The structure of AtALD1 from the flowering plant Arabidopsis thaliana was solved at a resolution of 2.3 Å. The structural analysis supports previous biochemical evidence for differential expression and distinct functions of AtALD1 and AtDAP-AT.
Keywords: LPC/CSU analysis, DAP-AT, PLP binding, Arabidopsis thaliana
Abstract
Diaminopimelate aminotransferase (DAP-AT) is an enzyme in the lysine-biosynthesis pathway. Conversely, ALD1, a close homologue of DAP-AT in plants, uses lysine as a substrate in vitro. Both proteins require pyridoxal-5′-phosphate (PLP) for their activity. The structure of ALD1 from the flowering plant Arabidopsis thaliana (AtALD1) was solved at a resolution of 2.3 Å. Comparison of AtALD1 with the previously solved structure of A. thaliana DAP-AT (AtDAP-AT) revealed similar interactions with PLP despite sequence differences within the PLP-binding site. However, sequence differences between the binding site of AtDAP-AT for malate, a purported mimic of substrate binding, and the corresponding site in AtALD1 led to different interactions. This suggests that either the substrate itself, or the substrate-binding mode, differs in the two proteins, supporting the known in vitro findings.
1. Introduction
The Arabidopsis thaliana aberrant growth and death 2 protein (AGD2) was originally recognized in association with pathogen resistance and was subsequently identified as the diaminopimelate aminotransferase (DAP-AT) enzyme involved in lysine synthesis (Rate & Greenberg, 2001 ▶; Song, Lu & Greenberg, 2004 ▶). A homologue was subsequently found and named AGD2-like defence response protein 1 (ALD1; Song, Lu, McDowell et al., 2004 ▶). DAP-AT and ALD1 homologues are present in many plant species. ALD1-deficient mutations show reduced production of salicylic acid and have increased susceptibility to disease (Song, Lu & Greenberg, 2004 ▶; Song, Lu, McDowell et al., 2004 ▶). As A. thaliana ALD1 efficiently removes NH3 from Lys and transaminates it into an unknown compound (Song, Lu & Greenberg, 2004 ▶; Song, Lu, McDowell et al., 2004 ▶), the defence response may possibly be regulated by a secondary metabolite that is derived from Lys catabolism. In vitro studies showed A. thaliana ALD1 to possess aminotransferase activity, which is distinct from the DAP-AT enzyme in the direction of action, and that its most preferable substrate is Lys (Song, Lu & Greenberg, 2004 ▶; Song, Lu, McDowell et al., 2004 ▶).
Pyridoxal-5′-phosphate (PLP) dependent enzymes catalyze a wide variety of diverse biochemical reactions in cells. All of these reactions have two initial steps in common (Toney, 2005 ▶; Cerqueira et al., 2011 ▶): the formation of an enzyme–PLP complex followed by the covalent attachment of the PLP cofactor to the ∊-amino group of a lysine in the active site. PLP-dependent enzymes have overlapping cofactor-binding and substrate-binding sites. Many enzyme–PLP complex structures have been solved and analyzed (see, for example, Shimon et al., 2007 ▶; Watanabe et al., 2007 ▶; Dobson et al., 2011 ▶). The structure of the substrate-binding sites is more difficult to study as enzyme–substrate complexes are usually reactive and short-lived. In many cases, complexes with surrogate compounds that mimic the substrate binding are used, such as the binding of malate to A. thaliana DAP-AT (Watanabe et al., 2007 ▶).
The structures of the PLP-dependent DAP-ATs from A. thaliana (AtDAP-AT), Chlamydia trachomatis (CtDAP-AT) and Chlamydomonas reinhardtii (CrDAP-AT) have been published (Watanabe et al., 2007 ▶, 2008 ▶, 2011 ▶; Dobson et al., 2011 ▶). Comparative sequence analysis of these proteins demonstrates approximately 60, 50 and 40% sequence identity to ALD1, respectively. However, the structure of ALD1 has not been determined to date. Here, we report the structure of A. thaliana ALD1 (AtALD1; UniProt ID Q9ZQI7) at a resolution of 2.3 Å. A comparison of the AtALD1 structure with that of AtDAP-AT (both containing PLP in their cofactor-binding sites) revealed that despite differences in the residues within the PLP-binding site, the interaction with PLP is very similar in the two structures. However, differences in the residues within the malate-binding site of AtDAP-AT suggest different substrate interactions in AtDAP-AT and AtALD1. The solved structure of AtALD1 can be exploited to understand the substrate specificity of this protein.
