The Thermofluor assay constitutes a quick and easy-to-perform high-throughput method with which it is possible to identify protein-stabilizing buffer compositions or small-molecule additives.
Keywords: thermal shift assay, Thermofluor, protein buffer cocktails, differential scanning fluorimetry, protein unfolding, small-molecule additives
Abstract
The stability and homogeneity of a protein sample is strongly influenced by the composition of the buffer that the protein is in. A quick and easy approach to identify a buffer composition which increases the stability and possibly the conformational homogeneity of a protein sample is the fluorescence-based thermal-shift assay (Thermofluor). Here, a novel 96-condition screen for Thermofluor experiments is presented which consists of buffer and additive parts. The buffer screen comprises 23 different buffers and the additive screen includes small-molecule additives such as salts and nucleotide analogues. The utilization of small-molecule components which increase the thermal stability of a protein sample frequently results in a protein preparation of higher quality and quantity and ultimately also increases the chances of the protein crystallizing.
1. Introduction
Although many biological macromolecules have been crystallized over the past few decades, the crystallization of a new macromolecule usually requires screening against a large number and a large variety of crystallization cocktails (Rupp, 2009 ▶). The chances of identifying successful crystallization conditions are usually higher when the macromolecule exhibits a higher level of purity, chemical and conformational homogeneity and stability. In order to assess the stability and monodispersity of a sample, various quality-control techniques are available such as size-exclusion chromatography, mass spectrometry, static and dynamic light scattering, circular-dichroism spectroscopy and analytical ultracentrifugation (Geerlof et al., 2006 ▶). Another high-throughput approach is the fluorescence-based thermal-shift assay (Thermofluor), which is also referred to as differential scanning fluorimetry (DSF; Niesen et al., 2007 ▶). The Thermofluor assay was originally developed for drug-discovery applications since it allows the rapid screening of molecules from compound libraries that bind to and therefore stabilize the protein of interest (Pantoliano et al., 2001 ▶; Senisterra et al., 2006 ▶; Niesen et al., 2007 ▶). In addition, the Thermofluor method is often used in everyday laboratory work in order to screen for optimized buffer conditions by varying the pH, buffer molecules and small-molecule additives. It has been demonstrated a number of times that increased thermal stability is correlated with increased structural order and decreased protein flexibility. This is often accompanied by increased conformational homogeneity of the protein sample (Ericsson et al., 2006 ▶; Vedadi et al., 2006 ▶; Nettleship et al., 2008 ▶). Consequently, the identification of an optimized buffer cocktail or additive may have beneficial effects for protein-production procedures (Mezzasalma et al., 2007 ▶) and may also increase the chances of the protein crystallizing.
The Thermofluor assay makes use of an environmentally sensitive dye, e.g. a hydrophobic fluorophore, which allows one to distinguish between folded and unfolded states of a protein. In the ideal case, no fluorescence is observed at low temperature because the protein is completely and correctly folded and no hydrophobic areas are exposed. Upon an increase in temperature the protein starts to unfold and hydrophobic areas become exposed. The fluorophore can now bind to these areas and fluorescence occurs. For data analysis, the fluorescence intensity may be plotted as a function of temperature. The resulting curve is ideally sigmoidal and can be used to estimate the apparent melting temperature T m of the protein of interest, which corresponds to the midpoint of the transition curve. Strictly speaking, the T m value determined by Thermofluor corresponds to the temperature at which hydrophobic areas of the protein become exposed. This is generally accepted to be a good approximation of the actual melting temperature of the protein (Senisterra et al., 2006 ▶), although methods such as differential scanning calorimetry (DSC) are specifically geared towards determination of the actual melting temperature.
Here, we present and discuss a novel 96-condition screen for Thermofluor experiments. The screen is based on a previously published screen (Nettleship et al., 2008 ▶) and has undergone a number of rounds of development and improvement. Since the screen is suitable for a wide variety of protein samples, it may turn out to be useful as a starting screen for Thermofluor experiments with the goal of the identifying optimized buffer conditions for protein crystallization.
2. Development of the Thermofluor screen
The original screen (Nettleship et al., 2008 ▶) was built up of two equally sized parts (each consisting of 48 conditions): a ‘buffer screen’ and an ‘additive screen’. The buffer screen contained 12 buffers covering a pH range from 4.0 to 9.5. These buffers were combined with NaCl at concentrations of up to 500 mM. The additive screen contained nucleotide cofactors, metal salts, sugars, reducing agents and other chemicals.
