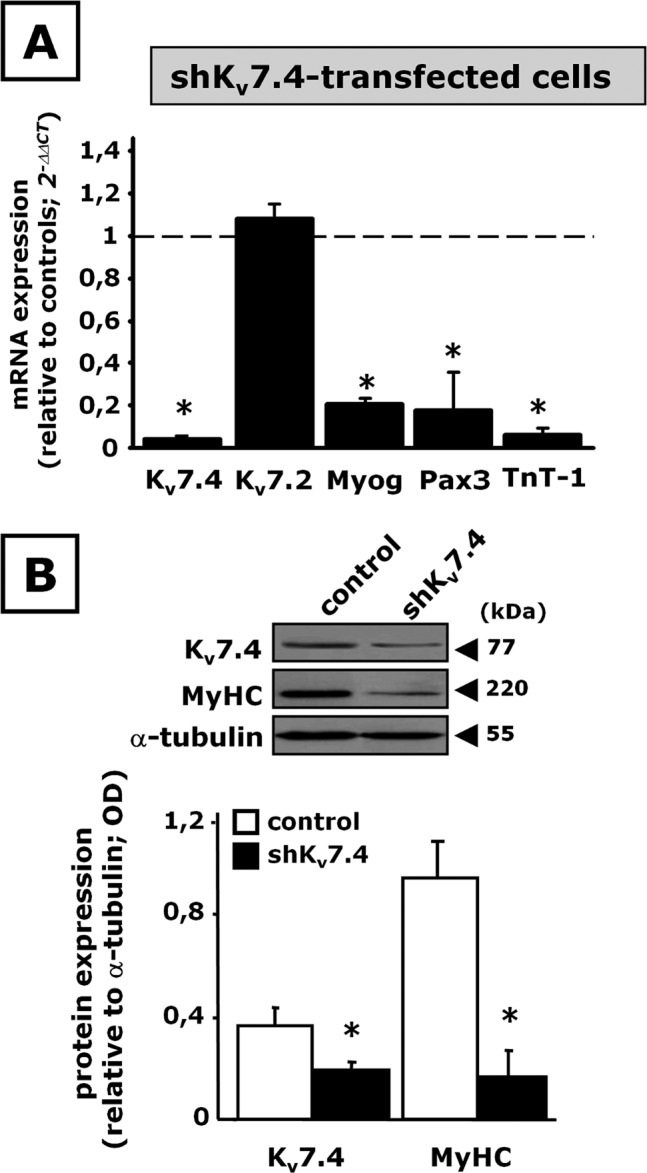
Effect of Kv7.4 gene silencing on C2C12 cell differentiation. C2C12 myoblasts transfected with pLKO.1-scramble (control) or pLKO.1-shKv7.4 (shKv7.4) plasmids were exposed to DM for 48 h. (A) Kv7.4, Kv7.2, Myog, Pax3, and TnT-1 mRNA expression levels by qPCR. (B) Western blot analysis for Kv7.4 and MyHC protein expression on total lysates from control- or shKv7.4-tranfected cells. Top, a representative blot for Kv7.4, MyHC, and α-tubulin. The approximate molecular mass for each of these proteins (expressed in kDa) is shown on the right. Bottom, bar graph showing the quantification of the averaged OD values for the Kv7.4 and MyHC bands, normalized to the OD value for α-tubulin, in both control-transfected (open columns) and shKv7.4-transfected (solid columns) C2C12 cells. *p ≤ 0.05 vs. respective control; data from three separate transfections for each experimental condition (scramble or shKv7.4 plasmid).
To assess whether Kv7.4 silencing affected myogenesis beside protein markers, and to evaluate whether differentiation was suppressed or delayed, the expression of MyHC was monitored at various time points (2, 3, 4, and 7 d) in immunocytochemical experiments. The results showed that, particularly at later time points (after 4–7 d), silencing of Kv7.4 impaired myotube formation; in fact, Kv7.4-silenced cells exposed to differentiating conditions did not increase their cell size and displayed an impaired multinucleation (Figure 2A), resulting in a marked decrease in the cell fusion index (Figure 2B).
FIGURE 2:
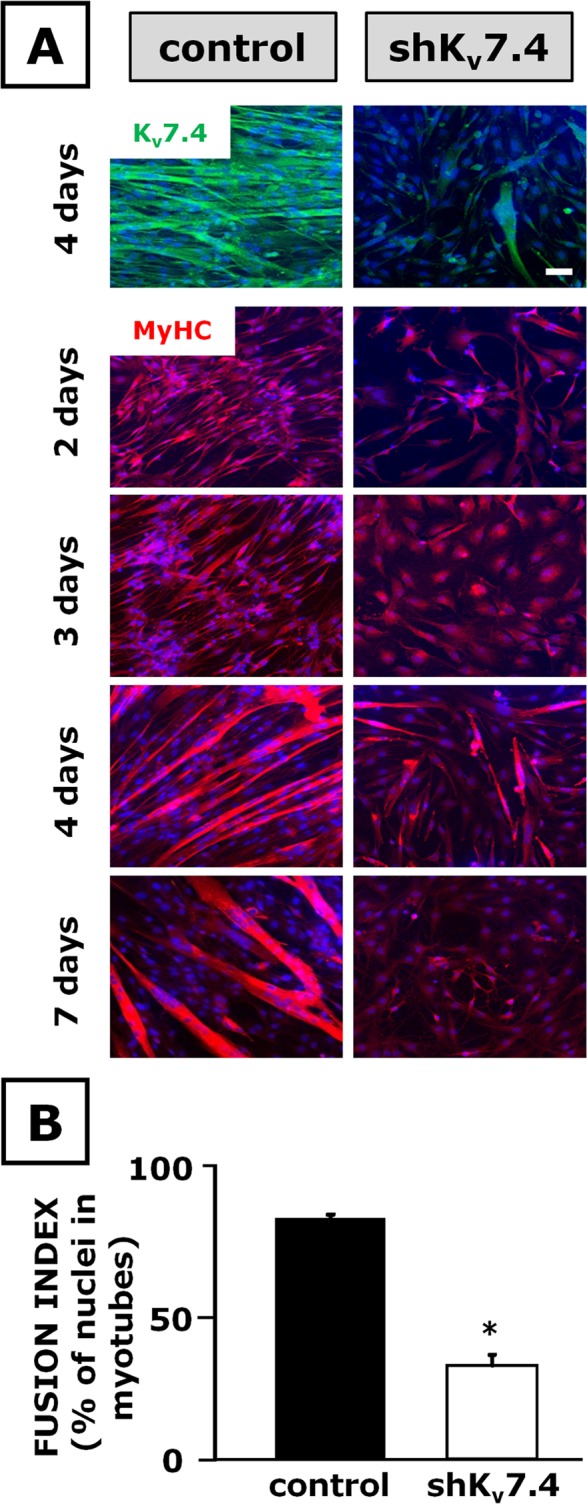
Immunocytochemical analysis of the effects of Kv7.4 gene silencing in C2C12 cells. C2C12 myoblasts transfected with pLKO.1-scramble (control) or pLKO.1-shKv7.4 (shKv7.4) plasmids were exposed to DM for the time indicated on the left. (A) Kv7.4 (green pseudocolor) and MyHC (red pseudocolor) immunofluorescence in control (left) and shKv7.4-transfected cells (right); nuclei stained with Hoechst are shown in blue pseudocolor. Scale bar, 100 μm. (B) Fusion index calculated in both control-transfected (solid column) and shKv7.4-transfected (open column) C2C12 cells exposed to DM for 4 d. *p ≤ 0.05 vs. control-transfected cells. Values are from independent fusion index measurements performed in 15 microscopic fields randomly selected from four coverslips for each experimental condition (scramble or shKv7.4 transfection).
Pharmacological experiments revealed that 10,10-bis(4-pyridinylmethyl)-9(10H)-anthracenone (XE-991), a rather specific inhibitor of channels formed by Kv7 subunits (Wang et al., 1998; Zaczek et al., 1998), when used at 30 μM was able to impede myogenesis, as revealed by the reduced expression of Myog and TnT-1 in C2C12 myotubes; consistent with previous results (Iannotti et al., 2010), lower drug concentrations (0.6–10 μM) were ineffective (Supplemental Figure S3).