2. Materials and methods
2.1. Protein expression and purification
The AtALD1 gene (encoding amino acids 21–456) was cloned into pET28-TEVH (Peleg & Unger, 2008 ▶). This clone lacked the N-terminal amino acids corresponding to possible plastid-targeting signals. The protein was fused at its N-terminus to a 6×His tag separated by a TEV cleavage site and expressed in Escherichia coli BL21 (DE3) cells. A bacterial culture was grown at 310 K in 5 l of LB medium containing kanamycin (30 µg ml−1). When the absorbance of the culture reached A 600 = 0.6, protein expression was induced by the addition of 100 µM isopropyl β-d-1-thiogalactopyranoside (IPTG). Continued growth was allowed for an additional 18 h at 288 K. The bacteria were lysed by sonication in a solution consisting of 50 mM Tris–HCl pH 7.2, 500 mM NaCl, 1 mM PMSF, Protease Inhibitor Cocktail Set III, EDTA-Free (Calbiochem), DNase (1 µg ml−1) and lysozyme (40 U per millilitre of culture). The lysate was clarified by centrifugation at 20 000g for 20 min at 277 K and the supernatant was applied onto an Ni–NTA column (HiTrap Chelating HP, GE Healthcare) equilibrated with a buffer solution consisting of 50 mM Tris–HCl pH 7.5, 0.2 M NaCl, 5 mM imidazole. AtALD1 was eluted with the equilibrating buffer supplemented with 250 mM imidazole and applied onto a gel-filtration column (HiLoad 16/60 Superdex 200 prep grade, GE Healthcare) equilibrated with 50 mM Tris–HCl pH 7.0, 0.1 M NaCl. Peak elution samples were pooled and 1 mM PLP and 10%(w/v) glycerol were added before the sample was concentrated to 6 mg ml−1 protein for crystallization screening.
2.2. Crystallization, data collection and refinement
Crystals of AtALD1 obtained under oil using the microbatch method and an Oryx6 robot (Douglas Instruments Ltd, East Garston, Hungerford, Berkshire, England) diffracted to 2.3 Å resolution at best. Crystals of AtALD1 in the presence of PLP were grown from a solution consisting of 100 mM sodium citrate tribasic dihydrate, 2.2 mM Fos-Choline-8 fluorinated, 18% polyethylene glycol 3350. The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 57.30, b = 89.85, c = 180.97 Å.
A complete data set was collected to 2.3 Å resolution at 100 K from a single crystal on beamline ID23-1 (wavelength 0.9724 Å) at the European Synchrotron Radiation Facility (ESRF). The diffraction images were indexed and integrated using the MOSFLM program (Leslie & Powell, 2007 ▶) and the integrated reflections were scaled using the SCALA program (Evans, 2006 ▶).
Structure-factor amplitudes were calculated using TRUNCATE from the CCP4 program suite (French & Wilson, 1978 ▶). Details of the data collection are described in Table 1 ▶. The structure was solved by molecular replacement with the program Phaser (McCoy, 2007 ▶) using the refined structure of ll-diaminopimelate aminotransferase from A. thaliana (PDB entry 3ei6; Watanabe et al., 2008 ▶) as a model. All steps of atomic refinement were carried out with the REFMAC5 program from the CCP4 suite (Murshudov et al., 2011 ▶). The model was built into 2mF obs − DF calc and mF obs − DF calc maps using the Coot program (Emsley & Cowtan, 2004 ▶). Refinement weights were optimized. In the early stages of refinement noncrystallographic symmetry was restrained and in later stages it was gradually released, followed by a concomitant decrease in R free. Refinement movements were only accepted when they produced a decrease in the R free value. In later rounds of refinement water molecules were built into peaks greater than 3σ in mF obs − DF calc maps. The AtALD1 construct is composed of 436 amino-acid residues (21–456). The final model includes residues 35–440 in one monomer and 34–442 in the second monomer (no electron density was observed for residues 21–34 and 441–456 of the first monomer and residues 21–33 and 443–456 of the second monomer). The R free value was 24.09% (for 5% of reflections not used in refinement) and the R work value was 20.53% for all data to 2.3 Å resolution. The AtALD1 model was evaluated using the PROCHECK program (Laskowski et al., 1993 ▶). Details of the refinement statistics for the AtALD1 structure are described in Table 1 ▶. The coordinates and structure factors for AtALD1 have been deposited in the PDB under code 4fl0.
