Lamin A tail domains are SUMO1 modified at K420 (nuclear localization signal) and K486 (Ig-fold). K486 modification requires Ig-fold surface residues E460 and D461 and is reduced by familial partial lipodystrophy–causing mutations G465D and K486N. These results suggest novel mechanisms of functional control over lamin A in cells.
Abstract
Lamin filaments are major components of the nucleoskeleton that bind LINC complexes and many nuclear membrane proteins. The tail domain of lamin A directly binds 21 known partners, including actin, emerin, and SREBP1, but how these interactions are regulated is unknown. We report small ubiquitin-like modifier 1 (SUMO1) as a major new posttranslational modification of the lamin A tail. Two SUMO1 modification sites were identified based on in vitro SUMOylation assays and studies of Cos-7 cells. One site (K420) matches the SUMO1 target consensus; the other (K486) does not. On the basis of the position of K486 on the lamin A Ig-fold, we hypothesize the SUMO1 E2 enzyme recognizes a folded structure–dependent motif that includes residues genetically linked to familial partial lipodystrophy (FPLD). Supporting this model, SUMO1-modification of the lamin A tail is reduced by two FPLD-causing mutations, G465D and K486N, and by single mutations in acidic residues E460 and D461. These results suggest a novel mode of functional control over lamin A in cells.
INTRODUCTION
The small ubiquitin-like modifier (SUMO) family consists of four conserved ∼10-kDa proteins (SUMO1, SUMO2, SUMO3, SUMO4) that are covalently and reversibly attached to lysine residues on target proteins (Gareau and Lima, 2010). SUMOylation can regulate the localization, function, and interactions of target proteins and influences many cellular pathways, including nuclear import/export, transcription, apoptosis, cell cycle regulation, and protein stability (Geiss-Friedlander and Melchior, 2007). At the molecular level, SUMOylation can block binding to specific partners, confer binding to new partners bearing a “SUMO interaction motif” (SIM), or change protein conformation (Wilkinson and Henley, 2010). The enzymes responsible for SUMO conjugation, and many SUMOylated proteins, are located primarily in the nucleus (Gareau and Lima, 2010). For example, actin, a major component of the nucleoskeleton (Visa and Percipalle, 2010; Simon and Wilson, 2011), is modified by SUMO2 and SUMO3 as a mechanism for retention in the nucleus (Hofmann et al., 2009). Another nucleoskeletal protein, lamin A, is modified by SUMO2 (Zhang and Sarge, 2008).
Nuclear intermediate filaments formed by A- and B-type lamins are major components of the nucleoskeleton and are responsible for nuclear shape, assembly, and genome tethering (Dittmer and Misteli, 2011; Simon and Wilson, 2011; Gerace and Huber, 2012). Lamins also bind signaling and chromatin-regulatory proteins, support epigenetic regulation, and are involved in mechanotransduction, development, transcription, replication, and DNA-damage repair (Dechat et al., 2008; Wilson and Berk, 2010; Wilson and Foisner, 2010). In mammalian cells, LMNB1 and LMNB2 encode somatic lamins B1 and B2, respectively; LMNB2 also encodes the spermatocyte-specific lamin B3 (Dittmer and Misteli, 2011). Together B-type lamins are essential for embryogenesis in mice (Kim et al., 2011b), with distinct contributions to the developing brain (Takamori et al., 2007; Coffinier et al., 2010, 2011). The mammalian LMNA gene is alternatively spliced to generate somatic lamins A and C (and minor isoform AΔ10) and spermatocyte-specific lamin C2 (Dittmer and Misteli, 2011). The A-type lamins are not essential at the cellular level but influence many specific tissues during development (Dechat et al., 2010a; Dittmer and Misteli, 2011; Gerace and Huber, 2012), with particular roles in mechanosensitive gene expression (Lammerding et al., 2004) and pRb-dependent cell proliferation control (Dechat et al., 2010b). Mutations in LMNA cause at least 15 tissue-specific diseases (laminopathies), including Emery–Dreifuss muscular dystrophy, Dunnigan-type familial partial lipodystrophy (FPLD), and cardiomyopathy (LMNA mutations are frequent in heart transplant patients; Cowan et al., 2010), and multisystem disorders, including Hutchinson–Gilford progeria syndrome (HGPS; Worman, 2012). Disrupted nucleoskeletal organization and lamin A missense mutations (e.g., G411D, G631D) are also seen relatively frequently in patients with metabolic syndrome, suggesting this too is a laminopathy (Dutour et al., 2011). The molecular mechanisms and tissue specificity of these diseases are poorly understood.
Lamins are extensively posttranslationally modified. The lamin A precursor polypeptide is C-terminally farnesylated and carboxymethylated and then proteolytically cleaved at Y646 to generate mature lamin A (Dechat et al., 2010a). Mature lamin A can be acetylated (Choudhary et al., 2009), O-GlcNAcylated (Wang et al., 2010; Alfaro et al., 2012), or Ser/Thr/Tyr-phosphorylated (Eggert et al., 1993; Haas and Jost, 1993; Olsen et al., 2006, 2010; Pan et al., 2009; Wang et al., 2010; Rigbolt et al., 2011) at various positions or SUMOylated by SUMO2 in the rod domain (Zhang and Sarge, 2008) or SUMO3 in the tail domain (Galisson et al., 2011). The tail domain of mature lamin A, comprising residues 385–646, interacts with at least 21 specific partners, including actin (Simon et al., 2010), titin (Zastrow et al., 2006), emerin (Clements et al., 2000), and the transcription factor sterol response element–binding protein 1 (SREBP1; Lloyd et al., 2002). Motivated by predicted SUMOylation site(s) (see later discussion), we explored potential SUMO modification of the lamin A tail.