Retigabine (10 μM), a specific activator of channels formed by Kv7.2–Kv7.5 subunits, enhanced C2C12 cell in vitro differentiation (Iannotti et al., 2010); it is noteworthy that in DM-exposed Kv7.4-silenced cells, the effects of retigabine on C2C12 cell differentiation were completely abolished, as revealed by the loss of the ability of this Kv7 activator to increase the expression of TnT-1 (Figure 3A) and Pax3 (Figure 3B).
FIGURE 3:
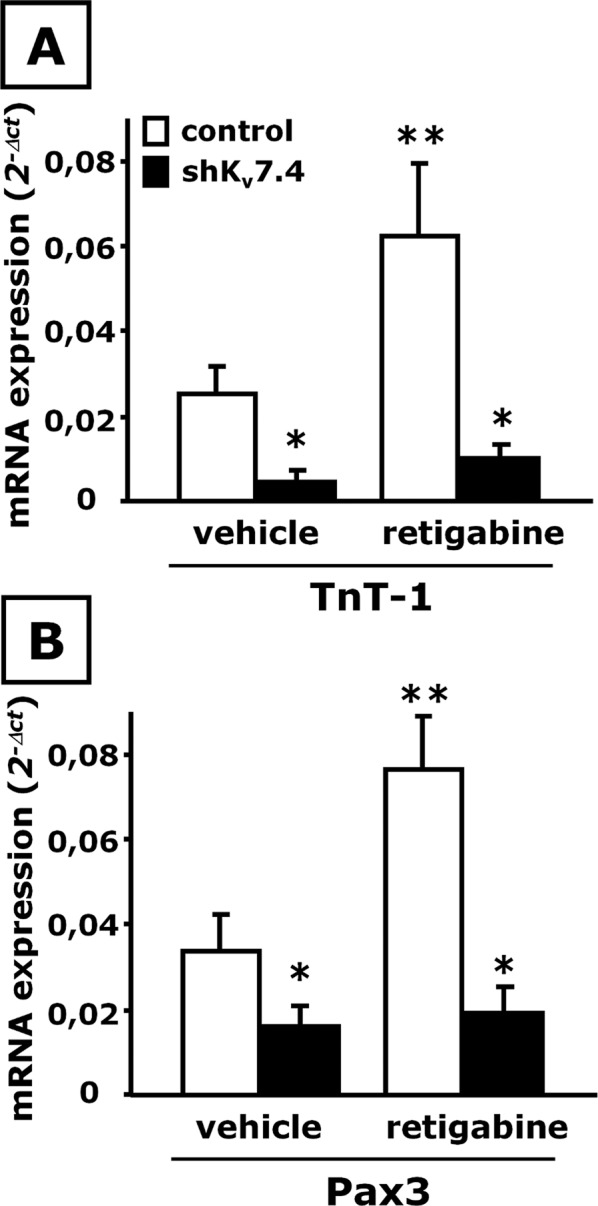
Effect of retigabine on skeletal muscle differentiation in shKv7.4-transfected C2C12 cells. mRNA expression analysis by qPCR for TnT-1 (A) and Pax3 (B) in control-transfected (open columns) or shKv7.4-transfected (solid columns) C2C12 cells exposed to DM for 48 h in the presence of vehicle (0.1% dimethyl sulfoxide) or retigabine (10 μM) as indicated. *p ≤ 0.05 vs. control-transfected C2C12 cells exposed to vehicle but not against each other; **p ≤ 0.05 vs. both control-transfected C2C12 cells exposed to vehicle and shKv7.4-transfected C2C12 cells exposed to retigabine; data from three separate transfections for each experimental condition (scramble or shKv7.4 plasmid).
Whereas skeletal muscle differentiation was dramatically impeded upon suppression of Kv7.4 expression, no significant changes in the transcript levels for Myog, Pax3, and TnT-1 (Figure 4A) or in MyHC expression (Figure 4B) were detected when Kv7.4 was overexpressed by transient transfection in C2C12 cells 1 d before DM exposure (48 h). Similarly, Kv7.4 transfection in proliferating C2C12 cells not subsequently exposed to DM also failed to induce the expression of TnT-1, Myog, and Pax3 (Supplemental Figure S4).
FIGURE 4:
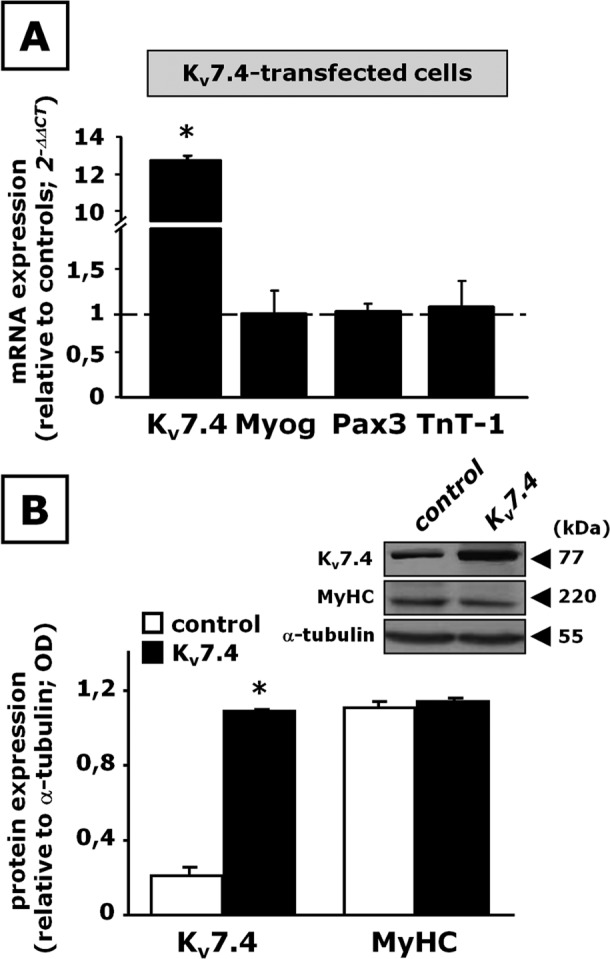
Effect of Kv7.4 overexpression on C2C12 cell differentiation. C2C12 myoblasts transfected with pcDNA3.1 (control) or pcDNA3.1- Kv7.4 (Kv7.4) were exposed to DM for 48 h. (A) Kv7.4, Myog, Pax3, and TnT-1 mRNA expression by qPCR. n = 3–6. (B) Western blot analysis for Kv7.4 and MyHC protein expression on total lysates from control- or Kv7.4-transfected cells. Top, a representative blot for Kv7.4, MyHC, and α-tubulin. The approximate molecular mass for each of these proteins (expressed in kDa) is shown on the right. Bottom, bar graph showing the quantification of the averaged OD values for the Kv7.4 and MyHC bands, normalized to the OD value for α-tubulin, in both control-transfected (open columns) and Kv7.4-transfected (solid columns) C2C12 cells. *p ≤ 0.05 vs. respective controls; data are from three separate transfections for each experimental condition (scramble or Kv7.4 plasmid).
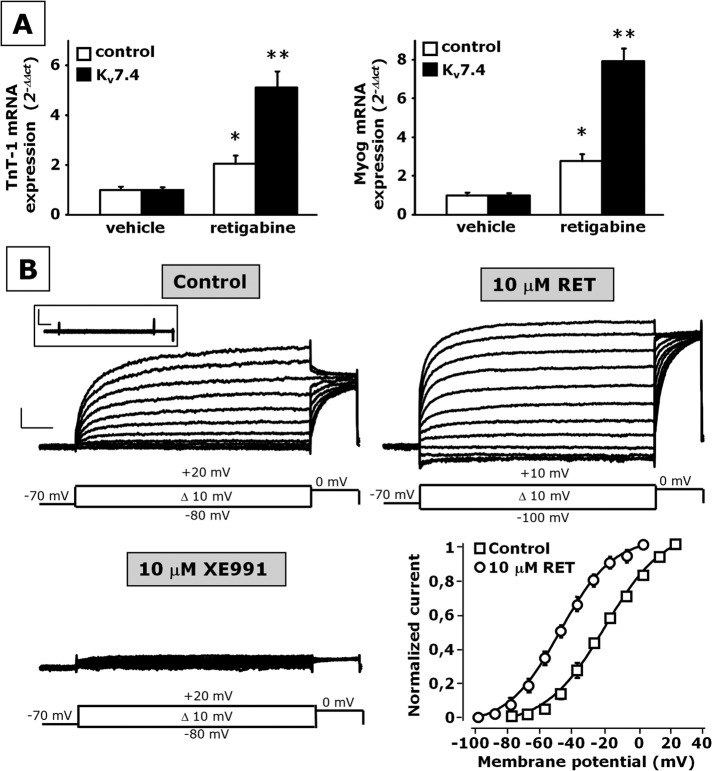
Although Kv7.4 hyperexpression per se did not modify skeletal myogenesis, Kv7.4-overexpressing cells showed an enhanced sensitivity to the differentiation-promoting effect of retigabine when compared with control cells; in fact, in Kv7.4-transfected cells, this neural Kv7 activator (10 μM) increased the transcript levels of TnT-1 and Myog by sixfold to eightfold; in control cells, TnT-1 (Figure 5A, left) and Myog (Figure 5A, right) were increased by twofold to threefold upon exposure to the same drug concentration.
