X-ray diffraction data were collected to 2.6 Å resolution from a crystal of the chicken MHC class I molecule BF2*1501. The crystal belonged to space group P3121, with unit-cell parameters a = 125.1, b = 125.1, c = 80.9 Å, and contained two molecules in the asymmetric unit. The Matthews coefficient and solvent content were calculated to be 2.08 Å3 Da−1 and 40.78%, respectively.
Keywords: chicken MHC class I molecules, MDV
Abstract
The chicken major histocompatibility complex (MHC) class I molecules named BF are strongly associated with Marek’s disease (MD). A single structure, that of chicken BF2*2101 from the B21 haplotype, which might provide resistance to MD, has been determined. However, little is known about other structures apart from BF2*2101. In order to provide further structures of chicken MHC class I molecules, BF2*1501 and chicken β2-microglobulin complexed with a nonapeptide (MDV-MEQRRR9) derived from Marek’s disease virus MEQ protein (MDV EcoRI Q fragment, residues 72–80) were assembled and crystallized. Diffraction data from the crystal were collected to 2.6 Å resolution; the crystal belonged to space group P3121, with unit-cell parameters a = 125.1, b = 125.1, c = 80.9 Å and two molecules in the asymmetric unit. The Matthews coefficient V M was 2.08 Å3 Da−1, with a calculated solvent content of 40.78%. These data will be helpful in obtaining insight into the structural basis of the involvement of BF2*1501 in MD.
1. Introduction
Major histocompatibility complex (MHC) class I molecules are membrane-surface glycoproteins that play a crucial role in the process of eliminating intracellular pathogens such as viruses and bacteria (Neefjes et al., 2011 ▶). The ternary peptide–MHC I–β2-microglobulin (β2m) complexes (pMHC I) are recognized by specific T-cell receptors (TCRs), with the help of the CD8 molecule on the surface of infected cells. The specific recognition is able to activate cytotoxic lymphocytes (CTL) and then lyse the infected cells (Garboczi et al., 1996 ▶). The MHC class I molecule comprises two chains: an α chain, whose extracellular portion is divided into three domains, α1, α2 and α3, and a light chain, β2m (Bjorkman et al., 1987 ▶).
Chicken major histocompatibility complex (chMHC) is also designated as a B locus, an erythrocyte antigen (Plachy et al., 1992 ▶). The B locus, the region that corresponds to the classical MHC in chickens, is much smaller and simpler than the typical mammalian MHC in terms of its genomic organization. Detailed analysis has revealed that the B locus is located on chromosome 16 and is composed of tightly linked polymorphic regions: BF (class I), BL (class IIβ) and BG (Kaufman et al., 1999 ▶). BF includes two genes, BF1, which is less expressed, and BF2, which is dominantly expressed in antigen-presenting cells. BF2 has been reported to be related to certain infectious pathogens. Peptide motifs for chMHC I of B21, B15 and B4 haplotypes have been detected by peptide pooling (Wallny et al., 2006 ▶). Moreover, the structure of the BF2*2101 from the B21 haplotype has been determined (Koch et al., 2007 ▶). The structure of BF2*2101 showed that a large central cavity in the peptide-binding groove might contribute to the resistance of the B21 haplotype in Marek’s disease (MD). In contrast to BF2*2101, BF2*1501 from the B15 haplotype is susceptible to Marek’s disease virus (MDV; Dalgaard et al., 2003 ▶). Therefore, determination of the structure of BF2*1501 may illustrate the sensitivity to MD.
MDV is a potent oncogenic herpesvirus which can elicit an acute lymphoproliferative disease within a few weeks and eventually leads to mass mortality of chickens. According to viral genome sequence and organization, MDV is classified as a member of the alphaherpesvirus subfamily (Ross, 1999 ▶). The MDV EcoRI Q fragment (MEQ) has been shown to be one of the most important proteins that can induce tumours (Liu et al., 1997 ▶). Since MDV is a highly cell-associated virus, T-cell-mediated response is more important than antibody/B-cell-mediated immune response (Parvizi et al., 2010 ▶). Here, we report the expression, refolding, purification and crystallization of chicken BF2*1501 with a nonapeptide (MDV-MEQRRR9) from the MDV MEQ (MDV EcoRI Q fragment, residues 72–80) protein.
