Abstract
The role of protein dynamics on different time scales in enzyme catalysis remains an area of active debate. The connection between enzyme dynamics on the femtosecond time scale and transition state formation has been demonstrated in human purine nucleoside phosphorylase (PNP) through the study of a mass-altered enzyme. Isotopic substitution in human PNP (heavy PNP) decreased the rate of on-enzyme chemistry but did not alter either the transition state structure or steady-state kinetic parameters. Here we investigate the underlying atomic motions associated with altered barrier crossing probability for heavy PNP. Transition path sampling was employed to illuminate the molecular differences between barrier crossing in light and heavy enzymes. The mass effect is apparent in promoting vibrations that polarize the N-ribosidic bond, and that promote the stability of the purine leaving group. These motions facilitate barrier crossing.
Keywords: purine nucleoside phosphorylase, kinetic isotope effect, promoting vibration, barrier crossing, transition path sampling, heavy enzyme
Computational studies using transition path sampling have supported the role of rapid protein dynamics in the catalytic mechanism of human purine nucleoside phosphorylase (PNP).1–3 PNP catalyzes the reversible phosphorolysis of 6-oxypurine nucleosides and deoxynucleosides to generate the corresponding purine base and α-D-ribose (or deoxyribose) 1-phosphate. This unbiased computational approach to study barrier passage predicted a ribocationic transition state fully consistent with that derived from experimental transition state analysis from kinetic isotope effects, and driven by local bond vibrational motions at the catalytic site (Fig. 1).3,4 These vibrational motions polarize the ribosidic bond to facilitate scission; rather than the more standard picture of this polarization arising from the approach of the attacking phosphate nucleophile. What has been lacking in the debate regarding the linkage of fs dynamics to catalysis is a definitive experimental test of these motions in catalysis.5–7
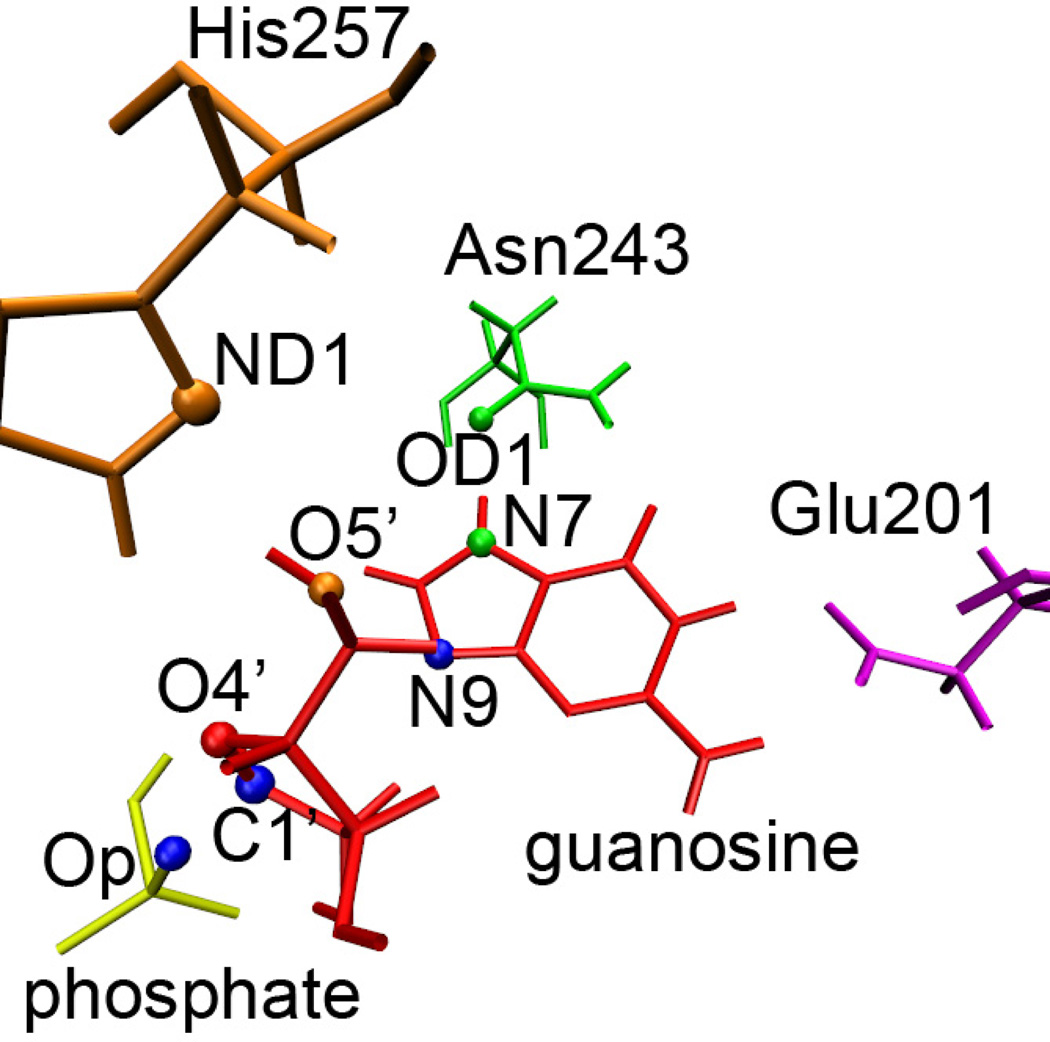
Figure 1.