FIGURE 5:
Effect of retigabine on skeletal muscle differentiation in Kv7.4-transfected C2C12 cells. (A) mRNA expression analysis by qPCR for TnT-1 (left) and Myog (right) in control-transfected (open columns) or Kv7.4 (solid columns) transfected C2C12 cells exposed to DM for 48 h in the presence of vehicle (0.1% dimethyl sulfoxide) or retigabine (10 μM), as indicated. *p ≤ 0.05 vs. control-transfected C2C12 cells exposed to vehicle but not against each other; **p ≤ 0.05 vs. both control-transfected C2C12 cells exposed to vehicle and Kv7.4-transfected C2C12 cells exposed to retigabine; data are from three separate transfections for each experimental condition (scramble or Kv7.4 plasmid). (B) Representative macroscopic current traces recorded from undifferentiated C2C12 transfected with Kv7.4 before and after application of 10 μM retigabine or 10 μM XE-991, as indicated. The voltage protocol is shown below each set of traces. The inset shows traces recorded from nontransfected control undifferentiated C2C12 cells. Current scale, 200 pA; time scale, 200 ms. Bottom right, steady-state activation curves for Kv7.4 channels before and after application of 10 μM retigabine. Continuous lines represent Boltzmann fits of the experimental data (data are from three to five cells for each experimental condition).
To investigate the mechanisms underlying the difference between Kv7.4 hyperexpression per se and retigabine on C2C12 cell differentiation, we performed patch-clamp recordings in the whole-cell configuration using the perforated-patch technique. Given that endogenous Kv7.4 currents in either C2C12 myoblasts or myotubes could not be resolved (see the inset in Figure 5B), we used C2C12 myoblasts overexpressing Kv7.4 channels by transient transfection for these experiments. Heterologously expressed Kv7.4 channels gave rise to voltage-dependent, outwardly-directed K+ currents having a threshold of activation around −50 mV and displaying slow activation and deactivation kinetics (Figure 5B, top left). Perfusion with retigabine (10 μM) enhanced the maximal current at depolarized potentials (≥0 mV), with a marked shift in the current activation threshold. As a result, during retigabine exposure, Kv7.4 currents were already significantly activated at the holding potential of −70 mV. In fact, hyperpolarizing pulses from −70 to −100 mV caused the channels to deactivate, generating inwardly-directed currents that relaxed toward zero (Figure 5B, top right). After washout of retigabine, exposure to the Kv7 blocker XE-991 (10 μM) largely suppressed the expressed Kv7.4 currents (Figure 5B, bottom left). Normalized current–voltage relationships revealed that retigabine left-shifted the midpoint potential of current activation by ∼25 mV (V1/2 was –24.5 ± 1.6 and −49.5 ± 5.2 mV in control and retigabine-exposed currents, respectively; p < 0.05; n = 3–5). No significant drug-induced changes were observed in the slope of the normalized current–voltage relationship (the k values were 16.3 ± 2.1 and 15.8 ± 0.7 in control and retigabine-exposed currents, respectively; p > 0.05; n = 3–5; Figure 5B, bottom right).
Changes in REST during skeletal myogenesis and effect of manipulating REST on myotube formation and Kv7.4 expression in C2C12 cells
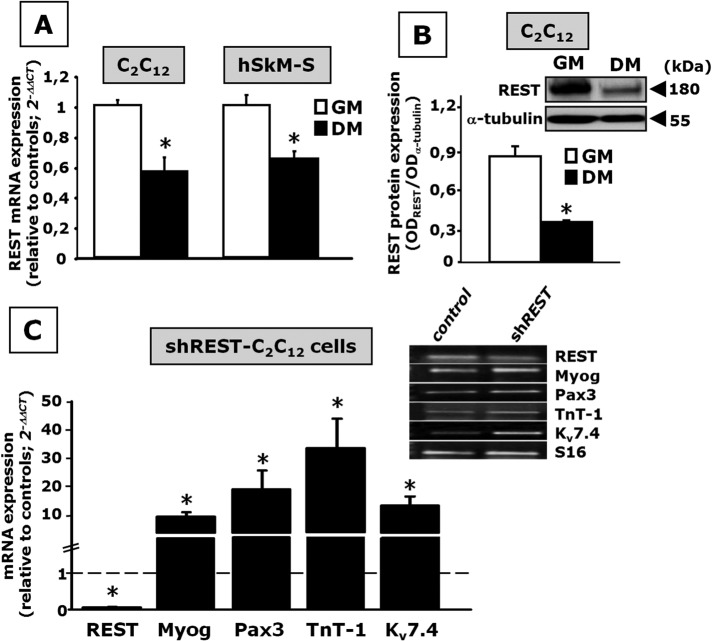
In both C2C12 and hSkM-S cells, REST is expressed in immature myoblasts, and its transcript (Figure 6A) and protein (Figure 6B) levels decline during myotube formation, showing an expression pattern opposite to that of Kv7.4. To evaluate whether changes in REST expression affect myogenesis and Kv7.4 expression levels, we transiently transfected C2C12 cells with a plasmid coding for a short hairpin RNA directed against REST transcripts (shREST) and then exposed them to DM. As shown in Figure 6C, in shREST-transfected C2C12 cells, REST mRNA levels were almost completely suppressed; in the same cells, qPCR analysis revealed that Myog, Pax3, and TnT-1 levels were markedly increased when compared with C2C12 cells differentiated after transfection with a control plasmid. In shREST-transfected C2C12 cells, also Kv7.4 mRNA levels were increased by ∼10-fold.
FIGURE 6:
Changes in REST levels during myotube formation and effect of REST gene silencing on C2C12 cell differentiation. (A) REST transcript levels in murine C2C12 and hSkM-S cells exposed to GM or to DM, as indicated. n = 3–6 plates of GM- or DM-exposed cells. (B) REST expression levels evaluated by Western blot analysis in proliferating myoblasts (GM) and in myotubes (DM). Top, a representative blot; the approximate molecular mass for REST and α-tubulin (expressed in kDa) is shown on the right. Bottom, quantification of the averaged OD values for the REST band, normalized to α-tubulin, in myoblasts (GM) and myotubes (DM). n = 3 plates of GM- or DM-exposed cells. *p ≤ 0.05 vs. respective control. (C) C2C12 myoblasts transfected with control or shREST plasmids were exposed to DM for 48 h. REST, Myog, Pax3, TnT-1, and Kv7.4 mRNAs expression levels were evaluated by qPCR. Data are from three to six separate transfections for both scramble and shREST plasmid. *p ≤ 0.05 vs. respective control. Top, a representative agarose gel electrophoresis of the qPCR products; the size of each band corresponds to the predicted amplicon size (Supplemental Table S1).
To investigate whether the suppression of REST was sufficient to trigger the differentiation program in proliferating C2C12 myoblasts, we evaluated the effect of REST silencing on the expression of Myog and TnT-1 also in C2C12 cells not exposed to DM. The results showed that REST silencing did not enhance skeletal muscle differentiation; moreover, in REST-silenced C2C12 myoblasts, Kv7.4 levels were also unaffected (Supplemental Figure 5).
On the other hand, REST overexpression largely prevented skeletal muscle differentiation in C2C12 cells, as revealed by the reduced mRNA levels for Myog, TnT-1, and Pax3 upon REST transfection (Figure 7A). A significant decrease of Kv7.4 mRNA levels was found in REST-overexpressing cells, whereas no changes were observed for Kv7.2. Western blot experiments also showed that REST-transfected cells displayed a marked decrease of MyHC and Kv7.4 protein levels (Figure 7B).