Over the years the original screen was modified and extended, reflecting the accumulated experience with the screen in practical terms. The Thermofluor screen presented here is also composed of a buffer screen (Table 1 ▶) and an additive screen (Table 2 ▶). The buffer screen was extended to 23 different buffer solutions covering a pH range from 4.0 to 10.0. These are used as such or in combination with 250 mM NaCl. This not only allows the effect of pH to be investigated but also the effect that different buffer chemicals may have on the stability of the protein. For the additive screen, different groups of chemicals have been defined. During the initial phase of the development of the additive screen all additive solutions were prepared in a basal buffer consisting of 50 mM Tris–HCl pH 7.5, 200 mM NaCl. However, since a variety of proteins have been found to be rather unstable in this buffer cocktail, the current additive screen is left unbuffered.
Table 1. Composition of the 48 buffer screen solutions.
The concentrations given are the final concentrations used during the experiment.
| Well | Buffer | Salt |
|---|---|---|
| A1 | Water | |
| A2 | 50 mM sodium citrate pH 4.0 | |
| A3 | 50 mM sodium acetate pH 4.5 | |
| A4 | 50 mM sodium citrate pH 5.0 | |
| A5 | 50 mM sodium citrate pH 5.5 | |
| A6 | 50 mM bis-tris pH 6.0 | |
| A7 | 50 mM sodium phosphate pH 6.0 | |
| A8 | 50 mM MES pH 6.2 | |
| A9 | 50 mM bis-tris propane pH 6.5 | |
| A10 | 50 mM ADA pH 6.5 | |
| A11 | 50 mM MES pH 6.7 | |
| A12 | 50 mM PIPES pH 6.7 | |
| B1 | 50 mM sodium phosphate pH 7.0 | |
| B2 | 50 mM MOPS pH 7.0 | |
| B3 | 50 mM HEPES pH 7.0 | |
| B4 | 50 mM HEPES pH 7.5 | |
| B5 | 50 mM Tris–HCl pH 7.5 | |
| B6 | 50 mM HEPES pH 8.0 | |
| B7 | 50 mM Tris–HCl pH 8.0 | |
| B8 | 50 mM Bicine pH 8.0 | |
| B9 | 50 mM Tris–HCl pH 8.5 | |
| B10 | 50 mM CHES pH 9.0 | |
| B11 | 50 mM CHES pH 9.5 | |
| B12 | 50 mM CHES pH 10.0 | |
| C1 | 250 mM NaCl | |
| C2 | 50 mM sodium citrate pH 4.0 | 250 mM NaCl |
| C3 | 50 mM sodium acetate pH 4.5 | 250 mM NaCl |
| C4 | 50 mM sodium citrate pH 5.0 | 250 mM NaCl |
| C5 | 50 mM sodium citrate pH 5.5 | 250 mM NaCl |
| C6 | 50 mM bis-tris pH 6.0 | 250 mM NaCl |
| C7 | 50 mM sodium phosphate pH 6.0 | 250 mM NaCl |
| C8 | 50 mM MES pH 6.2 | 250 mM NaCl |
| C9 | 50 mM bis-tris propane pH 6.5 | 250 mM NaCl |
| C10 | 50 mM ADA pH 6.5 | 250 mM NaCl |
| C11 | 50 mM MES pH 6.7 | 250 mM NaCl |
| C12 | 50 mM PIPES pH 6.7 | 250 mM NaCl |
| D1 | 50 mM sodium phosphate pH 7.0 | 250 mM NaCl |
| D2 | 50 mM MOPS pH 7.0 | 250 mM NaCl |
| D3 | 50 mM HEPES pH 7.0 | 250 mM NaCl |
| D4 | 50 mM HEPES pH 7.5 | 250 mM NaCl |
| D5 | 50 mM Tris–HCl pH 7.5 | 250 mM NaCl |
| D6 | 50 mM HEPES pH 8.0 | 250 mM NaCl |
| D7 | 50 mM Tris–HCl pH 8.0 | 250 mM NaCl |
| D8 | 50 mM Bicine pH 8.0 | 250 mM NaCl |
| D9 | 50 mM Tris–HCl pH 8.5 | 250 mM NaCl |
| D10 | 50 mM CHES pH 9.0 | 250 mM NaCl |
| D11 | 50 mM CHES pH 9.5 | 250 mM NaCl |
| D12 | 50 mM CHES pH 10.0 | 250 mM NaCl |
Table 2. Composition of the 48 additive screen solutions.
