Protein kinase D (PKD) is known to be involved in the fission of transport carriers at the Golgi complex. This study demonstrates that PKD is important for the cleavage of interstack Golgi connections in G2 of the cell cycle and thus entry of cells into mitosis.
Abstract
Before entering mitosis, the stacks of the Golgi cisternae are separated from each other, and inhibiting this process delays entry of mammalian cells into mitosis. Protein kinase D (PKD) is known to be involved in Golgi-to–cell surface transport by controlling the biogenesis of specific transport carriers. Here we show that depletion of PKD1 and PKD2 proteins from HeLa cells by small interfering RNA leads to the accumulation of cells in the G2 phase of the cell cycle and prevents cells from entering mitosis. We further provide evidence that inhibition of PKD blocks mitotic Raf-1 and mitogen-activated protein kinase kinase (MEK) activation, and, as a consequence, mitotic Golgi fragmentation, which could be rescued by expression of active MEK1. Finally, Golgi fluorescence recovery after photobleaching analyses demonstrate that PKD is crucial for the cleavage of the noncompact zones of Golgi membranes in G2 phase. Our findings suggest that PKD controls interstack Golgi connections in a Raf-1/MEK1–dependent manner, a process required for entry of the cells into mitosis.
INTRODUCTION
The Golgi ribbon is a continuous membranous system localized to the perinuclear area and has an essential role in lipid biosynthesis, protein modification, and secretory trafficking. The ribbon is composed of individual stacks of flattened cisternae that are laterally connected by membranous tubular bridges known as noncompact zones. During cell division, the Golgi complex disperses into vesicles to allow partitioning between daughter cells. The first step consists of the fragmentation of the noncompact zones of the Golgi ribbon. This occurs in the G2 phase of the cell cycle and results in the formation of isolated Golgi stacks. At the onset of mitosis, these isolated Golgi stacks are converted into scattered tubuloreticular elements and then further fragmented and dispersed throughout the cytoplasm, appearing as the Golgi haze. Golgi fragmentation is now known to be required for entry of cells into mitosis, suggesting a direct role for Golgi organelle architecture in G2/M checkpoint control (reviewed in Colanzi and Corda, 2007). Indeed, increasing evidence indicates that correct segregation of the Golgi complex is monitored by a “Golgi mitotic checkpoint.”
In recent years, several molecules involved in initial Golgi ribbon unlinking and further unstacking and vesiculation of Golgi membranes during mitosis have been identified. For example, Golgi fragmentation is inhibited via the functional block of the proteins BARS, Polo-like kinase, and Grasp65, resulting in cell cycle arrest at the G2 stage (Sütterlin et al., 2001, 2005; Colanzi et al., 2003b; Preisinger et al., 2005; Feinstein and Linstedt, 2007; Duran et al., 2008). BARS/CtBP3 is a protein with a dual role as a transcriptional corepressor in the nucleus and an involvement in vesicle trafficking by inducing fission of Golgi tubules (Hidalgo et al., 2004; Colanzi et al., 2007). How BARS fulfils its mitotic function is unresolved, but it has been proposed that BARS is regulated by phosphorylation to recruit protein cofactors involved in mitotic Golgi ribbon fission. The functions of Grasp65 and the structurally related Grasp55 protein are better understood. These proteins are important for the formation and/or maintenance of the tubules connecting the stacks within the Golgi ribbon. Mitotic phosphorylation of Grasp65 disrupted Grasp65 oligomerization, which is believed to be necessary for Golgi membrane linking (Wang et al., 2003, 2005). In mitosis, Grasp65 is phosphorylated on multiple sites by Cdk1 and Plk1 kinases (Lin et al., 2000; Wang et al., 2003; Preisinger et al., 2005; Sütterlin et al., 2005; Yoshimura et al., 2005). Of interest, expression of the wild-type Grasp65 C-terminus but not the phosphorylation-defective mutant caused a delay in mitotic entry, demonstrating the importance of phosphorylation in the regulation of Grasp65 function (Preisinger et al., 2005; Yoshimura et al., 2005). Similarly, multisite phosphorylation of Grasp55 downstream of mitogen-activated protein kinase kinase (MEK)–Erk appears to be required for Golgi fragmentation and mitotic progression (Duran et al., 2008; Feinstein and Linstedt, 2008; Xiang and Wang, 2010).
The best-characterized kinase cascade involved in mitotic Golgi fragmentation is the Raf-MEK-Erk pathway, playing a role before prophase in late G2. Initial in vitro reconstitution assays revealed a role for MEK1 but independence of Erk1/2 (Acharya et al., 1998). In line with this study was the observation that the first step of Golgi fragmentation, namely the breakup of perinuclear Golgi stacks into punctate structures, was mainly regulated by MEK, whereas the subsequent extensive dispersal of these structures was controlled by Cdk1/Cdc2 (Lowe et al., 1998b; Kano et al., 2000). Later MEK was shown to localize to Golgi membranes in mitotic cells (Colanzi et al., 2003b). Accordingly, inhibition of MEK1 by RNA interference or by using the MEK1/2-specific inhibitor U0126 delayed the passage of synchronized HeLa cells into M phase (Feinstein and Linstedt, 2007). Of importance, Raf kinases were shown to be upstream of MEK in mitotic Golgi fragmentation (Colanzi et al., 2003b).
Protein kinase D (PKD) is a family of serine/threonine–specific protein kinases comprising three structurally related members: PKD1, PKD2, and PKD3. PKD is recruited to the trans-Golgi network (TGN) by direct interaction of its cysteine-rich regions with diacylglycerol (DAG; Baron and Malhotra, 2002) and the GTPase Arf1 (Pusapati et al., 2010) and activated by PKCη-mediated phosphorylation (Diaz Anel and Malhotra, 2005). At the TGN, PKD is known to be involved in the fission of transport carriers en route to the cell surface (Liljedahl et al., 2001). It is intriguing that overexpression of constitutively active PKD is known to induce hypervesiculation of the Golgi, whereas kinase-dead PKD blocks vesicle fission, leading to the appearance of long, tubular structures (Bossard et al., 2007). Moreover, there is evidence that PKD activity is crucial for nocodazole-induced Golgi complex dispersal (Fuchs et al., 2009), pointing to a more general role of PKD in the maintenance of Golgi structure. In this study we provide evidence that PKD is required for mitotic entry of HeLa cells by driving the cleavage of Golgi interstack connections in the late G2 phase of the cell cycle. We further show that PKD signals through the Raf-1/MEK1 pathway to induce mitotic fragmentation of Golgi membranes.
RESULTS
Depletion of PKD induces an accumulation of HeLa cells in G2 stage
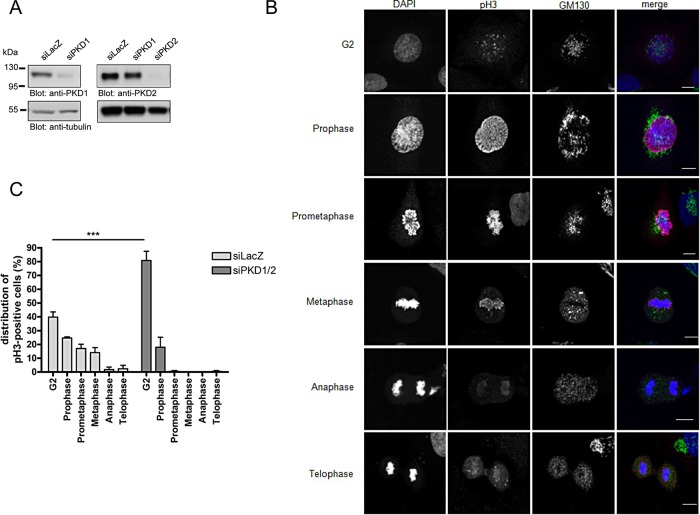
To analyze whether PKD is involved in cell cycle progression, we depleted PKD1 and PKD2 in asynchronous HeLa cells using specific small interfering RNAs (siRNAs; Peterburs et al., 2009). Western blot analyses revealed that ∼90% of the PKD1 and PKD2 proteins were removed, proving the efficiency and specificity of the siRNAs (Figure 1A). We then determined the mitotic index of HeLa cells lacking PKD1 and PKD2. We performed this as follows: HeLa cells were transfected with a control siRNA (siLacZ) or PKD1- plus PKD2-specific siRNAs and seeded on glass coverslips 48 h after transfection. Cells were fixed 24 h later and stained with an antibody specific for histone 3 phosphorylated at Ser-10 (pH3) to monitor the mitotic status of the transfected cells (Colanzi et al., 2007). Phosphorylation of histone 3 at Ser-10 starts at pericentromeric chromatin in late G2, is completed in prophase, and decreases upon exit from mitosis (Hendzel et al., 1997). Figure 1B shows the localization pattern of pH3 and the Golgi morphology during G2 and mitosis, which is in agreement with previous observations (Colanzi et al., 2007). In control cells, ∼11.3% were positive for pH3, whereas upon PKD1/2 depletion, slightly more cells were pH3 positive (12.3%), pointing to a marginal enrichment of cells in G2 and/or M phases of the cell cycle upon PKD1/2 knockdown in unsynchronized cells (unpublished data). However, detailed analysis and classification of the pH3-positive cells based on the localization pattern as shown in Figure 1B revealed that PKD1/2 depletion caused a dramatic accumulation of cells in G2 stage (87.7% in PKD-depleted cells vs. 33.2% in control cells; Figure 1C). Most of the remaining PKD1/2-depleted cells (8.7%) were in prophase, and later mitotic phases were hardly present, whereas all mitotic stages, including prophase, prometaphase, metaphase, anaphase, and telophase, were found in control cells (Figure 1C). To exclude off-targets effects and prove specificity of the siRNAs used, we performed this experiment with independent PKD1- and PKD2-specific siRNAs. Indeed, depletion of PKD1 and 2 using different siRNA tools also led to a dramatic accumulation of cells in G2 (60.8% in PKD-depleted cells vs. 32.4% in control cells; Supplemental Figure S1), thus confirming our previous results.