Substrate guanosine and active site residues in PNP involved at the transition state. The motion of His257 (orange) is known to compress O5′ (orange) toward the O4′ (red) and thereby polarize the glycosidic bond. The N-ribosidic bond is not shown here at the transition state, but would connect C1′ and N9 (blue) in the reactant. The reaction coordinate motion has C1′ migrating toward Op to form product. Asn243 and Glu201 neutralize and stabilize the leaving group.
Recently, one of us has published an experimental test that changes protein mode frequency without perturbing electrostatic properties of the enzyme.8,9 Human PNP was created in which all C, N and H were substituted with 13C, 15N, and 2H at non-exchangeable positions. This heavy PNP construct was termed a “Born-Oppenheimer enzyme” because while the altered mass of the system changes bond vibrational frequencies, and hence collective protein motions, these substitutions have no effect on the electronic potential energy surface that governs chemical reaction in the enzyme. While kinetic isotope effect measurements based on reactant isotope labels have long been used to probe chemical mechanism, such effects demonstrate the state change between free reactants and reactants at the transition state and provide no information on the involvement of protein motion on chemistry.10 Following purification in normal water, all exchangeable 2H is replaced by H, thus all H-bonds from exchangeable groups at the catalytic site are normal, and the only mass perturbation is from non-exchanged atoms with altered vibrational modes throughout the entire enzyme.9
This experimental construct is exactly what is needed to prove or disprove the proposition that vibrational modes in catalytic site chemistry are inherent parts of the chemical mechanism of an enzyme. We emphasize that the substrate molecules remain in their naturally occurring isotopic forms, and that all H-bonds between the enzyme and substrate are composed of normal isotopic H, so any changes in chemistry or observed protein dynamics is due solely to changes in the enzyme vibrational structure. The experiment, however, is an ensemble measurement that does not report on atomic level effects.
Heavy PNP exhibits a 9.9% increase in molecular mass relative to its unlabeled counterpart.9 The heavy enzyme was found to be unchanged from its normal counterpart in transition state structure and the steady state kinetic parameters, kcat and Km. As steady-state parameters for PNP are known to be linked to ms catalytic site conformational changes, it can be concluded that the motions linked to steady-state kinetics are diffusion-linked and independent of enzyme mass. However, the on-enzyme chemical rate constant for heavy PNP was slowed by approximately 30% for the phosphorolysis of guanosine, demonstrating a reduced probability of barrier-crossing for the chemistry step.9 This shows that the change in PNP mass has altered the approach to and passage over the chemical barrier as a result of the altered enzymatic bond vibrational frequencies, and so altered protein vibrational structure.
What was lacking was an atomistic picture of how the mass modulations effected protein dynamics. Transition path sampling (TPS)11 employing hybrid quantum mechanics/molecular mechanics (QM/MM) calculations was used to study the chemical barrier crossing of the reaction catalyzed by heavy PNP in comparison with light PNP using explicit solvent. Our previous transition path studies of PNP were limited to the use of implicit solvation.3 Given the nature of specific interactions of normal mass water solvent with the mass-altered protein, the most realistic computational model is desired, and so we now employ explicit solvation to explore barrier crossing in the Born-Oppenheimer enzyme.
The catalytic mechanism and nature of the transition state that we previously identified for PNP involved a promoting vibrations near the ribosyl ring to assist cleavage of the ribosidic bond followed by a conformational migration of the ribosyl cation to form the α-D-ribose 1-phosphate product.3 The results described in the present calculation provide insight into how the vibrational structure of the protein is altered by heavy atom substitution, and as a consequence, how mass-sensitive dynamics affects transition state barrier crossing and thus, explains the altered PNP catalytic efficiency.