Table 1. Data-collection and crystallographic refinement statistics for AtALD1.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Resolution range (Å) | 48.4–2.30 (2.42–2.30) |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 57.28, b = 89.87, c = 180.70 |
| No. of molecules in the asymmetric unit | 2 |
| No. of reflections measured | 544175 |
| No. of unique reflections | 42356 (6054) |
| R merge | 0.085 (0.350) |
| Completeness (%) | 99.9 (99.7) |
| Multiplicity | 12.8 (12.2) |
| 〈I〉/〈σ(I)〉 | 5.9 (1.8) |
| Refinement statistics | |
| Resolution (Å) | 48.4–2.30 |
| R work (%) | 20.53 |
| R free (%) | 24.09 |
| B factor (Å2) | |
| Overall | 39 |
| Protein | 39 |
| Ligand (PLP) | 32 |
| R.m.s.d. from ideal geometry | |
| Bond lengths (Å) | 0.016 |
| Bond angles (°) | 1.7 |
| Torsion angles (°) | 6.5 |
| Ramachandran plot (%) | |
| Most favoured | 92.6 |
| Additional favoured | 7.4 |
| Generously allowed | 0.0 |
| Disallowed | 0.0 |
2.3. Visualization and comparison of structures
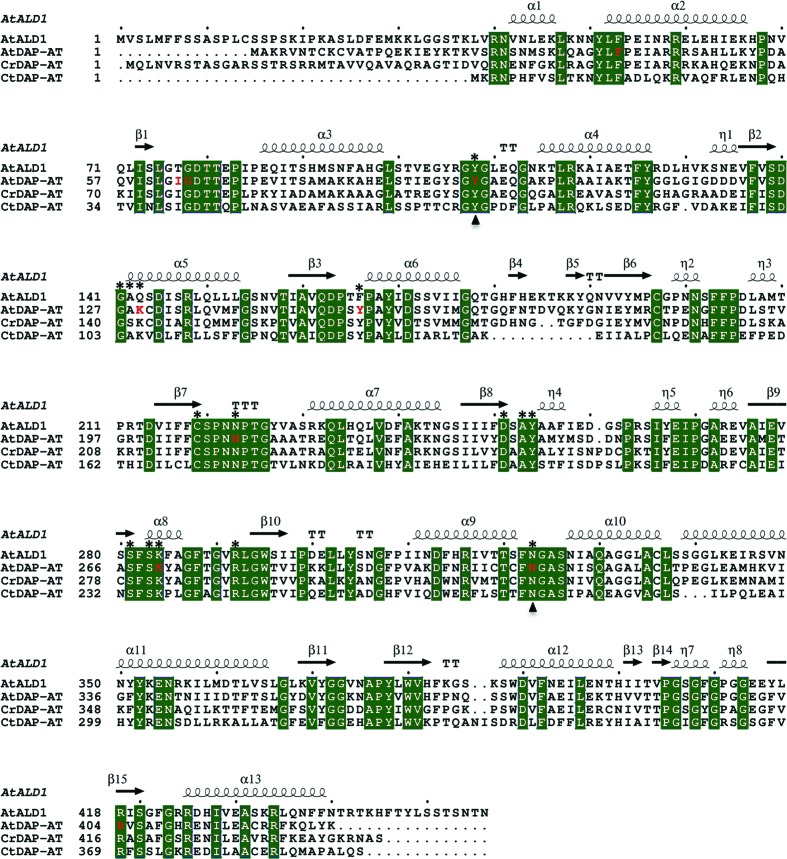
The multiple sequence alignment shown in Fig. 1 ▶ was performed using the Clustal Omega program (Sievers et al., 2011 ▶) and was visualized using ESPript (Gouet et al., 1999 ▶). The PyMOL software (DeLano, 2002 ▶) was used for molecular graphics and structural superimposition. For overall structure comparison, superimposition was performed using the Cα atoms of all aligned residues of the paired proteins. For comparison of the local structures of PLP- and malate-binding sites, the Cα atoms of first-sphere residues were used for superimposition.
