Abstract
Methods are presented through which one may isolate and identify novel bacterial DNA-binding proteins. Briefly, the DNA sequence of interest is affixed to beads, then incubated with bacterial cytoplasmic extract. Washes with buffers containing non-specific DNA and low salt concentrations will remove non-adhering and low-specificity DNA-binding proteins, while subsequent washes with higher salt concentrations will elute more specific DNA-binding proteins. Eluted proteins may then be identified by standard proteomic techniques.
Introduction
Bacteria and other organisms utilize specific DNA-binding proteins for a wide range of purposes, from regulation of gene transcription to DNA replication to maintenance of chromatin conformation. Although many DNA-binding proteins have been identified to date, they undoubtedly represent only a tiny fraction of such proteins produced by bacteria. Very often, a researcher will have evidence of a DNA-protein interaction, but without a clue as to the identity of the protein. To date, our laboratory has used the methods described herein to isolate and identify six proteins, most of which had not previously been thought to be DNA-binding proteins (unpublished results and Babb et al., 2006; Burns et al., 2010; Riley et al., 2009). In addition, there had been no reasons to assume a priori that any of these proteins would be associated with our DNAs of interest.
The methods described in this unit do not require any knowledge of the DNA-binding protein’s identity, or even the exact DNA sequence to which it binds. Ideally, the researcher will have some indication that a protein(s) is binding to the DNA of interest, such as electrophoretic mobility shift assay (EMSA) data. The methods then involve adhering the DNA of interest (“bait”) to macroscopic beads, incubating with bacterial cytoplasmic extract to fish out high-affinity DNA-binding proteins, then analyzing the bound proteins by proteomic methods. While the examples described are optimized for two different genera of spirochetes, these methods can be readily adapted for other bacteria. The only major modification one might need to make is the method of bacterial lysis, which needs to be determined empirically.
Strategic Planning
1.A Clean-Ultrapure Reagents
Materials used throughout the assay need to be free of contaminants. Avoiding nuclease contamination is critical, particularly in the absence of EDTA/EGTA. To minimize noise during mass spectrometry sample analysis, polyacrylamide gel reagents must be free of contaminants. Human keratin from skin is often the main source of contamination since it is virtually ubiquitous, so frequently changing gloves can improve results.
1.B DNA Probe
The size, purity/concentration, and overall probe design is paramount to the success of the assay.
1.BI Size
Due to the ability of one Streptavidin molecule to bind four biotin molecules, steric hindrance can be problematic. Small probes (<100bp) can cause the potential binding site(s) to be too close to Streptavidin beads and thus may also restrict protein binding. It’s the authors’ experience that a probe size of 125–425bp is optimal for assay success.
1.BII Purity/Concentration
DNA bait should be free of nucleases, to prevent degradation. Moreover, solutes should not be carried over from probe production procedures, as they may interfere with binding. Since the procedure takes advantage of a 5’ biotin moiety linked to the oligonucleotide used to amplify the probe via PCR, residual/unincorporated oligonucleotide may bind to the Streptavidin beads, reducing the amount of useful DNA bait bound to each bead. Therefore, it is critical to purify the initial template via gel electrophoresis prior to generating adequate amounts of probe. Probe concentration has been shown to be an exceptionally important aspect to the overall success of the assay. To ensure bead saturation, we recommend a probe concentration of 200–450ng/ul. This concentration can be achieved with the required volume by performing several 100ul PCR reactions using purified template DNA, pooling all reactions, and performing an ethanol precipitation (Jutras et al. 2010) or concentrating while performing buffer exchange using the appropriate Amicon/Microcon (Millipore). These methods will remove unincorporated, biotinylated oligonucleotides.
1.BIII Probe design
When constructing DNA bait for the affinity chromatography assay, one should consider the type of probe, the location of the biotin moiety, and the location of the potential binding site(s) of interest. Often promoters are the bait of interest, and thus a double stranded fragment of DNA is appropriate. However, one may have a ssDNA probe synthesized with a 5’ biotin modification. In the case of a dsDNA probe, bait is generated using PCR with one of the oligonucleotide primers modified at its 5’ end. Many companies that synthesize oligonucleotides can provide such modifications during the oligonucleotide synthesis. Since the biotin will interact with the Streptavidin bead, the biotin needs to be located on the 5’ end of an oligonucleotide. A ssDNA probe is not constrained to one particular end and may be modified at either the 3’ or 5’ end. Potential or hypothesized site(s) of interaction(s) should be relatively close to the middle of the probe to provide adequate availability for binding.
1.C In vitro Expression of DNA binding protein
Another aspect that requires some consideration prior to performing the assay is the relative expression profile of your potential target/ligand. Consider a situation where a hypothetical activator is thought to be required for the expression of a protein, which is upregulated during cold shock/stress. Then, one could simulate these in vitro culture conditions in order to potentially induce the expression of the activator. Should a repressor protein be involved in the regulatory network of this hypothetical protein, then comparing promoter pulldown assay results from lysates generated under inducing and non-inducing conditions may prove to be insightful.