We generated 105 and 99 reactive trajectories respectively for the light and heavy PNP enzymes, respectively. To eliminate correlation with the starting trajectories, only the last 40 reactive trajectories were collected for further analysis in both cases. These unbiased reactive trajectories form transition paths between reactants and products for the enzyme-catalyzed reactions. We report results from 10 of the last 40 reactive trajectories maximally separated in trajectory space – i.e. separated by 4 shooting moves.
Reaction Coordinates
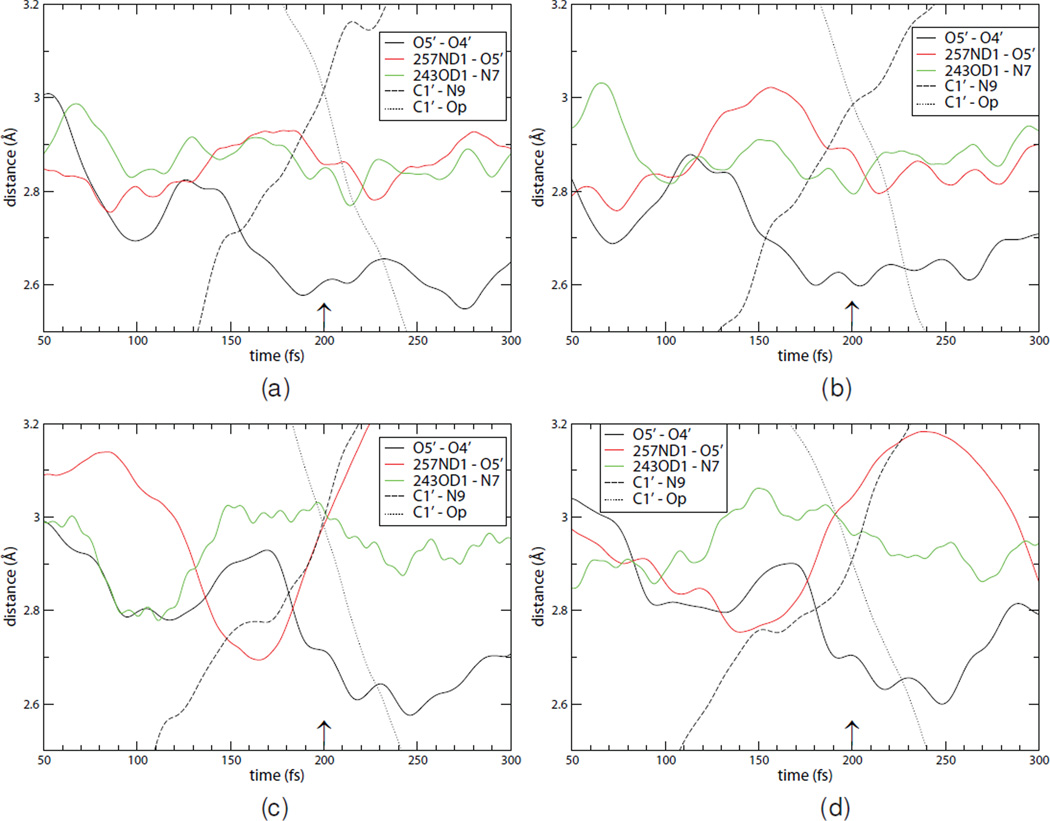
Time series of N-riboside bond-breaking (C1′–N9) and bond-forming (C1′–OP) distances were computed for reactive trajectories. As in our earlier reported calculations3 the barrier crossing event occurs in a sub-picosecond timescale for both light and heavy PNPs in reactive trajectories. During the reaction, resulting in N-ribosidic bond breaking, catalytic site residues and substrate oxygen atoms undergo regular dynamic motions. In the time series for the interaction of His257 ND1–O5′ and O5′–O4′ distances, the motion of these atom pairs are cooperative with the transition state formation. Two representative trajectories for light PNP demonstrate that ribosidic bond scission precedes formation of the α-D-ribose 1-phosphate product (Fig 2, panels a–b). The oscillatory nature of the O5′–O4′ motion, and the promoting vibration via the hydrogen bond contact of O5′ to His257 is less harmonic than was observed in implicit water.3 This difference is likely due to the damping presence of individual solvent molecule interactions. Two representative trajectories for the heavy enzyme, aligned so that the equivalence point for loss of the ribosidic bond and the bond length to the phosphoryl group assigned to occur at 200 fs, are also shown (Fig. 2, panels c–d).
Figure 2.
