Abstract
Background
The symbiotic interaction between leguminous plants and rhizobia involves two processes: bacterial infection, resulting in the penetration of bacteria in epidermal and cortical cells, and root nodule organogenesis. Root nodule symbiosis is activated by rhizobial signalling molecules, called Nodulation factors (NFs). NF perception induces the expression of several genes called early nodulins. The early nodulin N5 of Medicago truncatula is a lipid transfer protein that has been shown to positively regulate nodulation although it displays in vitro inhibitory activity against Sinorhizobium meliloti. The purpose of this work was to investigate the role of MtN5 by studying its spatial and temporal pattern of expression during the symbiotic interaction, also in relation to known components of the symbiotic signalling pathway, and by analysing the phenotypic alterations displayed by rhizobia-inoculated MtN5-silenced roots.
Results
We show here that MtN5 is a NF-responsive gene expressed at a very early phase of symbiosis in epidermal cells and root hairs. MtN5 expression is induced in vitro by rhizobial effector molecules and by auxin and cytokinin, phytohormones involved in nodule organogenesis. Furthermore, lipid signaling is implicated in the response of MtN5 to rhizobia, since the activity of phospholipase D is required for MtN5 induction in S. meliloti-inoculated roots. MtN5-silenced roots inoculated with rhizobia display an increased root hair curling and a reduced number of invaded primordia compared to that in wild type roots, but with no impairment to nodule primordia formation. This phenotype is associated with the stimulation of ENOD11 expression, an early marker of infection, and with the down-regulation of Flotillin 4 (FLOT4), a protein involved in rhizobial entry.
Conclusions
These data indicate that MtN5 acts downstream of NF perception and upstream of FLOT4 in regulating pre-infection events. The positive effect of MtN5 on nodule primordia invasion is linked to the restriction of bacterial spread at the epidermal level. Furthermore, MtN5 seems to be dispensable for nodule primordia formation. These findings provide new information about the complex mechanism that controls the competence of root epidermal cells for rhizobial invasion.
Keywords: Medicago truncatula, MtN5, Symbiosis, Sinorhizobium meliloti, Pre-infection stage, Root hair curling
Background
Plants belonging to the Leguminosae family have the ability to interact with rhizobia and produce a nitrogen-fixing organ, the root nodule. The symbiotic relationship starts with a molecular cross-talk between the two partners. Host-plant derived molecules are perceived by rhizobia and activate the synthesis of Nod factors (NFs), which, in turn, elicit a variety of biochemical responses in the root hair, including changes in ion fluxes, membrane depolarization, the oscillation of the cytosolic calcium and modification in the cytoskeleton [1-4] that lead to root hair deformation, to infection thread (IT) formation and eventually to the penetration of the bacteria into the epidermis [5].
NF perception relies on a pair of orthologous genes belonging to the LysM-family receptor-like kinases (LysM-RLK), NFP and LYK3 in M. truncatula and NFR1 and NFR5 in L. japonicus[6-8]. Closely related genes for NF perception were also found in other leguminous plants such as pea and soybean [9]. The perception of the NF through the LysM-RLKs activates a signalling pathway, termed the common symbiotic pathway [5], constituted in M. truncatula by DMI1 (coding for an ion channel), DMI2 (coding for a leucine-rich repeat receptor-like kinase), which are involved in generating the calcium oscillations [10-12], and DMI3 (coding for a calcium calmodulin protein kinase), which is responsible for the decoding of calcium spiking amplitude and frequency [5]. NF-induced early infection events also involve the activity of phospholipase C and D [13,14]. NF perception and the activation of the signalling cascade take place in the root epidermis. At the same time, pericycle and cortical cells re-enter the cell cycle and form a primordium from which a nodule meristem arises [15,16]. The primordium is invaded by rhizobia harboured inside infection threads between 48 and 96 h post root hair infection.
Cell division is mainly controlled by two crucial plant phytohormones, auxin and cytokinin, which regulate the progression of cells through the cell cycle [17]. Concentrations and the auxins to cytokinins ratio both play a pivotal role in determining whether and where cells are about to enter the mitotic phase in plants [18-20]. The reduction of auxin transport in rhizobia-inoculated [18,21-23] roots changes not only the auxin fluxes but also the auxin to cytokinin ratio at the site of nodule initiation. One of the L. japonicus spontaneous nodulation mutants carries an alteration in the Lotus Histidine Kinase 1 (LHK1) gene that acts as a cytokinins receptor [24], whilst the LHK1 loss-of-function mutants show a marked reduction in the number of primordia and mature nodules [25]. In M. truncatula the RNA interference (RNAi)-mediated down-regulation of Cytokinin Response1 (CRE1), the LHK1 ortholog, resulted in a marked reduction of the cortical cell division and in a block of the majority of the ITs at root hair level [26]. Thus cytokinin seems to be involved in the coordination of the epidermal and cortical pathways of nodulation most likely through the NODULE INCEPTION (NIN) gene [27,28]. Until now, NIN has been the supposed key gene in coordinating the NFs signalling and entry pathways [27,28].
Recent studies have suggested that membrane microdomains and associated proteins such as Flotillin2 (FLOT2), FLOT4 and M. truncatula SYMBIOTIC REMORIN 1 (MtSYMREM1), are involved in epidermal responses to rhizobia and play a role in IT formation [29,30].
The extensive analysis of the M. truncatula transcriptome showed that a group of small, cysteine-rich peptides are up-regulated during the establishment of symbiosis [31]. MtN5 is a nodulin gene that was identified by means of a differential screening approach and is expressed in mature nodules [32-34]. The sequence homology suggests that MtN5 belongs to the plant non-specific Lipid Transfer Protein (nsLTP) super-family characterized by an eight cysteine motif and the phylogenetic analysis demonstrates that it has high homology with Arabidopsis thaliana DIR1 and groups with plant nsLTP-like protein [34,35]. Like other proteins belonging to the plant ns-LTP super-family, MtN5 is able to bind lipids in vitro and to inhibit the growth of pathogens and symbionts [34]. The RNAi-mediated MtN5 suppression resulted in a marked reduction in the number of nodules developed on transgenic hairy roots suggesting that MtN5 is required for nodulation [34]. However, the stages of the nodulation pathway that MtN5 activity might be involved in are still unknown.
The aim of this study was to gain further insight into the function of MtN5 in the M. truncatula-S. meliloti interaction. Our data indicated that MtN5 is a NF-responsive gene expressed at a very early phase of legume-rhizobium interaction in the root hairs and in the epidermal cells. The phenotypic analysis of MtN5-silenced roots showed that MtN5 is implicated in limiting root hair curling and that this role is necessary for an efficient colonization of nodule primordia. MtN5 response to rhizobia is dependent on phospholipase D (PLD) activity and does not seem to require DMI1. Furthermore, FLOT4 induction is drastically reduced in MtN5-silenced roots. This study demonstrates that MtN5 is involved in the control of rhizobial infection by acting in two apparently contrasting ways: firstly by restricting the invasion at the root epidermis and secondly by promoting the infection in the root cortex.