FIGURE 1:
Depletion of PKD induces an accumulation of HeLa cells in G2. HeLa cells were transfected with siLacZ as a control or PKD1- and PKD2-specific siRNAs and cultured for 72 h. (A) Cells were lysed, and expression of PKD1 and PKD2 was analyzed by Western blot analysis using specific antibodies. Detection of tubulin served as a loading control. (B, C) Cells were fixed and stained with a GM130-specific antibody to visualize the Golgi, pHistone 3 (Ser-10)–specific antibody, and DAPI to visualize the nucleus. (B) Localization pattern of pH3 and GM130 (Golgi marker) during G2 and mitosis in control cells. Scale bar, 10 μm. (C) pH3-positive cells were further classified into the different cell cycle phases based on the specific pH3 localization pattern. Shown is the mean ± SEM of three independent experiments. Statistical analysis was performed using two-way analysis of variance (ANOVA) followed by a Bonferroni post test. ***p < 0.001.
Depletion of PKD induces a delay in G2/M transition
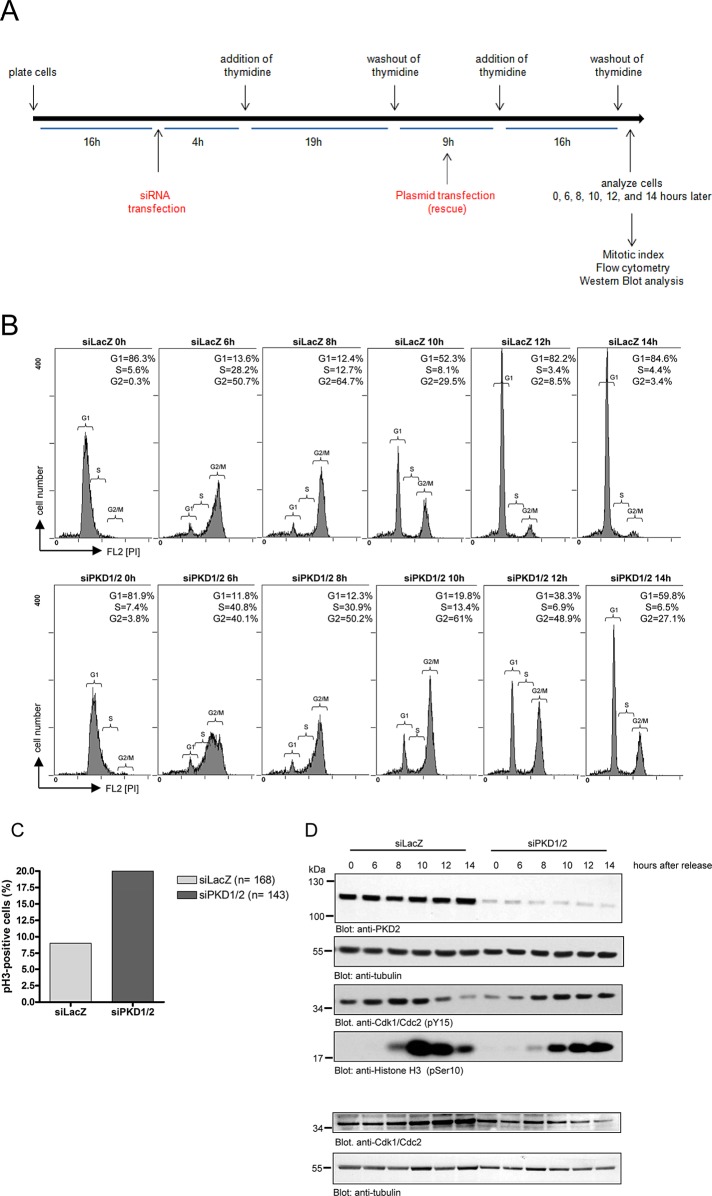
To further ascertain the involvement of PKD in mitotic entry and progression, we synchronized HeLa cells at the G1/S border using a double-thymidine block (Ma and Poon, 2011) according to the scheme shown in Figure 2A. In brief, HeLa cells were transfected with siLacZ or siPKD1 plus siPKD2 and cultured for 16 h, followed by incubation in growth medium containing thymidine for 19 h. Afterward, cells were released from the thymidine block (washout) and refed with growth medium for 9 h. Subsequently cells were subjected to the second thymidine block for an additional 16 h. After the second washout, cells were harvested at distinct time points (0, 6, 8, 10, 12, and 14 h), and cell cycle progression in siLacZ- and siPKD1/2-transfected cells was monitored by flow cytometry using propidium iodide staining (Figure 2B). We found that progression through S phase and into G2 phase was not altered in PKD1/2-depleted cells (Figure 2B, bottom). However, control cells progressed through G2/M phase much faster than did PKD1/2-depleted cells (Figure 2B, top). This is obvious from the fact that most of the PKD1/2-depleted cells were still in G2/M phase 10 and 12 h after thymidine release (61 and 48.9% in PKD1/2-depleted cells vs. 29.5 and 8.5% in control cells). Furthermore, whereas control cells finished G2/M phase 14 h after release, 27% of PKD1/2-depleted cells were still in G2/M phase. In a parallel approach, we analyzed the mitotic index of these cells using pH3 staining. In line with our previous results, we found that the amount of pH3-positive cells was dramatically increased in PKD1/2-depleted cells compared with control cells 14 h after release (20% in siPKD1/2 vs. 9% in siLacZ; Figure 2C). Thus depletion of PKD1/2 delayed passage through the G2 and M phases of the cell cycle after a thymidine block.
FIGURE 2:
Depletion of PKD induces a delay in mitotic entry. HeLa cells transfected with a control siRNA (siLacZ) or PKD1- and PKD2-specific siRNAs were synchronized at the G1/S border by a double-thymidine treatment and released for 0, 6, 8, 10, 12, and 14 h. (A) Scheme showing the experimental design. (B) Cells were fixed, stained with propidium iodide to visualize G1, S, and G2/M phases, and analyzed by flow cytometry. The percentage of cells in G1, S, and G2/M phases is indicated. Brackets indicate G1, S, and G2/M phases. (C) At 14 h after release, cells were fixed and stained with a pHistone 3 (Ser-10)–specific antibody and DRAQ5 to visualize the nucleus. The percentage of pH3-positive cells was quantified. n, total number of cells analyzed. Shown is a representative experiment (n = 2). (D) Cells were lysed, and phosphorylation of Cdk1/Cdc2 at tyrosine 15 and histone H3 at serine 10 was detected by Western blot analysis using specific antibodies. Detection of tubulin served as a loading control, and efficient knockdown was verified for PKD2. Lysates were also analyzed using a Cdk1/Cdc2–specific antibody to ensure that total protein levels were unchanged. Detection of tubulin verified equal loading. Shown is a representative experiment (n = 3).