Figure 1.
Multiple sequence alignment of AtALD1, AtDAP-AT, CrDAP-AT and CtDAP-AT. Sequence numbering corresponds to PDB entries 4fl0, 2z1z, 3qgu and 3asb, respectively. The secondary-structure elements of AtALD1 are labelled above the corresponding sequence. Residues that are conserved in all four proteins are shown as green blocks. Residues that are in contact with PLP in AtALD1 and AtDAP-AT, and correspondingly aligned residues in CrDAP-AT and CtDAP-AT, are marked by asterisks above the sequences. Residues that are in contact with malate in AtDAP-AT are marked in red. The PLP/malate-binding site spans the interface between the homodimer subunits A and B. Residues from subunit B are marked by triangles below the sequences.
LPC/CSU analysis (Sobolev et al., 1999 ▶) was used to define the protein–ligand and residue–residue interactions. A contact was considered to exist if the residue–ligand (or residue–residue) distance was less than 4.5 Å and the contact surface area was larger than 5 Å2. For analysis of PLP interactions, we compared the AtALD1 structure with the structure of AtDAP-AT from PDB entry 2z20 (Watanabe et al., 2007 ▶; both contain PLP). All other solved structures of AtDAP-AT are of the apo form, mutants or protein complexed with modified PLP or additionally complexed with malate. The solved structures of CtDAP-AT were also not used for comparison, as they are either of the apo form or covalently bound (not complexed) to PLP. The CrDAP-AT structure was only solved in the apo form. The PLP-binding sites in AtALD1 and AtDAP-AT are defined as the set of residues forming contacts with PLP. The PLP/malate-binding niche is defined as the residues of the overlapping PLP- and malate-binding sites in PDB entry 2z1z (Watanabe et al., 2007 ▶).
3. Results and discussion
3.1. Structural overview of AtALD1
The AtALD1 protein was crystallized with one homodimer per asymmetric unit and a V M of 2.35 Å3 Da−1, and its structure was solved (Table 1 ▶). A dimeric form is suggested by the apparent molecular weight of AtALD1 upon gel filtration (the peak eluted at 72 ml; Supplementary Fig. S11). Dimerization is also supported by analysis of the quaternary structure of AtALD1 using the PISA server (Krissinel & Henrick, 2007 ▶), which predicted that the dimeric form of AtALD1 would be the stable structure in solution. The root-mean-square deviation (r.m.s.d.) between the Cα atoms of the two subunits was calculated to be 0.46 Å. The crystal structure of each of the subunits consists of a large domain (LD) and a small domain (SD). The LD (Pro85–Ser338) consists of 254 residues and has an α–β–α sandwich fold. The LD includes both the PLP-binding site and the dimerization site. The SD is composed of the remaining residues: Gly34–Ile84 and Ser339–Thr441. The SD has an α/β architecture, with a β-sheet surrounded by two outer layers of α-helices [an animated Interactive 3D Complement (I3DC) is available in Proteopedia at http://proteopedia.org/w/Journal:Acta_Cryst_F:2].