Results
Prediction of regulatory motifs in the MtN5 promoter
Previous data demonstrated that MtN5 is precociously induced in S. meliloti-inoculated roots, is expressed in the root nodules and that its function is required for the successful symbiotic interaction between S. meliloti and M. truncatula[32-35]. With the aim of gaining a deeper insight into the regulation of MtN5 expression, the putative MtN5 promoter was analysed by means of an in silico approach for the detection of conserved responsive elements.
The sequence spanning 1.54 kb upstream of the ATG translation start codon of the MtN5 gene (Figure 1A; Additional file 1) was analysed by using the PLACE algorithm [36,37]. Along with common elements found in promoter sequences such as TATA and CAAT boxes, the consensus sequences of the organ-specific elements (OSE) OSE1ROOTNODULE and OSE2ROOTNODULE (AAAGAT and CTCTT, respectively), which are characteristic of promoters active in infected cells of root nodules [38,39], were found in the MtN5 promoter, although not canonically spaced (Figure 1A; Additional file 1). Moreover, the nucleotide sequence AATTT, termed Nodulation Responsive Element (NRE), was found to recur twice in the 590 bp upstream of the ATG initiation codon. This responsive element was demonstrated to be present in the promoter sequence of well characterized nodulin genes such as MtENOD11, MtNIN and ERN1 and to function as a cis-acting element targeted by the GRAS type NSP1 transcription factor [40]. The in silico analysis of the MtN5 promoter also highlighted the presence of motifs involved in the hormonal control of gene expression such as ARR1AT and AUXREPSIAA4, responsive to cytokinin and auxin respectively [41,42]. Interestingly the auxin response element AUXREPSIAA4 had been previously shown to be tissue specific and characteristic of those genes that are expressed in the root apical meristem of pea plants [42].
Figure 1.
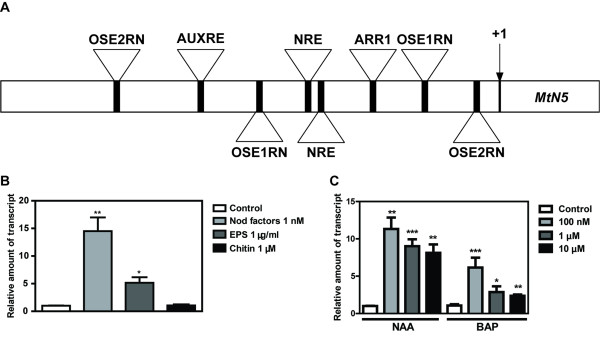
Bioinformatic analysis of the MtN5 promoter and expression of MtN5 in M. truncatula roots following treatments with bacteria-derived molecules and plant hormones. A. Schematic drawing of the putative MtN5 promoter and MtN5 open reading frame. The beginning of the ORF is identified by +1. The region analysed as the putative promoter encompasses 1.54 kb upstream the initiation codon. The following regulatory motifs are indicated: OSE1RN (OSE1ROOTNODULE; AAAGAT occurring in antisense orientation), OSE2RN (OSE2ROOTNODULE; AAGAG occurring in both sense and antisense orientations), NRE (AATTT), ARR1 (ARR1AT; GATT) and AUXRE (AUXREPSIAA4; GTCCCAT occurring in antisense orientation). B. M. truncatula roots were treated for 24 h with purified NFs (1 nM); for 48 h with EPS (1 μg/ml) and for 48 h with chitin (0.1 μM). C. M. truncatula roots were treated with α-naphtyl acetic acid (NAA) or with benzyl-amino-purine (BAP) at 100 nM, 1 μM and 10 μM for 48 h. The data were normalized to an internal actin control. The relative expression ratios were calculated using untreated roots as calibrator sample. The values reported are means ± SE (n=at least 3). Student’s t test was applied. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
MtN5 expression is induced by both rhizobia-derived molecules and plant hormones
On the basis of the motifs predicted by the PLACE algorithm [36], MtN5 expression was analyzed after treating the roots with both microorganism elicitor molecules and phytohormones. MtN5 expression is up-regulated after treatment with S. meliloti NFs [32 and this work], exhibiting an approximately 15-fold increase in NF treated roots compared with untreated roots (Figure 1B). However, a transient induction of MtN5 transcript level was also observed after inoculation with a S. meliloti strain defective in NF production [32]. To test whether other rhizobia-derived signals potentially contribute to MtN5 induction, we tested MtN5 expression in roots treated with exopolysaccharides (EPS) extracted from S. meliloti[43]. The roots were also treated with molecules like chitin oligomers (N,N’,N”,N”’-tetraacetylchitotetraose) that can be originated from the degradation of fungal cell wall [44]. In roots treated with 1 μg/ml EPS, the MtN5 transcript level increased about 5-fold as compared to the level in untreated roots, whilst MtN5 expression was not affected by chitin treatment (0.1 μM) (Figure 1B).
Considering that bioinformatic analysis of the MtN5 promoter predicts the presence of elements responsive to auxins and cytokinins, the phytohormones involved in the regulation of root nodules initiation and growth, the effects of auxin and cytokinin treatments on root MtN5 expression were evaluated. As reported in Figure 1C, both α-naphthyl acetic acid (NAA) and benzyl-amino-purine (BAP) supplied to roots can induce MtN5 expression in a wide range of concentrations (i.e. from 100 nM to 10 μM). Auxin displayed a similar ability to stimulate MtN5 expression at each of the concentrations tested whilst the effect of cytokinin was more pronounced at the lowest concentration used (100 nM) (Figure 1C). Interestingly, a stimulatory effect of auxin and cytokinin on MtN5 expression was detected in M. truncatula Jemalong in vitro-cultured leaf explants after a combined treatment with NAA and BAP [45,46].
The in silico analysis of the MtN5 promoter sequence and the experimental evidence suggest that MtN5 primarily responds to rhizobia-derived signals and can be regulated by hormones involved in the coordination of epidermal and cortical responses and in nodule formation.
MtN5 expression pattern during rhizobial infection and nodule development
To investigate the spatial and temporal pattern of MtN5 expression at the tissue and cell levels, MtN5 promoter activity was monitored in MtN5p::GUS transgenic roots with and without rhizobial inoculation. In non-inoculated roots, MtN5p::GUS is expressed at the root tip (Additional file 2). The β-glucuronidase (GUS) activity was also detected at the site of lateral root emergence and along the whole length of young lateral roots; as lateral roots get older, GUS activity was confined to the root tip (Additional file 2).