2. Materials and methods
2.1. Expression of BF2*1501 and chicken β2m proteins
BF2*1501 heavy-chain cDNA (GenBank accession No. L28958, residues 22–291) and chicken β2m (GenBank accession No. BAD22696, residues 22–119) were amplified by RT-PCR from a specific pathogen-free (SPF) chicken. The BF2*1501 and chicken β2m genes were ligated into the pET21a expression vector (Novagen) and transformed into Escherichia coli strain BL21 (DE3). The recombinant proteins were both expressed as inclusion bodies. When the bacteria reached an OD600 of about 0.6, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce protein expression. After 5 h, the cells were harvested by centrifugation at 6000g for 10 min. After lysis by sonication, the cells were centrifuged at 16 000g and the pellets were washed three times with buffer (20 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.5% Triton X-100). Finally, BF2*1501 heavy-chain and β2m inclusion bodies were dissolved in guanidinium chloride (Gua–HCl) buffer [6 M Gua–HCl, 50 mM Tris–HCl pH 8.0, 10 mM EDTA, 100 mM NaCl, 10%(v/v) glycerine, 10 mM DTT] to a concentration of 30 mg ml−1 (Chen et al., 2011 ▶).
2.2. Refolding and purification of the BF2*1501 complex
The protocol was essentially the same as described previously (Garboczi et al., 1992 ▶) with modification of the refolding temperature from 283 to 277 K, the refolding buffer volume from 200 to 500 ml and the molar ratio of heavy chain to light chain from 1:2 to 1:1 (Pan et al., 2011 ▶). The whole refolding process took about 24 h. 1 ml dissolved β2m inclusion bodies was first added gradually to 500 ml refolding buffer (100 mM Tris–HCl pH 8.0, 2 mM EDTA, 400 mM l-Arg, 0.5 mM oxidized glutathione, 5 mM reduced glutathione) and incubated at 277 K. After 8 h, 5 mg nonapeptide (RRRRKQTDY) dissolved in dimethyl sulfoxide (DMSO) was added to the solution. Half an hour later, 3 ml dissolved BF2*1501 heavy-chain inclusion bodies were added gradually into the solution and incubated at 277 K for a further 8 h. After refolding, the soluble portion was concentrated and purified by chromatography on a Superdex 200 16/60 HiLoad (GE Healthcare) size-exclusion column followed by Resource Q (GE Healthcare) anion-exchange chromatography.
2.3. Crystallization of the BF2*1501 complex
The purified protein was dialysed against crystallization buffer (20 mM Tris–HCl pH 8.0, 50 mM NaCl) and concentrated to 6 and 12 mg ml−1. The commercial kits Index, Crystal Screen, Crystal Screen 2 (Hampton Research, California, USA) consisting of pre-formulated solutions were used for crystallization. Two drops with 1 µl protein solution (6 and 12 mg ml−1) and 1 µl reservoir crystallization buffer were placed over a well containing 160 µl reservoir solution using a VDX plate (Hampton Research) at 291 K. 10 d later, crystals were obtained using Index solution No. 6 (0.1 M Tris–HCl pH 8.5, 2.0 M ammonium sulfate) at 291 K.
2.4. Data collection and processing
Diffraction data were collected on beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF; Shanghai, People’s Republic of China) at a wavelength of 0.97916 Å using an ADSC 315 imaging-plate detector. Before diffraction, the crystal was first immersed in reservoir solution containing 20%(v/v) glycerol as cryoprotectant for a few seconds and then flash-cooled in liquid nitrogen (Zhang et al., 2011 ▶). The collected intensities were indexed, integrated, corrected for absorption, scaled and merged using HKL-2000 (Otwinowski & Minor, 1997 ▶).