To corroborate this finding, we further analyzed the cell cycle–specific phosphorylation of Cdk1/Cdc2 in synchronized siLacZ- or siPKD1/2-transfected HeLa cells. Cdk1/Cdc2 is a cell cycle–dependent kinase that is activated upon dephosphorylation of a specific tyrosine residue (Tyr-15) at the G2/M boundary, controlling mitotic entry and progression (Timofeev et al., 2010). In control cells, Tyr-15 phosphorylation of Cdk1/Cdc2 strongly decreased 10 h after release, indicative of mitotic progression (Figure 2D). Of note, in cells lacking PKD1/2, Cdk1 was still phosphorylated 12 and 14 h after release, pointing to a delay in mitotic entry. This is corroborated by the phosphorylation pattern of histone 3 (Figure 2D). In control cells, histone 3 phosphorylation was first detectable 8 h after release, peaked at 10 h, and decreased 12 and 14 h after release. However, PKD1/2-depleted cells displayed a weaker and delayed histone 3 phosphorylation compared with control cells. Of note, histone 3 phosphorylation was still at a high level 14 h after release in PKD-depleted cells, which is in line with a delay in mitotic progression. To exclude that PKD-depleted cells show a delay in mitotic progression because of induction of apoptotic events, we analyzed HeLa cells for active caspase-3 expression. However, we could not detect cleaved caspase-3 in cells transfected with siLacZ or siPKD1/2 (Supplemental Figure S2).
PKD is upstream of the Raf–MEK1 signaling pathway in mitotic Golgi fragmentation
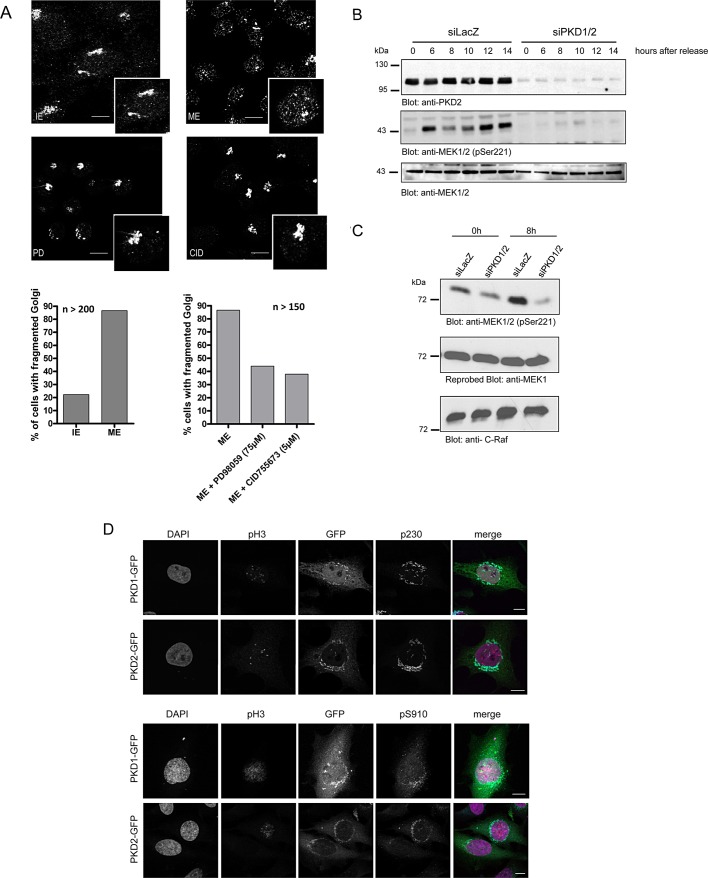
To ensure correct partitioning between daughter cells, Golgi membranes have to be dispersed during a two-step fragmentation process resulting in the complete vesiculation of the Golgi ribbon into small vesicles (Colanzi et al., 2003a). The first step of this fragmentation process occurs in G2 and is characterized by the cleavage of tubular bridges connecting adjacent stacks. The inhibition of proteins involved in ribbon cleavage leads to an arrest of the cells in G2 (Colanzi and Corda, 2007). To determine the impact of specific kinases in Golgi dispersal during mitosis, we used a “semi-intact assay” (Acharya et al., 1998). In this assay Golgi membrane organization of permeabilized interphase cells is monitored upon incubation with cellular mitotic extract. To analyze whether inhibition of PKD affects mitotic Golgi fragmentation, we applied mitotic and interphase extracts of HeLa cells to permeabilized HeLa cells and an ATP regeneration system at 32°C for 60 min. The samples were visualized by fluorescence microscopy using an anti–mannosidase II antibody. Microscopy images show that in 80% of the cells the interphase Golgi stacks stayed intact when incubated with interphase extract. However, upon incubation with mitotic extracts, Golgi membranes became fragmented in almost 90% of permeabilized cells (Figure 3A). It was demonstrated that the cytosolic protein mitogen-activated protein kinase kinase 1 (MEK1) is crucial for Golgi complex breakup, since the MEK1 inhibitor PD 98059 blocked fragmentation of Golgi stacks (Acharya et al., 1998). We therefore treated permeabilized cells with PD 98059 or the specific PKD inhibitor CID 755673 (Sharlow et al., 2008). In line with previous studies, inhibition of MEK1 strongly reduced the amount of cells harboring a fragmented Golgi down to 44% (Figure 3A). PKD inhibition was equally potent as inhibition by PD 98059, with ∼60% of permeabilized cells losing their ability to fragment Golgi membranes (Figure 3A). These data show that PKD is required for Golgi complex breakdown in HeLa cells during mitosis.
FIGURE 3:
PKD is upstream of the Raf–MEK1 signaling pathway in mitotic Golgi fragmentation. (A) HeLa cells were grown on coverslips and synchronized with thymidine for 8 h at 37°C. Cells were then permeabilized by digitonin treatment and washed with 1 M KCl in KHM. Afterward the cells were incubated with interphase extract (IE) or mitotic extract (ME) or mitotic extract pretreated with the MEK1 inhibitor PD 98059 (PD; 75 μM) and the PKD inhibitor CID 755673 (CID; 5 μM), respectively, and an ATP-regenerating system at 32°C for 1 h. The organization of Golgi membranes was monitored by fluorescence microscopy using a mannosidase II–specific antibody. Shown are representative images. Scale bar, 10 μm. Left graph, percentage of cells with dispersed Golgi membranes after incubation with interphase or mitotic extract (n = 200 cells/sample). Right graph, percentage of permeabilized cells after incubation with mitotic extract pretreated with PD 98059 or CID 755673 (n = 150 cells/sample). (B) HeLa cells transfected with a control siRNA (siLacZ) or PKD1- and PKD2-specific siRNAs were synchronized at the G1/S border by a double-thymidine treatment and released for the indicated times. Cells were lysed, and phosphorylation of MEK1 was detected by Western blot analysis using an antibody specific for phosphorylated serine 221. Lysates were also analyzed using a MEK1-specific antibody to ensure that total protein levels were unchanged. Successful knockdown of PKD2 was shown with a PKD2-specific antibody. Shown is a representative experiment (n = 3). (C) HeLa cells transfected with a control siRNA (siLacZ) or PKD1- and PKD2-specific siRNAs were synchronized at the G1/S border by a double-thymidine treatment and immediately harvested (0 h) or released for 8 h before analysis. Cells were lysed, and C-Raf kinase activity was determined as described in Materials and Methods. MEK1 phosphorylation was monitored using the pS221 antibody, and MEK1 and Raf-1 levels were visualized using specific antibodies as indicated. (D) HeLa cells transiently transfected with plasmids encoding PKD1-GFP or PKD2-GFP were synchronized at the G1/S border by aphidicolin treatment and released for 8 h before analysis. Cells were fixed and stained with antibodies specific for pH3 (red) and p230 (cyan, top) or pH3 and pS910 (cyan, bottom). Localization of PKD-GFP fusion proteins is shown in green. The nucleus was visualized with DAPI (pink). Scale bar, 10 μm.