1.D Lysate preparation and protein concentration
Depending on the bait DNA and DNA-binding protein, it can be difficult to provide conditions which are favorable for DNA-protein interactions. To that end, the buffer described below was developed to facilitate most DNA binding. However, a particular co-factor may be required for high affinity interactions, which is not present, or at insufficient levels. The trouble shooting section further addresses this issue. More often, many DNA binding proteins are expressed at very low levels in culture in relation to other molecules, which interact less specifically with DNA (Ball et al. 1992, Azam and Ishihama 1999). To ensure DNA binding proteins are not beyond the limits of detection, we often perform our assays with a substantial amount of culture. 107 cells/ml in a 250 ml- 1Liter of culture, which corresponds to 2.5–10 billion cells are routinely used for cell free extract preparations. The amount of DNA-binding protein present in the cytoplasmic extract will vary and is thus a step that will require some optimization. Due to the high amounts of proteins in many spirochetal culture media, estimating the protein concentration of such preparation is virtually impossible. Quantification of protein concentration from successful assays in other bacteria has ranged from 10ug–250ug/ml. This will increase the background, although it does not have any predictable effect on DNA binding protein specificity or activity. Below are three different methods for extracting soluble, functional DNA binding proteins.
Basic Protocol 1: Cell Lysis by Sonication with freeze thaw
Cell lysis using several freeze/thaw cycles followed by sonication is an efficient method for Prokaryotes, which lack inherently stable outer membranes. Furthermore, this method limits the physical and chemical stresses other procedures employ. As such, it is ideal for downstream applications where biologically functional products are desired.
Materials
Bacterial culture of interest
Nuclease-free water
Sonicator for disrupting bacterial cells
-
1.
Following harvest of bacterial culture via centrifugation, gently wash pellet with 5–10ml of ultrapure or nuclease free water.
Repeat washes two times, three total.
-
2.
Freeze at −80°C overnight, or longer. Use pelleted cells within 6 months.
-
3.
Slowly thaw pellet on ice. Return to −80°C for at least one hour, followed by thawing again on ice.
-
4.
Repeat for a total of 3 freeze thaw cycles.
-
5.
Once complete, resuspend the pellet in as little BS/THES buffer as possible.
This typically requires 1–3ml. It’s critical to produce a lysate that is as concentrated as possible while being sure to create a homogenous mixture. In doing so, you will be ensuring that potentially low level expressed proteins are concentrated while facilitating efficient sonication.
-
6.
Sonicate resuspended pellet on ice, at 10–40% amplitude for pulses of 10 seconds, followed by 1 minute off. Repeat for a total of five pulses.
Depending on the organism of interest, this step may be changed and/or optimized. Consult specific, published, sonication procedures for lysis of the prokaryotic cell of interest. In the authors’ experiences, it is preferable to proceed directly to Basic Protocol 2 on the same day that the lysate is made. Storage for one day at 4°C may be acceptable, but yield will generally be reduced. Do not freeze the lysate.
Alternate Protocol 1: Cell Lysis using BPER-II Extraction
Commercially available, mild, non-ionic detergents are effective at lysing prokaryotic cells while limiting down stream complications. By disrupting the hydrophobic interactions of the membrane(s), while maintaining secondary and tertiary structure, functional cytoplasmic proteins are released and often collected in the soluble fraction. Commercially available products that fall into this category include BPER-II (Thermo Scientific) and Bacterial PE LB (G-Biosciences); both of which are amendable to DNA-protein interaction assays.
Materials
Frozen cell pellets
Lysis buffer
-
1.
Thaw previously pelleted cells from −80°C on ice.
-
2.
Prepare lysis buffer by adding 200ul of BPER-II, 4.7ml of BS/THES buffer, and 100ul of Lysozyme (10U/ml).
-
3.
Vortex buffer and filter sterilize using a 0.2uM filter.
-
4.
Add buffer to pellet starting with 1ml and pipetting up and down to resuspend. Continue adding buffer, 250ul at a time as needed to create a homogenous mixture.
The amount will vary depending on the size of the pellet. Generally, 1–3ml should be sufficient.
-
5.
Once pellet is completely resuspended shake mixture at room temperature for 20–30 minutes.
Shaking can be accomplished using a standard Belly Dancer or an incubator capable of shaking cultures. 50–100 rpm is ideal.
-
6.
Transfer to 1.5ml microfuge tubes and centrifuge >17,000×g at 4°C for 30 minutes.
-
7.
Upon clearing lysates of cellular debris, transfer soluble fraction to fresh microfuge tube, store on ice, and proceed to step 1 of DNA affinity chromatography protocol.
Alternate Protocol 2: Cell Lysis by Vigorous Vortexing and Freeze Thaw Cycles
An alternative method for lysis of bacteria with relatively fragile cell membranes includes repeated snap freezing and vigorous vortexing. The protocol can be adapted to address specific requirements but the broad outline is given below.
Materials
Cell suspension
-
BS/THES buffer
-
1.
Following resuspension of pelleted cells in BS/THES buffer, snap freeze cells at −80°C for 15 min.
-
2.
Slowly thaw on ice.
-
3.
Vortex cells at high oscillation for 1 min followed by incubation on ice for 1 min. Repeat two more times, for a total of three vortex cycles.
-
4.
Freeze cell suspension again at −80°C for 15 min and repeat steps 2 and 3.
-
5.
Repeat step 4 one more time.
-
6.
Transfer suspension to 1.5 ml microfuge tubes and centrifuge at >/17,000 × g and 4°C for 30 minutes.