MtN5p::GUS transgenic roots showed localized induction of GUS activity as a consequence of S. meliloti inoculation. 3 hours post-inoculation (hpi), GUS activity was visible in the epidermis as localized spots and in the root hairs (Figure 2A-2C). At more advanced stages of infection (i.e. 24 hpi), the MtN5 promoter activity was detected in the root cortex, in close proximity to the central stele (2D).
Figure 2.
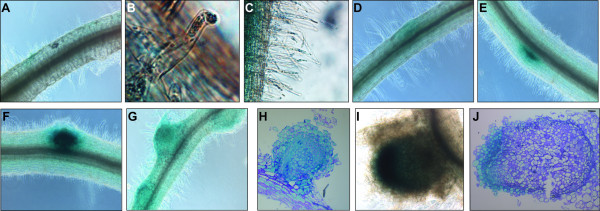
MtN5 promoter activity during rhizobial infection and nodule development. Representative expression patterns are shown. Localization of MtN5 expression in the root epidermis (A) and in the root hairs (B, C) 3 hours post-inoculation (hpi). (D) GUS staining detected in the root cortex at 24 hpi. MtN5 promoter activity in nodule primordia (E, F) and in young nodules (G, H). MtN5 promoter activity in fully developed root nodules (I, J).
During the early stages of nodule development, considerable activity of the MtN5 promoter could be observed at the boundary between the root cortex and the central stele (nodule primordia) (Figure 2E). As primordia grow and emerge from the root, the expression of the GUS reporter is detectable in the whole nodule (Figure 2F-2H). In fully developed root nodules, the promoter activity was predominantly localized in the distal zone (Figure 2I and 2J). The analysis of MtN5 promoter activity highlighted the presence of MtN5 expression during rhizobial infection in those cells that underwent structural changes and membrane rearrangements (i.e. root hairs) and in tissues showing a high cell division activity such as root meristems and nodule primordia.
The temporal and spatial pattern of MtN5 promoter activity indicates that MtN5 can be considered as an early marker of M. truncatula and S. meliloti interaction together with other epidermal nodulins such as RIP1 and ENOD11, which are putatively involved in cell wall modification before the formation of infection threads [47,48].
MtN5 influences ENOD11 response to rhizobia and is not required for NIN induction
In order to understand the relationships between MtN5 and other components of the early NFs signalling pathway, the expression of two early nodulins, ENOD11 and NIN was analysed in transgenic adventitious hairy roots carrying a hairpin construct (MtN5hp) for MtN5 silencing [34]. Composite plants were micro-flood inoculated with S. meliloti and roots were collected at different times after infection (3, 6, 12, and 72 hpi). The roots considered in this experiment were preselected for their transgenic status on the basis of the fluorescent signal deriving from the DsRED marker gene present in the T-DNA. MtN5 expression was induced very rapidly after S. meliloti inoculation (Figure 3A) in adventitious control roots, which were generated from the infection with Agrobacterium rhizogenes carrying an empty pRedRoot vector. In inoculated MtN5hp transgenic roots, MtN5 expression was significantly reduced at each time point considered compared to that in control roots (Figure 3A). The steady state level of the MtN5 transcript decreased on average by 70%, ranging from approximately 62% (6 hpi) to 80% (72 hpi).
Figure 3.
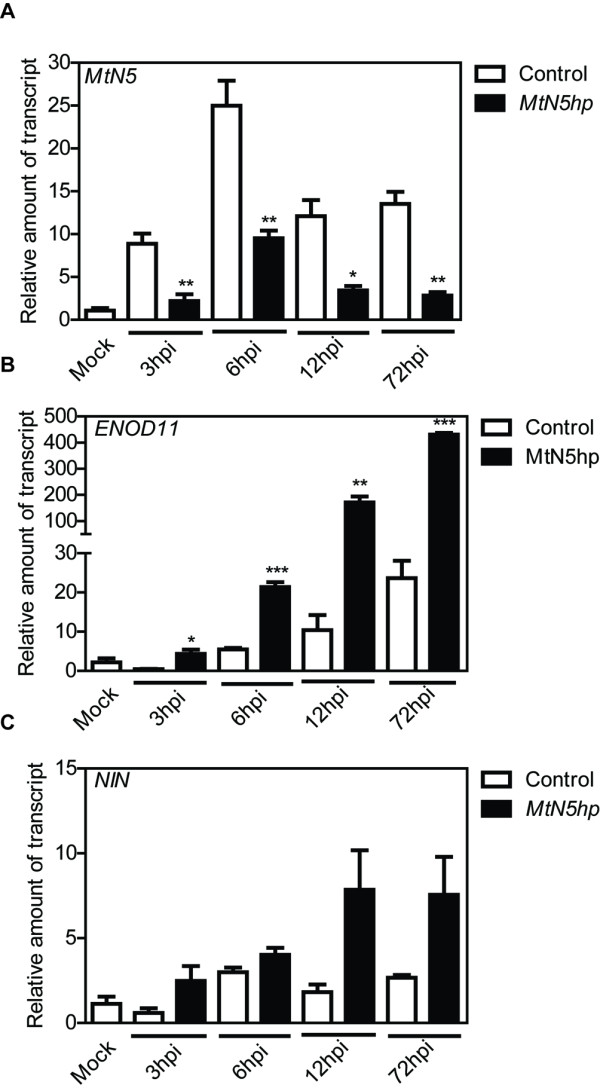
Time course expression analysis of MtN5, ENOD11 and NIN genes after rhizobial inoculation. The expression level of MtN5 (A), ENOD11 (B) and NIN (C) was assessed by qRT-PCR in adventitious hairy roots obtained by genetic transformation with A. rhizogenes harbouring a pRedRoot binary vector carrying either the MtN5hp construct or an empty T-DNA. The data were normalized to an internal actin control. The relative expression ratios were calculated using control mock-inoculated roots as calibrator sample. The values reported are means ± SE (n=at least 3). Student’s t test was applied. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The expression of ENOD11, an early marker of pre-infection and infection phases of rhizobial symbiosis, showed a progressive increase starting from 6 hpi in the control adventitious roots. In MtN5-silenced root tissues, ENOD11 was already strongly up-regulated at 3 hpi and its expression was significantly enhanced as compared to that in control roots in the following stages of infection (Figure 3B).
MtNIN is required for nodule primordia initiation having a role in the coordination of epidermal and cortical responses [28]. In control roots the expression of MtNIN was up-regulated starting from 6 hpi. MtN5 silencing did not significantly alter NIN mRNA steady state levels at each time point examined in the analysis (Figure 3C).