RESULTS
Because usually only a small percentage, at most, of endogenous SUMO substrates are modified at any given time (Johnson, 2004; Hay, 2005), we adapted a method used routinely in this field, namely transient coexpression assays (Sarge and Park-Sarge, 2009), to independently determine if lamin A is SUMO modified in vivo. Cos-7 cells were transiently cotransfected with full-length mature lamin A (myc tagged at the N-terminus [myc-lamin A]) plus histidine (His)-tagged SUMO1, His-SUMO2, or empty His vector (negative control). Whole-cell protein lysates were prepared 36 h after transfection, incubated with Ni2+ beads, and pelleted to affinity purify both free and protein-conjugated His-SUMO. Pelleted proteins were resolved by SDS–PAGE and Western blotted with antibodies specific for myc (Figure 1A). Unmodified myc-lamin A has a natural “His tag” (residues 563–566) that binds Ni2+ beads and served as the loading control (Figure 1A, myc-lamA, major ∼70-kDa band). An additional minor band at ∼125 kDa was detected weakly in cells that expressed myc-lamin A alone and might represent myc-lamin A that was modified by endogenous SUMO1. The ∼125-kDa signal was consistently greater in cells that also expressed either His-SUMO1 or His-SUMO2 (Figure 1A). We concluded that this ∼125-kDa band was SUMOylated lamin A. These results independently validated the previous report of SUMO2 modification of full-length lamin A (Zhang and Sarge, 2008) and further suggested novel lamin A modification by SUMO1.
FIGURE 1:
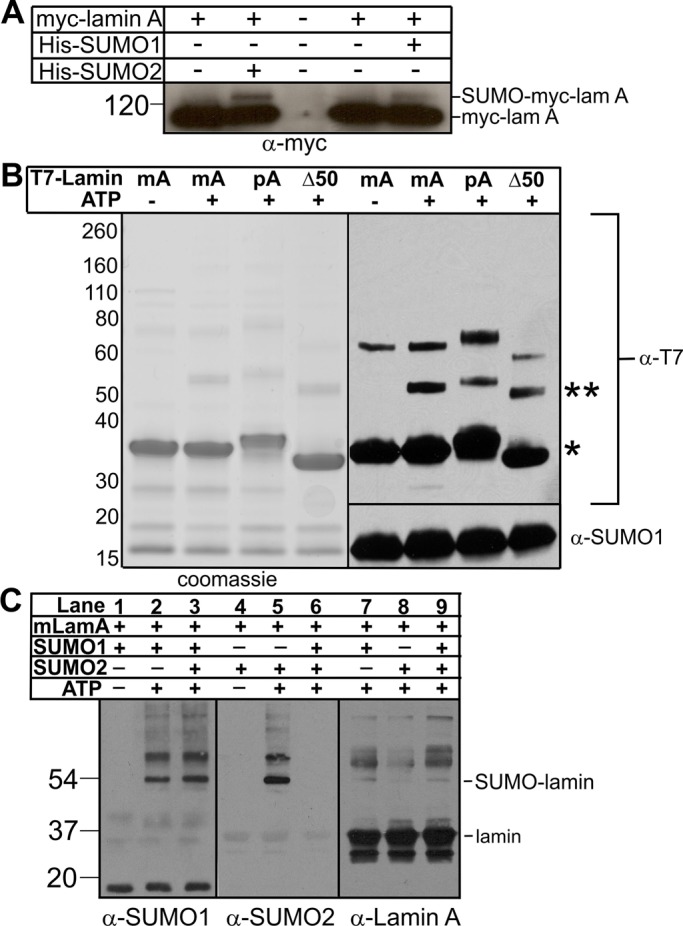
Modification of lamin A by SUMO1 or SUMO2 in vitro and in Cos-7 cells. (A) Exogenous mature myc-lamin A is modified by SUMO1 and SUMO2. Cos-7 cells were transfected to express the indicated constructs for 36 h, then lysed, incubated with Ni2+ beads to recover His-SUMO and lamin A (which has a natural His tag), resolved by SDS–PAGE, and immunoblotted with antibodies to myc (α-myc); n = 3. (B) Recombinant purified mature (mA), precursor (pA), or Δ50 (Δ50) lamin A tail-domain polypeptides were incubated with SUMO1, E1, and E2 with or without ATP for 3 h at 30°C. Reactions were resolved by SDS–PAGE in duplicate and either stained with Coomassie blue or immunoblotted first with antibodies to the T7 tag (α-T7) and then stripped and reprobed with antibodies to SUMO1 (α-SUMO1); n = 3. Unmodified lamin A tails are marked by an asterisk and SUMO1-modified lamin A tails by a double asterisk. (C) Mature lamin A tails were incubated with either SUMO1 or SUMO2 (or both) along with E1, E2, and ATP for 1 h at 30°C and then immunoblotted with antibodies specific for SUMO1 (α-SUMO1), SUMO2 (α-SUMO2), or lamin A tail (α-Lamin A); n = 3.
In vitro SUMOylation of purified lamin A tails
To specifically investigate lamin A tail SUMOylation, we incubated purified recombinant lamin A tail polypeptides (wild-type mature tail [mA] residues 394–646, wild-type precursor tail [pA] residues 394–664, and the HGPS-causing 50-residue-deleted precursor tail [Δ50]) with recombinant purified SUMO E1 activating enzyme, SUMO E2 conjugating enzyme, and SUMO1, with or without ATP. Reactions were stopped by adding SDS-sample buffer, resolved by SDS–PAGE, and either visualized by Coomassie blue or immunoblotted with antibodies specific for SUMO1 or the T7 tag on lamin tails (Figure 1B). Unmodified lamin A tails migrated at ∼34–36 kDa (Figure 1B; single asterisk). All three tails were efficiently modified by SUMO1 (∼52–54 kDa; Figure 1B; double asterisks), suggesting that residues 609–659, which are deleted in HGPS, contain no major SUMOylation sites. We also detected a minor slow-migrating (∼60- to 70-kDa) T7-tagged lamin A band in all reactions, independent of SUMOylation.
To determine whether lamin A tails were preferentially modified by SUMO1 versus SUMO2, we incubated mature wild-type lamin A tails with SUMO1, SUMO2, or both for 1 h, then resolved by SDS–PAGE and immunoblotted with antibodies specific for SUMO1 (Figure 1C, lanes 1–3), SUMO2 (Figure 1C, lanes 4–6), or lamin A (Figure 1C, lanes 7–9). In this competition assay, lamin A tail modification by SUMO1 was qualitatively unaffected by the presence of SUMO2 (Figure 1C, α-SUMO1, lane 2 vs. lane 3), suggesting a preference for SUMO1. Supporting this interpretation, modification of the lamin A tail by SUMO2 (Figure 1C, lane 5) was competed by the presence of SUMO1 (Figure 1C, α-SUMO2, lane 6). Thus, under these conditions, the E2 conjugating enzyme preferentially attached SUMO1 to the lamin A tail. Because our in vitro reactions lacked isopeptidases (enzymes that remove SUMO1 or SUMO2/3 and thereby influence preference indirectly in cells; Zhu et al., 2009), we speculate that lamin A tails might associate with SUMO noncovalently, with potentially higher affinity for SUMO1 than SUMO2, thereby increasing the probability that the SUMOylation machinery chooses SUMO1.