We next investigated whether PKD is upstream or downstream of active MEK1. MEK1 is specifically activated during mitosis in a Raf-1–dependent manner (Colanzi et al., 2003b). We therefore synchronized siLacZ- and siPKD1/2-transfected HeLa cells at the G1/S border using a double-thymidine block as depicted in Figure 2A. Cells were harvested at distinct time points after the second thymidine washout, and MEK1 phosphorylation was visualized using Western blot analysis with a pS221-MEK1–specific antibody, which reports Raf-induced activation of MEK (Alessi et al., 1994). In control cells MEK1 was phosphorylated at 6 h after release and stayed elevated (Figure 3B). It is striking that PKD1/2 depletion completely blocked MEK1 phosphorylation during mitosis (Figure 3B). We further analyzed whether Raf-1 activation was affected. To this end, we precipitated endogenous Raf-1 from lysates obtained from cells synchronized at the G1/S border (0 h) or released for 8 h. Precipitates were incubated with a glutathione S-transferase (GST)—tagged, kinase-inactive MEK1 in the presence of ATP for 30 min. MEK1 phosphorylation was then monitored using the pS221 antibody. Figure 3C demonstrates that in control cells Raf-1 kinase activity was strongly increased 8 h after release; however, depletion of PKD1/2 inhibited this increase completely. Of note, PKD did not directly phosphorylate Raf-1 in an in vitro kinase assay (unpublished data). Taken together, this indicates that PKD is upstream of the Raf-1/MEK1 signaling pathway in mitotic Golgi fragmentation. We further analyzed the localization of active PKD1 and PKD2 in G2 cells. To this end, we transiently transfected HeLa cells with PKD1–green fluorescent protein (GFP) or PKD2-GFP encoding expression plasmids and arrested cells in S phase by aphidicolin treatment for 16 h (Sütterlin et al., 2002). Subsequently cells were released from the S-phase block, and 8 h later cellular localization of active PKD-GFP during mitosis was examined by staining cells with anti-pS910, an antibody specific for phosphorylated PKD1 and PKD2 (Hausser et al., 2002). The cells were also stained with 4,6-diamino-2-phenylindole (DAPI), pH3 antibody, and an antibody to the Golgi protein p230 to identify the mitotic stage and visualize the organization of Golgi membranes, respectively (Figure 3D). Both PKD isoforms colocalized with the Golgi marker p230 in the G2 phase of the cell cycle (Figure 3D, top). Additional staining with the pS910-specific antibody revealed that Golgi-localized PKD is active in G2 (Figure 3D, bottom). These data prove that active PKD is present at Golgi membranes in G2.
Depletion of PKD inhibits mitotic Golgi fragmentation in intact cells
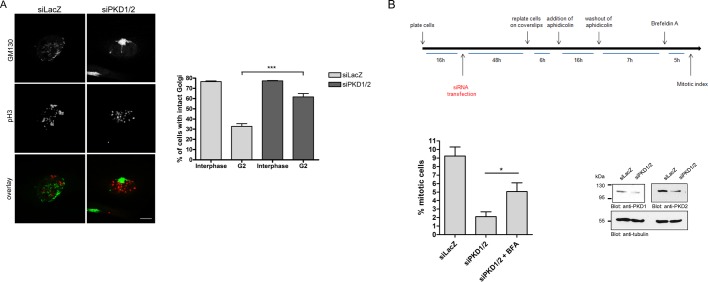
The first step in mitotic Golgi disassembly is the fragmentation of the noncompact zones of the Golgi ribbon. This occurs in G2 and results in the formation of isolated Golgi stacks (Feinstein and Linstedt, 2007). A functional block of proteins involved in this Golgi fragmentation step results in inhibition of the severing of the Golgi ribbon and arrest of the cell cycle at the G2 stage (Sütterlin et al., 2001; Hidalgo et al., 2004; Yoshimura et al., 2005; Feinstein and Linstedt, 2007). Because our results indicate that cells accumulate in G2 stage when PKD1 and 2 are depleted, we further investigated the morphology of the Golgi complex in this stage. To this end, we stained synchronized control and PKD1/2-depleted cells released for 8, 10, and 12 h with the pH3-specific antibody to visualize cells in late G2 stage and, in addition, with a GM130-specific antibody to visualize the Golgi complex. Figure 4A and Supplemental Figure S3 show representative images of pH3-positive late G2 cells, demonstrating ribbon cleavage of tubular bridges in control cells released for 8 h, whereas PKD1/2-knockdown G2 cells showed an intact Golgi at this time point. Quantification of >500 cells in late G2 in three independent experiments revealed that only 32.6% of control cells harbored an intact Golgi complex, whereas 61.7% of PKD1/2-depleted cells showed this Golgi phenotype (Figure 4A, graph). In interphase, the majority (∼77%) of control and PKD-depleted cells harbor an intact Golgi complex (Figure 4A, graph, and Supplemental Figure S3).
FIGURE 4:
Depletion of PKD inhibits mitotic Golgi fragmentation in intact cells. (A) HeLa cells transfected with the indicated siRNAs were synchronized at the G1/S border and released for 8 and 10 h. Cells were fixed and stained with antibodies specific for GM130 and pH3. The images show cells in G2 stage. Scale bar, 10 μm. Cells in interphase and late G2 were scored for their Golgi complex morphology. Golgi complexes were scored as fragmented when at least three clearly separated stacks were observed. The graph shows the percentages of cells in interphase and G2 stage harboring an intact Golgi complex. At least 150 cells (8- and 10-h release) were counted per sample and experiment; shown is mean ± SEM of three independent experiments. Statistical analysis was performed using one-way ANOVA followed by Tukey’s multiple comparison test (siLacZ G2 vs. siPKD1/2 G2: ***p < 0.001). (B) Top, scheme showing the experimental design. Bottom, HeLa cells were transfected with specific siRNAs and arrested in S phase with aphidicolin for 16 h. Cells were washed to remove the aphidicolin and placed in fresh medium. After 7 h, the cells were either left untreated or treated with brefeldin A (BFA) for an additional 5 h. The cells were then fixed and processed for immunofluorescence microscopy using anti-GM130 antibody to visualize the morphology of the Golgi apparatus. Cells were also stained with DAPI and anti–phospho histone H3 antibody. The mitotic index was determined as described in Figure 1. The data presented are mean ± SEM from four independent experiments. Statistical evaluation was performed using Student’s t test, *p < 0.05. Successful depletion of PKD1 and PKD2 was demonstrated by Western blot analysis using specific antibodies. Detection of tubulin served as a loading control.
We next asked whether the depletion of PKD blocks the cell cycle at the level of the Golgi complex or could also affect independent pathways. We thus analyzed whether artificial fragmentation of the Golgi by treatment with brefeldin A (BFA) could rescue mitotic progression in PKD-depleted cells (Sütterlin et al., 2002). We did this as follows: HeLa cells transfected with siLacZ or siPKD1/2 were arrested in S phase by aphidicolin treatment. Aphidicolin was removed, and 7 h later cells were treated with BFA for additional 5 h. The cells were then fixed and processed for fluorescence microscopy to visualize the organization of the Golgi complex and determine the mitotic index (Figure 4B, top). As described before, depletion of PKD reduced the number of mitotic cells compared with control cells (2.1% ± 0.6 in PKD-depleted cells vs. 9.2 ± 1% in control cells). However, in PKD-depleted cells treated with BFA, mitotic features were found in 5.1 ± 1% of the cells. These data show that the fragmentation of the Golgi complex with BFA alleviates the inhibition of Golgi fragmentation in PKD-depleted cells and, thus, the block in mitotic entry. Nevertheless, we cannot rule out that PKD controls G2 exit by further Golgi fragmentation–independent pathways.
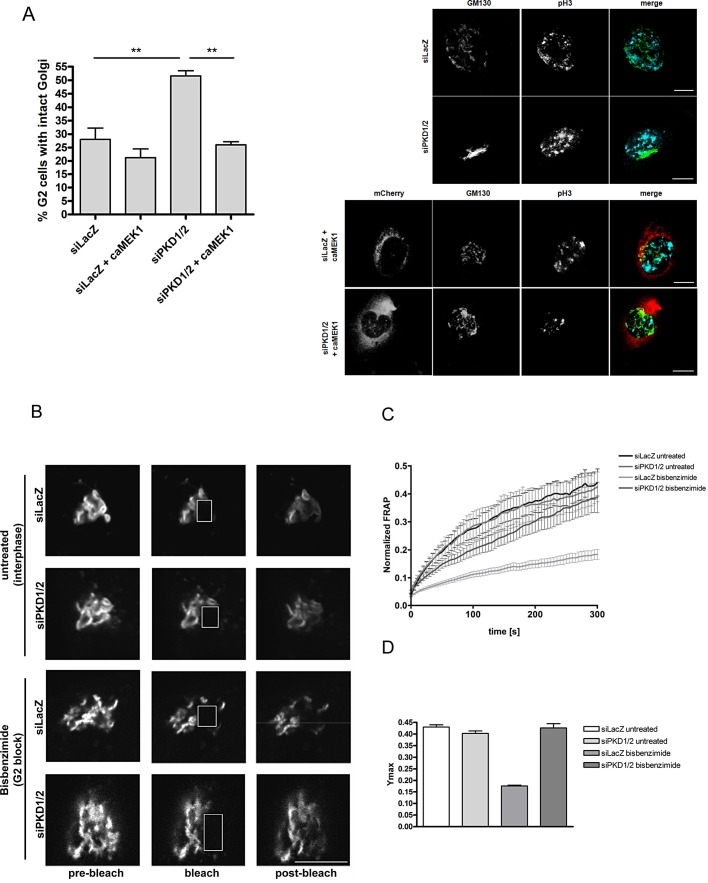
Our previous results indicate that PKD is upstream of the Raf-1/MEK pathway in mitotic Golgi fragmentation. We thus investigated whether a constitutively active (ca) form of MEK1 is able to rescue Golgi complex fragmentation and thus mitotic progression in PKD1/2-depleted cells. To this end, we synchronized control and PKD1/2-depleted cells, with or without overexpression of a constitutively active form of MEK1 fused to mCherry, at the G1/S border. Transfection with caMEK1 was done after releasing cells from the first thymidine block (see Figure 2A). After the release from the second thymidine block for 10, 12, and 14 h, cells were fixed and stained with the pH3- and GM130-specific antibodies to visualize cells in G2 and the Golgi complex, respectively. We specifically monitored Golgi complex morphology of control and PKD1/2 knockdown cells in G2 stage using fluorescence microscopy. Representative images are shown in Figure 5A. Quantification revealed that approximately only 28% of control cells showed an intact Golgi complex during late G2, whereas 51% of PKD-depleted cells showed this Golgi phenotype (Figure 5A). Remarkably, overexpression of caMEK1 restored Golgi fragmentation in PKD1/2-depleted cells to the control level (Figure 5A), demonstrating that MEK1 is downstream of PKD during mitotic Golgi fragmentation. However, caMEK1 overexpression in PKD-depleted cells was not sufficient to fully rescue progression through mitosis (unpublished data).