To date, the data available regarding the involvement of nodulin genes in the NFs signalling pathway place both ENOD11 and MtNIN downstream of the calcium spiking [13,28]. To analyze the relationship between MtN5 expression and the calcium oscillation we used the recently established Tnt1 transposon mutant collection of M. truncatula R108 [49] and searched for an insertion line in DMI1 gene, which is required for the generation of calcium spiking [10,50]. We identified the line NF4257, which carries a transposon insertion 155 nucleotides downstream of the translation initiation site of DMI1 gene. Plants were propagated and were subsequently screened for homozygous offsprings by polymerase chain reaction (PCR) (Additional file 3). In the NF4257 homozygous line, DMI1 expression was almost completely abolished (DMI1 steady state level was 20% as compared with wild type plants) (Additional file 3). In the S. meliloti-inoculated NF4257 mutant, the MtNIN induction was eliminated (Figure 4A and [28]), whilst MtN5 showed a wild-type behaviour (Figure 4B). This finding suggests that MtN5 activation might occur upstream of calcium spiking, although we cannot exclude a priori that other Tnt1 insertions, potentially present in the DMI1 homozygous mutant seedlings, could affect MtN5 expression in trans.
Figure 4.
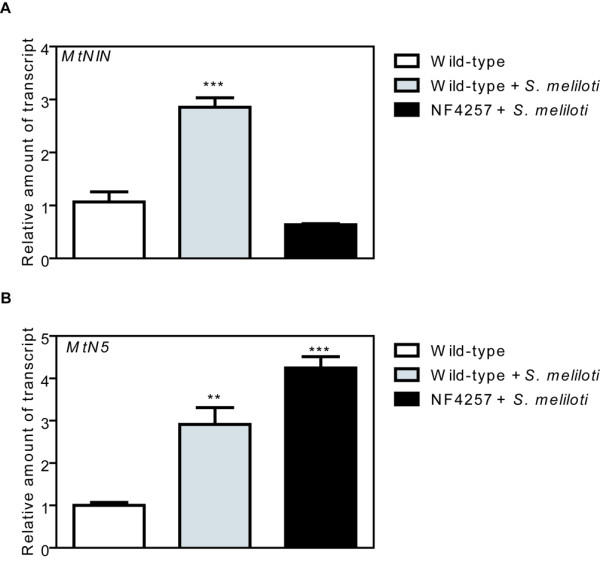
Expression level of NIN and MtN5 genes in M. truncatula DMI1 insertional mutant line. The expression level of NIN (A) and MtN5 (B) was assessed in M. truncatula R108 wild-type and DMI1 insertional mutant roots, both mock-inoculated and inoculated with S. meliloti. The data were normalized to an internal actin control. The relative expression ratios were calculated using mock-inoculated roots as calibrator sample. The values reported are means ± SE (n=3). Student’s t test was applied. **, P < 0.01; ***, P < 0.001.
PLD activity is required for the response of MtN5 to S. meliloti
Several pieces of research have demonstrated that the signalling cascade activated by NFs implicates, at an early stage, the intermediation of heterotrimeric G-proteins and small GTPases [51-54], which, in turn, stimulate the functionality of phospholipase C (PLC) and D (PLD) [51,53,55,56]. According to the current model, the products of hydrolysis generated by PLC (inositol trisphosphate and diacyl glycerol) stimulate the activity of ligand-gated calcium pumps causing the increase in the cytosolic Ca2+ concentration, whereas the PLD product (phosphatidic acid, PA) seems to be required for the onset of a kinase/phosphatase signalling cascade, that eventually leads to the activation of nodulin genes and to the reorganization of the cytoskeleton in preparation for the root hair inward growth [57].
With the aim of investigating the dependence of MtN5 induction on the two parallel pathways of the lipid signalling, a pharmacological approach was adopted, using specific inhibitors of PLC and PLD (neomycin and n-butanol, respectively). As already demonstrated, the application of both 100 μM neomycin and 68 mM n-butanol did not cause a significant loss of viability in root hair cells [13]. M. truncatula seedlings, pretreated with neomycin and subsequently inoculated with S. meliloti, showed a 13-fold up-regulation of MtN5 (Figure 5A) compared to untreated, control roots, whereas in the absence of the pharmacological treatment, inoculated roots displayed a 4-fold MtN5 induction indicating that neomycin did not repress MtN5 induction. Similarly, the treatment with the PLC inhibitor alone resulted in an approximately 9-fold increase of MtN5 expression. Analogous stimulatory effects were reported for other nodulin genes after treatment with PLC inhibitors and interpreted as the result of membrane trafficking alteration or microtubule reorganization [14,58]. These data suggest that the MtN5 induction due to rhizobial inoculation and to neomycin treatment are most probably independent effects (Figure 5A). The n-butanol competes for the phosphatidyl group acting as antagonist of PA production by PLD enzymatic activity [59]. Interestingly, S. meliloti-inoculated roots treated with the PLD inhibitor did not show a significant variation in MtN5 expression when compared to control roots indicating that the n-butanol treatment prevents MtN5 induction (Figure 5B). The treatment with tert-butanol, a butanol isomer, which does not act as a phosphatidyl group acceptor, did not hamper MtN5 induction (Figure 5C). On the whole, these results suggest that MtN5 response is dependent on PA production.
Figure 5.
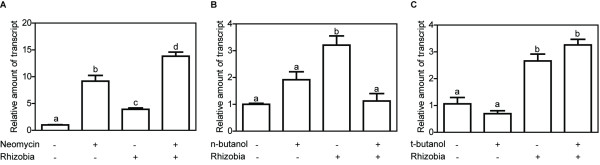
Effect of specific inhibitors of PLC and PLD (neomycin and n-butanol, respectively) on MtN5 expression. A. The relative mRNA level of MtN5 was assessed by qRT-PCR analysis in wild-type M. truncatula roots either untreated or treated with PLC agonist neomycin and inoculated with S. meliloti. B. qRT-PCR performed on wild-type M. truncatula roots either untreated or treated with PLD agonist n-butanol and inoculated with S. meliloti. C. qRT-PCR carried out on wild-type M. truncatula roots either untreated or treated with n-butanol isomer, t-butanol, and inoculated with S. meliloti. The expression data were normalized to an internal actin control. The relative expression ratios were calculated using untreated, mock-inoculated roots as calibrator sample. The data reported are means ± SE (n = at least 3). Common letters indicate no significant difference according to one-way ANOVA with the Bonferroni post-test.