In vitro and in vivo analysis of K-to-R–mutated lamin A tail polypeptides
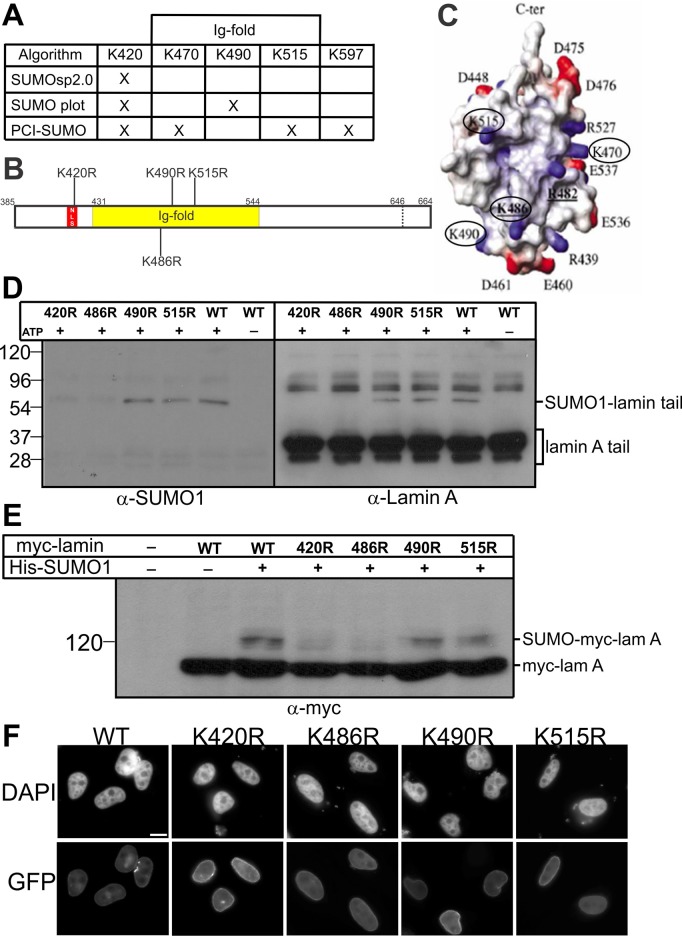
Three different algorithms (SUMOsp2.0, SUMO plot, and PCI-SUMO) were used to predict potential SUMOylation sites in the precursor lamin A tail (Figure 2A). All three predicted SUMOylation at residue K420, with additional sites predicted at K470, K490, K515, or K597 by a single algorithm. K420 is part of the nuclear localization signal (NLS) in lamin A (Dechat et al., 2010a; Dittmer and Misteli, 2011) and is therefore presumably surface exposed. Residues K470, K490, and K515 are exposed on the surface of the lamin A tail Ig-fold domain (Figure 2B, shaded region; Figure 2C shows atomic structure from Krimm et al., 2002), whereas K597 is located outside the Ig-fold in an area of undetermined structure. Further analysis focused on the predicted NLS and Ig-fold sites.
FIGURE 2:
Testing predicted SUMOylation sites in the lamin A tail. (A) SUMOylated lysine (K) sites predicted by each algorithm are marked by X. (B) Schematic diagram showing predicted SUMOylation sites and residue K486 in the lamin A tail; the NLS is red, and the Ig-fold (residues 436–552) is yellow. (C) Structure of the lamin A tail Ig-fold from Krimm et al. (2002); predicted SUMOylation residues K470, K490, K515, and “control” residue K486 are circled. (D) Mature wild-type or K-to-R–mutated lamin A tails were incubated with SUMO1, E1, and E2 with or without ATP for 1 h at 30°C. Reactions were resolved by SDS–PAGE and immunoblotted sequentially for SUMO1 (α-SUMO1) and then lamin A (α-lamin A); n = 3. (E) Mature wild-type or K-to-R–mutated full-length myc-lamin A was transfected into Cos-7 cells alone, or cotransfected with His-SUMO1, for 36 h. Whole-cell lysates were then incubated with Ni2+ beads to pellet His-SUMO1 (and lamin A), resolved by SDS–PAGE, and immunoblotted with antibodies to myc (α-myc); n = 3. (F) Mature wild-type or K-to-R–mutated full-length GFP-lamin A was transfected into HeLa cells for 24 h. Cells were visualized using DAPI or GFP autofluorescence. Scale bar, 10 μm.
We used site-directed mutagenesis to generate recombinant mature lamin A tails (residues 394–646) with single K-to-R mutations at K420, K470, K490, K515, or, as a predicted negative control, K486 (Figure 2B). The K470R polypeptide was expressed very poorly in bacteria and was not studied further. Each purified lamin A tail polypeptide was incubated in vitro in the presence of SUMO1 and ATP. Reactions were resolved by SDS–PAGE and first immunoblotted with antibodies specific for lamin A and then stripped and reprobed with antibodies to SUMO1 (Figure 2D). The K490R and K515R polypeptides were SUMOylated as efficiently as the wild-type lamin A tail (Figure 2D), suggesting that K490 and K515 were not involved in SUMOylation. However, the K420R and K486R polypeptides had consistently reduced or undetectable SUMOylation compared with wild type (Figure 2D), suggesting that K420 and K486 either were SUMO1 modification sites or required for SUMOylation of the lamin A tail. We were surprised by the K486R result, since this was not a predicted site.