-
7.
Transfer soluble fraction to fresh microfuge tubes, store on ice, and proceed to step 1 of the DNA affinity chromatography protocol.
-
1.
Basic Protocol 2A. DNA affinity chromatography: Borrelia burgdorferi & Leptospira interrogans
The following protocols have been optimized for purification of DNA-binding proteins from the spirochetes B. burgdorferi and L. Interrogans. The same procedures will probably work for any bacterium, but researchers will need to determine that empirically. See “Troubleshooting” for hints on optimization.
Materials
1.5ml microfuge tubes
M-280 Dynabeads (Invitrogen)
Magnetic Column: PolyATtract System 1000 (Promega)
THES Buffer
BS Buffer
Nuclease Free Water
TE
2X B/W buffer
Elution buffers 100mM–1M
Poly dI-dC or dA-dT
Nonspecific inhibitor DNA (Optional-See Trouble Shooting Section)
70°C Heat Block
Rocker/Belly Dancer
Spinning Rotor
Ice
Preparing Invitrogen Dynabeads M-280 Streptavidin
-
1.
Vigorously shake Dynabeads to resuspend in supplied preservative. Upon completion, beads should have a uniform, rust color.
-
2.
Add 200ul of beads per reaction to 1.5ml microfuge tubes.
One may desire to use less to conserve beads. As little as 100ul can be used, but this decreases the potential amount of biotinylated DNA bound and thus can adversely affect the reaction. Beads may be re-used, see Invitrogen instructions for doing so.
-
3.
Secure microfuge tube on a Promega PolyATract 1000 magnet to pull Dina beads down.
-
4.
Using a P200, remove the preservative without disrupting the beads and discard supernatant.
-
5.
Wash Beads with 500ul of 2X B/W buffer (recipe follows). Washing involves removing the microfuge tube from the magnet. Re-suspending the beads in the desired amount of wash buffer; tapping the tube to ensure re-suspension, and reapplying the tube to the magnet to pulldown the beads.
-
6.
Discard wash and repeat two more times (3 total).
Probe binding/Washing
-
7.
Following final wash, remove 2X B/W buffer, and resuspend in 190ul of 2X B/W buffer. This creates a 1X B/W buffer suitable for biotinylated probe DNA: Streptavidin binding. 190ul is used instead of 200ul since it is assumed that all 2X B/W buffer cannot be removed from previous wash.
-
8.
Add 200ul of 200–400ng/ul biotinylated probe DNA diluted in nuclease-free water. See “General considerations for probe design” for further details.
-
9.
Incubate while rolling at room temperature for 20 minutes. We prefer a Labnet Revolver since it is able to move liquid along all three axis of the microfuge tube. Alternatively, microfuge tubes affixed to “Bellydancers” or other rocking devices will work. However, it is critical that the mixture be constantly moving in the tube.
-
10.
Pull beads down by applying to magnet. Optional: Reserve a portion of the supernatant for later analysis.
-
11.
Repeat steps 8–10 using an equal volume (and concentration) of probe to ensure that the beads are saturated with DNA. Reserving supernatant for analysis is optional, but recommended.
-
12.
Wash the probe-bead complex with 400ul of TE (recipe follows). Reserve wash for analysis.
-
13.
Repeat washes twice (total of 3 washes).
This will dilute the high NaCl concentration in the binding/washing buffer in addition to removing any unbound DNA probe and potential contaminates.
Preparing DNA probe for lysate incubation
-
14.
To ensure that the DNA probe is in reactions conditions suitable for DNA-Protein interactions wash the probe-beads with 500ul of BS/THES Buffer.
-
15.
Repeat Wash once
-
16.
Wash with 500ul of BS/THES buffer supplemented with 10ug/ml of Poly dI-dC. This step will begin to introduce an excess of non-specific, randomly structured DNA-like molecules, which will effectively sequester non-specific DNA binding proteins. Alternative inhibition options are available, and are discussed below and in the trouble shooting section.
Bait-target/ligand binding
-
17.
Apply 200ul of BS/THES buffer to probe-bead complex along with 600–750ul of cleared lysate (supernatant). To provide an excess of non-specific competitor DNA add 25–100ug of Poly dI-dC. Roll for 30 minutes at room temperature or 4 °C (when repeated in step 19).
Amount of Poly dI-dC added should be optimized, more stringent conditions will result in a decrease in non-specific interactions. Although, depending on protein-probe affinity, structure of probe, concentration of required co-factors, etc the Poly dI-dC concentration can become inhibitory after binding capacity threshold is reached. Thus, it is important to optimize this step.
Other non-specific competitors that we have used with success are 1000X excess of PCR amplified, non-biotinylated DNA using an alternative genomic region. Sonicated/sheared genomic Escherichia coli is also suitable in some situations.
Incubations under room temperature or refrigerated conditions may increase or decrease the stability of protein-DNA complex. As such, this will require some optimization. It is often best to attempt room temperature incubation first. Alternatively, this step may be performed once at room temperature and once at 4 °C.
-
18.
Pull bead-probe-protein complex down using magnet and discard supernatant.
-
19.
Repeat step 17 and 18. Conditions can be varied (see text below 17).
-
20.
Wash bead-probe-protein complex with 500ul of BS/THES buffer supplemented with 10ug/ml Poly dI-dC.