3.2. Sequence and structure comparisons
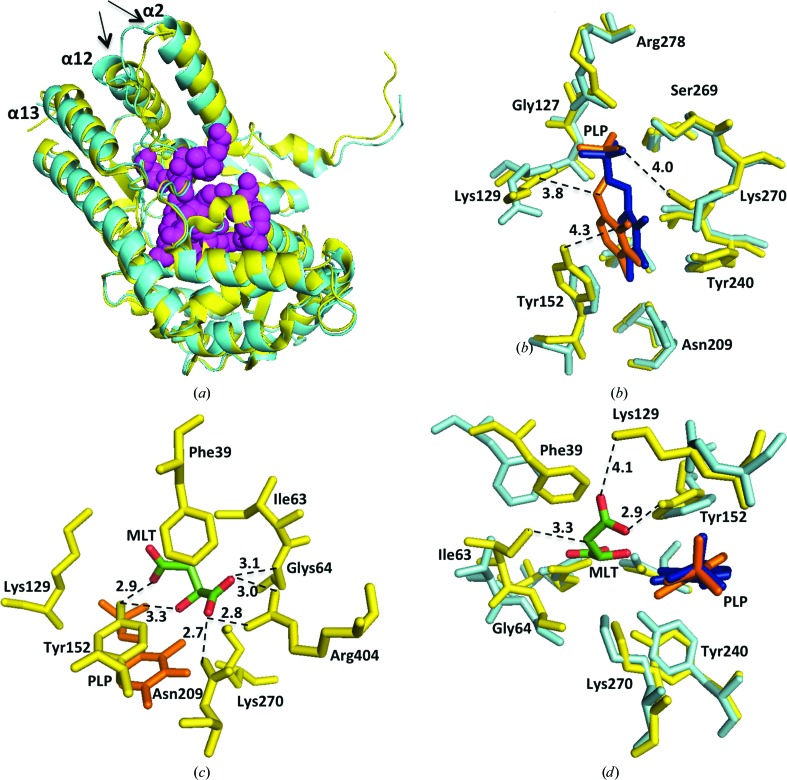
A multiple sequence alignment of AtALD1, AtDAP-AT, CrDAP-AT and CtDAP-AT is shown in Fig. 1 ▶. The four sequences are related: AtALD1 is more than 60% identical in sequence to AtDAP-AT and is about 50% and 40% identical to CrDAP-AT and CtDAP-AT, respectively. This similarity is also evident at the structural level. When superimposed, AtALD1 and AtDAP-AT (both containing PLP) have an r.m.s.d. of 1.2 Å (Fig. 2 ▶ a). The differences are larger between AtALD1 containing PLP and DAP-AT structures lacking the cofactor (r.m.s.d.s of 2.2 and 1.9 Å for AtALD1 versus CtDAP-AT and CrDAP-AT, respectively). This may arise from species differences, as a comparison of PLP-bound and apo forms of AtDAP-AT shows a very small difference in structure (r.m.s.d. of 0.3 Å), as expected for heterogroup binding (Najmanovich et al., 2000 ▶). On the other hand, malate binding in the substrate pocket causes a larger conformational change. A comparison of the PLP-bound and malate-bound form of AtDAP-AT with its PLP-bound form gives an r.m.s.d. of 1.5 Å. This can be rationalized, as binding at an active site is often accompanied by conformational changes because of the higher flexibility required for chemical activity (Tsou, 1993 ▶; Benkovic & Hammes-Schiffer, 2003 ▶).
Figure 2.
Structural comparison between AtALD1 and AtDAP-AT. (a) Superposition of AtALD1 (cyan) and AtDAP-AT (PDB entry 2z20; yellow) structures containing PLP. Helices α2, α12 and α13 are indicated (the arrows point to the C-termini of the helices). A CPK model of first-sphere residues forming the PLP/malate-binding niche (magenta) is shown. (b) Superposition of the PLP-binding site residues of AtALD1 (cyan) and AtDAP-AT (PDB entry 2z20; yellow). PLP complexed with AtDAP-AT (orange) and AtALD1 (blue) is shown. The bond distances discussed in the text are indicated. Only subunit A residues are presented. (c) Structure of the malate-binding site in AtDAP-AT (PDB entry 2z1z). Binding-site residues (yellow) and PLP (orange) are shown. Malate (MLT) is coloured by atom type (green, carbon; red, oxygen). Hydrogen-bond distances to malate are shown. Only subunit A residues are presented. (d) Superposition of the malate-binding site in AtDAP-AT (yellow) with aligned AtALD1 residues (cyan). PLP complexed with AtDAP-AT (orange) and AtALD1 (blue) is indicated. Contacts of malate (coloured by atom type: green, carbon; red, oxygen) in AtDAP-AT that are missing in the AtALD1 structure are shown. Only subunit A residues are shown.
3.3. Cofactor- and substrate-binding niche
The PLP- and malate-binding sites in AtDAP-AT strongly overlap (Watanabe et al., 2007 ▶). First-sphere residues in AtDAP-AT forming the PLP/malate-binding niche are marked in Fig. 1 ▶. 16 out of 19 binding-niche residues are conserved in all four proteins (AtALD1, AtDAP-AT, CrDAP-AT and CtDAP-AT). Three residues are not conserved in AtALD1. These are Thr77, Gln143 and Phe166, which correspond to Ile, Lys and Tyr, respectively, in the other three proteins. Fig. 2 ▶(a) shows the position of the PLP/malate-binding niche. The binding niche is situated close to the N-termini of helices α2 and α12 and is distant from the C-termini, which is the region of largest difference between the structures. Therefore, we conclude that there are no dramatic differences in the overall structure of the binding niche between AtALD1 and AtDAP-AT. Still, local differences could in principle strongly influence cofactor and/or substrate binding. This possibility is considered below.