To test the potential biological significance of these Lys residues, we transiently coexpressed myc-tagged, full-length mature lamin A (myc-lamin A; wild type or each K-to-R mutant) with His-SUMO1 or the empty His vector in Cos-7 cells. Whole-cell protein lysates were prepared 36 h after transfection, incubated with Ni2+ beads, and pelleted to affinity purify His-SUMO1 and both endogenous and myc-tagged lamin A due to its natural His tag (residues 563–566). Pelleted proteins were resolved by SDS–PAGE and immunoblotted with myc-specific antibodies (Figure 2E). Wild-type myc-lamin A and the K490R and K515R mutants were SUMO1 modified at similar levels in vivo (Figure 2E), consistent with our in vitro results (Figure 2D). Also consistently, the K420R and K486R mutations each substantially reduced lamin A SUMO1 modification in vivo (Figure 2E). Control fluorescence imaging of transfected HeLa cells showed that at low to medium expression levels, green fluorescent protein (GFP)-tagged, full-length mature lamin A polypeptides (wild type or each K-to-R mutant) localized as expected and did not grossly perturb nuclear morphology (Figure 2F). We concluded that the lamin A tail domain is modified both in vitro and in vivo by SUMO1 and that this modification targets at least two residues: consensus residue K420 and totally unexpected residue K486.
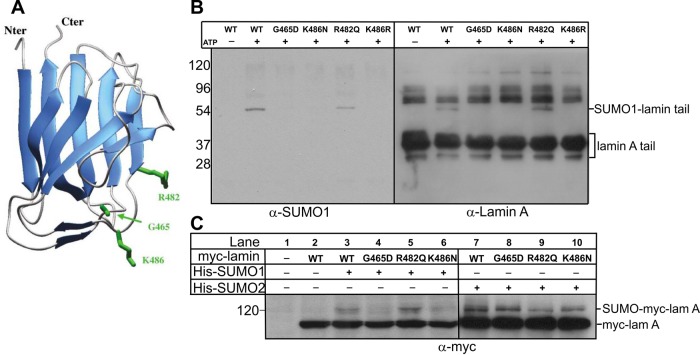
FPLD-causing R482Q, K486N, or G465D mutations and lamin A tail SUMOylation
For most laminopathies, disease-causing missense mutations map throughout the lamin A molecule (Dittmer and Misteli, 2011). Of the 24 different residues mutated in FPLD, 16 are located in the tail (Shackleton et al., 2000; Speckman et al., 2000; Haque et al., 2003; Dittmer and Misteli, 2011; Le Dour et al., 2011), and of these 16, 7 (R439, G465, R471, R482, K486, I497, K515E; see later discussion) map to the Ig-fold surface (Dhe-Paganon et al., 2002; Krimm et al., 2002). Our evidence revealed one (K515) as nonessential for SUMO1 modification and another (K486) as critical for SUMO1 modification. This suggested that FLPD disease might arise, at least in part, by disrupted SUMO1 modification of the lamin A tail. We first tested this hypothesis for FPLD-linked mutations G465D, R482Q, and K486N (Figure 3A) in vitro by SUMO1 modification of recombinant lamin A tail residues 385–646 (wild type, G465D, R482Q, or K486N), resolving by SDS–PAGE and sequential immunoblotting for SUMO1 and then lamin A (Figure 3B). The G465D and K486N mutations each reduced lamin A modification by SUMO1 (Figure 3B), whereas R482Q and wild-type tails were modified to similar extents (Figure 3B).
FIGURE 3:
Effects of FPLD-causing mutations on lamin A modification by SUMO1 or SUMO2 in vitro and in vivo. (A) Structure of the lamin A tail Ig-fold from Krimm et al. (2002) indicating three residues (G465, R482, K486) in which mutations cause FPLD. (B) Mature lamin A tails (wild type or FPLD mutated) were incubated with SUMO1, E1, and E2 with or without ATP for 1 h at 30°C. Reactions were resolved by SDS–PAGE and immunoblotted sequentially for SUMO1 (α-SUMO1) or lamin A (α-lamin A); n = 3. (C) Mature myc-lamin A (wild type or FPLD mutated) was cotransfected into Cos-7 cells with His-SUMO1 or His-SUMO2 (or neither) for 36 h. Whole-cell lysates were then incubated with Ni2+ beads to pellet His-SUMO1 (and lamin A), resolved by SDS–PAGE, and immunoblotted with antibodies to myc (α-myc); n = 3.
To determine whether these disease-causing mutations also affected lamin A SUMOylation in cells, we cotransfected Cos-7 cells with myc-lamin A (wild type, G465D, R482Q, or K486N) plus either His-SUMO1 or His-SUMO2. Consistent with the biochemical results (Figure 3B), myc-lamin A bearing either G465D or K486N reduced modification by SUMO1 in cells relative to wild-type myc-lamin A (Figure 3C, lanes 4 and 6 vs. lane 3). Also consistently, the R482Q mutation did not reduce SUMO1 modification in cells (Figure 3C, lane 5 vs. lane 3). SUMOylation by SUMO2 in vivo was not grossly affected by these FPLD-causing mutations, with one exception: R482Q appeared to slightly reduce modification by SUMO2 (Figure 3C, lanes 7–10).
SUMOylation-recognition surface motif in the lamin A tail
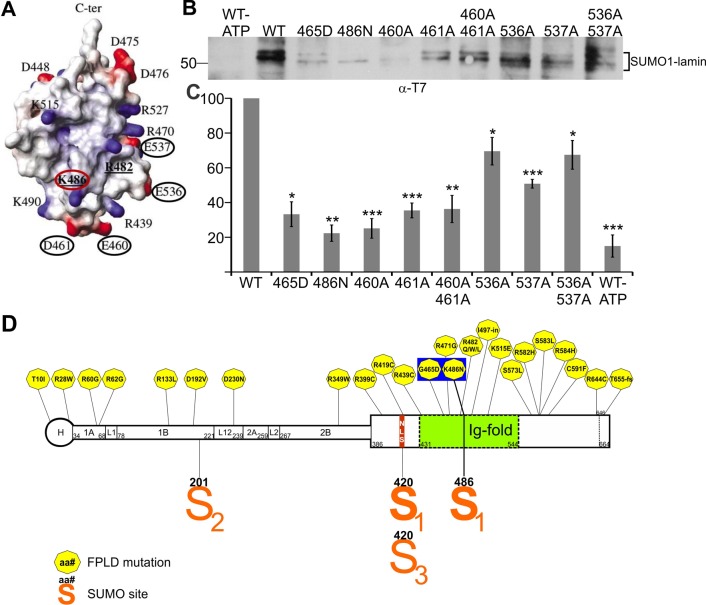
The foregoing experiments identified K486 as either SUMOylated or required for SUMOylation of the lamin A tail. However, this residue did not match any known SUMOylation consensus motif, all of which have negatively charged (acidic) residues near the modified lysine. To solve this conundrum, we considered whether acidic residues were provided by polypeptide folding. Indeed, four acidic residues are surface exposed near K486 in the Ig-fold structure as shown in Figure 4A: E460 and D461 (directly below K486), and E536 and E537 (close to K486 but on a different side). We hypothesized that one or more of these acidic residues provide the SUMOylation-recognition motif for K486.