MtN5 influences root hair curling and primordia invasion
The microscopic observations of MtN5hp hairy roots inoculated with a rhizobium strain harbouring the hemA::LacZ reporter gene revealed a significantly higher number - about twice that of control roots - of root hair curling events (Figure 6A). However, the 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-Gal) staining of rhizobia highlighted that the number of bacteria-colonized primordia was significantly reduced (by about 80%) in MtN5hp roots as compared to that in the control samples (Figure 6A) and, as already reported, the composite plants bearing MtN5hp silenced roots also showed a reduction in the number of fully developed nodules (by about 80%). Nonetheless, the total number of nodule primordia did not vary significantly between MtN5-silenced and control roots (Figure 6A) nor were any phenotypic alterations observed in curled root hairs such as excessive curling or root hair deformation (Additional file 4). It was also apparent that nodule primordia and mature nodules of MtN5hp plants were not dissimilar in morphology to those produced in control roots (Additional file 4). On the basis of these data, the transcript levels of M. truncatula Flotillin 4 (FLOT4), a gene involved in the bacterial entry pathway, were compared in MtN5-silenced roots and control roots. FLOT4 showed itself to be readily induced in control roots after rhizobial inoculation (3 hpi), reached a maximum of expression at 6 hpi and decreased afterwards. In MtN5hp roots infected with the symbiont, a drastic decrease of FLOT4 expression (between 70% and 90%) was detected between 3 and 12 hpi (Figure 6B).
Figure 6.
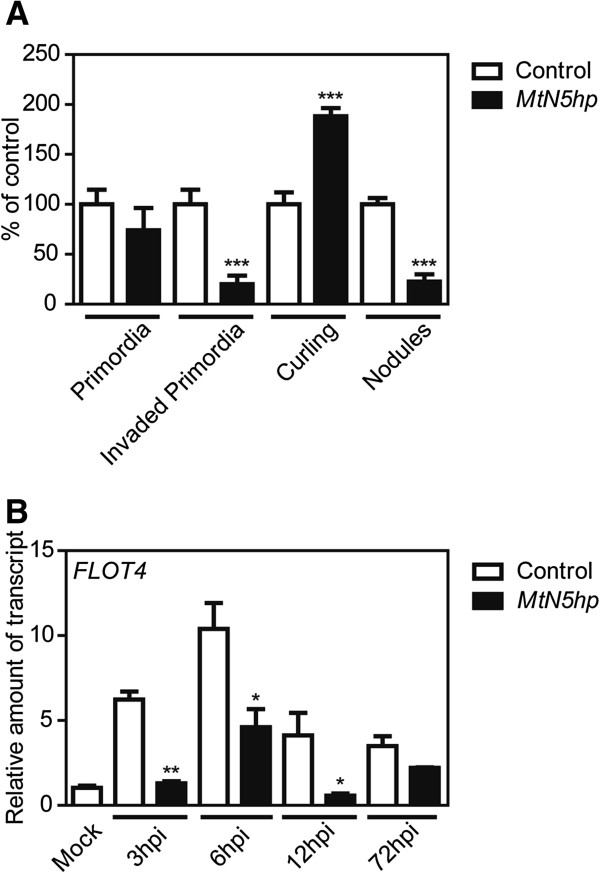
Phenotypical analysis of inoculated MtN5hp roots and FLOT4 expression. A. Number of root hair curling events, total and invaded primordia and mature nodules in MtN5hp roots inoculated with S. meliloti. The data reported are means ± SE (n=40) calculated as percentage relatively to control inoculated roots. Student’s t test was applied. ***, P < 0.001. B. The expression level of FLOT4 was assessed by qRT-PCR in adventitious hairy roots obtained by genetic transformation with A. rhizogenes harbouring a pRedRoot binary vector carrying either the MtN5hp construct or an empty T-DNA (control). The expression data were normalized to an internal actin control. The relative expression ratios were calculated using control mock-inoculated roots as calibrator sample. The values reported are means ± SE (n=3). Student’s t test was applied. *, P < 0.05; **, P < 0.01.
Discussion
The early nodulin MtN5 was identified by means of a subtractive hybridization approach [32] and, according to the sequence homology, was annotated as a putative Lipid Transfer Protein [32,33]. The in vitro characterization of MtN5 recombinant protein highlighted that it has the ability to bind lipid and counteract the growth of microorganisms [34] suggesting a possible involvement in the limitation of the rhizobial invasion. However, the functional study based on both silencing and over-expression approaches showed that MtN5 function is required for a successful establishment of the symbiosis [34].
To shed light on the molecular role of MtN5 in M. truncatula-S. meliloti symbiosis, we initially focused on the regulation of MtN5 expression. The data presented in this work confirm that the induction of MtN5 is a NF-responsive event, but also show that MtN5 expression is probably controlled by several additional signals, either rhizobia-derived effectors or phytohormones, auxin and cytokinins. These observations are consistent with the finding that MtN5 is induced at different stages of symbiotic interaction from pre-infection to mature nodules and also with the constitutive expression in root tips of principal and lateral roots. The rhizobial infection does not modify MtN5 expression at the root tip level, but it triggers very early the induction of MtN5 in the epidermal tissues (3 hpi) and in the root cortex (24 hpi). Thus we propose that MtN5 can be regarded as an early molecular marker of M. truncatula and S. meliloti interaction.
The detection of MtN5 promoter activity in root hairs and in localized spots of the epidermis supports the idea that MtN5 might be involved in pre-infection or/and infection events. Pre-infection responses to rhizobia are accompanied by the induction of the marker gene ENOD11 in the root epidermis, whereas in later phases its expression is confined to the invaded zones of the roots [48]. The transcription factor MtNIN seems to regulate ENOD11 activity contributing to the restriction of its expression to the S. meliloti-responsive zone of the root [28]. In inoculated MtN5-silenced roots, the ENOD11 induction is not prevented. Indeed, ENOD11 expression is strongly stimulated in MtN5-silenced roots compared with that in control roots, indicating that MtN5 function is required for controlling ENOD11 expression. In addition, MtN5 down-regulation does not inhibit NIN transcription suggesting that the mis-regulation of ENOD11 in MtN5-silenced roots is independent of the NIN effect on ENOD11 transcription. The stimulation of ENOD11 expression in MtN5-silenced roots could also be interpreted as an indirect consequence of the impaired rhizobial infection.
The phenotypic analysis of MtN5-silenced roots confirms a role of this nodulin in pre-infection stages of the symbiosis; in fact, although abnormalities in root hair curling were not observed, the number of curled root hairs was significantly greater in comparison to that in control roots. An enhanced responsiveness of root hairs to rhizobia was observed in several infection-defective leguminous plants [28,60-62], which could suggest the presence of a mechanism that represses root cell competence in wild type plants [61]. From our data we can infer that MtN5 might be implicated in the mechanism that limits root hair curling.