2.5. Comparison of BF2*1501 with other class I molecules
The amino-acid sequence of BF2*1501 was aligned with those of typical BF2*2101, human, mouse, bovine, horse and swine MHC class I molecules using DNAMAN v.5.2.2 (Raiola et al., 2004 ▶). The GenBank accession numbers are as follows: BF2*2101, AY234769; BoLA-N*01301, CAA56909; ELA_7-6, AY225155; SLA-1*1502, HQ909439; H-2Kb, V00746; HLA-A2, U56825.
3. Results and discussion
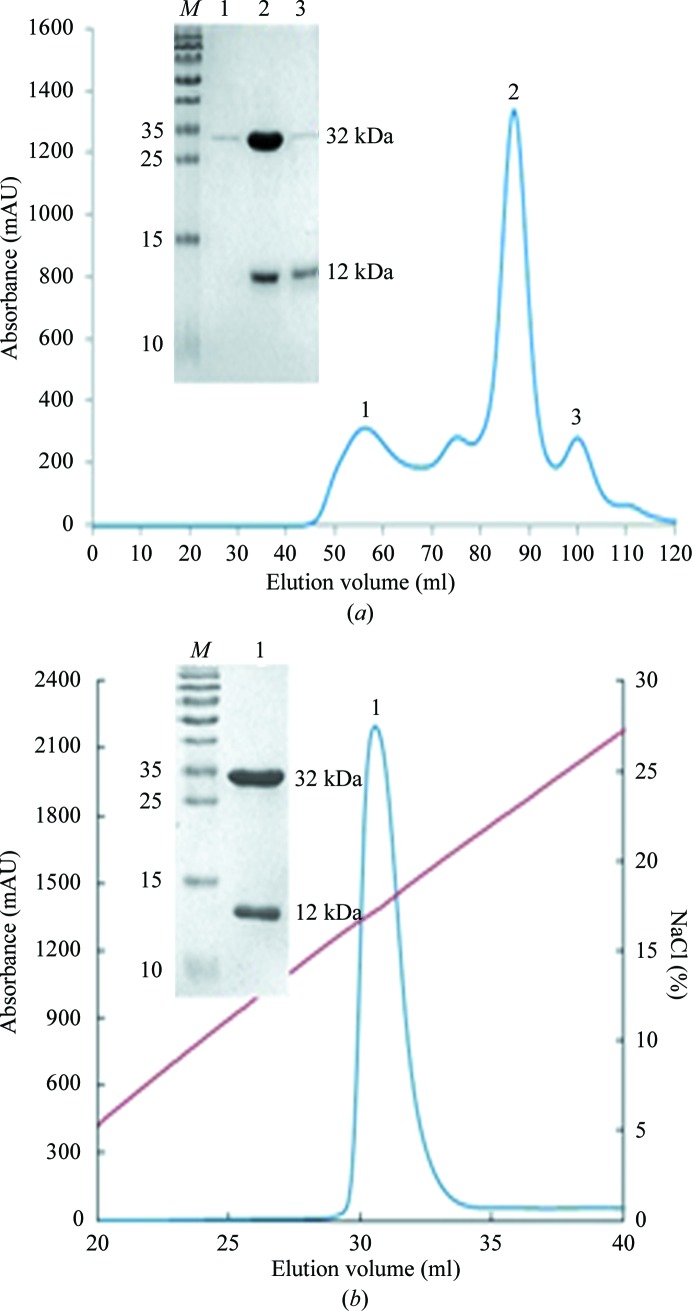
In each case, about a 10% yield of the stable complex was obtained by refolding in vitro. The refolded proteins were first purified by Superdex 200 16/60 HiLoad size-exclusion chromatography (GE Healthcare; Fig. 1 ▶ a). The profile revealed three peaks, which were aggregated BF2*1501 heavy chain (peak 1), the correctly refolded complex (peak 2, ∼44 kDa) and excess β2m (peak 3). The collected complex was further purified by Resource Q anion-exchange chromatography. The specific peak appeared at a NaCl concentration of 16–20% (Fig. 1 ▶ b). SDS–PAGE analysis showed two bands corresponding to the expressed molecular weights of BF2*1501 (∼32 kDa) and chicken β2m (∼12 kDa) (see inset in Fig. 1 ▶).
Figure 1.