FIGURE 4:
Effects of alanine substitutions at acidic residues near K486. (A) Structure of the lamin A tail Ig-fold from Krimm et al. (2002) indicating K486 (red circle) and acidic residues E460, D461, E536, and E537 (black circles) tested for potential relevance to lamin A tail recognition by the E2 enzyme. (B) Mature lamin A tails (wild type or K-to-A substituted, 0.3 μg/μl) were incubated with SUMO1, E1, and E2 with or without ATP for 1 h at 30°C. Reactions were resolved by SDS–PAGE and immunoblotted for the T7 tag. (C) Quantification of B by densitometry of SUMOylated lamin bands (bracket) relative to the wild-type lamin A tail (n ≥ 3; bars indicate SEM). Differences were significant as determined by Student’s t test relative to wild-type lamin A tail: *p < 0.05, **p < 0.01, ***p < 0.005. (D) Schematic diagram of the lamin A molecule showing locations of FPLD-causing mutations (hexagons) and residues modified by SUMO1 (S1), SUMO2 (S2), or SUMO3 (S3). Residues G465 and K486, noted in the text as located on the “bottom front” side of the Ig-fold, are highlighted.
To test this model, we analyzed in vitro SUMOylation of T7-tagged lamin A tails (residues 385–646) bearing a single Ala substitution at E460, D461, E536, or E537 or the double mutation E460A/D461A or E536A/E537A. For these experiments, reactions were resolved by SDS–PAGE using 3-(N-morpholino)propanesulfonic acid buffer (instead of 2-(N-morpholino)ethanesulfonic acid buffer, as in Figure 1B) to improve band resolution, and blots were probed with more sensitive antibodies to the T7 tag on lamin A (not lamin A antibodies) to ensure equal recognition of mutated lamin tail polypeptides. Results were quantified by densitometry as a percentage of the SUMOylated wild-type T7-lamin A signal (Figure 4C). This analysis revealed at least three ∼50.2- to 54.5-kDa (presumably singly modified) lamin-SUMO1 bands; we speculate that SDS–PAGE migration might be slightly different, depending on which site is modified. The E460A, D461A, and double E460A/D461A mutations each reduced lamin A modification by SUMO1 by 65–80%, similar to the effect of G465D or K486N (Figure 4C; n ≥ 3). This result supported the hypothesis that E460 and D461 provide acidic-residue support for lamin A recognition by the SUMOylation machinery. The other tested mutations (E536A, E537A, E536A/E537A) reduced SUMO1 modification by 30–50% (Figure 4C; n ≥ 3), suggesting that these acidic residues are less important than E460 and D461 but nevertheless contribute, either to E2 enzyme recognition of lamin A or to the speculative noncovalent association of lamin A with SUMO1.
DISCUSSION
We discovered that the lamin A tail domain is modified, both in vitro and in Cos-7 cells, by SUMO1. This modification, as well as its locations in the lamin molecule (NLS and Ig-fold domain of the lamin A tail) and disease implications, are all distinctly different from the previously reported SUMO2 modification of the lamin A coiled-coil “rod” domain (Zhang and Sarge, 2008). SUMO1 modification of the lamin A tail targets at least two residues: highly predicted residue K420 and totally unexpected residue K486. Of importance, lamin C residues 1–566 are identical to lamin A, suggesting that lamin C might also be modified by SUMO1.
Like other SUMO targets (Gareau and Lima, 2010; Wilkinson and Henley, 2010), SUMO1 modification of lamin A/C is likely to be dynamic. For example, residue 420 can alternatively be modified by SUMO3 (Galisson et al., 2011) or ubiquitin (Kim et al., 2011a) and is surrounded by known phosphorylation sites (Malik et al., 2009; Olsen et al., 2010). Residue 486 has one known alternative modification (ubiquitin; Kim et al., 2011a). SUMOylation at this site might also be dynamically regulated by phosphorylation of nearby surface-exposed Ig-fold residues (Olsen et al., 2010; Kim et al., 2011a; Simon and Wilson, in press).
We propose that SUMO1 modification of K420 within the NLS might inhibit lamin A/C binding to partners such as cyclin D3 or core histones that require an unmodified NLS (Taniura et al., 1995; Zastrow et al., 2004; Mariappan et al., 2007). We further propose that SUMO1 modification of K420 might inhibit lamin A/C binding to α-importin as a potential mechanism for controlling the assembly of A-type lamin filaments after mitosis (Adam et al., 2008). Alternative modification of K420 by SUMO3 in HEK293 cells (Galisson et al., 2011) adds further interest to this site. We found that lamin A tails were modified in vitro by SUMO2 in the absence of SUMO1 but at much lower levels than by SUMO1. Thus, given a choice, the SUMOylation machinery preferentially attached SUMO1 to the lamin A tail in vitro. The mechanism and consequences of this preference for SUMO1 are unknown and will be important to determine in future.
We focused on residue K486, which was critical for SUMO1 modification but located in a region that lacked a canonical linear SUMOylation consensus motif. Our findings support the hypothesis that negatively charged residues required for SUMO-E2 enzyme recognition are provided by the three-dimensional conformation of the Ig-fold domain. SUMOylation machinery recognition and modification of K486 can be explained by a surface consensus “patch” formed by acidic residues E460 and D461 (directly “beneath” K486), each of which was critical for SUMO1 modification. Two other acidic residues, E536 and E537, may provide backup recognition sites since single and double E536A and E537A mutations caused mild defects. To our knowledge this is the first evidence of a “conformational” consensus site for SUMOylation.