FIGURE 5:
Depletion of PKD affects the cleavage of the noncompact zones in G2. (A) HeLa cells cultured on collagen-coated coverslips were synchronized at the G1/S border and transfected with siLacZ or siPKD1/2. One sample of each setup was additionally transfected with a caMEK1- plasmid fused to mCherry. Cells were released for 10, 12, and 14 h, fixed, and stained with antibodies specific for GM130 and pH3. Left, cells in late G2 were scored for their Golgi complex morphology. The graph displays the results of the quantification of cells in G2 stage harboring an intact Golgi complex (percentage of total G2 cells). At least 35 cells per sample and experiment were analyzed. Shown is mean ± SEM of three independent experiments. Statistical evaluation was performed using one-way ANOVA followed by Tukey’s multiple comparison test. **p < 0.01. Right, representative images showing Golgi complex morphology of control cells (siLacZ), PKD1/2-depleted cells (siPKD1/2), and siRNA-transfected cells expressing caMEK1-mCherry (siLacZ + caMEK1 and siPKD1/2 + caMEK1) in G2 stage. Color code: GM130 (green), pH3 (cyan), caMEK1-mCherry (red). Scale bar, 10 μm. (B, C) HeLa cells stably transfected with ManII-GFP and transiently transfected with siLacZ or siPKD1/2 were left untreated (interphase) or treated with bisbenzimide (G2 block). The region marked with a rectangle was photobleached, and FRAP was measured over time. (B) Representative images illustrating the Golgi complex before (prebleach), directly after (bleach). and during recovery (postbleach). The bleached areas are delineated by white-bordered rectangles. Scale bar, 10 μm. (C) The curves show the normalized FRAP kinetics of interphase cells and G2 cells over time. Fluorescence recovery in bleached areas was monitored every 2 s. Curves of fluorescence intensities were normalized to nonbleached areas. Shown is mean ± SEM of two independent experiments (10 cells/sample per experiment). (D) Ymax (maximum fluorescence recovery) was determined by fitting the curves shown in C to a one-phase exponential equation.
Finally, we analyzed whether PKD is crucial for the cleavage of the noncompact zones of the Golgi ribbon in G2, using a quantitative approach based on fluorescence recovery after photobleaching (FRAP) of Golgi-resident enzymes (Colanzi et al., 2007). These enzymes diffuse along the length of the Golgi ribbon, as revealed by their fast FRAP. Hence, severing of tubular connections between stacks blocks diffusion of resident enzymes and thus impairs FRAP. HeLa cells stably transfected with the Golgi-resident enzyme mannosidase II fused to GFP (Man II-GFP) were either left untreated or accumulated in G2 stage with the topoisomerase-I inhibitor bisbenzimide (Colanzi et al., 2007). A region corresponding to half of the Golgi complex was bleached by repeated laser illumination at high intensity, and the FRAP of ManII-GFP was examined over time. Representative images are shown in Figure 5B. In Figure 5C, the normalized FRAP curves represent the mean ± SEM of 20 individual FRAP curves per sample from two independent experiments (10 FRAP curves per experiment). In untreated control and PKD1/2-knockdown cells, recovery of ManII-GFP fluorescence reached a high level after 300 s, consistent with an intact Golgi ribbon (Figure 5C). In bisbenzimide-treated control cells, however, FRAP of ManII-GFP did not reach the plateau that was seen in untreated cells, indicating that the noncompact zones were cleaved and thus continuous diffusion of enzymes was interrupted (Figure 5C). By contrast, PKD1/2-depleted cells treated with bisbenzimide demonstrated FRAP comparable to that seen in untreated cells, proving that PKD1/2-knockdown cells harbor an intact Golgi ribbon in G2 stage. Calculation of the maximum fluorescence recovery (Ymax) is plotted in Figure 5D. Compared with untreated control cells, 41% recovery is reached in bisbenzimide-treated control cells, whereas in those lacking PKD, recovery is almost complete (99%). Our results thus clearly show that PKD is specifically required for severing of the noncompact zones of the Golgi complex in G2, which is a prerequisite for cells to pass the G2/M Golgi checkpoint.
DISCUSSION
In this study we identify PKD as a regulator of Golgi interstack connections and a novel player in Golgi mitotic checkpoint control. Using a cellular reconstitution assay, we provide evidence that PKD is necessary and sufficient for the mitotic fragmentation of Golgi membranes. In intact cells we further show that PKD is critical for Golgi fragmentation in the late G2 phase of the cell cycle and mitotic entry by activating the Raf-1/MEK1 pathway.
The importance of the Raf-1/MEK1 pathway in Golgi mitotic checkpoint control is underscored by the fact that expression of active MEK1 in PKD1/2-depleted cells rescued the block in Golgi membrane fragmentation and mitotic entry. The role of MEK in mitotic Golgi fragmentation has been clarified. Inhibition of MEK1 by the use of specific inhibitors and depletion of MEK1 by RNA interference caused cells to arrest in the G2 stage of the cell cycle (Feinstein and Linstedt, 2007). Dispersal of Golgi membranes by brefeldin A treatment or Grasp65 depletion abrogated the requirement for MEK1, indicating that the Golgi ribbon cleavage is the essential step for mitotic entry (Feinstein and Linstedt, 2007). Similarly, we show that brefeldin A treatment rescues mitotic entry in PKD-depleted cells. A specific function for MEK1 in the Golgi mitotic checkpoint is further supported by the identification of its Golgi-localized effector, Erk1c, which shows increased expression during G2 and mitosis (Shaul and Seger, 2006; Shaul et al., 2009). Depletion of Erk1c reduced mitotic Golgi fragmentation and mitotic progression (Shaul and Seger, 2006). A putative downstream target of this Raf-1/MEK1/Erk1c pathway is the Golgi stacking protein Grasp55 (Feinstein and Linstedt, 2008). The functional block of the Golgi matrix components Grasp65 (Sütterlin et al., 2005) and Grasp55 (Duran et al., 2008), respectively, resulted in inhibition of the severing of the Golgi ribbon and arrest of the cell cycle at the G2 stage. Neither Grasp55 nor Grasp65 is phosphorylated by PKD in vitro (unpublished data), and BARS does not contain a PKD consensus motif. The known PKD substrates at the Golgi complex PI4KIIIβ, CERT, and OSBP are involved in lipid metabolism and trafficking (for a review see Olayioye and Hausser, 2012). By phosphorylating and regulating these substrates, PKD drives the fission of Golgi-derived transport carriers to maintain Golgi secretory function (Hausser et al., 2005; Fugmann et al., 2007; Nhek et al., 2010). However, in mitotic cells, overall secretory traffic is shunted (Kreiner and Moore, 1990; Lowe et al., 1998a). Indeed, both PI4KIIIβ and CERT show reduced localization to Golgi membranes in mitotic cells (Godi et al., 1999; Chandran and Machamer, 2008), pointing toward an alternative PKD-dependent mechanism that drives Golgi fragmentation in G2.
The molecular mechanism by which MEK1 is activated in late G2 stage of the cell cycle is unclear. Raf-1 is immediately upstream of MEK1 (Colanzi et al., 2003b); however, mitotic Raf-1 activation was found to be Ras independent (Ziogas et al., 1998) but Pak1 dependent (Zang et al., 2001, 2002). Here we show that PKD is upstream of Raf-1 and MEK1 in the Golgi mitotic checkpoint, which is in line with our previous observation that PKD can stimulate mitogen-activated protein kinase signaling through Raf-1 (Hausser et al., 2001). Because PKD does not directly phosphorylate Raf-1, the question of how the signal is transferred from PKD to Raf-1 remains unresolved. PKD was recently identified to phosphorylate and activate Pak4 (Spratley et al., 2011), and Pak kinases are known to phosphorylate and activate Raf-1 during mitosis (Zang et al., 2001, 2002). It is therefore tempting to speculate that PKD signals through intermediary kinases such as Paks to activate the Raf-1/MEK1 pathway. Recent global mapping of PKD-dependent phosphorylation events upon nocodazole-induced Golgi fragmentation may support the identification of mitotic PKD downstream targets (Franz-Wachtel et al., 2012).
