The expression, purification, characterization and crystallization of GK2848, a carbonic anhydrase from G. kaustophilus, are described. The crystals diffracted to a resolution of 2.70 Å.
Keywords: hypothetical proteins, carbonic anhydrases, left-handed β-helix, Geobacillus kaustophilus
Abstract
GK2848, a hypothetical protein from the thermophilic organism Geobacillus kaustophilus, was cloned and overexpressed in Escherichia coli. The protein was purified to homogeneity using Ni–NTA affinity-column and gel-filtration chromatography. The purified protein was crystallized using the sitting-drop vapour-diffusion method. The crystals diffracted to a resolution of 2.70 Å and belonged to the orthorhombic space group P21212. GK2848 bears sequence homology to carbonic anhydrases of various bacterial species, indicating that it belongs to the carbonic anhydrase family of proteins. A subsequent carbonic anhydrase activity assay of GK2848 using the Wilbur–Anderson method confirmed its function as a carbonic anhydrase. A preliminary structure solution was obtained by molecular replacement using MOLREP. Mutation and biochemical characterization of the protein are in progress. The structure and functional analysis of GK2848 might provide valuable information on a novel class of carbonic anhydrases, as none of its homologous structures have been characterized.
1. Introduction
Geobacillus kaustophilus, a moderate thermophile, grows in aquatic environments and can withstand temperatures between 278 and 351 K (Takami et al., 2004 ▶). The organism consists of a 3.54 Mb chromosome and encodes several thermophilic proteins which have been useful in industrial processes (Takami et al., 2004 ▶). GK2848, a conserved hypothetical protein from the genome of G. kaustophilus, has a molecular mass of about 20 kDa and is composed of 182 amino acids.
GK2848 exhibits significant sequence homology to carbonic anhydrases from various bacterial species, indicating that it belongs to the carbonic anhydrase family of proteins. Carbonic anhydrases are zinc-containing enzymes that are present in organisms of all kingdoms and known for their physiological role in interconverting CO2 and HCO3 −; they are divided into at least five classes (α, β, γ, δ and ∊) based on their occurrence (Alber & Ferry, 1994 ▶). Several structures are available in the literature for the α class (Lindskog, 1997 ▶) and β class of enzymes (Zimmerman & Ferry, 2008 ▶). However, only two structures have been reported for the γ class of carbonic anhydrases. The α, β and γ classes of carbonic anhydrases show no significant sequence identity to one another and display distinct folds.
In this study, we report the crystallization, characterization and preliminary crystallographic analysis of GK2848 from G. kaustophilus. Interestingly, none of the close sequence homologues of GK2848 annotated as carbonic anhydrase have been structurally and functionally characterized. Therefore, the crystal structure of GK2848 will help us gain insights into the function and mechanism of GK2848 as a carbonic anhydrase. To our knowledge, this is the first report of a carbonic anhydrase from G. kaustophilus.
2. Materials and methods
2.1. Cloning, expression and purification of GK2848
The gene encoding the hypothetical protein (GK2848) from G. kaustophilus (GenBank accession No. BAD77133.1) was amplified from genomic DNA by PCR using the primers 5′-GGA ATT CAT ATG ATT TAC CCA TAC AAA GGC AAA-3′ and 5′-GGA ATT GGA TCC TTA TTA CGG CAG CTC TTT TTT GTC GG-3′. The amplified fragment was cloned under the control of the T7 promoter of the Escherichia coli expression vector pET-HisTEV, with the tobacco etch virus (TEV) protease-recognition site, Glu–Asn–Leu–Tyr–Phe–Gln–Gly, instead of the thrombin-recognition site of the pET-15b vector (Novagen, Madison, Wisconsin, USA). The expression vector was transformed into the E. coli BL21-CodonPlus (DE3)-RIL (Stratagene) strain and GK2848 protein was overexpressed at mid-log phase by the addition of 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside). The cells (35.6 g) were collected by centrifugation, washed with 50 ml buffer A (20 mM Tris–HCl pH 8.0) containing 0.5 M NaCl, 5 mM 2-mercaptoethanol and 1 mM phenylmethylsulfonyl fluoride and resuspended in 50 ml of the same buffer. The cells were then disrupted by sonication for 10 min in a chilled water bath and the cell lysate was incubated at 343 K for 13 min. The sample was centrifuged at 15 000g for 30 min and the supernatant was applied onto a HisTrap HP5 column (GE Healthcare Biosciences, New Jersey, USA) pre-equilibrated with buffer A containing 0.5 M NaCl and 20 mM imidazole. The protein was eluted with a linear gradient of 0.02–0.5 M imidazole and all the fractions were analysed by SDS–PAGE. The sample containing GK2848 was then loaded onto a HiLoad 16/60 Superdex 200 column (GE Healthcare Biosciences) pre-equilibrated with buffer A containing 0.5 M NaCl and 20 mM imidazole. The eluted fractions containing GK2848 were collected and treated with TEV protease at 303 K for 1 h. The protein sample was applied onto a HisTrap HP5 column (GE Healthcare Biosciences) pre-equilibrated with buffer A containing 0.5 M NaCl and 20 mM imidazole. The flowthrough fraction was collected and the purified protein was loaded onto a HiPrep 26/10 desalting column, which was eluted with buffer A containing 0.2 M NaCl. The sample-containing fractions were pooled and dialysed against the buffer (0.02 M Tris–HCl, 20 mM NaCl and 20 mM imidazole). The concentration of the protein used for crystallization (26 mg ml−1) and subsequent assays was measured using a UV spectrophotometer (A 280) and assuming a calculated absorption coefficient of 0.723 (ProtParam; Gasteiger et al., 2005 ▶).