3.4. PLP binding-site structure
The structure of AtALD1 incorporates PLP situated in a binding site formed by the two monomers (Supplementary Table S1a). The interaction of PLP with the protein is predominately aromatic–aromatic and hydrophilic, involving 13 residues from subunit A and two residues from subunit B.
We compared the PLP interactions in the AtALD1 and AtDAP-AT structures. The overall binding-site structure and position of PLP is similar in both enzymes (Fig. 2 ▶ b and Supplementary Tables S1a and S1b). However, there are two residues (out of 15) that are not conserved: Lys129 and Tyr152 in AtDAP-AT versus Gln143 and Phe166, respectively, in AtALD1. While PLP contacts the backbone N and Cβ atoms of Lys129, it is likely that the difference in side chain between Lys and Gln does not significantly influence the positions of these atoms and their contact area. Furthermore, PLP only has a minor contact area (of about 1 Å2) with the side-chain O atom of Tyr152. Therefore, the replacement of Tyr by Phe at this position is also not likely to significantly influence PLP binding.
Comparison of the data in Supplementary Tables S1(a) and S2(b) reveals that there are two relatively large differences in PLP contact area between AtDAP-AT and AtALD1: Asn209 in AtDAP-AT corresponding to residue Asn223 in AtALD1, and Tyr240 in AtDAP-AT corresponding to Tyr254 in AtALD1. Yet, the positions of these two residues in the two structures relative to PLP are similar and the changes in contact area are the result of small reorientations of nearby residues (mainly Tyr152 and Lys270). Overall, PLP is almost completely buried in both structures (6% solvent-accessible surface area in the AtDAP-AT structure and 8% in the AtALD1 structure). While the reorientation of Lys270 leads to the loss of a putative water-mediated hydrogen bond (distance of 4.0 Å), all other putative hydrogen bonds in the two structures are identical. We conclude that as the interactions that are responsible for PLP binding in the two structures are very similar, AtALD1, as AtDAP-AT, is a PLP-dependent enzyme.
3.5. Putative substrate-binding site
Can AtALD1 bind similar substrates as AtDAP-AT? This was addressed by analyzing the interaction of malate with AtDAP-AT (PDB entry 2z1z), comparing the structure of the malate-binding site in AtDAP-AT with the site formed by the corresponding residues in AtALD1 and speculating on the malate interactions if malate were placed in the same position in AtALD1. Malate is a small hydrophilic molecule containing nine non-H atoms, five of which are O atoms that form seven hydrogen bonds to AtDAP-AT. The ten amino-acid residues contacting malate in AtDAP-AT are listed in Supplementary Table S2(a) and are illustrated in Fig. 2 ▶(c). Eight are from subunit A and two are from subunit B. These ten residues are conserved in the DAP-AT sequences (Fig. 1 ▶). AtALD1 shows differences at three positions: Thr77, Gln143 and Phe166 versus Ile63, Lys129 and Gln143 in AtDAP-AT. A superposition of the residues forming the malate-binding site in AtDAP-AT with the corresponding residues of AtALD1 is presented in Fig. 2 ▶(d). Despite some rearrangement at the site, most contacts are conserved (Supplementary Tables S2a and S2b). However, we can see at the three positions that differ that malate would lose some or all of its contacts in AtALD1. It would lose a hydrophobic contact with the side chain of Ile63 and, probably more importantly, the hydrophobic surface area would become available to the solvent. While Lys129 and Tyr152 contact both PLP and malate (Supplementary Tables S1c and S2a), the replacement of these residues would only affect malate binding. Indeed, replacement of Tyr152 by Phe would lead to the loss of a strong side-chain hydrogen bond to malate (length 2.9 Å) and replacement of Lys129 by Gln would lead to the loss of a putative water-mediated hydrogen bond (length 4.1 Å) to malate. In summary, as opposed to AtDAP-AT, AtALD1 is not likely to bind malate. Therefore, the two enzymes most probably differ in substrate specificity.