PKD activity was found to increase during mitosis, and active PKD associated with centrosomes, spindles, and the midbody in rat intestinal epithelial IEC-18 cells (Papazyan et al., 2008). However, the question remains of how PKD is locally activated during G2/M transition. Treatment of cells with nocodazole, which disrupts the microtubule cytoskeleton and promotes the formation of Golgi ministacks, was shown to activate PKD (Fuchs et al., 2009). In addition, activation of RhoA has been shown to activate PKD (Cowell et al., 2009), and nocodazole-induced stimulation of Raf-1 is augmented by coexpression of Pak1/2 (Zang et al., 2001). Of interest, expression of constitutively active RhoA was recently reported to promote the fragmentation of the Golgi complex into ministacks (Zilberman et al., 2011), and inactivation of Rho GTPases by toxin B treatment delayed Aurora-A activation and histone H3 phosphorylation (Ando et al., 2007). It thus appears that the profound cytoskeletal rearrangements occurring at mitotic onset, emanating from the centrosome as the microtubule-organizing center, are linked to Golgi structural organization at the level of PKD. Taken together, our results provide evidence for a novel role of PKD in Golgi mitotic checkpoint control by acting upstream of Raf-1/MEK1. Our data further emphasize the importance of PKD in the maintenance of the structural integrity of the Golgi complex.
MATERIALS AND METHODS
Plasmids, siRNAs, reagents, and antibodies
The plasmid encoding constitutively active MEK1 fused to mCherry was obtained from Addgene (plasmid 31880; Cambridge, MA; Covassin et al., 2009). Plasmids encoding PKD1 and PKD2 fused to GFP have been described previously (Hausser et al., 2005). Commercially available antibodies used were anti-PKD2 rabbit polyclonal antibody (Calbiochem, Darmstadt, Germany), anti-PKD1 C-20 and anti-Raf-1 C12 rabbit polyclonal antibodies (Santa Cruz Biotechnology, Heidelberg, Germany), anti–tubulin α mouse monoclonal antibody (Neomarkers, Fremont, CA), anti–phospho-H3 (Ser-10) rabbit polyclonal antibody (Sigma-Aldrich, Munich, Germany), anti–phospho-MEK1 (S221) rabbit polyclonal antibody and anti–caspase-3 rabbit monoclonal antibody 8G10 (both from Cell Signaling, Beverly, MA), anti-Cdk1/Cdc2 and anti–phospho-Cdk1/Cdc2 (Tyr-15) rabbit polyclonal antibody and anti-GM130 and anti-p230 mouse monoclonal antibodies (BD Biosciences, Heidelberg, Germany), and anti-MEK1 rabbit polyclonal antibody (Millipore, Schwalbach, Germany). The antibody specific for phosphorylation of PKD1 at serine 910 is described elsewhere (Hausser et al., 2002). The rabbit polyclonal mannosidase II antibody was provided by Vivek Malhotra (Center of Genomic Regulation, Barcelona, Spain). Secondary antibodies used were goat anti–mouse and anti–rabbit immunoglobulin G (IgG), Alexa 488, Alexa 546, or Alexa 633 coupled (Invitrogen, Darmstadt, Germany), and goat anti–mouse and anti–rabbit IgG horseradish peroxidase (HRP) coupled (Dianova, Hamburg, Germany). siRNAs specific for LacZ, human PKD1, and human PKD2 were obtained from Eurofins MWG Operon (Ebersberg, Germany) and have been described in detail elsewhere (Peterburs et al., 2009). ON-Target plus Smartpools (human PRKD1, gene ID: 5587; and human PRKD2, gene ID: 25865) and the ON-Target plus NON-targeting control were obtained from Dharmacon (Thermo Fisher Scientific, Waltham, MA). The MEK1-specific inhibitor PD98059 was obtained from Cell Signaling, the PKD specific inhibitor CID755673 was from Tocris Bioscience (Bristol, United Kingdom), and brefeldin A was from Calbiochem.
Cell culture
HeLa cells were obtained from the American Type Culture Collection (Manassas, VA), and HeLa cells stably transfected with Mannosidase II fused to GFP were obtained from Vivek Malhotra. Cells were maintained in RPMI 1640 or DMEM supplemented with 10% fetal bovine serum. HeLa cells were transfected with siRNA and ON-Target plus Smartpools using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. Plasmid transfection was performed using TransIT-HeLaMONSTER reagent (MirusBio, Madison, WI) according to the manufacturer’s instructions.
Cell synchronization at the G1/S border
HeLa cells were synchronized at the G1/S border using a double-thymidine block or aphidicolin treatment. In brief, cells grown on Petri dishes or coverslips were transfected with specific siRNAs and 24 h later incubated in growth medium containing thymidine (Sigma-Aldrich) at a final concentration of 2 mM for 19 h. Afterward, cells were released from the thymidine block by washing with serum-free medium and were refed with growth medium for 9 h. Subsequently cells were subjected to the second thymidine block for an additional 16 h. After the second release, cells were harvested at distinct time points. In case of rescue experiments, plasmid transfection of HeLa cells was performed during the first release. In case of aphidicolin treatment, transfected HeLa cells were incubated in medium supplemented with aphidicolin (5 μg/ml; Calbiochem) for 16 h. Afterward, cells were released from the S-phase arrest by washing with serum-free medium and were refed with growth medium. Cells were harvested 10 h later. BFA treatment (2 μM) was performed 7 h after release, and cells were incubated another 5 h.
Flow cytometry
Trypsinized cells were pelleted, washed in cold phosphate-buffered saline (PBS), and resuspended in ice-cold ethanol while vortexing. Cells were incubated overnight at 4°C. The next day ethanol was removed by centrifugation and cells were washed using cold PBS and incubated with 50 μg/ml propidium iodide (Invitrogen) for 30 min. Cells were analyzed with a Cytomics FC500 instrument (Beckman Coulter, Krefeld, Germany). Data were plotted with CXP software (Beckman Coulter). Ten thousand events were analyzed for each sample.
Preparation of mitotic and interphase extracts
HeLa cells grown to 70% confluency were treated with thymidine (2 mM) for 10–12 h, causing the arrest of cells in S phase. Subsequently cells were washed three times with PBS and incubated with nocodazole (500 ng/ml) overnight to arrest them in mitosis. Mitotic cells were detached from the dishes by a “mitotic shake-off” procedure, leaving the nonmitotic cells attached. Two washing steps with ice-cold PBS and one with mitotic extract buffer (15 mM 1,4-piperazinediethanesulfonic acid, pH 7.2, 50 mM KCl, 10 mM MgCl2, 20 mM β-mercaptoethanol, 20 mM β-glycerophosphate, 15 mM ethylene glycol tetraacetic acid [EGTA], 10.5 mM spermidine, 0.2 mM spermine, and 1 mM dithiothreitol [DTT] plus complete protease inhibitors [Roche Diagnostics, Mannheim, Germany]) were performed. Cells were then resuspended in 2× volume of mitotic extract buffer. After swelling on ice for 10 min, cells were homogenized using a 24-gauge needle. To yield the supernatant termed “mitotic extract,” cells were centrifuged in a table top ultracentrifuge using a TLS55 rotor for 45 min at 48,000 rpm. The supernatant was harvested in aliquots, immediately frozen in liquid nitrogen, and stored at −80°C. For preparation of interphase extracts, confluent HeLa cells were washed and harvested with a cell scraper. Subsequent procedures were similar to those described for mitotic extract preparation. Both extracts contained protein concentrations in the range of 5–10 mg/ml.