-
21.
Pull down beads and discard supernatant.
-
22.
Repeat steps 20–21 4 times (5 washes total).
-
23.
Wash with 500ul of BS/THES buffer
-
24.
Pull down beads and discard supernatant
-
25.
Repeat once (2 washes total).
Eluting target/ligand
-
26.
Add 120ul of 100mM NaCl Elution buffer (recipe follows)
-
27.
Roll at room temperature for 3–5 minutes.
-
28.
Pull down beads and SAVE elution
-
29.
Repeat steps 26–28 with increasing NaCl concentration in each elution. Typically, 6 elutions are preformed: 100mM NaCl, 200mM NaCl, 300mM NaCl, 500mM NaCl, 750mM NaCl, 1M NaCl. Keep elutions on ice or at 4 °C until SDS PAGE. In the event of a long time lapse between legs of the experiment, protein elutions should be frozen at −20°C.
-
30.
Add 35ul of ultrapure, nuclease free water to beads-probe mixture following final elution. Incubate at 70°C for 10 minutes.
-
31.
Pull down beads and save supernatant.
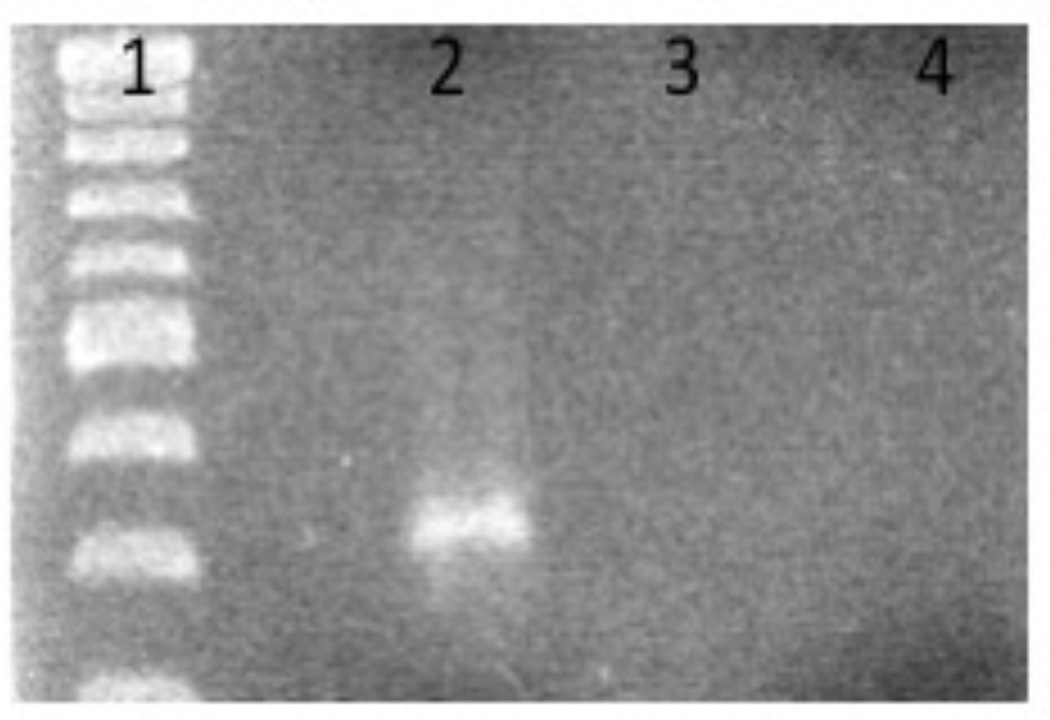
An incubation at 70°C will break the streptavidin-biotin interaction (Holmberg et al. 2005) eluting the DNA probe. This elution can be analyzed via agarose gel electrophoresis along with previous samples collected (steps 10, 12–13) to determine the efficiency of each step (Fig. 1). This final step is critical since it indicates that the DNA probe was the bait during each cytoplasmic extract incubation and confirms that the beads alone were not the target of bound proteins. To further confirm this one may run a control experiment that lacks biotinylated DNA (replace with nuclease free water).
Figure 1.
1% Agarose Gel electrophoresis following DNA affinity chromatography. Lane 1: Molecular Weight Marker. Lane 2 Eluted probe following assay completion using a 200 ng/ul starting probe concentration. Lane 3 Dynabeads alone following DNA affinity chromatography. Lane 4 Elution following DNA affinity chromatography using 85 ng/ul of probe.
Basic Protocol 3. Visualization and Identification of DNA binding proteins
Successful identification of DNA binding protein(s) is contingent on a careful separation of the eluted proteins. The standard method of discontinuous gel electrophoresis is used for the separation of eluates (Laemmli, 1970). In this system, sodium dodecylsulfate (SDS) imparts a constant negative charge-to-mass ratio to proteins via the negative sulfate group, thus enabling their separation based on molecular size. A reducing agent, such as Beta-mercaptoethanol, is typically included to prevent disulfide bond formation. For a detailed protocol on one-dimensional SDS gel electrophoresis readers are referred to Gallagher, 2006 and Appendix 3M). However, some important considerations are discussed below:
-
1.