FIGURE 7:
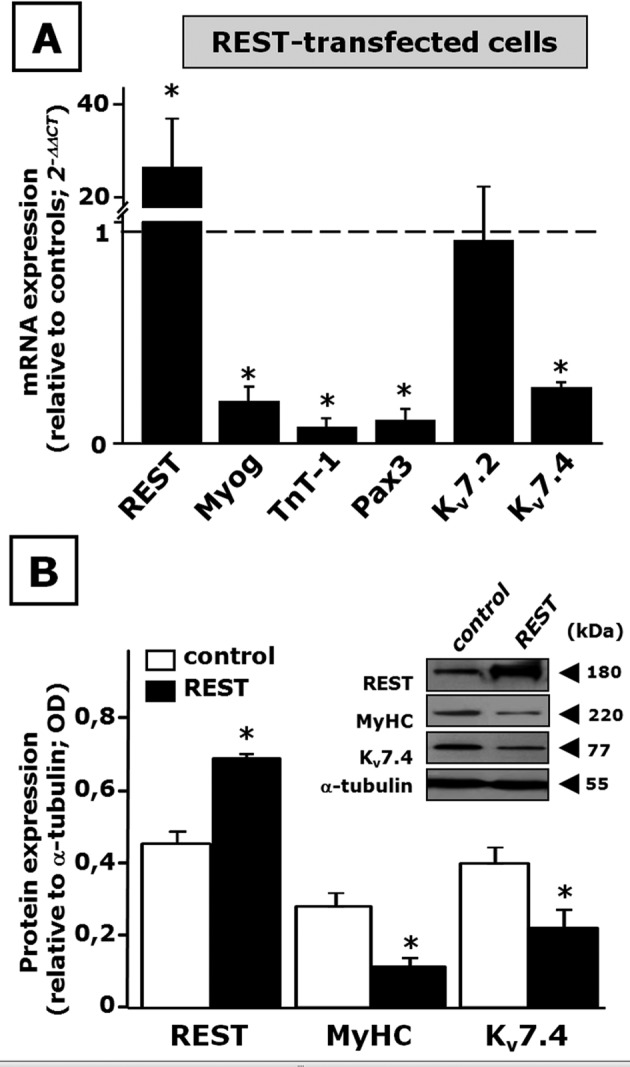
Effect of REST overexpression on C2C12 cell differentiation. (A) REST, Myog, TnT-1, Pax3, Kv7.2, and Kv7.4 mRNA expression levels in C2C12 myoblasts transfected with REST. (B) Western blot analysis for REST, MyHC, and Kv7.4 protein expression in total lysates from control- or REST-transfected cells. The inset shows a representative blot for REST, MyHC, Kv7.4, and α-tubulin. The approximate molecular mass for each of these proteins (expressed in kDa) is shown on the right. The bar graph reports the quantification of the averaged OD values for each band, normalized to α-tubulin, in both control-transfected (open columns) and REST-transfected (solid columns) C2C12 cells. *p ≤ 0.05 vs. respective control. Data are from three to six separated transfections for each experimental condition (scramble or REST plasmid).
Differential binding of REST to RE1-rich regions in the Kv7.4 gene during C2C12 cell differentiation
Members of the Kv7 gene family are under direct transcriptional regulation by REST (Mucha et al., 2010). Therefore we next investigated whether a similar mechanism might be responsible for the reciprocal changes in the expression levels of REST and Kv7.4 observed during skeletal myogenesis. To this aim, we performed a bioinformatic analysis of putative RE1 consensus sites for REST binding in the area surrounding the transcription start site (TSS) of the Kv7.4 genes from various mammalian species, including the beginning of the first intron. Given the well-known sequence variability in the RE1 consensus (Bruce et al., 2004), and particularly the presence of a 0- to 11-nucleotide (nt) spacer between the two more-conserved half-site regions (Johnson et al., 2008), the input sequence for this bioinformatic screening was rather degenerated, to gain information about most, if not all, potential REST-binding sites in the Kv7.4 gene region investigated (see Materials and Methods for further details). The results revealed a large number of potential RE1 sites, some of which clustered within four regions of high sequence similarity among Kv7.4 genes from human, rat, and mouse (Figure 8A). To test whether REST effectively bound to these regions and whether such binding was modified during in vitro myogenesis, we performed ChIP experiments in proliferating or differentiated C2C12 cells. REST-immunoprecipitated DNA was then used in qPCR experiments using primers specifically able to amplify each of these four regions. The results revealed that REST effectively bound to all four regions in myoblasts and that the occupancy of sites 1, 3, and 4 was greatly reduced during C2C12 cell differentiation into myotubes; no significant difference in REST binding between myoblasts and myotubes was found in region 2 (Figure 8B).
FIGURE 8:
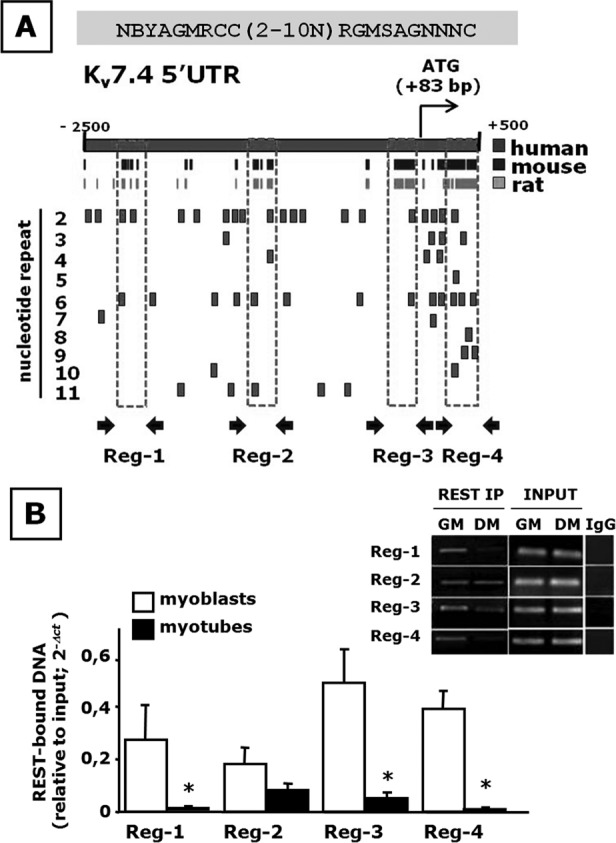
Bioinformatic and ChIP analysis of putative RE1 sites in the Kv7.4 gene. (A) A schematic representation of ∼2500 nt upstream and of ∼ 500 nt downstream from the TSS in the human Kv7.4 gene. The black and light gray boxes correspond to regions of high sequence homology between the mouse and the human genes, and the rat and the human genes, respectively. RE-1 sites identified are shown as small squares in the lower part. RE1 sites are indicated relative to the number of nucleotides in the spacer region separating the hemihalves of the RE1 canonical sequence, as indicated on the left. Regions identified as 1–4, indicated by the broken boxes, correspond to RE1-containing gene regions of high sequence homology among species. (B) REST occupancy of Kv7.4 RE1 sites evaluated by ChIP. Average data of the relative amount of the REST-immunoprecipitated DNA in both myoblasts and myotubes are shown in the bar graph; data are normalized relative to the input DNA, using the 2−ΔCt formula. Data are from three to six separate experiments in GM- or DM-exposed C2C12 cells. *p ≤ 0.05 vs. respective control. The inset shows a representative agarose gel electrophoresis of the qPCR products obtained from REST-immunoprecipitated DNA from both C2C12 myoblasts (open columns; GM) and myotubes (solid columns; DM). Control lanes corresponding to amplification products obtained using DNA inputs or immunoglobulin G–immunoprecipitated DNA as a template for each primer pairs are also shown.
DISCUSSION
Kv7.4 as a crucial regulator of skeletal muscle differentiation
Skeletal muscle development is a multifactorial, highly controlled process involving the coordinated regulation of a large number of genes that allow proliferating myoblasts to withdraw from the cell cycle and fuse in ordered arrays of large, multinucleated myotubes, which further differentiate into mature muscle fibers (Walsh and Perlman, 1997).