The critical role of K486 in SUMO1 modification of the lamin A tail is particularly interesting since the K486N mutation causes FPLD (Lloyd et al., 2002). We found that K486N and the nearby FPLD-causing mutation G465D both significantly decreased lamin A tail modification by SUMO1 in vitro and in cells. Of importance, neither mutation affected modification of lamin A by SUMO2 in vivo. Another FPLD-causing mutation in the lamin A/C Ig-fold, R482Q, did not significantly affect modification by SUMO1 but showed a slight decrease in SUMO2 modification in vivo. These findings suggested three conclusions: 1) since R482 is not involved in SUMO1 modification of lamin A/C, it might cause disease by a different mechanism (potentially involving SUMO2), 2) K486 is either directly modified by SUMO1 or required for this modification, and 3) G465 is required for lamin A tail recognition by the SUMOylation machinery.
The molecular mechanisms of FPLD are unknown. One study showed that the FPLD-causing G465D, R482W, and K486N mutations each weakly decreased lamin A binding to SREBP1 (residues 1–463; full-length transcription factor) by 25–40% in vitro (Lloyd et al., 2002). This weakened binding suggested, but did not prove, that SREBP1 contacts this region of the Ig-fold. For example, in a separate study, R482W-mutated and wild-type lamin A tails bound similarly well to a smaller SREBP1 fragment (residues 227–487) in vitro (Duband-Goulet et al., 2011). Most previous studies of FPLD patient cells focused on mutations at residue R482 (Broers et al., 2005; Boguslavsky et al., 2006; Bidault et al., 2011). Whether G465D, K486N, or FPLD-causing mutations elsewhere in lamin A/C (Figure 4D) cause disease by similar or distinct mechanisms is an important question for future work. However, our findings suggest that residues located on the “bottom front” of the Ig-fold (G465, K486, E460, D461; Figure 4D) significantly disrupt SUMO1 modification of the lamin tail. On the other hand, our tested mutations located in other regions of the Ig-fold (R482, K490, K515, E536, E537; Figure 4A), did not significantly affect SUMO1 modification of the lamin tail. We therefore propose that in patients with mutations on the “bottom front” of the Ig-fold, FPLD might arise from defective SUMO1 modification, whereas in patients with mutations elsewhere in the Ig-fold or other domains, FPLD arises by a different mechanism.
These findings are significant because they suggest two mechanisms by which SUMO1 might normally contribute to lamin A/C filament regulation in living cells. First, SUMO1 modification at K420 is predicted to block partners that require the unmodified NLS, whereas K486 modification is predicted to block partners (potentially including SREBP1) that require the unmodified Ig-fold (Figure 5). Whether SUMO1 modification of lamin A/C affects the binding or transcriptional activity of SREBP1 is unknown. Second, SUMO1 modification has the potential to recruit or stabilize novel FPLD-relevant partner(s) bearing the SIM motif. In FPLD patients with G465D- or K486N-mutated lamin A/C, we predict both types of partner—SREBP1 and/or other Ig-fold-binding proteins (Zastrow et al., 2004), and hypothetical SIM-containing partner(s)—might be misregulated (Figure 5). These models suggest new ways to think about FPLD disease and explore the differential regulation of lamin A/C function in specific tissues.
FIGURE 5:
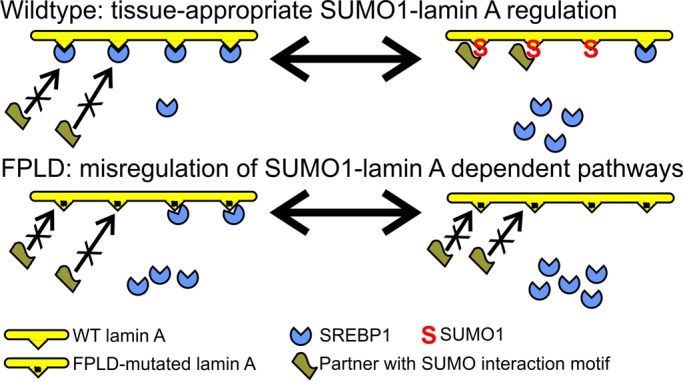
Proposed models for SUMO1 regulation of lamin A tails. Proposed mechanisms by which SUMO1 modification of lamin A tails might regulate 1) tissue-appropriate binding or release of SREBP1 (or other partners) and 2) recruitment of hypothetical SIM-containing partner(s) relevant to adipose tissue. We propose that both types of interaction are misregulated in a subset of FPLD patients.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis
Human lamin A tail constructs in pET23b (Novagen, Rockland, MA), which places a His6 tag at the C-terminus and a T7 tag at the N-terminus, for the wild-type mature lamin A tail (residues 394–646) and pre–lamin A tail (residues 394–664) were described previously (Simon et al., 2010). Wild-type FLAG-lamin A mutants R482Q and K486N in pSVK3 were kindly provided by Howard Worman (Columbia University, New York, NY). Wild-type GFP-lamin A in pEGFP-C1 was kindly provided by Kris Dahl (Carnegie Mellon University, Pittsburgh, PA). Single missense mutations in either full-length wild-type lamin A or the lamin A tail were generated by PCR mutagenesis using the primers shown in Supplemental Table S1.
Myc-tagged full-length lamin A or lamin A tail constructs were generated by cloning each wild-type or mutant lamin A into the pCMV-myc vector (Clontech, Mountain View, CA). The SUMO1 and SUMO2 cDNAs were generated by reverse transcription-PCR (RT-PCR) as described (Tatham et al., 2001) using total HeLa cell RNA as template. SUMO cDNAs were cloned into the pcDNA3.1/His vector (Invitrogen, Carlsbad, CA) for expression in mammalian cells. Ubc9 (E2 enzyme) and cDNAs encoding human His-Uba2 and Aos1 (subunits of the E1 enzyme) were kindly provided by Michael Matunis (Johns Hopkins Bloomberg School of Public Health, Baltimore, MD).
Recombinant lamin tails, SUMO1, SUMO2, E1, and E2 enzymes
Recombinant His-tagged lamin tail peptides and His-tagged SUMO1, SUMO2, and human E1 (subunits Uba2 and Aos1) were each expressed separately in Escherichia coli BL-21 and purified using nickel-nitriloacetic acid (NTA)–agarose per manufacturer instructions (Qiagen Valencia, CA). Lamin tail peptides were stored in buffer (50 mM NaHPO4, pH 8.0, 300 mM NaCl, 100 mM imidazole, 0.5 mM phenylmethylsulfonyl fluoride) at −80°C. Glutathione S-transferase (GST)–tagged E2 was expressed in E. coli BL-21 and purified using glutathione–Sepharose 4B (GE Healthcare Bio-Sciences, Piscataway, NJ), and GST was cleaved using factor Xa per manufacturer’s instructions (GE Healthcare Bio-Sciences).