Percentage of the gels- For an efficient separation of small DNA binding proteins we recommend using 15% or 20% gels. Lower percentage gels may result in inefficient separation of close molecular size species.
-
2.
Purity of the reagents- All buffers/reagents used for casting gels should be free of any contaminants. We recommend that buffers for casting gels should be made fresh and filter sterilized. Similarly, a new stock of reagents should be used for casting gels for this purpose, as contaminated reagents may adversely affect down-stream protein identification by mass spectrometry (MS). Wells should be rinsed with 1 X SDS-running buffer to remove any residual ammonium persulfate (APS) that may affect down-stream applications. To further eliminate excess radicals supplied by APS, gels may be casted and left, covered, in moist paper towel at 4°C overnight.
-
3.
Cleanliness of the equipment- If glass plates are used for casting gels, they should be cleaned with laboratory detergent and wiped with 70% ethanol soaked paper tissue.
-
4.
Staining of gels- The location of proteins in SDS-polyacrylamide gels can be detected by SYPRO-Ruby (Molecular Probes, Eugene, OR) as per manufacturer’s recommendations or by silver staining. Though both stains have comparable sensitivities, SYPRO-Ruby has advantages over silver staining in terms of decreased hands-on time, less toxic, and better consistency.
-
5.
Visualization of stained gels- Prior to visualization of SYPRO stained gels, UV-transilluminator should be thoroughly cleaned with 70% ethanol and water. Alternatively, gels can be placed on a clean glass plate and then visualized on a UV-transilluminator.
Reagent and Solutions
THES Buffer
50mM Tri HCl (pH 7.5)
10mM EDTA
20% Sucrose (mass/vol)
140mM NaCl
0.7% Protease Inhibitor Cocktail II (sigma) (vol/vol)
0.1% Phosphatase Inhibitor Cocktail II (sigma) (vol/vol)
5× BS Buffer *
50mM HEPES
25mM CaCl2
250mM KCl
60% Glycerol
*Additional salts may be added to buffer if one suspects that a cofactor is required for binding. Examples include Zn, Fe, Mn, etc. Typical final concentrations of additional salts in the 5X BS Buffer is 25mM–100mM, however some optimization will be required
BS/THES BINDING WASHING BUFFER
44.3% THES Buffer
20.0% BS Buffer
35.7% Nuclease Free Water
i.e.: 13.3ml THES Buffer: 6ml 5XBS Buffer: 10.7ml nuclease-free water
Final concentration of all components in BS/THES Binding Washing Buffer
22mM Tris HCl (pH 7.5)
4.4mM EDTA
8.9% Sucrose (mass/vol)
62mM NaCl
0.3% Protease Inhibitor
0.04% Phosphatase Inhibitor
10mM HEPES
5mM CaCl2
50mM KCl
12% Glycerol
Filter Sterilize Final Mixture using 0.2uM filter
2 × B/W Buffer
10mM Tris HCl (pH 7.5)
1mM EDTA
2M NaCl
Elution Buffer
25mM Tris HCl
Varying [NaCl]: 100mM, 200mM, 300mM, 500mM, 750mM, 1M NaCl.
COMMENTARY
Background Information
Transcription, DNA recombination, DNA replication and modification are all processes that require the interaction of proteins with nucleic acids. Experimental approaches to study such associations have taken advantage of the biochemical and biophysical properties of nucleotides/sides. A variety of interfering and protecting gel footprinting assays are invaluable for examining specific interactions. Other techniques include EMSA, ChIP-chip/seq, solid discontinuous phase transcription factor binding assays, circular dichroism, electron microscopy, crystallography, and crosslinking. However, most often the aforementioned assays require the identification of a candidate protein prior to downstream experimentation. There are a variety of methods to identify these molecules, but we have found that a promoter pulldown/DNA affinity chromatography is a fast, reliable, and reproducible method for doing so. The extent of the bait is not limited to promoters and can be used with downstream/transcribed regions.
This procedure utilizes an ionic titration to disrupt protein-nucleic acids interactions at a concentration, which exceeds the empirical Na2+ concentration within the cell. In the described protocol Van der Waals, Hydrogen (Seeman, Rosenberg, Rich 1976) bonding, and hydrophobic interactions are not directly taken into account; each of which being important in DNA-protein interactions. Indeed, H-bonds account for the majority of interactions between proteins and dsDNA (LeJune et al. 2005). An alternative elution method could incorporate aspects of these types of associations by altering pH or temperature in addition to ionic conditions.
The conformational topology of DNA in vitro and in vivo differs significantly (summarized in Peters and Maher 2010). Furthermore, the cytoplasmic microenvironment is littered with molecules, which influence local and distal interactions in addition to DNA structure, often referred to as molecular crowding (Miyoshi and Sugimoto 2008). The latter points illustrate the difficulties associated with simulating these conditions in vitro. Practical examples are those DNA binding proteins that recognize specific DNA topology rather than nucleic acids sequence (Hadjfrangiskout and Koechler 2008) Therefore, many relevant interactions may not occur in vitro and thus somewhat limits the promoter pulldown/DNA affinity chromatography assay efficacy. Although, many sequence specific DNA binding protein instigate interaction via the unique information displayed by the base edge of each nucleotide or directly with phosphate atoms in the DNA backbone (LeJune et al. 2005). These types of proteins should be efficiently purified via the above assay.