Purification of the refolded BF2*1501 complex (BF2*1501, chicken β2m and the epitope MDV-MEQRRR9) by FPLC Superdex 200 16/60 HiLoad gel-filtration and Resouce-Q anion-exchange chromatography (GE Healthcare). (a) Gel-filtration profile of the refolded products. Peak 1 corresponds to the aggregated BF2*1501 heavy chain, peak 2 corresponds to the correctly refolded complex (∼44 kDa) and peak 3 corresponds to excess chicken β2m. Inset, reduced SDS–PAGE gel (15%) for peaks 1, 2 and 3. Lane M contains molecular-mass markers (labelled in kDa). (b) Anion-exchange chromatography profile of the refolded products. Peak 1 corresponds to correctly refolded complex, which was eluted at an NaCl concentration of 16–20%. Inset, reduced SDS–PAGE gel (15%) for peak 1. Lane M contains molecular-mass markers (labelled in kDa).
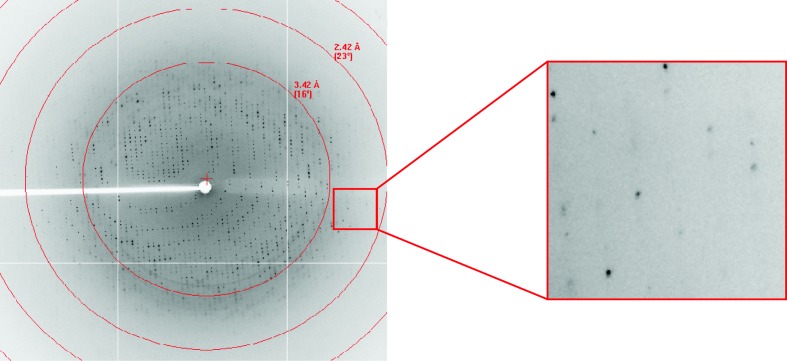
After purification and concentration, the BF2*1501–MDV-MEQRRR9 complex was set up for crystallization screening. Many single crystals appeared using Index solution No. 6 (0.1 M Tris–HCl pH 8.5, 2.0 M ammonium sulfate) at 291 K after 10 d. After optimization, a single crystal was used for diffraction (Fig. 2 ▶) that grew from a solution consisting of 0.1 M Tris–HCl pH 8.5, 1.9 M ammonium sulfate at 291 K after 10 d. The crystal belonged to space group P3121, with unit-cell parameters a = 125.1, b = 125.1, c = 80.9 Å, and diffracted to 2.6 Å resolution (Fig. 3 ▶, Table 1 ▶). The Matthews coefficient value V M was 2.08 Å3 Da−1, with a calculated solvent content of 40.78%.
Figure 2.
Photograph of the crystal used for diffraction analysis.
Figure 3.
Diffraction pattern of the chicken MHC I molecule. The high-resolution diffraction spots are highlighted in the box.
Table 1. X-ray diffraction data and processing statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P3121 |
| Unit-cell parameters (Å) | a = 125.1, b = 125.1, c = 80.9 |
| Volume of the unit cell (Å3) | 1096701.6 |
| Resolution range (Å) | 50.00–2.60 (2.60–2.69) |
| Total No. of reflections | 205783 |
| No. of unique reflections | 24077 |
| Completeness (%) | 100.0 (100.0) |
| Average I/σ(I) | 22.119 (5.194) |
| R merge † (%) | 11.7 (54.0) |
| Average multiplicity | 9.0 (9.1) |
R
merge = 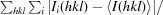
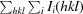 , where I
i(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity from multiple measurements.
, where I
i(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity from multiple measurements.
A multiple amino-acid sequence alignment of BF2*1501 with other known MHC class I molecules was performed. A deletion of five amino acids was a notable difference between chicken MHC class I molecules and those from other species. Otherwise, chicken BF2 molecules share very low homology with those from other species. BF2*1501 has 42, 42.8, 42.8, 43.9 and 43.9% sequence identity to the swine, bovine, equine, mouse and human class I molecules, respectively. However, different haplotypes in chicken share high homology with each other. BF2*1501 has 90.7% sequence identity to BF2*2101. The results suggest that there is an obvious species difference between chicken MHC class I molecules and their bovine, equine, mouse and human homologues.