An intricate network of reciprocal regulation among myogenic regulatory factors controls the early phases of myogenesis (Martin, 2003). This highly coordinated expression of myogenic factors at various stages of skeletal muscle development is strictly dependent on changes in membrane potential; in particular, membrane hyperpolarization underlain by several classes of K+ channels (Wieland and Gong, 1995; Fischer-Lougheed et al., 2001) is known to trigger the expression of myogenin and myocyte-enhancer factor-2, thus exerting a permissive role on the Ca2+-dependent initiation of the skeletal muscle differentiation program (Konig et al., 2004).
We previously showed that significant changes in the expression levels of some members of the Kv7 subclass of voltage-gated K+ channels, most notably Kv7.4, accompany the myoblast-to-myotube transition in murine C2C12 cells (Iannotti et al., 2010), with similar changes also occurring in human primary myoblasts. To investigate the role of Kv7.4 in skeletal myogenesis, expression levels of this gene were down-regulated or up-regulated by transfection techniques. Transfection with the shKv7.4 plasmid specifically suppressed Kv7.4 mRNA expression in proliferating C2C12 myoblasts, with no effect on other Kv7 members; this result argues in favor of the specificity of the silencing plasmid for its target sequence and shows that compensatory mechanisms among members of the Kv7 subfamily are not taking place. Silencing of Kv7.4 in proliferating C2C12 myoblasts prevents their ability to initiate the differentiation program, suggesting a permissive role for this K+ channel subunit in skeletal myogenesis. Kv7.4-silenced cells exposed to differentiating conditions for up to 7 d did not increase their cell size and displayed an impaired multinucleation, resulting in a marked decrease in cell fusion index.
Pharmacological results corroborate the hypothesis that Kv7.4 subunits play a crucial role in skeletal myogenesis. In fact, in Kv7.4-silenced cells, the effects of the Kv7 activator retigabine on C2C12 myogenesis were abolished, arguing in favor of Kv7.4 subunits being a major target for the drug-induced differentiation-promoting effects. On the other hand, channels formed by different Kv7 subunits display a differential sensitivity to the M-type current inhibitor XE-991; in fact, whereas homomeric or heteromeric Kv7.2 or Kv7.3 channels display an IC50 of ∼1 μM (Wang et al., 1998), currents carried by Kv7.4 and Kv7.5 channels display IC50s of 5.5 μM (Søgaard et al., 2001) and 65–75 μM (Schroeder et al., 2000; Yeung et al., 2008), respectively. Therefore the observation that XE-991 reduced C2C12 cells differentiation only when used at relatively high concentrations (30 μM) suggests a major role for Kv7.4 subunits, possibly in heteromeric configuration with Kv7.5 subunits (Schroeder et al., 2000; Bal et al., 2008; Roura-Ferrer et al., 2008), in skeletal muscle differentiation in vitro.
Kv7.4 overexpression in differentiating C2C12 cells failed to accelerate myotube formation, possibly suggesting that the levels of Kv7.4 attained during myogenesis were sufficient to allow the achievement of the differentiated state. On the other hand, Kv7.4-overexpressing cells showed an enhanced sensitivity to the differentiation-promoting effects of retigabine when compared with control cells. Unfortunately, we failed to record native Kv7 currents in C2C12 cells, possibly because of their small single-channel conductance and opening probabilities (Li et al., 2004; Miceli et al., 2009) or of their location in membrane microdomains, which might escape electrophysiological detection (Roura-Ferrer et al., 2012). Nevertheless, the present electrophysiological results in C2C12 cells overexpressing Kv7.4 channels showed that retigabine exposure caused functional consequences on channel behavior that could not be reproduced by Kv7.4 overexpression. In fact, retigabine, in addition to increasing the maximal conductance, also caused a marked leftward shift in the current activation threshold. Thus, it seems possible to speculate that retigabine exposure would increase Kv7.4 participation in membrane potential control in C2C12 cells, and the resulting hyperpolarization would favor skeletal myogenesis; these effects would be amplified in Kv7.4-overexpressing cells, thereby explaining their higher sensitivity to the differentiation-promoting effects of retigabine.
It should be noticed that mouse models of Kv7.4 deficiency, although reproducing some features of the auditory impairment observed in humans affected with DFNA2 (Kharkovets et al., 2006) and showing abnormal mechanosensitivity (Heidenreich et al., 2011), lacked a visible phenotype and displayed normal growth and development; however, no specific investigation on skeletal muscle structure and function has been carried out in these animals, and the possibility exists that other K+ channels might compensate in vivo for the loss of the myogenesis-promoting effects of Kv7.4.
REST expression changes during myogenesis
In nerve cells, a decrease in REST levels releases the repression of hundreds of specific genes, thus orchestrating the acquisition of the differentiated phenotype (D’Alessandro and Meldolesi, 2010). Rat L6 myoblasts express high levels of REST activity (Chong et al., 1995; Seth and Majzoub, 2001); in these cells, the inhibition of REST derepresses non-muscle–specific proteins, such as the brain type II voltage-gated sodium channel (Chong et al., 1995).
Our results show that the expression levels of REST inversely correlated with the differentiation stage in both C2C12 cells and human primary myoblasts, being lower at later developmental stages. Moreover, REST level modulation strongly regulates skeletal muscle differentiation, and high levels of REST contribute to the suppression of the differentiation program in proliferating skeletal muscle cells. However, the observation that REST suppression in C2C12 myoblasts grown under proliferating conditions did not enhance the expression of skeletal muscle–specific genes suggests that, in the absence of other molecular triggers, REST suppression alone is not sufficient to sustain the acquisition of the myogenic program. Similar hypotheses have been also put forward to explain REST-induced regulation of genes involved in neuronal differentiation (Majumder, 2006).
REST-induced regulation of Kv7.4 expression during myogenesis
Kv7.2 and Kv7.3 genes are direct targets for regulation by REST in sensory cells of the dorsal root ganglia (DRG; Mucha et al., 2010), and REST-induced down-regulation of Kv7.2 contributes to DRG overexcitability during neuropathic pain states (Rose et al., 2011). Our results show that REST levels are inversely related also to those for Kv7.4. In fact, Kv7.4 transcript levels were significantly reduced in REST-transfected C2C12 cells, whereas they were markedly enhanced upon REST silencing. Potential REST-binding RE1 sites in the Kv7.4 gene were identified by bioinformatics analysis; these clustered within four regions of high sequence homology among human, rat, and mouse species located between ∼2500 nt upstream and ∼500 nt downstream from the TSS of the Kv7.4 gene. ChIP experiments revealed that REST effectively bound to these four regions, and, more important, that REST-binding efficiency was decreased in myotubes when compared with myoblasts at three of them. These results suggest that a reduction in REST binding to RE1 sites in the Kv7.4 gene during skeletal muscle differentiation might enhance Kv7.4 transcription, allowing this K+-channel gene to exert its permissive role on myogenesis
Tissue-specific regulation of Kv7 genes by REST
As already introduced, Mucha et al. (2010) identified an RE1 site, conserved among various species, located in the first intron of the Kv7.2 gene, ∼10 kb downstream of the first exon. Bioinformatic analysis of the corresponding position of all Kv7 gene members revealed that the RE-1 sequence identified in Kv7.2 also had high matching scores in Kv7.3 and Kv7.5 genes but low scores in Kv7.1 and Kv7.4; electromobility shift assays confirmed the ability of REST to bind to this RE1 site in Kv7.2, Kv7.3, and Kv7.5 but not in Kv7.1 or Kv7.4 genes. In the present experiments, given the strong sequence variability in the potential RE1 sites (Johnson et al., 2008), we used a consensus sequence more degenerated than that used in Mucha et al. (2010). This strategy allowed the identification of regulatory regions distinct from those of Mucha et al. (2010). Taken together, these data are strongly suggestive of a marked tissue-specific transcriptional regulation of the different Kv7 gene family members. In fact, Kv7.2 transcript levels in DRGs seem to be highly regulated by REST (Mucha et al., 2010); by contrast, in C2C12 cells, Kv7.2 transcript levels appear insensitive to changes in REST levels. Moreover, a decrease in REST levels during myogenesis contributes to the enhanced expression of Kv7.4 in C2C12 cells, whereas, despite the low levels of REST, mature DRG neurons express low (Heidenreich et al., 2011) or undetectable (Passmore et al., 2003) Kv7.4 levels. A complex cross-talk among families of factors acting as activators, including Sp1 (Mucha et al., 2010), thyroid hormone receptors (Winter et al., 2006), and Pit1 (Mustapha et al., 2009), or repressors, such as REST (Mucha et al., 2010), of Kv7 gene transcription is likely responsible for such phenomenon. Indeed, several Sp1 sites are also present in the REST-binding regions identified here in the Kv7.4 gene.