Time series from reactive trajectories from the transition path ensembles aligned to the crossing points of ribosidic bond breakage and ribose-1 phosphate formation for both light (panels a and b) and heavy (panels c and d) enzymes. The time series demonstrate that the reaction occurs as previously proposed via scission of the ribosidic bond assisted by a promoting vibration, followed by stabilization of the leaving group with another promoting vibration. The trajectories are aligned so that 200 fs (marked with an arrow) is chosen as the transition point. The heavy enzyme shows significantly delayed timing of these promoting vibration events relative to the transition point as compared to the naturally occurring mass variant.
In the first segment of the PNP reaction coordinate, compression of the O4′–O5′ distance near the crossing region leads to loss of the N-ribosidic bond at the transition state, yielding a fully formed ribocation without significant bond participation of the attacking phosphate nucleophile (Fig. 2). Dynamic motion linked to the ribocation feature of the transition state formation is initiated by interactions with His257, hydrogen bonded to the 5′-hydroxyl group, in a motion that pushes O5′ toward the ribosyl ring to cause compression of the O5′–O4′ distance. This ballistic motion results in the His257-ND1 to O5′ distance increasing as O5′ is moving toward O4′, a motion contributing to formation of the ribocation transition state. Compression of the O5′, O4′ oxygen atoms causes electrons from the ribosyl group to migrate into the purine from the C1′–N9 bond. Electron addition to the purine would create an unfavorable anionic leaving group. However, compression of Asn243 with N7 to approximately 2.8 Å at the transition point protonates N7 and neutralizes the charge to assist formation of the purine leaving group. In addition to the interaction with Asn243, Glu201 and a nearby water molecule stabilizes electron delocalization into the purine ring. As barrier crossing for human PNP requires concerted ribocation formation and leaving group activation, the TPS analysis reported here considers both interactions.
Formation of the ribocation is associated with a more planar ribosyl group via a C2′-endo to C3′-endo conformational change. Following transition state formation, the reaction coordinate is completed as the ribosyl group continues its geometric change to migrate the C1′ anomeric carbon toward the phosphate, resulting in formation of α-D-ribose 1-phosphate in a process termed “nucleophilic displacement by electrophile migration”.12 We note that the choice of the ribosidic bond breaking and phosphoryl bond forming to be a ?reaction coordinate? is not meant to imply that these along form the set of motions needed for reaction. In fact that this is not the case is exactly the point to this letter. They are simply chosen as convenient physical means to demark reaction progress.
Mass alters promoting vibrations
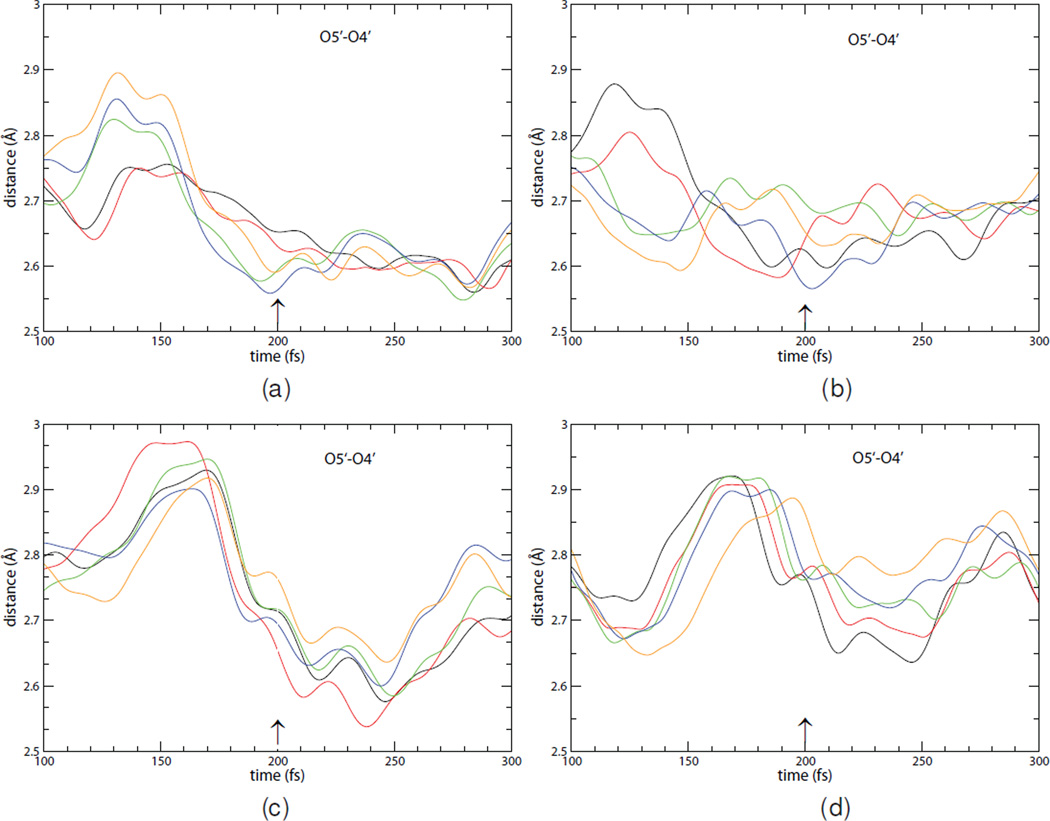