The concentrations given are the final concentrations used during the experiment.
| Well | Additive 1 | Additive 2 |
|---|---|---|
| E1 | 50 mM sodium chloride | |
| E2 | 100 mM sodium chloride | |
| E3 | 150 mM sodium chloride | |
| E4 | 200 mM sodium chloride | |
| E5 | 400 mM sodium chloride | |
| E6 | 600 mM sodium chloride | |
| E7 | 800 mM sodium chloride | |
| E8 | 1000 mM sodium chloride | |
| E9 | 2.5%(v/v) glycerol | |
| E10 | 5%(v/v) glycerol | |
| E11 | 10%(v/v) glycerol | |
| E12 | 15%(v/v) glycerol | |
| F1 | 20%(v/v) glycerol | |
| F2 | 3% D-glucose | |
| F3 | 3% sucrose | |
| F4 | 10 mM magnesium chloride | |
| F5 | 10 mM calcium chloride | |
| F6 | 5 mM manganese chloride | |
| F7 | 5 mM nickel chloride | |
| F8 | 5 mM iron(III) chloride | |
| F9 | 5 mM zinc chloride | |
| F10 | 5 mM EDTA | |
| F11 | 100 mM potassium chloride | |
| F12 | 100 mM lithium chloride | |
| G1 | 50 mM sodium bromide | |
| G2 | 50 mM sodium iodide | |
| G3 | 100 mM sodium formate | |
| G4 | 100 mM sodium acetate | |
| G5 | 100 mM sodium malonate | |
| G6 | 100 mM sodium tartrate | |
| G7 | 100 mM sodium nitrate | |
| G8 | 100 mM sodium thiocyanate | |
| G9 | 100 mM sodium sulfate | |
| G10 | 100 mM ammonium sulfate | |
| G11 | 100 mM ammonium chloride | |
| G12 | 100 mM imidazole pH 7.6 | |
| H1 | 500 mM imidazole pH 7.6 | |
| H2 | 2 mM AMP | 5 mM magnesium chloride |
| H3 | 2 mM ADP | 5 mM magnesium chloride |
| H4 | 2 mM AMPPcP | 5 mM magnesium chloride |
| H5 | 2 mM dCMP | 5 mM magnesium chloride |
| H6 | 2 mM dGMP | 5 mM magnesium chloride |
| H7 | 2 mM GDP | 5 mM magnesium chloride |
| H8 | 2 mM TMP | 5 mM magnesium chloride |
| H9 | 2 mM NAD | 5 mM magnesium chloride |
| H10 | 30 mM L-arginine | 30 mM L-glutamate |
| H11 | 5 mM dithiothreitol | |
| H12 | 5 mM β-mercaptoethanol |
The roles of the different groups of chemicals in the additive screen are as follows. Conditions E1–E8 (Table 2 ▶) are included to analyze protein stability in the presence of NaCl in the concentration range 50–1000 mM. This allows investigation of whether low- or high-salt concentrations in the purification procedure are tolerated by the protein. Conditions E9–F3 are used to analyze the effect of glycerol, d-glucose and sucrose, which are frequently encountered as protein-stabilizing agents. In conditions F4–G11 different metal salts are tested. EDTA was also included in this group to assess the effect of the absence of divalent metal ions on protein stability. Imidazole is commonly used for the elution of proteins from immobilized metal-affinity chromatography (IMAC) columns. Two concentrations of imidazole (conditions G12 and H1) are included to assess protein stability at high imidazole concentrations. If the stability is found to be significantly reduced at high imidazole concentrations, care should be taken to remove the imidazole as soon as possible after elution of the protein, e.g. by dilution, dialysis or size-exclusion chromatography. Conditions H2–H9 contain various nucleotide cofactors supplemented with 5 mM magnesium chloride, which often assists in nucleotide-cofactor binding. Finally, the two amino acids Glu and Arg (Golovanov et al., 2004 ▶) as well as two different reducing agents constitute the last group of the screen (conditions H10–H12).