Conclusion
The present results suggest that REST-regulated Kv7.4 channels play a crucial role in the induction and/or maintenance of the differentiated state in skeletal myotubes and highlight REST as a crucial regulator of skeletal muscle differentiation fate. In view of the described dysregulation of REST-target genes in myofibrillar myopathies (Barrachina et al., 2007), the novel mechanism described here may be a relevant target for intervention against skeletal muscle diseases characterized by abnormal repair and differentiation.
MATERIALS AND METHODS
Cell culture
C2C12 myoblasts were cultured and differentiated as previously described (Iannotti et al., 2010). Briefly, proliferating C2C12 cells were propagated in a growth medium (GM) composed of DMEM (Life Technologies Italia, Milan, Italy) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin plus 50 μg/ml streptomycin, and 1% l-glutamine (Invitrogen, Milan, Italy) in a humidified atmosphere of 95% air/5% CO2 at 37°C. Myotube differentiation was achieved upon exposure of proliferating C2C12 cells to a lower (0.1%) FBS concentration, plus 5 μg/ml insulin and 5 μg/ml apotransferrin (differentiation medium) for 2–7 d, according to the experimental protocol. Human primary myoblasts (hSKM-S; A12555, Invitrogen) were cultured in GM and differentiated upon exposure to DMEM plus 2% horse serum, according to the manufacturer’s instructions.
Cell transfection, RNA extraction, and quantitative real-time PCR
C2C12 cells were plated in 60-mm culture dishes; the day after plating, cells were transfected with the plasmids of interest using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions, together with a plasmid encoding for the enhanced green fluorescent protein (Clontech, Mountain View, CA) to assess transfection efficiency. Total cDNA in the transfection mixture was kept constant at 8 μg. For qPCR and Western blot experiments, 24 h after transfection C2C12 cells were exposed to DM for 2 d. In time-course immunofluorescence assays, in order to prolong transcript suppression of the Kv7.4 by the silencing strategy, we exposed shKv7.4 or scramble-transfected C2C12 cells to selection media containing 1.5 μg/ml puromycin for 2 d; after this period, cells were plated on poly-l-lysine–coated coverslip, allowed to adhere for further 24 h, and then exposed to DM (puromycin supplemented) for different times (Mancini et al., 2011). Total RNA isolation, purification, and cDNA synthesis were performed as described (Iannotti et al., 2010). qPCR was carried out in a real-time PCR system (7500 fast; Applied Biosystems, Monza, Italy) using the SYBR Green detection technique and specific primers (Supplemental Table S1). Samples were amplified simultaneously in triplicate in a one-assay run with a nontemplate control blank for each primer pair to control for contamination or primer-dimer formation, and the cycle threshold (Ct) value for each experimental group was determined. A housekeeping gene (the ribosomal protein S16) was used as an internal control to normalize the Ct values, using the 2−ΔCt formula; differences in mRNA content between groups were as expressed as 2−ΔΔCt, as previously described (Winer et al., 1999).
Immunofluorescence
C2C12 cells were washed twice in ice-cold phosphate-buffered saline (PBS) and fixed with 2% paraformaldehyde for 15 min at room temperature (RT; 20–22°C). Cells were treated with 0.1 M glycine for 5 min and incubated overnight at 4°C with the following primary antibodies: mouse anti-MyHC (1:100; Sigma-Aldrich, Milan, Italy) and rabbit anti-Kv7.4 (1:100; Abcam, Cambridge, United Kingdom). Cells were then washed for 30 min with PBS, incubated for 1 h at RT with anti-mouse conjugated to Cy3 or anti-rabbit conjugated to Alexa Fluor 488 (1:100; Jackson ImmunoResearch, Newmarket, United Kingdom), and counterstained with Hoechst 33258 (0.2 μg/ml, 10 min at RT; Sigma-Aldrich) to visualize cells nuclei. All antibodies and Hoechst 33258 were diluted in PBS containing 0.1% Triton X-100 and 1% bovine serum albumin. Coverslips were mounted in Fluoromount (Sigma-Aldrich) and analyzed using a Zeiss LSM 510 Meta argon/krypton laser scanning confocal microscope (Carl Zeiss, Jena, Germany). Images were acquired using the multitrack system to avoid cross-talk among channels. The excitation/emission settings were as follows: 430/505–550 nm for Alexa Fluor 488 and 543/560–615 nm for CY3. Images were confocally captured using a 20× objective (PlanApochromat, numerical aperture, 1.4) pinhole, <1 Airy unit. Each image was acquired eight times, and the signal averaged to improve the signal-to-noise ratio. The fusion index was defined as the percentage of nuclei inside the myotubes; myotubes were identified as MyHC-positive cells containing three or more nuclei, as previously described (Mancini et al., 2011).
Whole-cell electrophysiology
Currents from C2C12 cells were recorded at room temperature (20–22°C) 1 d after transfection with an Axopatch 200A (Molecular Devices, Union City, CA) using the perforated whole-cell configuration of the patch-clamp technique, with glass micropipettes of 3–5 MΩ resistance. The extracellular solution contained (mM): 138 NaCl, 2 CaCl2, 5.4 KCl, 1 MgCl2, 10 glucose, and 10 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4, with NaOH. The pipette (intracellular) solution contained (mM): 140 KCl, 2 MgCl2, 10 ethylene glycol tetraacetic acid, 10 HEPES, 5 Mg-ATP, pH 7.3–7.4, with KOH. Nystatin (120–240 μg/ml) was added to this solution immediately before each recording session. The pCLAMP software (version 10.0.2; Molecular Devices) was used for data acquisition and analysis. To generate conductance–voltage curves, the cells were held at −70 mV and then depolarized for 1.5 s from −100/−80 to 0/+20 mV in 10-mV increments, followed by an isopotential pulse at 0 mV of 350-ms duration. The current values recorded at the beginning of the 0-mV pulse were normalized and expressed as a function of the preceding voltages. The data were fitted to a Boltzmann equation of the following form: y = max/[1 + exp(V1/2 − V)/k], where V is the test potential, V1/2 is the half-activation potential, and k is the slope factor.
Bioinformatic analysis and chromatin immunoprecipitation assays
Kv7.4 sequence alignments and potential RE1 consensus sites identification were performed using Lasergene software (DNASTAR, Madison, WI). The sequence NBYAGMRCC(2-10N)RGMSAGNNNC was used to identify RE1 sites in the Kv7.4 gene (from−3360 to +1950 relative to the TSS), where N = A, C, G, or T; B = C, G, or T; Y = C or T; M = A or C; R = A or G; and S = C or G. Each putative RE1 site identified showed a homology score of >85%. ChIP assays were performed following published protocols (Formisano et al., 2007), with slight modifications. Briefly, C2C12 cells were cross-linked in 1% formaldehyde for 20 min at room temperature. The cross-linking reaction was stopped by adding glycine to a final concentration of 0.125 M. The cells were washed two times in cold PBS containing protease inhibitors (Sigma-Aldrich), and cell lysates were prepared and sonicated to generate chromosomal DNA in the range of 400–600 base pairs. Sonicated DNA was precleared with salmon sperm DNA/protein G-agarose 50% gel slurry for 1 h at 4°C and immunoprecipitated overnight at 4°C with rabbit polyclonal antibodies directed against the N-terminus of REST (REST-N; 1:300; Santa Cruz Biotechnology, Santa Cruz, CA). After washes and elution, immunoprecipitated DNA was released from histones (65°C for 4 h in 5 M NaCl), treated with proteinase K (1 h at 45°C), purified by phenol-chloroform extraction and ethanol precipitation, and finally resuspended in PCR-grade water. Immunoprecipitated DNA was analyzed by qPCR (40–45 cycles) using primers amplifying each of the four RE1-rich regions identified (Supplemental Table S1). DNA immunoprecipitated by anti–rabbit immunoglobulin G antibodies (GE Healthcare Europe, Milan, Italy) was used as a negative control. In addition, for each primer pair, nonimmunoprecipitated DNA was used as input for normalization. All ChIP assays were performed in triplicate for at least three different biological preparations.