A detailed analysis of multiple individual reactive trajectories is needed to understand the atomic level cause of lowered barrier crossing probability, as each crossing will differ in atomic detail. The reaction is initiated via a compression mode of three oxygens, those of O5′, O4′ in the ribosyl group and the OP from the phosphate nucleophile. The electronegativity of the oxygen atoms in close proximity polarizes the ribosidic bond. Analysis of statistically independent reactive trajectories was used to calculate this distance as the transition state is traversed (Fig. 3, only 5 trajectories per panel are presented for clarity, with panels a–b corresponding to the native enzyme and panels c–d corresponding to the heavy enzyme). The O4′–O5′ distance achieves a minimum of 2.56–2.60 Å for native PNP and occurs 7–11 fs before the transition state is reached at 200 fs. This ribosyl group bond compression happens just before the transition state occurs, and can be considered an initiating event resulting from polarization of the ribosidic bond. In the heavy enzyme, a broader range of O4′–O5′ minima are found at 2.54–2.72 Å and crucially, the compression minimum is reached 35–55 fs after the transition state is formed at 200 fs. At the transition state for heavy PNP the O5′–O4′ distance is larger, in the range of 2.75 to 2.80 ÅQuantum chemical calculations have established that loss of the ribosidic bond is exquisitely sensitive to the O4′–O5′ distance and small variations are important in the catalytic efficiency of PNP.13 All trajectories shown are reactive trajectories, thus these compression distances, together with leaving group interactions (see below) are sufficient to cause reaction. Weaker O4′–O5′ compression is one factor expressed in the reduced catalytic efficiency for heavy PNP. These changes are due solely to changes in the vibrational structure of the enzyme, and this work establishes the way fs vibrational modes interact with the substrate. As substrate is unchanged in the light and heavy enzymes, and exchangeable protons in the reaction coordinate (for example those on His257) are unchanged, the altered probability of barrier crossing originates purely from the mass-dependent vibrational states of the enzyme protein.
Figure 3.
These panels demonstrate the effect of heavy atom replacement on the promoting vibration that modulates the O5-O4 distance in 10 reactive trajectories in the light (panels a and b) and the heavy enzyme (panels c and d.) The results from 5 different trajectories are depicted in each panel. As in the previous figure, the trajectories are aligned so that 200 fs is the transition point (marked with an arrow.) There is clearly a significant delay in the promoting vibration relative to the transition point in the heavy enzyme.
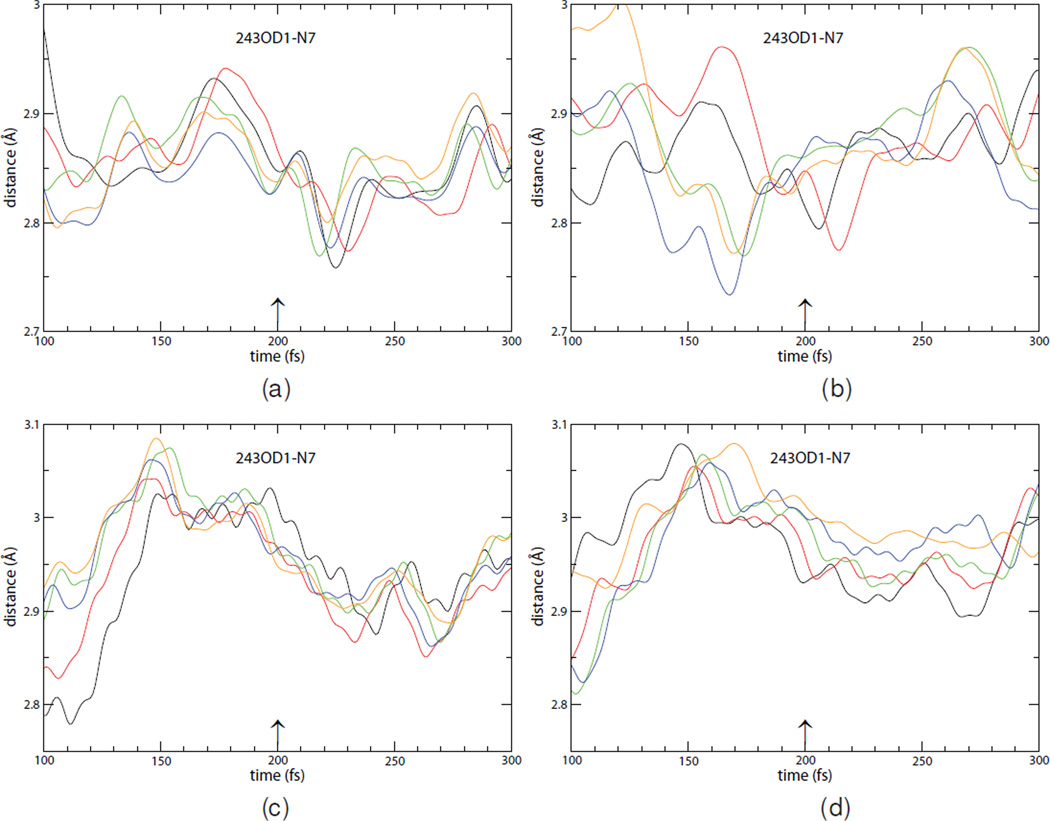
Although the His257-O5′–O4′ mode is important to barrier crossing, leaving group interactions are also essential for catalysis. Asn243 is important in stabilizing the protonated N7 in the purine base for leaving group interactions. This proton originates from solvent water and neutralizes charge on the purine base to favor barrier crossing.12 These interactions should also occur near the transition state to be maximally effective for barrier crossing. In native PNP the minimum range for this interaction is 2.77–2.80 Å and it occurs 6–22 fs after the transition state (Fig. 4, panels a–b.) In the heavy enzyme, the distance is 2.86–2.94 Å and this minimum is achieved later, 63–85 fs after the transition state (Fig. 4, panels c–d.) Similar to His257-mediated compression of the oxygen stack, there is greater variability, greater time-lag and therefore weaker coordination of this leaving group interaction to barrier crossing. Chemistry occurs for PNP when the ribocation and leaving group interactions are coordinated. We hypothesize that the 30% loss of catalytic efficiency in heavy PNP results from an increase in non-reactive trajectories caused by loss of coordination in promoting vibrations causing polarization of the scissile bond and stabilization of the leaving group.