The whole screen may be prepared in a 96 deep-well master block and stored at 253 K. However, multiple rounds of freezing and thawing may have adverse effects on some of the substances in the screen. It is therefore recommended to reformat the freshly prepared screen into 96-well thin-wall PCR plates, which are later directly used for the Thermofluor experiment, to seal them with low-cost sealing foil and to store the ready-to-use plate at 253 K for one-time use.
3. The Thermofluor experiment
For the Thermofluor experiment, no prior knowledge of the protein properties is required. Every standard real-time PCR machine (e.g. a MyiQTM Thermocycler equipped with a Single Color Real-Time PCR Detection System from Bio-Rad) is suitable for this experiment. The assay is performed using a 96-well thin-wall PCR plate (e.g. from Bio-Rad or Roche). The total reaction volume is 25 µl and the plate is set up on ice or in a cold room. Either a ready-to-use plate, which was previously prepared and stored at 253 K, is thawed for the experiment or the plate is freshly prepared. For the buffer screen, 20 µl of the screening solution is pipetted into the wells of the plate, ideally using a multi-channel pipette. Accordingly, for the additive screen the wells are filled with 15 µl of the screening solution and 5 µl water or 5× buffer (for instance, the most stabilizing buffer identified from the buffer screen or an alternative buffer system such as that used during the protein-purification procedure). A 5000× SYPRO Orange stock solution in DMSO (Invitrogen) is diluted 1:100 in water to yield a tenfold working solution, which can be stored in the dark at 277 K for several hours. The protein concentration in the purified protein solution should be in the range 20–100 µM. However, the concentration may be lower for higher molecular-weight samples. 2.5 µl protein solution and 2.5 µl of the tenfold SYPRO Orange working solution are added to the 20 µl well solution. The plates are sealed with Optical Quality Sealing Tape (e.g. from Bio-Rad) and centrifuged at 4000 rev min−1 for 2 min. For the Thermofluor experiment the plates are then heated from 277 to 368 K in increments of 0.5–1 K in a thermocycler equipped with a single-colour real-time PCR detection system (scan rate of 1 K min−1). The lid of the thermocycler is heated to 378 K to avoid condensation effects during the experiment. Fluorescence changes in the wells of the plate are monitored simultaneously using excitation and emission wavelengths of 492 and 516 nm, respectively. The whole experiment only takes about 1–2 h. It is also possible to start the Thermofluor experiment at a higher temperature than 277 K, e.g. 293 K; however, a lower starting temperature is recommended in order to ensure that proteins with a low melting temperature also exhibit clear baseline fluorescence before protein unfolding is initiated.
4. Use of the screen
The Thermofluor screen presented in this work may be used in multiple ways. Firstly, all 96 conditions can be tested simultaneously. However, it needs to be emphasized again that the additive screen lacks a buffer system; this might result in unstable protein, leading to low-quality fluorescence curves. Therefore, it is recommended that the additive screen is used in conjunction with a buffer system of choice (see §3). Another option would be to split the screen into the two halves. The buffer screen is first investigated in order to identify the most promising buffer composition; then, in a second experiment the most stabilizing buffer may be used in the additive screen.
It is also clear that with a coarse screen such as that described here it is not possible to identify ‘the best’ buffer composition in one step. Further protein-specific fine-tuning may be required to achieve this. Furthermore, for some proteins it may initially be necessary to also screen against the nature and the concentration of the dye used in order to yield optimal results.
5. Data evaluation
5.1. Data analysis
Data analysis can be performed using a Boltzmann model (Ericsson et al., 2006 ▶). However, in everyday laboratory work the approach, which is frequently used, is just a simple qualitative test. Its evaluation is simply based on visual inspection of superposed curves, which allows identification of the condition with the highest apparent melting temperature.