Their helical shape corresponding to the distinct endoflagellum/axial fibril classically defines spirochetes. Further distinguishing characteristics include an outer sheath, a protoplasmic cylinder, and varying peptidoglycan complexity. However, while structurally complex, spirochetes are relatively fragile making their lysis straightforward. In contrast, the morphological ultrastructure of other prokaryotes may create their own unique challenges associated with cytoplasmic protein isolation. The above cell lysis procedures highlight merely a few of many potential methods for effective membrane rupture and cytoplasmic protein isolation, which may require some optimization dependent on the organism of interest. However, we contend that the DNA affinity chromatography aspect of the assay is a broad scale platform, lending itself to a variety of applications.
Trouble Shooting
As previously described, in vitro assays have their limitations. Simulating inducing or repressing conditions in culture may increase the overall success of identifying regulatory proteins. In accordance with the former, improving conditions for some DNA binding proteins may require the supplementation of a specific co-factor. The addition of Fe2+, Zn3+, or Mn2+ (Ma et al. 2011) could facilitate target protein binding to DNA bait via the significant effects co-factors exert on protein structure and function (Zhou et al. 2007).
Should the procedure fail, checking the DNA bait concentration before and after, in addition to assuring DNA integrity, is highly recommended. A simple way to do so is spectrophotometrically using a Nanodrop (Thermo Scientific) or a more complete analysis can be performed via Bioanalyzer analysis (Agilent). We contend that a high starting probe concentration is essential to successful purification (Fig. 1). It is also feasible that further alterations be necessary to the DNA bait. Based on the nature of the probe and location of potential binding site(s) altering the position of the biotin moiety may increase success. By switching the tether to the downstream portion of the bait, binding may increase by decreasing steric hindrance, alleviating torsional stress, or preventing aberrant probe interactions.
The addition of Glycerol and sucrose in the binding buffer typically increases protein stability and protects against protein denaturation by providing an optimal microenvironment of hydration around the protein (Timasheff 1993). The concentration of Glycerol and Sucrose can be varied or substituted for the other to increase potential stability (Ruan et al 2003), which will be target-dependent and difficult to predict in advance.