The host plant controls the rhizobial invasion with different strategies [63,64]. For instance, the induction of pathogen defence genes is one of the early events detected during symbiosis and it has been indicated as one of the mechanisms involved in IT abortion [33,65,66]. At later stages, the expression of these defence genes usually declines probably as a consequence of bacteria recognition by the host plant mediated also by molecular signals, e.g. exopolisaccharides, produced by rhizobia [33,65,67]. Furthermore, host plants limit the number of nodules through a mechanism termed autoregulation of nodulation that implicates the intervention of shoot receptor kinases [63,64]. MtN5 belongs to the non specific LTP family that includes apoplastic proteins with inhibitory activity against pathogens and itself possesses the capacity to limit S. meliloti growth in vitro[34]. It has also been suggested that MtN5 could belong to a group of LTPs having a role in plant-microbe interaction signalling [35]. MtN5 might act by limiting the stable attachment and/or proliferation of rhizobia at the surface of epidermal cells/root hairs, by modulating, for instance, either the perception or the activity of rhizobia-derived signal molecules. Besides the increased curling, MtN5-silenced roots showed a reduction in the number of invaded root nodules, indicating that MtN5 could also participate, either directly or indirectly, in the regulation of bacterial invasion. If this is so, the limitation of curling events might favour the penetration of bacteria at a restricted number of infected spots.
Several experimental observations are consistent with the existence of a parallel NF-mediated signalling pathway that operates in root hair curling and bacterial entry acting in concert with the common symbiotic pathway [25,68]. Furthermore, pharmacological studies have demonstrated that lipid signalling is implicated in both root hair deformation and IT initiation [14]. The response of MtN5 to rhizobium is dependent on PLD activity and we obtained a first indication that it might be independent of DMI1, a component of the common symbiotic pathway. Further investigations are needed to prove the independence of MtN5 induction from Ca2+ spiking and to ascertain whether MtN5 is a component of the parallel NF-mediated pathway that controls root hair curling and/or rhizobial entry. It was recently discovered that a member of the flotillin family, FLOT4, which is up-regulated during early symbiotic events and localized in membrane microdomains in infected root tips, plays a role in IT initiation and elongation [29]. Our observations indicate that MtN5 acts upstream with respect to FLOT4, supporting the idea that MtN5 participates in the events preceding IT development.
Bacterial infection occurring at the epidermis level is coordinated with the cortical process that promotes nodule formation and development [19]. The formation of nodule primordia requires the perception of Ca2+ oscillations by calcium-activated kinase CCaMK (DMI3) and the activation of cytokinin signalling. In addition, the phytohormone auxin participates both locally and systemically in the regulation of nodule organogenesis [22,69]. Both auxin and cytokinins induce MtN5 expression in vitro. Furthermore, in S. meliloti inoculated roots the MtN5 transcript localizes in the inner cortex at a site where cellular divisions take place to form the nodule primordium. These findings would suggest the possibility that MtN5 participates in nodule organogenesis. However, we observed that the total number of nodule primordia in MtN5-silenced roots did not differ from that measured in wild type roots. Therefore, MtN5 does not seem to be required for nodule primordia initiation. This is consistent with the finding that MtN5 functions downstream or more likely independently of MtNIN (Figure 3C).
Even though nodule initiation appears not to be impaired, the number of invaded primordia was markedly reduced in MtN5-silenced roots. Thus the decreased number of mature nodules in MtN5-silenced roots could be the consequence of the curtailed capacity of the rhizobia to invade the primordia. It has in fact been reported that bacterial invasion can be crucial for the maintenance of nodule developmental program [70]. On the other hand, we cannot exclude that MtN5 might have a direct role in promoting the maintenance of nodule primordia. Further investigations are necessary to clarify this point.
Conclusions
The data presented here demonstrate that MtN5 participates in the molecular events occurring at the epidermis after NF perception and PLD activation and before root hair invasion and that its function is dispensable for nodule initiation but required for nodule invasion. Plants lipid transfer proteins are usually small, secreted, basic proteins, characterized by the presence of a hydrophobic cavity that enables the interaction with lipid molecules. Members of the plant LTP superfamily display a wide range of biological activities such as a defensive role against pathogens, deposition of cuticular wax, modification of cell walls and pollen tube guidance [71,72]. Based on sequence similarity, MtN5 and A. thaliana DIR1, an nsLTP implicated in pathogen defence systemic signalling [73] have been assigned to a sub-group of ns-LTP putatively involved in lipid-mediated signalling [35]. The lily LTP SCA and the A. thaliana SCA, which are implicated in pollen tube growth, have also been put forward as signal transducers [72,74].
We propose that MtN5 plays a role in the process that regulates the competence of epidermal cells for rhizobial infection. Thus MtN5 would be part of the machinery that gives the host control over the mutualistic partner in order to preserve plant fitness. To explore the hypothesis that MtN5 functions as a signal transducer in these processes, the identification of MtN5 interacting proteins or ligands would be necessary.
Methods
Bacterial strains
Sinorhizobium meliloti 1021 [75] was grown at 28°C in LBMC medium (10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl, 2.6 mM MgSO4, 2.6 mM CaCl2) supplemented with streptomycin 200 μg/ml. Agrobacterium rhizogenes ARqua1 [76] was grown at 28°C in TY medium (5 g/l tryptone, 3 g/l yeast extract, 6 mM CaCl2, pH 7.2) supplemented with streptomycin 100 μg/ml.
Plant growth and rhizobial inoculation
Medicago truncatula cv. Jemalong seeds were sterilized and germinated as already described [77]. For time course assay and for GUS detection experiments, M. truncatula seedlings were placed in square Petri plates, containing slanted BMN agar medium [78] supplemented with 0.1 μM L-α-2-Aminoethoxyvinyl glycine (AVG). The plates were kept vertically in a growth chamber at 25°C and 10-h light/14-h dark regimen. After seven days of nitrogen starvation, the seedlings were micro-flood inoculated as previously described [34]. Briefly, bacteria were grown overnight and suspended in 10 mM MgSO4. Microflood inoculation was performed by placing five drops (0.5 μl) of bacterial suspension on the surface of the root. For qRT-PCR experiments, control samples (mock-inoculated) were treated with the same volume of 10 mM MgSO4.
GUS constructs and histochemical staining
The reporter construct was prepared by fusing together the MtN5 promoter and the intronless coding region of E. coli β-glucuronidase (uidA), as already described [79]. Briefly, 1.5 kb-long sequence upstream of the transcription initiation site was amplified from wild-type plants by means of PCR with the following primers: 5′-GAATTCCACAATCTCTTTCTTTCTCG-3′ and 5′-GGATCCCTGGTTCTAGTTTACTATAT-3′. The PCR fragments were sub-cloned and checked by sequencing. The MtN5 promoter was cloned upstream the 1.812 kb sequence of GUS coding region into a pBIN19 derivative vector, harbouring the nptII gene, coding for the kanamycin resistance, under the transcriptional control of nos promoter [80]. The two transcriptional cassettes (MtN5promoter::GUS and kanamycin resistance) were placed convergently. The resulting chimeric gene was mobilized into A. rhizogenes ARquaI, which was used to obtain plants bearing genetically modified silenced roots. The histochemical GUS staining was performed as previously described [81]. Images were taken with a Leica DM2500 microscope equipped with a DFC420C digital camera (Leica Microsystems, Wetzlar, Germany). The GUS stained nodules were embedded, after fixation and dehydratation, in Technovit 7100 (HeraeusKulzer, Wehrheim, Germany) according to the manufacturer’s instructions. Sections (6 μm thick) were prepared and stained with 0.05% toluidine blue. The slides were observed with a Zeiss Axiophot D1 microscope (http://www.zeiss.com/) and images were recorded with an Axiocam MRc5 camera (Zeiss) using the Axiovision program (version 4.1).