Western blots
C2C12 cells were washed twice in cold PBS and lysed with lysis solution (150 mM NaCl, 1 mM EDTA, pH 7.4, 10 mM Tris-HCl, pH 8, 1% SDS, and protease inhibitors). Lysates were sonicated for 40–60 s and then boiled for 5 min in Laemmli SDS loading buffer, loaded (100–150 μg) on 8% SDS–PAGE, and then transferred to a polyvinylidene fluoride membrane. Filters were incubated overnight at 4°C with the following antibodies: rabbit anti-REST (1:400; Millipore, Temecula, CA); mouse anti-Kv7.4 (1:500; Neuromab, Davis, CA); and mouse anti-MyHC (1:1000; Millipore). An anti α-tubulin antibody (1:5000; Sigma-Aldrich) was used to check for equal protein loading. Reactive bands were detected by chemiluminescence (ECL or ECL-plus; Amersham Bioscience, Piscataway, NJ). Images were analyzed on a ChemiDoc station with Quantityone software (Bio-Rad, Segrate, Italy).
Drugs and other reagents
Retigabine was from ASTA Medica (Radebeul, Germany) or Valeant Pharmaceuticals (Aliso Viejo, CA); XE-991 was from Tocris Bioscience (Bristol, United Kingdom). The expression plasmid pcDNA3.1-REST was kindly provided by R. Suzanne Zukin (Department of Neuroscience, Albert Einstein College of Medicine, New York, NY). The shRNA lentiviral plasmid designed to silence Kv7.4 gene expression (pKLO.1-shKv7.4; 5′-CCGGCCGTTCTGTCAGGATTCTGAACTCGAGTTCAGAATCCTGACAGAACGGTTTT-3′) was from Sigma-Aldrich (Ref. Seq. XM_143960). The nonsense pLKO.1-puro plasmid containing a scrambled sequence was obtained from Addgene (Cambridge, MA). The shRNA lentiviral plasmid (psMagic2-amp) encoding a short sequence 5′-GCGCCCAGATATTTACAGTTCAAATTAGTGAAGCCACAGATGTAATTTGAACTGTAAATATCTGGA TGT-3′) directed against REST mRNA (shREST), as well as the scrambled plasmid used as control, were from Open Biosystems (Huntsville, AL) and were kindly provided by Lucio Pastore (Department of Medical Biotechnology, University of Naples Federico II, Naples, Italy).
Statistics
Data are expressed as mean ± SEM of the given number of experiments (n); for qPCR experiments, each data point was performed in triplicate. Data sets were compared by use of matched Student’s t tests or, if necessary, with one-way analysis of variance, followed by the Newman-Keuls test. Statistically significant differences were accepted when p was ≤0.05.
Supplementary Material
Acknowledgments
We acknowledge the contributions of Natascia Guida (Division of Pharmacology, Department of Neuroscience, University of Naples Federico II, Naples, Italy) for initial help with ChIP experiments, Vincenzo Romaniello (Division of Pharmacology, Department of Neuroscience, University of Naples Federico II) for initial help with qPCR experiments, and Antonio Porcellini (Department of General Pathology, Faculty of Science, University of Naples Federico II) and Silvia Bartollino (Department of Medicine and Health Science, University of Molise, Campobasso, Italy) for bioinformatic tools and software. This study was supported by grants from the European Union (E-Rare 2007, EUROBFNS), the Fondazione Telethon Italy (GGP07125), the Fondazione San Paolo–IMI (Project Neuroscience), Regione Molise (Convenzione AIFA/Regione Molise), and the Italian Ministry for University and Research (PRIN 2009).
Abbreviations used:
- ChIP
chromatin immunoprecipitation
- Ct
cycle threshold
- DFNA2
nonsyndromic autosomal-dominant hearing loss type 2
- DM
differentiation medium
- DRG
dorsal root ganglia
- FBS
fetal bovine serum
- GM
growth medium
- MyHC
myosin heavy chain
- Myog
myogenin
- PBS
phosphate-buffered saline
- qPCR
quantitative PCR
- REST
repressor element-1 silencing transcription factor
- TnT-1
troponinT-1
- TSS
transcription start site
- XE-991
10,10-bis(4-pyridinylmethyl)-9(10H)-anthracenone
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-12-1044) on December 14, 2012.
REFERENCES
- Bal M, Zhang J, Zaika O, Hernandez CC, Shapiro MS. Homomeric and heteromeric assembly of KCNQ (Kv7) K+ channels assayed by total internal reflection fluorescence/fluorescence resonance energy transfer and patch clamp analysis. J Biol Chem. 2008;283:30668–30676. doi: 10.1074/jbc.M805216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrachina M, Moreno J, Juvés S, Moreno D, Olivé M, Ferrer I. Target genes of neuron-restrictive silencer factor are abnormally up-regulated in human myotilinopathy. Am J Pathol. 2007;171:1312–1323. doi: 10.2353/ajpath.2007.070520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AM, Tonks NK. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science. 1997;278:1288–1291. doi: 10.1126/science.278.5341.1288. [DOI] [PubMed] [Google Scholar]
- Bijlenga P, Liu JH, Espinos E, Haenggeli CA, Fischer-Lougheed J, Bader CR, Bernheim L. T-type alpha 1H Ca2+ channels are involved in Ca2+ signaling during terminal differentiation (fusion) of human myoblasts. Proc Natl Acad Sci USA. 2000;97:7627–7632. doi: 10.1073/pnas.97.13.7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlenga P, Occhiodoro T, Liu JH, Bader CR, Bernheim L, Fischer-Lougheed J. An ether-à-go-go K+ current, Ih-eag, contributes to the hyperpolarization of human fusion-competent myoblasts. J Physiol. 1998;512:317–323. doi: 10.1111/j.1469-7793.1998.317be.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Göttgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci USA. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramírez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Cooper E. Human myoblast fusion requires expression of functional inward rectifier Kir2.1 channels. J Cell Biol. 2001;153:677–686. doi: 10.1083/jcb.153.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro R, Meldolesi J. In PC12 cells, expression of neurosecretion and neurite outgrowth are governed by the transcription repressor REST/NRSF. Cell Mol Neurobiol. 2010;30:1295–1302. doi: 10.1007/s10571-010-9602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioretti B, Pietrangelo T, Catacuzzeno L, Franciolini F. Intermediate-conductance Ca2+-activated K+ channel is expressed in C2C12 myoblasts and is downregulated during myogenesis. Am J Physiol Cell Physiol. 2005;289:C89–C96. doi: 10.1152/ajpcell.00369.2004. [DOI] [PubMed] [Google Scholar]
- Fischer-Lougheed J, Liu JH, Espinos E, Mordasini D, Bader CR, Belin D, Bernheim L. Human myoblast fusion requires expression of functional inward rectifier Kir2.1 channels. J Cell Biol. 2001;153:F9–F12. doi: 10.1083/jcb.153.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formisano L, Noh KM, Miyawaki T, Mashiko T, Bennett MV, Zukin RS. Ischemic insults promote epigenetic reprogramming of mu opioid receptor expression in hippocampal neurons. Proc Natl Acad Sci USA. 2007;104:4170–4175. doi: 10.1073/pnas.0611704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich M, Lechner SG, Vardanyan V, Wetzel C, Cremers CW, De Leenheer EM, Aránguez G, Moreno-Pelayo MA, Jentsch TJ, Lewin GR. KCNQ4 K(+) channels tune mechanoreceptors for normal touch sensation in mouse and man. Nat Neurosci. 2011;15:138–145. doi: 10.1038/nn.2985. [DOI] [PubMed] [Google Scholar]
- Iannotti FA, Panza E, Barrese V, Viggiano D, Soldovieri MV, Taglialatela M. Expression, localization, and pharmacological role of Kv7 potassium channels in skeletal muscle proliferation, differentiation, and survival after myotoxic insults. J Pharmacol Exp Ther. 2010;332:811–820. doi: 10.1124/jpet.109.162800. [DOI] [PubMed] [Google Scholar]