As described in the notes above, varying the type and concentration of competitor DNA will change the amount of background or stringency of the assay. Different non-specific inhibitors include: Poly dA-dT/dI-dC, vortex-sheared and/or sonicated E. coli DNA, Salmon sperm DNA, genomic DNA from bacteria of interest, PCR amplified DNA from bacterial DNA of interest. These considerations should be assessed by the investigator, and pursued in the event initial experimentation fails.
Anticipated Results
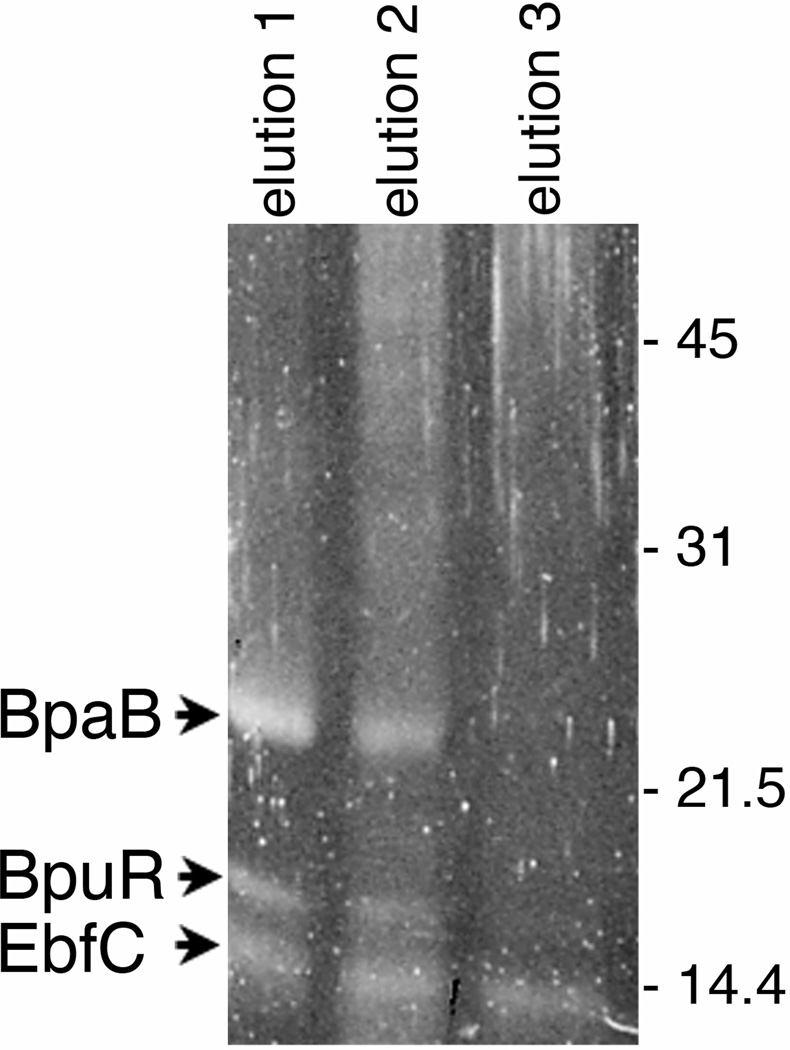
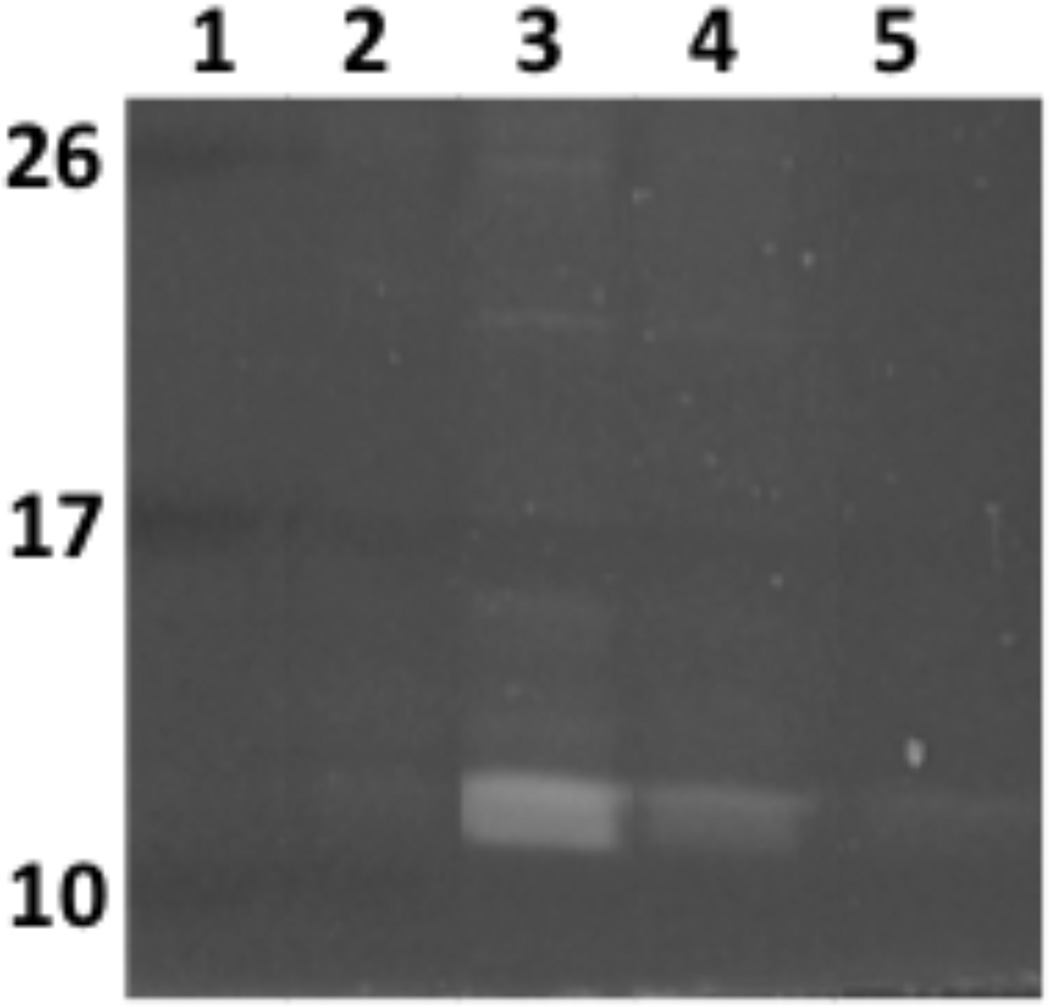
Typically, proteins that elute at higher salt concentrations are those which have the highest affinity for the DNA bait. Electrostatic interactions are not the only pertinent attractant forces with respect to protein-nucleic acid interactions. Hydrogen bonding, hydrophobic interactions and Van der Waals forces are also relevant and thus, gradually increasing the temperature or pH may also result in the isolation of high affinity protein. Focusing on electrostatic interactions; lower NaCl concentrations (<300mM) are not typically relevant in terms of high affinity interactions. However, it is often a good idea to separate these elutions using SDS PAGE to provide a baseline of background noise and will be dependent on cytoplasmic extract concentration to ensure that the reactions yielded proteins. In the authors’ experiences, high affinity interactions elute at >500mM NaCl (Fig. 2 and 3). Moreover, bands present in lower concentrations that continue to elute at higher salt concentrations typically indicates a highly abundant protein. In this regard, having lower salt concentration elution as a means for comparison is always helpful (Fig. 4). Another result that we often observe is several high molecule weight species eluting a various salt concentrations. Our previous analysis have often identified these as subunits of DNA and RNA polymerase, which in our view is a positive result, reflecting the fact that DNA binding cell machinery are interacting with probe DNA. Indeed, an excess of salt may result in co-purification of RNA polymerase subunits since there exists a surplus of these molecules, which will dissociate at relevant sodium concentrations (Kontur et al. 2010). Figure 2 shows initial results that led to the identification of three B. burgdorferi DNA-binding proteins that we subsequently verified as having high relative affinities for erp Operator DNA. Results of studies to identify L. interrogans DNA-binding proteins are shown in Fig. 3.
Figure 2.