Plants inoculated with S. meliloti bearing the pXLGD4 plasmid, containing the constitutive hemA::LacZ gene, were stained as previously described [82]. Whole root samples were mounted on glass slides with coverslips and observed with a Leica DM2500 microscope equipped with a DFC420C digital camera (Leica Microsystems, Wetzlar, Germany).
Plant transformation
Root transformation with A. rhizogenes ARqua1 was performed as previously described [83]. Plants infected with ARqua1 were kept in square Petri dishes containing Fåhraeus Modified Medium (FMM) for about three weeks. When the binary vector employed in the transformation was pBIN19 (MtN5p::GUS construct), the FMM was supplemented with kanamycin 50 μg/ml for transformants selection. For MtN5 silencing, the MtN5hp construct was cloned into the pRedRoot binary vector, as previously described [34]. Transformed roots were checked using a Leica MZ16F fluorescence microscope with the following filter setting for DsRED1 detection: 541-551 nm bandpass excitation filter and 590 nm long-pass emission filter. Composite plants were micro-flood inoculated in Petri dishes and kept vertically in the growth chamber at 25°C and 10-h light/14-h dark regimen, as previously described [34].
Exopolysaccharide extraction
The extraction of exopolysaccharide (EPS) from S. meliloti 1021 was performed as previously described [43]. The polysaccharides were dissolved in 10% NaCl and the concentration of the preparation was checked using the phenol-H2SO4 method [84].
Plant treatments
To test the effects of plant hormones on MtN5 gene expression, M. truncatula seeds were scarified and sterilized as described above and then germinated on solid FMM medium. 7-day-old seedlings were moved to square Petri dishes containing slanted FMM agar medium supplemented with either α-naphthyl acetic acid (NAA) or benzyl-amino-purine (BAP) at different concentrations. Plants were kept vertically in the growth chamber at 25°C with a regimen of 10 h of light and 14 h of darkness for 48 h. Root apparatuses were collected, frozen in liquid nitrogen and stored at -80°C until RNA extraction.
For the treatments with Nod Factors (NFs), chitin tetramers (N,N’,N”,N”’-tetraacetylchitotreaose) (Carbosynth, UK) and exopolysaccharides (EPS), 7-day-old seedlings were transferred to 50 ml test tubes containing liquid FMM supplemented with the chosen concentration of the effector and kept in the growth chamber at 25°C with a regimen of 10 h of light and 14 h of darkness. The treatment with NFs was carried out for 24 h and the treatments with EPS and chitin tetramers were performed for 48 h.
For the pharmacological treatments seven-day-old M. truncatula seedlings were placed on Petri dishes containing BMN medium supplemented with both 0.1 μM AVG and the pharmacological effectors, and treated for 24 h. Neomycin (Sigma) was prepared as 10 mM aqueous solution and n-butanol and tert-butanol were diluted in sterile water just before use [13]. The plants were then transferred onto fresh BMN medium supplemented only with AVG and micro-flood inoculated with S. meliloti, as previously described. The root tissues were collected 4 hpi and MtN5 expression was checked by quantitative RT-PCR (qRT-PCR) analysis.
Quantitative RT-PCR
The qRT-PCR analyses were carried out as already described [85]. The nucleotide sequences of the primers used for the qRT-PCR are reported in Additional file 5. The pairs of primers used to analyse the expression of MtN5 in hairy roots were specifically chosen at the 3′ end of the transcript to avoid the amplification of sequences derived from the hp construct itself.
Genomic DNA extraction and insertional mutant characterization
The M. truncatula genomic DNA was prepared from 100 mg of leaves as previously described (http://medicago.org/documents/Protocols/dna.html).
The genetic characterization of the M. truncatula R108 NF4257 mutant line which, besides other insertions, harbours a transposon insertion in DMI1 gene, was carried out by means of a PCR-based approach. The genomic DNA was used as template in a PCR reaction containing two primers annealing on the DMI1 coding sequence (DMI1_for: 5′-ATCCTTGGCTGGAGTGACAAATTG-3′; DMI1_rev: 5′-CTGATCTGCATTTTCGTCCGCAGC-3′) and a third primer complementary to the Tnt1 sequence (Tntail1: 5′-TATGCAAAGAACTTGTCGGCATGC-3′) [49] (Additional file 3). The discrimination between heterozygous and homozygous plants for the insertion in the DMI1 gene was carried out on the basis of the number and the size of the amplicons obtained following PCR reaction.
Statistical analysis
The mean values ± SE are reported in the figures. Statistical analyses were conducted using a Student’s t test or a one–way ANOVA with Bonferroni post-test, as appropriate.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YP carried out the nodulation experiments and wrote the manuscript; YP, BM performed the molecular analyses; SM performed the microscopic analysis of the root nodules; TP coordinated the study and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Nucleotide sequence of the putative MtN5 promoter and MtN5 open reading frame. The beginning of the ORF is identified by +1. The region analysed as the putative promoter encompasses 1.54 kb upstream the initiation codon. TATA box and CAAT box are double underlined and underlined with dashed line, respectively. The other motifs highlighted are listed as follow: OSE1ROOTNODULE (AAAGAT occurring in antisense orientation), OSE2ROOTNODULE (AAGAG occurring in both sense and antisense orientations), NRE (AATTT) , ARR1AT (AGATT) and AUXREPSIAA4 (GTCCCAT occurring in antisense orientation).
MtN5 promoter activity in non-inoculated M. truncatula root tissue. A. Representative GUS staining pattern in M. truncatula transgenic adventitious roots harbouring the MtN5::GUS construct. Insets show the representative GUS staining pattern at the lateral root apex (B) and at the lateral root emergence (C), respectively.
Description of NF4257 insertional mutant and determination of DMI1 expression in the mutated background. A. Schematic drawing of the Tnt1 insertion and representation of the oligonucleotides used for the genetic characterization of the insertional line. B. qRT-PCR analysis of DMI1 expression in M. truncatula R108 wild-type and NF4257 roots inoculated with S. meliloti. The expression data were normalized to an internal actin control. The relative expression ratio was calculated using inoculated wild type roots as calibrator sample. The values reported are means ± SE (n=3). Student’s t test was applied. **, P < 0.01.