2.2. Crystallization and data collection
Crystallization screening was carried out using the Index kit (Hampton Research) and the experiments were performed in sitting drops using the vapour-diffusion technique. Drops consisting of 1 µl protein solution and 1 µl well solution were equilibrated against 100 µl well solution at 293 K. The best diffracting crystals appeared in around 20 d (Fig. 1 ▶) and were obtained in a condition consisting of 30%(v/v) pentaerythritol ethocylate, 0.05 M ammonium sulfate, 0.05 M bis-tris pH 6.5.
Figure 1.
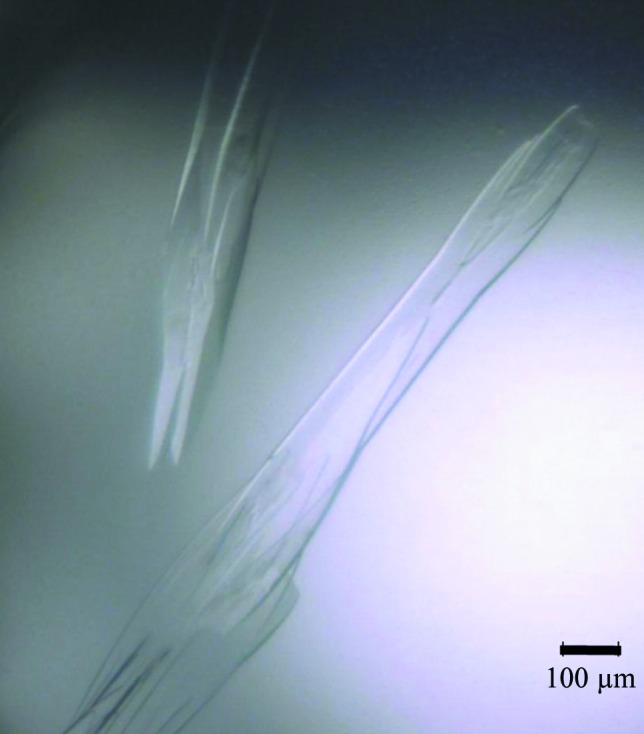
GK2848 crystals grown in 30%(v/v) pentaerythritol ethocylate, 0.05 M ammonium sulfate, 0.05 M bis-tris pH 6.5 at 293 K.
Prior to data collection, the crystal was soaked in a cryoprotectant solution [reservoir solution containing 50%(v/v) Paratone-N and 50%(v/v) paraffin oil] for a few seconds and flash-cooled in a nitrogen-gas stream at 100 K. A complete single data set was obtained on the RIKEN structural genomics beamline I (BL26B2) at SPring-8, Hyogo, Japan. The data set was processed to 2.7 Å resolution using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶). The data-collection statistics are given in Table 1 ▶.
Table 1. Data-collection statistics for GK2848.
Values in parentheses are for the highest-resolution shell.
| Wavelength (Å) | 1.000 |
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 137.98, b = 58.70, c = 94.39 |
| No. of molecules per asymmetric unit | 3 |
| Solvent content (%) | 61.36 |
| Resolution range (Å) | 50.0–2.70 (2.80–2.70) |
| Unique reflections | 20754 |
| Multiplicity | 5.7 (4.5) |
| Completeness (%) | 99.2 (95.8) |
| 〈I/σ(I)〉 | 14.0 (2.9) |
| R merge † | 0.093 (0.226) |
R
merge = 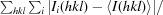
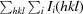 , where 〈I(hkl)〉 is the mean of the intensity measurements I
i(hkl) and the summation extends over all reflections.
, where 〈I(hkl)〉 is the mean of the intensity measurements I
i(hkl) and the summation extends over all reflections.
2.3. Carbonic anhydrase assay
The carbonic anhydrase activity of GK2848 was assayed electrometrically using a modification of the Wilbur–Anderson method (Wilbur & Anderson, 1948 ▶). The assay was carried out at 277 K by the addition of 0.01 mg protein to 6 ml chilled 20 mM Tris–HCl buffer pH 8. The reaction was initiated by adding 6.0 ml ice-cold CO2 saturated water. The time required for the pH of the buffer to drop from 8.0 to 6.0 was measured. The activity of GK2848 was calculated using the equation WAU (Wilbur–Anderson units) = 10(t 0/t − 1) per mg of protein, where t is the time required for the pH change when the test sample is present and t 0 is the time required for the pH change for the buffer without the test sample. Bovine carbonic anhydrase II (Sigma, St Louis, USA) was used as a positive control. Bovine serum albumin (BSA) and laminin-binding protein Lmb of Streptococcus agalactiae were used as negative controls.