Figure 4.
These panels are as in Fig. 3 except they show the effects of mass substitution in 10 reactive trajectories in the light (panels a and b) and the heavy enzyme (panels c and d) in leaving group interactions. The results from 5 different trajectories are depicted in each panel. As in the previous figure, the trajectories are aligned so that 200 fs is the transition point. Again, as in the previous figure, mass substitution causes a significant shift in timing of the maximum effect of this stabilizing promoting vibration.
The results of this QM/MM transition path sampling study comparing barrier crossing catalyzed by native and heavy PNPs shows the probability of forming the transition state is altered by increased catalyst mass. Heavy isotope replacement of C, N and non-exchangeable H atoms in PNP affects the intrinsic protein motion, which, in turn, couples directly to transition state formation and barrier crossing in this enzyme catalyzed reaction.
Given the modest decrease in barrier crossing with heavy PNP, it is expected that the changes in dynamic range of motion are also subtle. The same types of promoting vibrations that are involved in ribocation formation (His257–O5′ coupling) and leaving group stabilization (Asn243–N7 coupling) are present in heavy and light enzymes but their efficiency is modified. Transition state activation via these promoting vibrations requires simultaneous formation of the ribocation and leaving group interactions. We are able to identify a decreased correlation of these promoting vibrations, with maximum compression of the polarizing oxygen stack occurring some femtoseconds after ribocation formation. In addition, the average compression distance of the polarizing oxygen stack in the heavy enzyme is greater than these distances in the native enzyme.
Another major interaction for barrier crossing involves stabilization of the leaving group. In this interaction, there are also longer overall distances between enzyme and reactants in all reactive trajectories for heavy PNP. As indicated above, the minimum distance, corresponding to the maximum leaving group interaction, is reached after ribocation formation has occurred. Clearly these suboptimal dynamic interactions still allow for reaction to occur since all observed TPS ensembles are comprised of reactive events. The dynamic changes in vibrational structure found in the Born-Oppenheimer enzyme disrupt the approach to and departure from the transition state region, making the process less efficient. PNP enzyme function, resulting in the successful catalysis of this reaction, combines coupled features that have evolved in the context of their natural vibrational frequencies. The small perturbation induced by heavy isotope replacement slightly alters the dynamic features, causing less frequent coordination of the multiple dynamic events that lead to barrier crossing.