5.2. Non-ideal curve shape
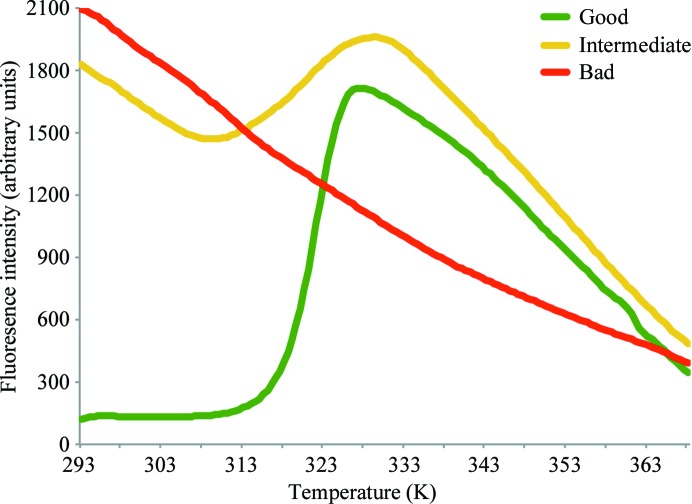
In practice, the unfolding curves obtained from a Thermofluor experiment will mostly not exhibit an ideal sigmoidal shape. Fig. 1 ▶ depicts three possible outcomes. A good unfolding curve has no or only low fluorescence intensity before unfolding is initiated, followed by a sharp transition (green curve in Fig. 1 ▶). After reaching the maximum fluorescence a decay in fluorescence intensity is observed, which might be explained by thermal motion of the fluorescent dye and/or protein aggregation.
Figure 1.
Different shapes of protein-unfolding curves in a Thermofluor experiment. Examples of unfolding curves of good (green), intermediate (yellow) and bad (red) quality are presented. The unfolding curve of good quality can be used for fitting procedures using the Boltzmann model (Ericsson et al., 2006 ▶), resulting in this case in an apparent T m value of 321.8 K.
The fluorescent dye SYPRO Orange may exhibit high background fluorescence below the transition temperature, which is often accompanied by a broadened transition to a non-interpretable curve shape (yellow curve in Fig. 1 ▶). This happens if the protein is already partially unfolded, e.g. owing to a sub-optimal initial buffer composition or protein:SYPRO Orange ratio or if the protein exhibits hydrophobic patches on its surface, e.g. owing to a missing binding partner. Such curves are usually not useful for fitting procedures using the Boltzmann model. However, by careful visual inspection of broadened curves a stabilizing buffer can often still be identified which might result in better melting curves in subsequent Thermofluor experiments (Crowther et al., 2009 ▶).
For some proteins, no useful unfolding transition at all can be obtained (red curve in Fig. 1 ▶) because the protein is already unfolded at the beginning of the experiment and hence no further transition takes place. This observation is made for some proteins in the extreme regions of the pH buffer screen (e.g. pH 4.0 and 10.0). However, if the buffer used for protein purification is so unfavourable, it can be envisaged that the whole Thermofluor screen will not yield any useful results. In such cases different initial buffer systems should be tried, although it may not be apparent from the Thermofluor assay which one to choose. Moreover, no unfolding transition is also observed when the analyzed protein does not have a hydrophobic core. This is, for instance, the case for some DNA-binding proteins.
5.3. When is a shift significant?
The effect of different compounds on the shift of the melting temperature cannot be predicted and the shift can range from as little as only a few degrees to several tens of degrees. The accuracy of the measurement is correlated with the shape of the unfolding curve (Crowther et al., 2009 ▶) and hence can be considered as a protein-dependent factor. If desired, the precision of the measured value can be determined by performing multiple independent Thermofluor experiments. All changes larger than the determined error can be considered as being significant.
5.4. Negative shift in melting temperature
Obviously, for most proteins the goal of the experiment is the identification of an increased melting temperature in order to obtain a protein sample with higher stability and hopefully homogeneity. However, it is also feasible to look for the opposite: considering the very high melting temperatures of more than 363 K observed for some proteins from thermophilic microorganisms, it may also be worth identifying a buffer system which is destabilizing. In such a buffer system, protein solubility would be decreased and consequently the chance of crystallization may be enhanced.
Moreover, information about a negative shift can be used during protein-production procedures, e.g. destabilization of the protein in high imidazole or extremely high- or low-salt concentrations.