Semi-intact assay
The semi-intact assay was performed according to Acharya et al. (1998). In brief, HeLa cells grown on fibronectin-coated coverslips to 90% confluency were treated with 2 mM thymidine for 8 h. The cells were washed with KHM buffer (25 mM HEPES, pH 7.2, 125 mM potassium acetate, 2.5 mM magnesium acetate) at room temperature, shifted to ice, and washed again with cold KHM buffer. Permeabilization was done by incubation with 30 mg/ml digitonin in KHM buffer for 90 s at room temperature. After digitonin treatment, semi-intact cells were washed with 1 M KCl-KHM buffer to remove cytosolic proteins, followed by a final washing step using KHM buffer. Coverslips with semi-intact cells were inverted on a 50-μl drop of a reaction mixture on a sheet of Parafilm and incubated in a 32°C water bath for 1 h. The reaction mixture contained mitotic or interphase extract together with an ATP-regenerating system (100 mM ATP, 100 mM UTP, 200 mM creatine phosphate, 2.76 mg/ml creatine kinase). After incubation, cells were fixed with 4% paraformaldehyde, and the Golgi complex was visualized using a rabbit mannosidase II–specific antibody, followed by an Alexa 488–coupled anti-rabbit antibody. The Golgi state was monitored using confocal laser scanning microscopy.
Immunofluorescence and microscopy
Transfected HeLa cells were grown on collagen-coated coverslips, washed with PBS, fixed in 4% paraformaldehyde at room temperature for 15 min, washed, permeabilized with 0.1% Triton X-100 (5 min, room temperature), and blocked with blocking buffer (5% fetal bovine serum in PBS) for 30 min. The cells were incubated with the primary antibodies diluted in blocking buffer (1 μg/ml) for 2 h, washed, incubated with secondary antibodies diluted in blocking buffer for 1 h, washed, mounted in Fluoromount G (Southern Biotechnology, Birmingham, AL), and analyzed on a confocal laser scanning microscope (LSM 710; Zeiss, Jena, Germany). Nuclei were stained by incubation with 2.5 μM DRAQ5 (Biostatus Limited, Shepshed, United Kingdom) in PBS for 15 min before mounting or by mounting in ProLong Gold antifade reagent supplemented with 1 μg/ml DAPI (Invitrogen). DAPI was excited with the 405 nm line of the argon laser, and fluorescence was detected at 415–485 nm. Alexa 488 and GFP were excited with the 488 nm line of the argon laser, and fluorescence was detected at 490–550 nm. Alexa 546 was excited with the 561 nm line of a diode-pumped solid-state laser, and fluorescence was detected at 560–620 nm. Alexa 633 and DRAQ5 were excited with the 633 nm line of a helium–neon laser, and fluorescence was detected at 640–750 nm. Cells were imaged with an EC Plan-Neofluar 40.0×/1.3 oil differential interference contrast (DIC) or a Plan Apochromat 20×/0.8 DIC objective lens. Images were processed with Photoshop (Adobe, San Jose, CA).
Golgi FRAP analysis
To investigate membrane continuity, we transiently transfected HeLa–Man II-GFP cells grown on glass-bottom dishes with siRNAs. At 48 h posttransfection, half of the samples were incubated with 40 μg/ml bisbenzimide (Sigma-Aldrich) for 18 h, and the other cells were left untreated. Subsequently the diffusion mobility of the Man II-GFP protein in living cells was imaged at room temperature using a laser scanning confocal microscope. Cells were imaged with a Plan Apochromat 63×/1.4 DIC M27 objective lens. An initial prebleach image was taken, followed by bleaching the Golgi region of interest five times with high-intensity laser light (488 nm line, 80% laser power). Recovery of fluorescence in the bleached area was measured over time by scanning every 2 s. Fluorescence intensity values were normalized to the unbleached region. Fitting of curves was performed with a one-phase exponential equation (Y = Ymax[1 − exp(−K*X)]; Prism, GraphPad Software, La Jolla, CA).
Protein extraction of cells
Whole-cell extracts were obtained by solubilizing cells in hot protein sample buffer (95°C). After 10 min of incubation at 95°C, extracts were placed on ice and centrifuged (16,000 × g, 15 min, and 4°C). Samples were subjected to SDS–PAGE.
Western blotting
Equal amounts of proteins were separated by SDS–PAGE and blotted onto nitrocellulose membranes (Pall, Dreieich, Germany). After blocking with 0.5% blocking reagent (Roche Diagnostics), filters were probed with specific antibodies. Proteins were visualized with HRP-coupled secondary antibodies using the ECL detection system.
Raf-kinase assay
Synchronized HeLa cells released for 0 and 8 h were lysed in RIPA buffer (50 mM HEPES, pH 7.4, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM NaF, and 5 mM EDTA plus complete protease inhibitors and PhoSTOP [Roche Diagnostics]). For immunoprecipitation, equal amounts of proteins were incubated with a Raf-1–specific antibody overnight at 4°C. Immune complexes were collected with protein G–agarose (KPL, Gaithersburg, MD) and washed four times with kinase buffer (20 mM 3-(N-morpholino)propanesulfonic acid, pH 7.2, 5 mM EGTA, 25 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM DTT). The Raf-1 kinase reaction was carried out for 30 min at 37°C in 30 μl kinase buffer. The reaction was started by addition of a kinase buffer mixture containing 5 mM ATP, 18.75 mM MgCl2 and 1 μg of purified inactive GST-MEK1 (Millipore). To terminate the reaction, SDS-sample buffer was added, and the samples were resolved by SDS–PAGE, blotted onto nitrocellulose, and analyzed by Western blotting.
Supplementary Material
Acknowledgments
We appreciate the excellent technical assistance of G. Link. The work was supported by grants from the German Cancer Aid (DKH 107545 and 109576) to A.H. and M.A.O. M.A.O. and T.B. are supported by the Heisenberg and Emmy Noether programs of the Deutsche Forschungsgemeinschaft, respectively. J. Villeneuve is supported by a long-term European Molecular Biology Organization fellowship.
Glossary
Abbreviations used:
- MEK1
mitogen-activated protein kinase kinase 1
- PKD
protein kinase D
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-03-0198) on December 14, 2012.
The authors declare that they have no competing financial interests.
REFERENCES
- Acharya U, Mallabiabarrena A, Acharya JK, Malhotra V. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell. 1998;92:183–192. doi: 10.1016/s0092-8674(00)80913-7. [DOI] [PubMed] [Google Scholar]
- Alessi DR, Saito Y, Campbell DG, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall CJ, Cowley S. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. EMBO J. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando Y, Yasuda S, Oceguera-Yanez F, Narumiya S. Inactivation of Rho GTPases with Clostridium difficile toxin B impairs centrosomal activation of Aurora-A in G2/M transition of HeLa cells. Mol Biol Cell. 2007;18:3752–3763. doi: 10.1091/mbc.E07-03-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron CL, Malhotra V. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science. 2002;295:325–328. doi: 10.1126/science.1066759. [DOI] [PubMed] [Google Scholar]
- Bossard C, Bresson D, Polishchuk RS, Malhotra V. Dimeric PKD regulates membrane fission to form transport carriers at the TGN. J Cell Biol. 2007;179:1123–1131. doi: 10.1083/jcb.200703166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran S, Machamer CE. Acute perturbations in Golgi organization impact de novo sphingomyelin synthesis. Traffic. 2008;9:1894–1904. doi: 10.1111/j.1600-0854.2008.00810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A, Corda D. Mitosis controls the Golgi and the Golgi controls mitosis. Curr Opin Cell Biol. 2007;19:386–393. doi: 10.1016/j.ceb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Colanzi A, Hidalgo CC, Persico A, Cericola C, Turacchio G, Bonazzi M, Luini A, Corda D. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 2007;26:2465–2476. doi: 10.1038/sj.emboj.7601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A, Sütterlin C, Malhotra V. Cell-cycle-specific Golgi fragmentation: how and why. Curr Opin Cell Biol. 2003a;15:462–467. doi: 10.1016/s0955-0674(03)00067-x. [DOI] [PubMed] [Google Scholar]