The results we describe are fully consistent with mass-dependent femtosecond vibrations being central to the function of enzymes. We again emphasize that the experimental results demonstrate that only rapid vibrations are effected in the enzyme. Two critical examples of these alterations have been identified in this letter. These local catalytic-site promoting vibrations are part of the reaction coordinate. Others have questioned the role of dynamics in enzymatic function.5–7 Other groups question whether it is possible that such rapid motions can be linked to chemistry since enzymatic catalytic cycles have timescales in the millisecond range. The combination of the experimental results demonstrating mass-dependent catalytic effects and the present computational identification of altered motions linked to barrier crossing establishes the importance of these effects. Enzymes have been optimized through evolution to function with the naturally occurring isotopic elements. Enzyme structures have been optimized to hunt efficiently through configuration space for catalytically competent conformations based on the bond vibrational modes of the natural isotopic compositions. Mass perturbations in this hunt decrease the efficiency of the search; and thus decrease the rate of on-enzyme chemistry. The unique combination of experimental and computational approaches to catalysis with mass-altered PNP shows in this specific example, how catalytic efficiency is influenced by femtosecond bond vibrational modes.
Computational Methods
The crystal structure of the trimeric human PNP in complex with the transition state analogue, Immucillin-H, and phosphate (PDB code 1RR6) was used to build the simulation models. CHARMM (version 35b1)14 was used for simulations. The catalytic site residue Glu201 was deprotonated at neutral pH to stabilize the purine base. His257 was also modeled as the neutral form with a proton at NE leaving ND to interact with 5′-hydroxyl of ribose ring.15 The transition state (TS) inhibitor was modified to the guanosine substrate by replacing atoms at N4 with O4 and C9 with N9. Atom N7 of the purine base was protonated because transition state analysis has established N7 protonation to N7H before reaching the transition state.4 For heavy PNP, the atomic masses of C, N and nonexchangeable H of the amino acids were modified with heavy isotopes. Exhangeable protons are identified as solvent exposed ionizable protons. This is of course a manual replacement process. Simulations for light and heavy PNPs were performed separately following the same protocol.
All crystallographic waters were retained. In addition, the system was solvated with a water sphere of 60 Å radius and neutralized with counterions, then followed with minimization, heating and equilibration with Langevin Dynamics.
For QM/MM simulations, the QM region consisted of 40 atoms including the protonated N-guanosine and moieties. The PM3 semiempirical methodi was used to calculate the electronic structure of the QM atoms. PM3 was chosen because of the reactive phosphate group in the QM region. The remaining atoms of the protein, ions and solvent water molecules were in the MM region. There were no covalent bonds between the atoms of the QM and the MM regions.
In the MM region, the CHARMM27 all-atom force field was used for protein and ion species. Explicit water molecules were represented by the TIP3P model.16 For the MM atoms, the SHAKE algorithm was used to constrain all bonds involving hydrogen atoms. The time step for integration was 1 fs. The system was minimized using 150 steps of steepest descent (SD) followed by 6,000 steps of adopted basis Newton-Raphson (ABNR) minimization. The temperature of the system was slowly increased to 300 K within 70 ps, and this temperature was held for the rest of the simulations. Harmonic constraints were applied to the system and gradually released during the minimization and heating steps. After the heating phase, the system was equilibrated with velocities assigned from a Gaussian distribution every 100 steps at 300 K for another 10 ps and then dynamically equilibrated for a final 150 ps. This final structure was used as a starting configuration for TPS study.
Transition path sampling (TPS)17,18 was used to simulate the rare chemical event of catalysis. We,19,20 and others,21,22 have previously reported extensively on its use in enzymatic reactions.
In order to map the reaction path, three non-overlapping regions need to be defined in the configuration space, specifically, reactants, products and intermediate states. The order parameters were defined by the C1′–N9 bond-breaking and C1′–OP bonding-forming distances, such that the reactant region contains all configurations with C1′–N9 bond length < 1.7 Å and C1′–OP bond length > 1.8 Å, while the product region contains all configurations with C1′–N9 bond length > 1.7 Å and C1′–OP bond length < 1.8 Å.
Acknowledgement
The authors acknowledge the support of the National Institutes of Health Grant GM068036.