6. Results and discussion
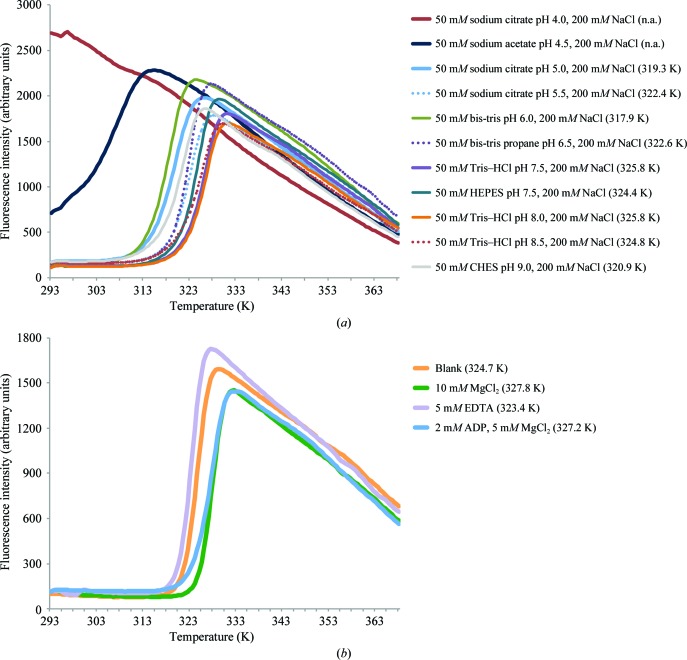
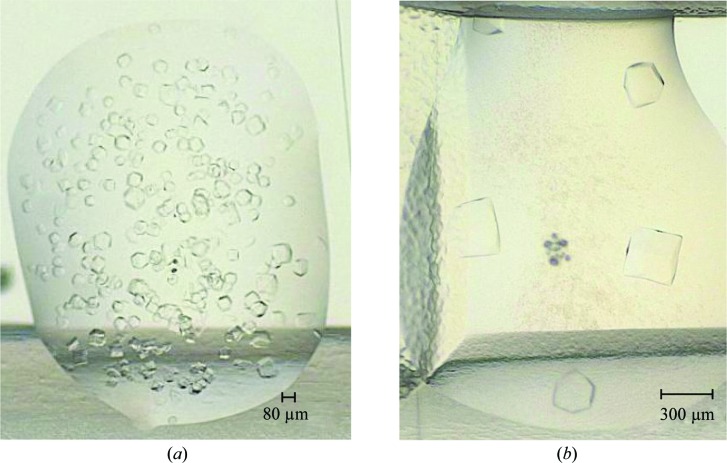
In recent years the screen presented in this work, or earlier versions of it, have been used for buffer optimization of more than a dozen proteins, of which more than 50% were successfully crystallized. One example is the enzyme Mtb-DapD, a tetrahydrodipicolinate-N-succinyltransferase from M. tuberculosis, the successful crystallization and X-ray structure of which have been described previously (Schuldt et al., 2008 ▶, 2009 ▶). Looking at results from a Thermofluor assay performed using DapD, it seems obvious that the Thermofluor assay is potentially very useful in order to identify conditions in which the protein is stable and therefore crystallizable (Figs. 2 ▶ and 3 ▶). For instance, DapD is significantly less stable in HEPES buffer than in Tris–HCl although both have the same pH value (Fig. 2 ▶ a). Moreover, MgCl2 was identified as a stabilizing agent (Fig. 2 ▶ b). Subsequently, DapD was successfully crystallized from two different crystallization conditions. The first condition was based on a precipitant solution which contained 200 mM MgCl2 (Schuldt et al., 2008 ▶). In the second condition, the precipitant solution contained a high concentration of sodium acetate pH 7.0. Here, addition of Mg2+ significantly increased the crystal size from about 80 to 300 µm (Fig. 3 ▶; Reinhard et al., unpublished work). In retrospect, this behaviour is nicely explained by the crystal structure of DapD obtained from the MgCl2 crystallization condition (Schuldt et al., 2009 ▶), which exhibits two tightly coordinated magnesium ions along the threefold axis of the DapD homotrimer.
Figure 2.
Thermofluor-based protein-unfolding curves of Mtb-DapD. The values in parentheses are T m values as determined using the Boltzmann model (Ericsson et al., 2006 ▶). A selection of protein-unfolding curves obtained in the buffer screen (a) and the additive screen (b) is shown. The buffer system used for the additive screen was 50 mM Tris–HCl pH 7.5, 200 mM NaCl. The Thermofluor experiment was prepared as described in §3 using 50 µM protein. The experiment was performed from 293 to 368 K in increments of 0.5 K. Please note that in a deviation from the screen described in this manuscript, the NaCl concentration of the screen was 200 mM instead of 250 mM. (a) The most stable protein sample was obtained in 50 mM Tris–HCl pH 7.5 or 8.0 supplemented with 200 mM NaCl. In 50 mM sodium citrate pH 4.0, 200 mM NaCl, DapD is unfolded at the beginning, resulting in a non-interpretable curve. (b) The additive screen showed that the T m value for DapD was increased by about 3 K in the presence of MgCl2, whereas the removal of divalent metal ions using EDTA resulted in destabilization. Adenosine-based nucleotides also result in a slight stabilization of DapD, probably because DapD uses succinyl-CoA as a cofactor, which exhibits an ADP moiety, and/or because of the presence of MgCl2 in the nucleotide conditions.