In vitro SUMOylation assays and immunoblotting
Each purified recombinant lamin A tail polypeptide was incubated with purified recombinant SUMO1 or SUMO2 plus recombinant-purified E1, E2, and 100 mM ATP as described (Desterro et al., 1998). Reactions were stopped by adding SDS-sample buffer, resolved by SDS–PAGE, transferred to nitrocellulose, and probed using antibodies specific for the lamin A tail (NCL-Lamin A, Novocastra; Leica Microsystems, Buffalo Grove, IL; raised against human lamin residues 598–611; 1:2000 dilution), c-Myc (9E10, Santa Cruz Biotechnology, Santa Cruz, CA; 1:5000), T7 tag (69048-3, Novagen; 1:10,000), SUMO1 (either SC-9060, Santa Cruz Biotechnology, 1:2000; or α-SUMO1, Enzo Life Sciences, Plymouth Meeting, PA; 1:4000), or SUMO2 (α-SUMO2; Enzo Life Sciences; 1:4000). Secondary antibodies were horseradish peroxidase–coupled anti-mouse and anti-rabbit (GE Healthcare, Chalfont St Giles, United Kingdom; 1:10,000).
In vivo SUMOylation assay
Cos-7 cells were transfected with myc-tagged full-length mature lamin A (wild type or each indicated mutant) plus His-SUMO1, His-SUMO2, or the empty His vector as control. Cells were lysed 36 h after transfection, and His-tagged proteins (including lamin A, which has a natural His tag) were affinity enriched under denaturing conditions using nickel NTA-agarose beads (Qiagen) as described previously (Hofmann et al., 2009). After addition of SDS-sample buffer, samples were resolved by SDS–PAGE, transferred to nitrocellulose, probed using antibodies specific for c-Myc (9E10, Santa Cruz Biotechnology; 1:5000), and detected using horseradish peroxidase–coupled anti-mouse secondary antibodies (GE Healthcare; 1:10,000).
HeLa transfections and microscopy
Approximately 60,000 HeLa cells were seeded on glass coverslips (Fisher Scientific, Waltham, MA) and transfected with 3 μg of DNA using LT1 transfection reagent (Roche, Indianapolis, IN). At 24 h after transfection, cells were fixed 15 min in 3% formaldehyde, permeabilized for 20 min in phosphate-buffered saline (PBS)/0.2% Triton X-100, and then blocked for 1 h in PBS/3% bovine serum albumin. DNA was stained using 4′,6-diamidino-2-phenylindole (DAPI), and GFP fluorescence was directly visualized using a Nikon Eclipse E600 equipped with a Nikon Plan APO 60×/numerical aperture 1.40 oil objective (Nikon, Melville, NY). Images were acquired with a Q Imagine Retiga Exi 12-bit digital camera using IP Lab software from Scanalytics (Spectra Services, Ontario, NY).
Supplementary Material
Acknowledgments
We gratefully acknowledge Howard Worman and Kris Dahl for lamin constructs, Michael Matunis for E1 and E2 plasmids and insightful discussions, Jason M. Berk for valuable discussions and comments on the manuscript, and research funding from the Department of Defense CDMRP (PC111523 to W.A.H.) and the National Institutes of Health (RO1 048646 to K.L.W.).
Abbreviations used:
- FPLD
familial partial lipodystrophy
- GFP
green fluorescent protein
- GST
glutatione S-transferase
- HGPS
Hutchinson-Gilford progeria syndrome
- LINC
links the nucleoskeleton and cytoskeleton
- SIM
SUMO interaction motif
- SREBP1
sterol response element binding protein 1
- SUMO
small ubiquitin-like modifier
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-07-0527) on December 14, 2012.
REFERENCES
- Adam SA, Sengupta K, Goldman RD. Regulation of nuclear lamin polymerization by importin alpha. J Biol Chem. 2008;283:8462–8468. doi: 10.1074/jbc.M709572200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro JF, et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc Natl Acad Sci USA. 2012;109:7280–7285. doi: 10.1073/pnas.1200425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidault G, Vatier C, Capeau J, Vigouroux C, Bereziat V. LMNA-linked lipodystrophies: from altered fat distribution to cellular alterations. Biochem Soc Trans. 2011;39:1752–1757. doi: 10.1042/BST20110675. [DOI] [PubMed] [Google Scholar]
- Boguslavsky RL, Stewart CL, Worman HJ. Nuclear lamin A inhibits adipocyte differentiation: implications for Dunnigan-type familial partial lipodystrophy. Hum Mol Genet. 2006;15:653–663. doi: 10.1093/hmg/ddi480. [DOI] [PubMed] [Google Scholar]
- Broers JL, Kuijpers HJ, Ostlund C, Worman HJ, Endert J, Ramaekers FC. Both lamin A and lamin C mutations cause lamina instability as well as loss of internal nuclear lamin organization. Exp Cell Res. 2005;304:582–592. doi: 10.1016/j.yexcr.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Clements L, Manilal S, Love DR, Morris GE. Direct interaction between emerin and lamin A. Biochem Biophys Res Commun. 2000;267:709–714. doi: 10.1006/bbrc.1999.2023. [DOI] [PubMed] [Google Scholar]
- Coffinier C, Chang SY, Nobumori C, Tu Y, Farber EA, Toth JI, Fong LG, Young SG. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc Natl Acad Sci USA. 2010;107:5076–5081. doi: 10.1073/pnas.0908790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol Biol Cell. 2011;22:4683–4693. doi: 10.1091/mbc.E11-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan J, Li D, Gonzalez-Quintana J, Morales A, Hershberger RE. Morphological analysis of 13 LMNA variants identified in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:6–14. doi: 10.1161/CIRCGENETICS.109.905422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol. 2010a;2:a000547. doi: 10.1101/cshperspect.a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Gesson K, Foisner R. Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring Harb Symp Quant Biol. 2010b;75:533–543. doi: 10.1101/sqb.2010.75.018. [DOI] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S, Werner ED, Chi YI, Shoelson SE. Structure of the globular tail of nuclear lamin. J Biol Chem. 2002;277:17381–17384. doi: 10.1074/jbc.C200038200. [DOI] [PubMed] [Google Scholar]