- Colanzi A, Sütterlin C, Malhotra V. RAF1-activated MEK1 is found on the Golgi apparatus in late prophase and is required for Golgi complex fragmentation in mitosis. J Cell Biol. 2003b;161:27–32. doi: 10.1083/jcb.200208099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covassin LD, Siekmann AF, Kacergis MC, Laver E, Moore JC, Villefranc JA, Weinstein BM, Lawson ND. A genetic screen for vascular mutants in zebrafish reveals dynamic roles for Vegf/Plcg1 signaling during artery development. Dev Biol. 2009;329:212–226. doi: 10.1016/j.ydbio.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell CF, Yan IK, Eiseler T, Leightner AC, Doppler H, Storz P. Loss of cell-cell contacts induces NF-kappaB via RhoA-mediated activation of protein kinase D1. J Cell Biochem. 2009;106:714–728. doi: 10.1002/jcb.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Anel AM, Malhotra V. PKCeta is required for beta1gamma2/beta3gamma2- and PKD-mediated transport to the cell surface and the organization of the Golgi apparatus. J Cell Biol. 2005;169:83–91. doi: 10.1083/jcb.200412089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran JM, Kinseth M, Bossard C, Rose DW, Polishchuk R, Wu CC, Yates J, Zimmerman T, Malhotra V. The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol Biol Cell. 2008;19:2579–2587. doi: 10.1091/mbc.E07-10-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein TN, Linstedt AD. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol Biol Cell. 2007;18:594–604. doi: 10.1091/mbc.E06-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein TN, Linstedt AD. GRASP55 regulates Golgi ribbon formation. Mol Biol Cell. 2008;19:2696–2707. doi: 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz-Wachtel M, Eisler SA, Krug K, Wahl S, Carpy A, Nordheim A, Pfizenmaier K, Hausser A, Macek B. Global detection of protein kinase D-dependent phosphorylation events in nocodazole-treated human cells. Mol Cell Proteomics. 2012;11:160–170. doi: 10.1074/mcp.M111.016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs YF, Eisler SA, Link G, Schlicker O, Bunt G, Pfizenmaier K, Hausser A. A Golgi PKD activity reporter reveals a crucial role of PKD in nocodazole-induced Golgi dispersal. Traffic. 2009;10:858–867. doi: 10.1111/j.1600-0854.2009.00918.x. [DOI] [PubMed] [Google Scholar]
- Fugmann T, Hausser A, Schoffler P, Schmid S, Pfizenmaier K, Olayioye MA. Regulation of secretory transport by protein kinase D-mediated phosphorylation of the ceramide transfer protein. J Cell Biol. 2007;178:15–22. doi: 10.1083/jcb.200612017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- Hausser A, Link G, Bamberg L, Burzlaff A, Lutz S, Pfizenmaier K, Johannes FJ. Structural requirements for localization and activation of protein kinase C mu (PKC mu) at the Golgi compartment. J Cell Biol. 2002;156:65–74. doi: 10.1083/jcb.200110047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser A, Storz P, Hubner S, Braendlin I, Martinez-Moya M, Link G, Johannes FJ. Protein kinase C mu selectively activates the mitogen-activated protein kinase (MAPK) p42 pathway. FEBS Lett. 2001;492:39–44. doi: 10.1016/s0014-5793(01)02219-0. [DOI] [PubMed] [Google Scholar]
- Hausser A, Storz P, Martens S, Link G, Toker A, Pfizenmaier K. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat Cell Biol. 2005;7:880–886. doi: 10.1038/ncb1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Hidalgo CC, Bonazzi M, Spano S, Turacchio G, Colanzi A, Luini A, Corda D. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305:93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- Kano F, Takenaka K, Yamamoto A, Nagayama K, Nishida E, Murata M. MEK and Cdc2 kinase are sequentially required for Golgi disassembly in MDCK cells by the mitotic Xenopus extracts. J Cell Biol. 2000;149:357–368. doi: 10.1083/jcb.149.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiner T, Moore HP. Membrane traffic between secretory compartments is differentially affected during mitosis. Cell Regul. 1990;1:415–424. doi: 10.1091/mbc.1.5.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljedahl M, Maeda Y, Colanzi A, Ayala I, Van Lint J, Malhotra V. Protein kinase D regulates the fission of cell surface destined transport carriers from the trans-Golgi network. Cell. 2001;104:409–420. doi: 10.1016/s0092-8674(01)00228-8. [DOI] [PubMed] [Google Scholar]
- Lin CY, Madsen ML, Yarm FR, Jang YJ, Liu X, Erikson RL. Peripheral Golgi protein GRASP65 is a target of mitotic polo-like kinase (Plk) and Cdc2. Proc Natl Acad Sci USA. 2000;97:12589–12594. doi: 10.1073/pnas.220423497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M, Nakamura N, Warren G. Golgi division and membrane traffic. Trends Cell Biol. 1998a;8:40–44. doi: 10.1016/s0962-8924(97)01189-6. [DOI] [PubMed] [Google Scholar]
- Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998b;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- Ma HT, Poon RY. Synchronization of HeLa cells. Methods Mol Biol. 2011;761:151–161. doi: 10.1007/978-1-61779-182-6_10. [DOI] [PubMed] [Google Scholar]
- Nhek S, Ngo M, Yang X, Ng MM, Field SJ, Asara JM, Ridgway ND, Toker A. Regulation of oxysterol-binding protein Golgi localization through protein kinase D-mediated phosphorylation. Mol Biol Cell. 2010;21:2327–2337. doi: 10.1091/mbc.E10-02-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye MA, Hausser A. Integration of non-vesicular and vesicular transport processes at the Golgi complex by the PKD-CERT network. Biochim Biophys Acta. 2012;1821:1096–1103. doi: 10.1016/j.bbalip.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Papazyan R, Doche M, Waldron RT, Rozengurt E, Moyer MP, Rey O. Protein kinase D isozymes activation and localization during mitosis. Exp Cell Res. 2008;314:3057–3068. doi: 10.1016/j.yexcr.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterburs P, Heering J, Link G, Pfizenmaier K, Olayioye MA, Hausser A. Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 2009;69:5634–5638. doi: 10.1158/0008-5472.CAN-09-0718. [DOI] [PubMed] [Google Scholar]
- Preisinger C, Korner R, Wind M, Lehmann WD, Kopajtich R, Barr FA. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusapati GV, Krndija D, Armacki M, von Wichert G, von Blume J, Malhotra V, Adler G, Seufferlein T. Role of the second cysteine-rich domain and Pro275 in protein kinase D2 interaction with ADP-ribosylation factor 1, trans-Golgi network recruitment, and protein transport. Mol Biol Cell. 2010;21:1011–1022. doi: 10.1091/mbc.E09-09-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharlow ER, Giridhar KV, Lavalle CR, Chen J, Leimgruber S, Barrett R, Bravo-Altamirano K, Wipf P, Lazo JS, Wang QJ. Potent and selective disruption of protein kinase D functionality by a benzoxoloazepinolone. J Biol Chem. 2008;283:33516–33526. doi: 10.1074/jbc.M805358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul YD, Gibor G, Plotnikov A, Seger R. Specific phosphorylation and activation of ERK1c by MEK1b: a unique route in the ERK cascade. Genes Dev. 2009;23:1779–1790. doi: 10.1101/gad.523909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul YD, Seger R. ERK1c regulates Golgi fragmentation during mitosis. J Cell Biol. 2006;172:885–897. doi: 10.1083/jcb.200509063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratley SJ, Bastea LI, Doppler H, Mizuno K, Storz P. Protein kinase D regulates cofilin activity through p21-activated kinase 4. J Biol Chem. 2011;286:34254–34261. doi: 10.1074/jbc.M111.259424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C, Hsu P, Mallabiabarrena A, Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- Sütterlin C, Lin CY, Feng Y, Ferris DK, Erikson RL, Malhotra V. Polo-like kinase is required for the fragmentation of pericentriolar Golgi stacks during mitosis. Proc Natl Acad Sci USA. 2001;98:9128–9132. doi: 10.1073/pnas.161283998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeev O, Cizmecioglu O, Settele F, Kempf T, Hoffmann I. Cdc25 phosphatases are required for timely assembly of CDK1-cyclin B at the G2/M transition. J Biol Chem. 2010;285:16978–16990. doi: 10.1074/jbc.M109.096552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Yoshioka K, Barr FA, Lowe M, Nakayama K, Ohkuma S, Nakamura N. Convergence of cell cycle regulation and growth factor signals on GRASP65. J Biol Chem. 2005;280:23048–23056. doi: 10.1074/jbc.M502442200. [DOI] [PubMed] [Google Scholar]
- Zang M, Hayne C, Luo Z. Interaction between active Pak1 and Raf-1 is necessary for phosphorylation and activation of Raf-1. J Biol Chem. 2002;277:4395–4405. doi: 10.1074/jbc.M110000200. [DOI] [PubMed] [Google Scholar]
- Zang M, Waelde CA, Xiang X, Rana A, Wen R, Luo Z. Microtubule integrity regulates Pak leading to Ras-independent activation of Raf-1. Insights into mechanisms of Raf-1 activation. J Biol Chem. 2001;276:25157–25165. doi: 10.1074/jbc.M100152200. [DOI] [PubMed] [Google Scholar]
- Zilberman Y, Alieva NO, Miserey-Lenkei S, Lichtenstein A, Kam Z, Sabanay H, Bershadsky A. Involvement of the Rho-mDia1 pathway in the regulation of Golgi complex architecture and dynamics. Mol Biol Cell. 2011;22:2900–2911. doi: 10.1091/mbc.E11-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziogas A, Lorenz IC, Moelling K, Radziwill G. Mitotic Raf-1 is stimulated independently of Ras and is active in the cytoplasm. J Biol Chem. 1998;273:24108–24114. doi: 10.1074/jbc.273.37.24108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.