Structure determination of BF2*1501 will be useful for epitope vaccine design for Marek’s disease and other pathogens. Structure determination and refinement are currently under way.
Acknowledgments
This work was supported by grants from the National Key Basic Research Program of China (973 Program, grant No. 2013CB835302) and the Key National Natural Science Foundation of China (grant No. 31230074). We thank Professor George F. Gao and Dr Jianxun Qi (Institute of Microbiology, Chinese Academy of Sciences) for helpful suggestions.
References
- Bjorkman, P. J., Saper, M. A., Samraoui, B., Bennett, W. S., Strominger, J. L. & Wiley, D. C. (1987). Nature (London), 329, 506–512. [DOI] [PubMed]
- Chen, R., Qi, J., Yao, S., Pan, X., Gao, F. & Xia, C. (2011). Acta Cryst. F67, 1633–1636. [DOI] [PMC free article] [PubMed]
- Dalgaard, T. S., Højsgaard, S., Skjødt, K. & Juul-Madsen, H. R. (2003). Scand. J. Immunol. 57, 135–143. [DOI] [PubMed]
- Garboczi, D. N., Ghosh, P., Utz, U., Fan, Q. R., Biddison, W. E. & Wiley, D. C. (1996). Nature (London), 384, 134–141.
- Garboczi, D. N., Hung, D. T. & Wiley, D. C. (1992). Proc. Natl Acad. Sci. USA, 89, 3429–3433. [DOI] [PMC free article] [PubMed]
- Kaufman, J., Milne, S., Göbel, T. W., Walker, B. A., Jacob, J. P., Auffray, C., Zoorob, R. & Beck, S. (1999). Nature (London), 401, 923–925. [DOI] [PubMed]
- Koch, M., Camp, S., Collen, T., Avila, D., Salomonsen, J., Wallny, H. J., van Hateren, A., Hunt, L., Jacob, J. P., Johnston, F., Marston, D. A., Shaw, I., Dunbar, P. R., Cerundolo, V., Jones, E. Y. & Kaufman, J. (2007). Immunity, 27, 885–899. [DOI] [PubMed]
- Liu, J.-L., Lee, L. F., Ye, Y., Qian, Z. & Kung, H.-J. (1997). J. Virol. 71, 3188–3196. [DOI] [PMC free article] [PubMed]
- Neefjes, J., Jongsma, M. L., Paul, P. & Bakke, O. (2011). Nature Rev. Immunol. 11, 823–836. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pan, X., Qi, J., Zhang, N., Li, Q., Yin, C., Chen, R., Gao, F. & Xia, C. (2011). Acta Cryst. F67, 568–571. [DOI] [PMC free article] [PubMed]
- Parvizi, P., Abdul-Careem, M. F., Haq, K., Thanthrige-Don, N., Schat, K. A. & Sharif, S. (2010). Anim. Health Res. Rev. 11, 123–134. [DOI] [PubMed]
- Plachy, J., Pink, J. R. & Hála, K. (1992). Crit. Rev. Immunol. 12, 47–79. [PubMed]
- Raiola, A., Camardella, L., Giovane, A., Mattei, B., De Lorenzo, G., Cervone, F. & Bellincampi, D. (2004). FEBS Lett. 557, 199–203. [DOI] [PubMed]
- Ross, N. L. (1999). Trends Microbiol. 7, 22–29. [DOI] [PubMed]
- Wallny, H. J., Avila, D., Hunt, L. G., Powell, T. J., Riegert, P., Salomonsen, J., Skjødt, K., Vainio, O., Vilbois, F., Wiles, M. V. & Kaufman, J. (2006). Proc. Natl Acad. Sci. USA, 103, 1434–1439. [DOI] [PMC free article] [PubMed]
- Zhang, N., Qi, J., Pan, X., Chen, Z., Li, X., Gao, F. & Xia, C. (2011). Acta Cryst. F67, 888–891. [DOI] [PMC free article] [PubMed]