- Johnson R, et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharkovets T, Dedek K, Maier H, Schweizer M, Khimich D, Nouvian R, Vardanyan V, Leuwer R, Moser T, Jentsch TJ. Mice with altered KCNQ4 K+ channels implicate sensory outer hair cells in human progressive deafness. EMBO J. 2006;25:642–652. doi: 10.1038/sj.emboj.7600951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig S, Hinard V, Arnaudeau S, Holzer N, Potter G, Bader CR, Bernheim L. Membrane hyperpolarization triggers myogenin and myocyte enhancer factor-2 expression during human myoblast differentiation. J Biol Chem. 2004;2:28187–28196. doi: 10.1074/jbc.M313932200. [DOI] [PubMed] [Google Scholar]
- Lerche C, Scherer CR, Seebohm G, Derst C, Wei AD, Busch AE, Steinmeyer K. Molecular cloning and functional expression of KCNQ5, a potassium channel subunit that may contribute to neuronal M-current diversity. J Biol Chem. 2000;275:395–400. doi: 10.1074/jbc.M002378200. [DOI] [PubMed] [Google Scholar]
- Lesage F, Attali B, Lazdunski M, Barhanin J. Developmental expression of voltage-sensitive K +channels in mouse skeletal muscle and C2C12 cells. FEBS Lett. 1992;310:162–166. doi: 10.1016/0014-5793(92)81320-l. [DOI] [PubMed] [Google Scholar]
- Li Y, Gamper N, Shapiro MS. Single‑channel analysis of KCNQ K+ channels reveals the mechanism of augmentation by a cysteine‑modifying reagent. J Neurosci. 2004;24:5079–5090. doi: 10.1523/JNEUROSCI.0882-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie AR, Byron KL. Cardiovascular KCNQ (Kv7) potassium channels: physiological regulators and new targets for therapeutic intervention. Mol Pharmacol. 2008;74:1171–1179. doi: 10.1124/mol.108.049825. [DOI] [PubMed] [Google Scholar]
- Maljevic S, Wuttke TV, Seebohm G, Lerche H. KV7 channelopathies. Pflugers Arch. 2010;460:277–288. doi: 10.1007/s00424-010-0831-3. [DOI] [PubMed] [Google Scholar]
- Mancini A, Sirabella D, Zhang W, Yamazaki H, Shirao T, Krauss RS. Regulation of myotube formation by the actin-binding factor drebrin. Skelet Muscle. 2011;1:36. doi: 10.1186/2044-5040-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PT. Role of transcription factors in skeletal muscle and the potential for pharmacological manipulation. Curr Opin Pharmacol. 2003;3:300–308. doi: 10.1016/s1471-4892(03)00050-x. [DOI] [PubMed] [Google Scholar]
- Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5:1929–1935. doi: 10.4161/cc.5.17.2982. [DOI] [PubMed] [Google Scholar]
- Miceli F, Cilio MR, Taglialatela M, Bezanilla F. Gating currents from neuronal K(V)7.4 channels: general features and correlation with the ionic conductance. Channels. 2009;3:274–283. [PubMed] [Google Scholar]
- Mucha M, Ooi L, Linley JE, Mordaka P, Dalle C, Robertson B, Gamper N, Wood IC. Transcriptional control of KCNQ channel genes and the regulation of neuronal excitability. J Neurosci. 2010;30:13235–13245. doi: 10.1523/JNEUROSCI.1981-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustapha M, Fang Q, Gong TW, Dolan DF, Raphael Y, Camper SA, Duncan RK. Deafness and permanently reduced potassium channel gene expression and function in hypothyroid Pit1dw mutants. J Neurosci. 2009;29:1212–1223. doi: 10.1523/JNEUROSCI.4957-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore GM, et al. KCNQ/M currents in sensory neurons: significance for pain therapy. J Neurosci. 2003;23:7227–7236. doi: 10.1523/JNEUROSCI.23-18-07227.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose K, Ooi L, Dalle C, Robertson B, Wood IC, Gamper N. Transcriptional repression of the M channel subunit Kv7.2 in chronic nerve injury. Pain. 2011;152:742–754. doi: 10.1016/j.pain.2010.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roura-Ferrer M, Solé L, Martínez-Mármol R, Villalonga N, Felipe A. Skeletal muscle Kv7 (KCNQ) channels in myoblast differentiation and proliferation. Biochem Biophys Res Commun. 2008;369:1094–1097. doi: 10.1016/j.bbrc.2008.02.152. [DOI] [PubMed] [Google Scholar]
- Roura-Ferrer M, Solé L, Oliveras A, Villarroel A, Comes N, Felipe A. Targeting of Kv7.5 (KCNQ5)/KCNE channels to surface microdomains of cell membranes. Muscle Nerve. 2012;45:48–54. doi: 10.1002/mus.22231. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Schroeder BC, Hechenberger M, Weinreich F, Kubisch C, Jentsch TJ. KCNQ5, a novel potassium channel broadly expressed in brain, mediates M-type currents. J Biol Chem. 2000;275:24089–24095. doi: 10.1074/jbc.M003245200. [DOI] [PubMed] [Google Scholar]
- Seth KA, Majzoub JA. Repressor element silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) can act as an enhancer as well as a repressor of corticotropin-releasing hormone gene transcription. J Biol Chem. 2001;276:13917–13923. doi: 10.1074/jbc.M007745200. [DOI] [PubMed] [Google Scholar]
- Soldovieri MV, Miceli F, Taglialatela M. Driving with no brakes: molecular pathophysiology of Kv7 potassium channels. Physiology (Bethesda) 2011;26:365–376. doi: 10.1152/physiol.00009.2011. [DOI] [PubMed] [Google Scholar]
- Søgaard R, Ljungstrøm T, Pedersen KA, Olesen SP, Jensen BS. KCNQ4 channels expressed in mammalian cells: functional characteristics and pharmacology. Am J Physiol Cell Physiol. 2001;280:C859–866. doi: 10.1152/ajpcell.2001.280.4.C859. [DOI] [PubMed] [Google Scholar]
- Tsevi I, Vicente R, Grande M, López-Iglesias C, Figueras A, Capellà G, Condom E, Felipe A. KCNQ1/KCNE1 channels during germ-cell differentiation in the rat: expression associated with testis pathologies. J Cell Physiol. 2005;202:400–410. doi: 10.1002/jcp.20132. [DOI] [PubMed] [Google Scholar]
- Villalonga N, Martínez-Mármol R, Roura-Ferrer M, David M, Valenzuela C, Soler C, Felipe A. Cell cycle-dependent expression of Kv1.5 is involved in myoblast proliferation. Biochim Biophys Acta. 2008;1783:728–736. doi: 10.1016/j.bbamcr.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Walsh K, Perlman H. Cell cycle exit upon myogenic differentiation. Curr Opin Genet Dev. 1997;7:597–602. doi: 10.1016/s0959-437x(97)80005-6. [DOI] [PubMed] [Google Scholar]
- Wang HS, Pan Z, Shi W, Brown BS, Wymore RS, Cohen IS, Dixon JE, McKinnon D. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science. 1998;282:1890–1893. doi: 10.1126/science.282.5395.1890. [DOI] [PubMed] [Google Scholar]
- Wieland SJ, Gong QH. Modulation of a potassium conductance in developing skeletal muscle. Am J Physiol. 1995;268:C490–C495. doi: 10.1152/ajpcell.1995.268.2.C490. [DOI] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Winter H, et al. Thyroid hormone receptors TRalpha1 and TRbeta differentially regulate gene expression of Kcnq4 and prestin during final differentiation of outer hair cells. J Cell Sci. 2006;119:2975–2984. doi: 10.1242/jcs.03013. [DOI] [PubMed] [Google Scholar]
- Yeung SY, Lange W, Schwake M, Greenwood IA. Expression profile and characterisation of a truncated KCNQ5 splice variant. Biochem Biophys Res Commun. 2008;371:741–746. doi: 10.1016/j.bbrc.2008.04.129. [DOI] [PubMed] [Google Scholar]
- Zaczek R, Chorvat RJ, Saye JA, Pierdomenico ME, Maciag CM, Logue AR, Fisher BN, Rominger DH, Earl RA. Two new potent neurotransmitter release enhancers, 10,10-bis(4-pyridinylmethyl)-9(10H)-anthracenone and 10,10-bis(2-fluoro-4-pyridinylmethyl)-9(10H)-anthracenone: comparison to linopirdine. J Pharmacol Exp Ther. 1998;285:724–730. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.