References
- 1.Núñez S, Antoniou D, Schramm VL, Schwartz SD. Promoting vibrations in human PNP: a molecular dynamics and hybrid quantum mechanical/molecular mechanical study. J. Am. Chem. Soc. 2004;126:15720–15729. doi: 10.1021/ja0457563. [DOI] [PubMed] [Google Scholar]
- 2.Saen-oon S, Ghanem M, Schramm V, Schwartz S. Remote mutations and active site dynamics correlate with catalytic properties of purine nucleoside phosphorylase. Biophys. J. 2008;94:4078–4088. doi: 10.1529/biophysj.107.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saen-oon S, Quaytman S, Schramm V, Schwartz S. Atomic detail of chemical transformation at the transition state of an enzymatic reaction. Proc. Natl. Acad. Sci. USA. 2008;105:16543–16548. doi: 10.1073/pnas.0808413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewandowicz A, Schramm VL. Transition state analysis for human and plasmodium falciparum purine nucleoside phosphorylases. Biochemistry. 2004;43:1458–1468. doi: 10.1021/bi0359123. [DOI] [PubMed] [Google Scholar]
- 5.Pisliakov AV, Cao J, Kamerlin SCL, Warshel A. Enzyme millisecond conformational dynamics do not catalyze the chemical step. Proc. Natl. Acad. Sci. USA. 2009;78:1339–1375. doi: 10.1073/pnas.0909150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamerlin SC, Warshel A. At the dawn of the 21st century: is dynamics the missing link for understanding enzyme catalysis? Proteins. 2010;78:1339–1375. doi: 10.1002/prot.22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammes-Schiffer S, Benkovic S. Relating protein motion to catalysis. Ann. Rev. Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 8.Silva R, Murkin A, Schramm V. Femtosecond dynamics coupled to chemical barrier crossing in a Born-Oppenheimer enzyme. Proc. Natl. Acad. Sci. USA. 2011;108:18661–18665. doi: 10.1073/pnas.1114900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kipp DR, Silva RG, Schramm VL. Mass-dependent bond vibrational dynamics influence catalysis by HIV-1 protease. J. Am. Chem. Soc. 2011;133:19358–19361. doi: 10.1021/ja209391n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schramm V. Enzymatic transition states, transition-state analogs, dynamics, thermodynamics, and lifetimes. Ann. Rev. Biochem. 2011;80:703–732. doi: 10.1146/annurev-biochem-061809-100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolhuis P, Chandler D, Dellago C, Geissler P. Transition path sampling: throwing ropes over mountain passes, in the dark. Annu. Rev. Phys. Chem. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 12.Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, et al. Transition state structure of PNP and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- 13.Núñez S, Wing C, Antoniou D, Schramm VL, Schwartz SD. Insight into catalytically relevant correlated motions in human purine nucleoside phosphorylase. J. Phys. Chem. A. 2006;110:463–476. doi: 10.1021/jp051277u. [DOI] [PubMed] [Google Scholar]
- 14.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: the biomolecular simulation program. J. Comp. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirschi JS, Arora K, III, CLB, Schramm VL. Conformational dynamics in human purine nucleoside phosphorylase with reactants and transition-state analogues. J. Phys. Chem. B. 2010;114:16263–16272. doi: 10.1021/jp108056s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen W, Chandrasekhar J, Madura J, Impey R, Klein M. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 17.Dellago C, Bolhuis P, Csajka F, Chandler D. Transition path sampling and the calculation of rate constants. J. Chem. Phys. 1998;108:1964–1977. [Google Scholar]
- 18.Dellago C, Bolhuis P, Chandler D. Efficient transition path sampling: Application to Lennard-Jones cluster rearrangements. J. Chem. Phys. 1999;108:9236–9245. [Google Scholar]
- 19.Basner JE, Schwartz SD. How enzyme dynamics helps catalyze a reaction, in atomic detail: a transition path sampling study. J. Am. Chem. Soc. 2005;127:13822–13831. doi: 10.1021/ja043320h. [DOI] [PubMed] [Google Scholar]
- 20.Quaytman S, Schwartz S. Reaction coordinates of an enzymatic reaction revealed by transition path sampling. Proc. Natl. Acad. Sci. USA. 2007;104:12253–12258. doi: 10.1073/pnas.0704304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crehuet R, Field MJ. A transition path sampling study of the reaction catalyzed by the enzyme chorismate mutase. J. Phys. Chem. B. 2007;111:5708–5718. doi: 10.1021/jp067629u. [DOI] [PubMed] [Google Scholar]
- 22.Escobedo FA, Borrero EE, Araque JC. Transition path sampling and forward flux sampling. Applications to biological systems. J. Phys. Condend. Matter. 2009;21:333101. doi: 10.1088/0953-8984/21/33/333101. [DOI] [PubMed] [Google Scholar]