- Dittmer TA, Misteli T. The lamin protein family. Genome Biol. 2011;12:222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duband-Goulet I, et al. Subcellular localization of SREBP1 depends on its interaction with the C-terminal region of wild-type and disease related A-type lamins. Exp Cell Res. 2011;317:2800–2813. doi: 10.1016/j.yexcr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutour A, et al. High prevalence of laminopathies among patients with metabolic syndrome. Hum Mol Genet. 2011;20:3779–3786. doi: 10.1093/hmg/ddr294. [DOI] [PubMed] [Google Scholar]
- Eggert M, Radomski N, Linder D, Tripier D, Traub P, Jost E. Identification of novel phosphorylation sites in murine A-type lamins. Eur J Biochem. 1993;213:659–671. doi: 10.1111/j.1432-1033.1993.tb17806.x. [DOI] [PubMed] [Google Scholar]
- Galisson F, Mahrouche L, Courcelles M, Bonneil E, Meloche S, Chelbi-Alix MK, Thibault P. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol Cell Proteomics. 2011;10:M110 004796. doi: 10.1074/mcp.M110.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Gerace L, Huber MD. Nuclear lamina at the crossroads of the cytoplasm and nucleus. J Struct Biol. 2012;177:24–31. doi: 10.1016/j.jsb.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M, Jost E. Functional analysis of phosphorylation sites in human lamin A controlling lamin disassembly, nuclear transport and assembly. Eur J Cell Biol. 1993;62:237–247. [PubMed] [Google Scholar]
- Haque WA, Oral EA, Dietz K, Bowcock AM, Agarwal AK, Garg A. Risk factors for diabetes in familial partial lipodystrophy, Dunnigan variety. Diabetes Care. 2003;26:1350–1355. doi: 10.2337/diacare.26.5.1350. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hofmann WA, Arduini A, Nicol SM, Camacho CJ, Lessard JL, Fuller-Pace FV, de Lanerolle P. SUMOylation of nuclear actin. J Cell Biol. 2009;186:193–200. doi: 10.1083/jcb.200905016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011a;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011b;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm I, et al. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure. 2002;10:811–823. doi: 10.1016/s0969-2126(02)00777-3. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dour, et al. A homozygous mutation of prelamin-A preventing its farnesylation and maturation leads to a severe lipodystrophic phenotype: new insights into the pathogenicity of nonfarnesylated prelamin-A. J Clin Endocrinol Metab. 2011;96:E856–862. doi: 10.1210/jc.2010-2234. [DOI] [PubMed] [Google Scholar]
- Lloyd DJ, Trembath RC, Shackleton S. A novel interaction between lamin A and SREBP1: implications for partial lipodystrophy and other laminopathies. Hum Mol Genet. 2002;11:769–777. doi: 10.1093/hmg/11.7.769. [DOI] [PubMed] [Google Scholar]
- Malik R, Lenobel R, Santamaria A, Ries A, Nigg EA, Korner R. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J Proteome Res. 2009;8:4553–4563. doi: 10.1021/pr9003773. [DOI] [PubMed] [Google Scholar]
- Mariappan I, Gurung R, Thanumalayan S, Parnaik VK. Identification of cyclin D3 as a new interaction partner of lamin A/C. Biochem Biophys Res Commun. 2007;355:981–985. doi: 10.1016/j.bbrc.2007.02.060. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Pan C, Olsen JV, Daub H, Mann M. Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol Cell Proteomics. 2009;8:2796–2808. doi: 10.1074/mcp.M900285-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK. Detection of proteins sumoylated in vivo and in vitro. Methods Mol Biol. 2009;590:265–277. doi: 10.1007/978-1-60327-378-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic “network of networks.”. Nat Rev Mol Cell Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- Simon DN, Wilson KL. Partners and post-translational modifications of nuclear lamins. Chromosoma. 2013 doi: 10.1007/s00412-013-0399-8. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Zastrow MS, Wilson KL. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus. 2010;1:264–272. doi: 10.4161/nucl.1.3.11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckman RA, Garg A, Du F, Bennett L, Veile R, Arioglu E, Taylor SI, Lovett M, Bowcock AM. Mutational and haplotype analyses of families with familial partial lipodystrophy (Dunnigan variety) reveal recurrent missense mutations in the globular C-terminal domain of lamin A/C. Am J Hum Genet. 2000;66:1192–1198. doi: 10.1086/302836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori Y, Tamura Y, Kataoka Y, Cui Y, Seo S, Kanazawa T, Kurokawa K, Yamada H. Differential expression of nuclear lamin, the major component of nuclear lamina, during neurogenesis in two germinal regions of adult rat brain. Eur J Neurosci. 2007;25:1653–1662. doi: 10.1111/j.1460-9568.2007.05450.x. [DOI] [PubMed] [Google Scholar]
- Taniura H, Glass C, Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol. 2010;2:a000620. doi: 10.1101/cshperspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL, Berk JM. The nuclear envelope at a glance. J Cell Sci. 2010;123:1973–1978. doi: 10.1242/jcs.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL, Foisner R. Lamin-binding proteins. Cold Spring Harb Perspect Biol. 2010;2:a000554. doi: 10.1101/cshperspect.a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ. Nuclear lamins and laminopathies. J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zastrow MS, Flaherty DB, Benian GM, Wilson KL. Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J Cell Sci. 2006;119:239–249. doi: 10.1242/jcs.02728. [DOI] [PubMed] [Google Scholar]
- Zastrow MS, Vlcek S, Wilson KL. Proteins that bind A-type lamins: integrating isolated clues. J Cell Sci. 2004;117:979–987. doi: 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Sarge KD. Sumoylation regulates lamin A function and is lost in lamin A mutants associated with familial cardiomyopathies. J Cell Biol. 2008;182:35–39. doi: 10.1083/jcb.200712124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Goeres J, Sixt KM, Bekes M, Zhang XD, Salvesen GS, Matunis MJ. Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol Cell. 2009;33:570–580. doi: 10.1016/j.molcel.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.