3. Results and discussion
GK2848 was expressed in E. coli and the recombinant protein was purified using Ni–NTA affinity and gel-filtration chromatography. GK2848 was purified to homogeneity and appeared to run as a single band on SDS–PAGE with a molecular weight of 19.96 kDa. A PSI-BLAST analysis of GK2848 against the nonredundant protein database identified 400 entries, with sequence identity ranging from 95 to 58%. This included putative acetyltransferases and carbonic anhydrases from Geobacillus, Bacillus and Anoxibacillus species (Fig. 2 ▶). A BLAST search of the sequence of GK2848 against the Protein Data Bank to retrieve homologous structures revealed that GK2848 belonged to the left-handed β-helix family of proteins (Choi et al., 2008 ▶), with the closest homologue among them being a putative acetyltransferase from B. cereus (PDB entry 1xhd; Midwest Center for Structural Genomics, unpublished work). The homologues also included two γ-class carbonic anhydrases, Cam from Methanosarcina thermophila and Cap from Pyrococcus horikoshii (Jeyakanthan et al., 2008 ▶); however, both share low sequence identities of 38.5 and 22.5%, respectively, with GK2848. Only Cam has been extensively characterized and has been shown to exist as a homotrimer (Alber & Ferry, 1994 ▶, 1996 ▶; Iverson et al., 2000 ▶). To find out whether GK2848 possesses carbonic anhydrase activity or not, a carbonic anhydrase assay of the purified protein using the Wilbur–Anderson method (Wilbur & Anderson, 1948 ▶) was carried out. GK2848 was found to have a specific activity of 187.96 WAU per milligram. No detectable activity was found using samples of BSA and Lmb, which confirmed that GK2848 belongs to the carbonic anhydrase family.
Figure 2.
Multiple sequence alignment of GK2848 with its homologous sequences from G. thermodenitrificans (95%), B. methanolicus (81%), B. megaterium (81%), B. azotoformans (81%), B. cereus (79%), B. weihenstephanensis (80%) and B. thuringiensis (80%). The highly conserved residues are shown as red blocks. Sequence comparison shows that nearly 62% of the residues of GK2848 are highly conserved in homologous species. None of the close homologues of GK2848 have been characterized as carbonic anhydrase and the structural information is also not available except for B. cereus protein.
Good diffraction-quality crystals of GK2848 were obtained as described above. The crystals belonged to the orthorhombic space group P21212, with unit-cell parameters a = 137.98, b = 58.70, c = 94.39 Å, α = β = γ = 90°, and the crystal data were processed to 2.7 Å resolution using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶).
Assuming a molecular weight of 20 kDa for the protein and three molecules in the asymmetric unit, the resultant Matthews coefficient (V M; Matthews, 1968 ▶) was calculated to be 3.18 Å3 Da−1, with a solvent content of 61.36%. A preliminary solution of the structure was obtained by molecular replacement using MOLREP (Vagin & Teplyakov, 2010 ▶), with the structure of a putative acetyltransferase from B. cereus (1xhd) as the search model. As expected, three molecules were found in the crystallographic asymmetric unit. Further structural, mutational and biochemical characterization of GK2848 for carbonic anhydrase activity is in progress. Since none of the close sequence homologues of GK2848 have been characterized as carbonic anhydrases, the structural and functional information of GK2848 will provide additional insights into the mechanism of this ancient enzyme.
Acknowledgments
The authors thank C. Kuroishi for cloning of the gk2848 gene. This work was supported by the RIKEN Structural Genomic/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan. PR thanks the Department of Biotechnology (DBT), Government of India, for providing a fellowship.
References
- Alber, B. E. & Ferry, J. G. (1994). Proc. Natl Acad. Sci. USA, 91, 6909–6913. [DOI] [PMC free article] [PubMed]
- Alber, B. E. & Ferry, J. G. (1996). J. Bacteriol. 178, 3270–3274. [DOI] [PMC free article] [PubMed]
- Choi, J. H., Govaerts, C., May, B. C. H. & Cohen, F. E. (2008). Proteins, 73, 150–160. [DOI] [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Iverson, T. M., Alber, B. E., Kisker, C., Ferry, J. G. & Rees, D. C. (2000). Biochemistry, 39, 9222–9231. [DOI] [PubMed]
- Jeyakanthan, J., Rangarajan, S., Mridula, P., Kanaujia, S. P., Shiro, Y., Kuramitsu, S., Yokoyama, S. & Sekar, K. (2008). Acta Cryst. D64, 1012–1019. [DOI] [PubMed]
- Lindskog, S. (1997). Pharmacol. Ther. 74, 1–20. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Takami, H., Takaki, Y., Chee, G. J., Nishi, S., Shimamura, S., Suzuki, H., Matsui, S. & Uchiyama, I. (2004). Nucleic Acids Res. 32, 6292–6303. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wilbur, K. M. & Anderson, N. G. (1948). J. Biol. Chem. 176, 147–154. [PubMed]
- Zimmerman, S. A. & Ferry, J. G. (2008). Curr. Pharm. Des. 14, 716–721. [DOI] [PubMed]