In summary, there is considerable structural similarity between the AtALD1 and DAP-AT proteins; however, their modes of function appear to differ. Residue differences at the malate-binding site of AtDAP-AT and the resulting changes in the putative interaction at the corresponding malate-binding positions in AtALD1 lead us to conclude that the substrate specificity of AtALD1 is essentially different from those of the solved DAP-AT structures. Thus, the structural analysis supports the biochemical evidence (Song, Lu & Greenberg, 2004 ▶; Song, Lu, McDowell et al., 2004 ▶) for differential expression and distinct functions of AtALD1 and AtDAP-AT.
Supplementary Material
PDB reference: AtALD1, 4fl0
Supplementary material file. DOI: 10.1107/S1744309112050270/gx5211sup1.pdf
Acknowledgments
We thank Professor Joel Sussman for helpful discussions and Alexander Berchansky for producing the animated interactive three-dimensional analysis in Proteopedia. The protein expression and structure determination were carried out at the Israel Structural Proteomics Center (ISPC) supported by the Divadol and Dumbitz Foundations. This research was also supported by a grant from the Israeli Science Foundation.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: GX5211).
References
- Benkovic, S. J. & Hammes-Schiffer, S. (2003). Science, 301, 1196–1202. [DOI] [PubMed]
- Cerqueira, N. M. F. S. A., Fernandes, P. A. & Ramos, M. J. (2011). J. Chem. Theor. Comput. 7, 1356–1368. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Dobson, R. C., Girón, I. & Hudson, A. O. (2011). PLoS One, 6, e20439. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- French, S. & Wilson, K. (1978). Acta Cryst. A34, 517–525.
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Leslie, A. G. W. & Powell, H. R. (2007). Evolving Methods for Macromolecular Crystallography, edited by R. J. Read & J. L. Sussman, pp. 41-51. Dordrecht: Springer.
- McCoy, A. J. (2007). Acta Cryst. D63, 32–41. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Najmanovich, R., Kuttner, J., Sobolev, V. & Edelman, M. (2000). Proteins, 39, 261–268. [DOI] [PubMed]
- Peleg, Y. & Unger, T. (2008). Methods Mol. Biol. 426, 197–208. [DOI] [PubMed]
- Rate, D. N. & Greenberg, J. T. (2001). Plant J. 27, 203–211. [DOI] [PubMed]
- Shimon, L. J., Rabinkov, A., Shin, I., Miron, T., Mirelman, D., Wilchek, M. & Frolow, F. (2007). J. Mol. Biol. 366, 611–625. [DOI] [PubMed]
- Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D. & Higgins, D. G. (2011). Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed]
- Sobolev, V., Sorokine, A., Prilusky, J., Abola, E. E. & Edelman, M. (1999). Bioinformatics, 15, 327–332. [DOI] [PubMed]
- Song, J. T., Lu, H. & Greenberg, J. T. (2004). Plant Cell, 16, 353–366. [DOI] [PMC free article] [PubMed]
- Song, J. T., Lu, H., McDowell, J. M. & Greenberg, J. T. (2004). Plant J. 40, 200–212. [DOI] [PubMed]
- Toney, M. D. (2005). Arch. Biochem. Biophys. 433, 279–287. [DOI] [PubMed]
- Tsou, C.-L. (1993). Science, 262, 380–381. [DOI] [PubMed]
- Watanabe, N., Cherney, M. M., van Belkum, M. J., Marcus, S. L., Flegel, M. D., Clay, M. D., Deyholos, M. K., Vederas, J. C. & James, M. N. G. (2007). J. Mol. Biol. 371, 685–702. [DOI] [PubMed]
- Watanabe, N., Clay, M. D., van Belkum, M. J., Cherney, M. M., Vederas, J. C. & James, M. N. G. (2008). J. Mol. Biol. 384, 1314–1329. [DOI] [PubMed]
- Watanabe, N., Clay, M. D., van Belkum, M. J., Fan, C., Vederas, J. C. & James, M. N. G. (2011). J. Mol. Biol. 411, 649–660. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: AtALD1, 4fl0
Supplementary material file. DOI: 10.1107/S1744309112050270/gx5211sup1.pdf