DNA-affinity chromatography using erp Operator DNA and B. burgdorferi cytoplasmic lysate. Following extensive washing, proteins that eluted with increasing concentrations of NaCl were separated by SDS-PAGE and stained with Sypro Ruby. Proteins eluted at 500mM, 750 mM and 1M NaCl are shown.
Figure 3.
L. interrogans cytoplasmic proteins purified by affinity chromatography using lenA 5′ noncoding DNA as bait. Proteins in elutions 1, 2, 3, and 4 were eluted with NaCl at concentrations of 200, 350, 500, 750, and 1 M, respectively. Proteins were separated by SDS-PAGE and visualized with SYPRO-Ruby. Numbers on the right indicate positions of molecular mass standards.
Acknowledgements
Research described in this protocol was funded by US NIH grants R01-AI044254 and R21-AI078111, both awarded to B. Stevenson.
Literature Cited
- Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- Babb K, Bykowski T, Riley SP, Miller MC, DeMoll E, Stevenson B. Borrelia burgdorferi EbfC, a novel, chromosomally-encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete’s resident cp32 prophages. J. Bacteriol. 2006;188:4331–4339. doi: 10.1128/JB.00005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball AA, Osuna R, Ferguson KC, Johnson RC. Dramatic changes in FIS levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns LH, Adams CA, Riley SP, Jutras BL, Bowman A, Chenail AM, Cooley AE, Haselhorst LA, Moore AM, Babb K, Fried MG, Stevenson B. BpaB, a novel protein encoded by the Lyme disease spirochete's cp32 prophages, binds to erp Operator 2 DNA. Nucleic Acids Res. 2010;38:5443–5455. doi: 10.1093/nar/gkq284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR. One-dimensional SDS gel electrophoresis of proteins. Curr. Protoc. Mol. Biol. 2006;75:10.2A.1–10.2A.37. doi: 10.1002/0471142727.mb1002as75. [DOI] [PubMed] [Google Scholar]
- Hadjifrangiskou M, Koehler TM. Intrinsic curvature associated with the coordinately regulated anthrax toxin gene promoters. Microbiology. 2008;154:2501–2512. doi: 10.1099/mic.0.2007/016162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J, Uhlen M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperature. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- Jutras BL, Liu Z, Brissette CA. Simultaneous isolation of Ixodidae and bacterial (Borrelia spp.) genomic DNA. Curr. Protocols Microbiol. 2010 doi: 10.1002/9780471729259.mc01e02s19. 1E.2. [DOI] [PubMed] [Google Scholar]
- Kontur WS, Capp MW, Gries TJ, Saecker RM, Record MT. Probing DNA binding, DNA Opening and Assembly of a Downstream Clamp/Jaw in E. Coli RNA Polymerase- λPR Promoter Complexes Using Salt and the Physiological Anion Glutamate. Biochemistry. 2010;49:4361–4373. doi: 10.1021/bi100092a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lejeune D, Delsaux N, Charloteaux B, Thomas A, Brasseur R. Protein-nucleic acid recognition: statistical analysis of atomic interactions and influence of DNA structure. Proteins. 2005;61:258–271. doi: 10.1002/prot.20607. [DOI] [PubMed] [Google Scholar]
- Ma Z, Lee JW, Helmann JD. Identification of altered function alleles that affect Bacillus subtilis PerR metal ion selectivity. Nucleic Acids Res. 2011;39:5036–5044. doi: 10.1093/nar/gkr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi D, Sugimoto N. Molecular crowding effects on structure and stability of DNA. Biochimie. 2008;90:1040–1051. doi: 10.1016/j.biochi.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Peters JP, Maher LJ. DNA curvature and flexibility in vitro and in vivo. Quarterly Rev. Biophys. 2010;43:23–63. doi: 10.1017/S0033583510000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley SP, Bykowski TT, Cooley AE, Burns LH, Babb K, Brissette CA, Bowman A, Rotondi M, Miller MC, DeMoll E, Lim K, Fried MG, Stevenson B. Borrelia burgdorferi EbfC defines a newly-identified, widespread family of bacterial DNA-binding proteins. Nucleic Acids Res. 2009;37:1973–1983. doi: 10.1093/nar/gkp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan K, Chunhe X, Tingting L, Li J, Lange R, Balny C. The thermodynamic analysis of protein stabilization by sucrose and glycerol against pressure-induced unfolding The typical example of the 33-kDa protein from spinach photosystem II. European J. Biochem. 2003;270:1654–1661. doi: 10.1046/j.1432-1033.2003.03485.x. [DOI] [PubMed] [Google Scholar]
- Schlax PJ, Capp MW, Record MT. Inhibition of transcription initiation by lac repressor. J. Mol. Biol. 1995;245:331–350. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Nat. Acad. Sci. USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timasheff SN. The control of protein stability and association by interactions with water: how do solvent affect these processes? Annual Review of Biophysics and Biomolecular Structure. 1999;22:67–97. doi: 10.1146/annurev.bb.22.060193.000435. [DOI] [PubMed] [Google Scholar]
- Zhou H, Shatz W, Purdy MM, Fera N, Dahlquist FW, Reich NO. Long-range structural and dynamical changes induced by cofactor binding in DNA methyltransferase M.HhaI. Biochemistry. 2007;46:7261–7268. doi: 10.1021/bi602662e. [DOI] [PubMed] [Google Scholar]