Figure 3.
Crystals of Mtb-DapD obtained in the absence (a) and presence (b) of 10 mM MgCl2. The crystals grew in the presence of a high concentration of sodium acetate pH 7.0 (Reinhard et al., unpublished work). The protein buffer was 20 mM Tris–HCl pH 8.0, 200 mM NaCl, 3 mM DTT. As shown previously, the enzymatic activity of Mtb-DapD is enhanced in the presence of MgCl2 (Schuldt et al., 2009 ▶).
7. Summary and outlook
The Thermofluor assay provides a quick and easy approach for identifying an optimized buffer composition for a protein of interest. In day-to-day laboratory work many cases were encountered in which optimization of the protein buffer had a positive effect on the quality, quantity and crystallizability of a protein sample. Consequently, it is strongly recommended that a Thermofluor experiment should be performed for every new protein target that is being worked on. However, just like every other experimental method, the Thermofluor method does not guarantee success and has its limitations.
The Thermofluor screen presented in this work provides a general starting point for the identification of optimized buffer conditions and is suitable for a very wide variety of protein samples. Modifications of the screen are easy to perform in order to assess, for instance, more protein-specific properties such as the binding of certain ligands and inhibitors. Considering how easy it is to perform the experiment and also the small amount of protein sample that is required to perform a Thermofluor screen, it may be anticipated that this approach will play an even more important role in future structural biology projects.
Acknowledgments
We would like to thank Dr Matthew Groves from the EBML Hamburg Outstation for discussions.
References
- Crowther, G. J. et al. (2009). J. Biomol. Screen. 14, 700–707. [DOI] [PMC free article] [PubMed]
- Ericsson, U. B., Hallberg, B. M., DeTitta, G. T., Dekker, N. & Nordlund, P. (2006). Anal. Biochem. 357, 289–298. [DOI] [PubMed]
- Geerlof, A. et al. (2006). Acta Cryst. D62, 1125–1136. [DOI] [PMC free article] [PubMed]
- Golovanov, A. P., Hautbergue, G. M., Wilson, S. A. & Lian, L.-Y. (2004). J. Am. Chem. Soc. 126, 8933–8939. [DOI] [PubMed]
- Mezzasalma, T. M., Kranz, J. K., Chan, W., Struble, G. T., Schalk-Hihi, C., Deckman, I. C., Springer, B. A. & Todd, M. J. (2007). J. Biomol. Screen. 12, 418–428. [DOI] [PubMed]
- Nettleship, J. E., Brown, J., Groves, M. R. & Geerlof, A. (2008). Methods Mol. Biol. 426, 299–318. [DOI] [PubMed]
- Niesen, F. H., Berglund, H. & Vedadi, M. (2007). Nature Protoc. 2, 2212–2221. [DOI] [PubMed]
- Pantoliano, M. W., Petrella, E. C., Kwasnoski, J. D., Lobanov, V. S., Myslik, J., Graf, E., Carver, T., Asel, E., Springer, B. A., Lane, P. & Salemme, F. R. (2001). J. Biomol. Screen. 6, 429–440. [DOI] [PubMed]
- Rupp, B. (2009). Biomolecular Crystallography: Principles, Practice, and Application to Structural Biology. New York: Garland Science.
- Schuldt, L., Weyand, S., Kefala, G. & Weiss, M. S. (2008). Acta Cryst. F64, 863–866. [DOI] [PMC free article] [PubMed]
- Schuldt, L., Weyand, S., Kefala, G. & Weiss, M. S. (2009). J. Mol. Biol. 389, 863–879. [DOI] [PubMed]
- Senisterra, G. A., Markin, E., Yamazaki, K., Hui, R., Vedadi, M. & Awrey, D. E. (2006). J. Biomol. Screen. 11, 940–948. [DOI] [PubMed]
- Vedadi, M. et al. (2006). Proc. Natl Acad. Sci. USA, 103, 15835–15840.