Root hair deformations and nodule primordia in MtN5-silenced and control roots. A-B. Representative micrographs of root hair curling in M. truncatula roots inoculated with S. meliloti. A. Transgenic adventitious root bearing the MtN5hp gene construct. B. Transgenic adventitious root bearing an empty T-DNA (Control sample). C-D. Representative micrographs of root nodule primordia in M. truncatula roots inoculated with S. meliloti carrying the pXLGD4 plasmid. The presence of rhizobia within the primordia was highlighted through the staining for the β-galactosidase activity. C. Nodule primordium generated on transgenic adventitious root bearing the MtN5hp gene construct. D. Nodule primordium generated on transgenic adventitious root bearing an empty T-DNA (Control sample).
Primers used for RT-PCR. List of the oligonucleotides used as primers in the qRT-PCR experiments.
Contributor Information
Youry Pii, Email: youry.pii@univr.it.
Barbara Molesini, Email: barbara.molesini@univr.it.
Simona Masiero, Email: simona.masiero@unimi.it.
Tiziana Pandolfini, Email: tiziana.pandolfini@univr.it.
Acknowledgements
We thank Sharon R. Long for generously gifting the purified Nod Factors and Pascal Gamas for kindly providing S. meliloti harbouring pXLGD4 plasmid. We are grateful to Thomas Ott for critical reading of the manuscript.
References
- Timmers ACJ, Auriac MC, Truchet G. Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development. 1999;126:3617–3628. doi: 10.1242/dev.126.16.3617. [DOI] [PubMed] [Google Scholar]
- Sieberer BJ, Timmers AC, Emons AM. Nod factors alter the microtubule cytoskeleton in Medicago truncatula root hairs to allow root hair reorientation. Mol Plant Microbe Interact. 2005;18:1195–1204. doi: 10.1094/MPMI-18-1195. [DOI] [PubMed] [Google Scholar]
- Cárdenas L, Thomas-Oates JE, Nava N, López-Lara IM, Hepler PK, Quinto C. The role of nod factor substituents in actin cytoskeleton rearrangements in Phaseolus vulgaris. Mol Plant Microbe Interact. 2003;16:326–334. doi: 10.1094/MPMI.2003.16.4.326. [DOI] [PubMed] [Google Scholar]
- de Ruijter NCA, Bisseling T, Emons AMC. Rhizobium Nod factors induce an increase in subapical fine bundles of actin filaments in Vicia sativa root hairs within minutes. Mol Plant Microbe Interact. 1999;12:829–832. doi: 10.1094/MPMI.1999.12.9.829. [DOI] [Google Scholar]
- Oldroyd GE, Downie AJ. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Bio. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- Gough C, Cullimore J. Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol Plant Microbe Interact. 2011;24:867–878. doi: 10.1094/MPMI-01-11-0019. [DOI] [PubMed] [Google Scholar]
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, Rosenberg C, Roe BA, Long SR, Dénarié J, Cook DR. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science. 2004;303:1364–1367. doi: 10.1126/science.1092986. [DOI] [PubMed] [Google Scholar]
- Esseling JJ, Lhuissier FG, Emons AM. A nonsymbiotic root hair tip growth phenotype in NORK-mutated legumes: implications for nodulation factor-induced signaling and formation of a multifaceted root hair pocket for bacteria. Plant Cell. 2004;16:933–944. doi: 10.1105/tpc.019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely BK, Lougnon G, Ané JM, Cook DR. The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant J. 2007;49:208–216. doi: 10.1111/j.1365-313X.2006.02957.x. [DOI] [PubMed] [Google Scholar]
- Charron D, Pingret JL, Chabaud M, Journet EP, Barker DG. Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 2004;136:3582–3593. doi: 10.1104/pp.104.051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg-Grossman S, Volpin H, Levine A. Root hair curling and Rhizobium infection in Medicago truncatula are mediated by phosphatidylinositide-regulated endocytosis and reactive oxygen species. J Exp Bot. 2007;58:1637–1649. doi: 10.1093/jxb/erm013. [DOI] [PubMed] [Google Scholar]
- Foucher F, Kondorosi E. Cell cycle regulation in the course of nodule organogenesis in Medicago. Plant Mol Biol. 2000;43:773–786. doi: 10.1023/A:1006405029600. [DOI] [PubMed] [Google Scholar]
- Roudier F, Fedorova E, Lebris M, Lecomte P, Györgyey J, Vaubert D, Horvath G, Abad P, Kondorosi A, Kondorosi E. The Medicago species A2-type cyclin is auxin regulated and involved in meristem formation but dispensable for endoreduplication-associated developmental programs. Plant Physiol. 2003;131:1091–1103. doi: 10.1104/pp.102.011122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E, Redondo-Nieto M, Kondorosi A. Ubiquitin-mediated proteolysis. To be in the right place at the right moment during nodule development. Plant Physiol. 2005;137:1197–1204. doi: 10.1104/pp.105.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Schlaman HR, Spaink HP, Sautter C, Rolfe BG, Djordjevic MA. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 1998;14:23–34. doi: 10.1046/j.1365-313X.1998.00090.x. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- Frugier F, Kosuta S, Murray JD, Crespi M, Szczyglowski K. Cytokinin: secret agent of symbiosis. Trends Plant Sci. 2008;13:115–120. doi: 10.1016/j.tplants.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Boot KJM, van Brussel AAN, Tak T, Spaink HP, Kijne JW. Lipochitin oligosaccharides from Rhizobium leguminosarum bv. viciae reduce auxin transport capacity in Vicia sativa subsp. nigra roots. Mol Plant Microbe Interact. 1999;12:839–844. doi: 10.1094/MPMI.1999.12.10.839. [DOI] [Google Scholar]
- van Noorden GE, Ross JJ, Reid JB, Rolfe BG, Mathesius U. Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 2006;140:1494–1506. doi: 10.1104/pp.105.075879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U. Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell. 2006;18:1617–1629. doi: 10.1105/tpc.105.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315:104–107. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315:101–104. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell. 2006;18:2680–2693. doi: 10.1105/tpc.106.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD. Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact. 2011;24:631–639. doi: 10.1094/MPMI-08-10-0181. [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007;144:324–335. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney CH, Long SR. Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc Natl Acad Sci USA. 2010;107:478–483. doi: 10.1073/pnas.0910081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Timmers T, Mbengue M, Moreau S, Hervé C, Tóth K, Bittencourt-Silvestre J, Klaus D, Deslandes L, Godiard L, Murray JD, Udvardi MK, Raffaele S, Mongrand S, Cullimore J, Gamas P, Niebel A, Ott T. A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA. 2010;107:2343–2348. doi: 10.1073/pnas.0913320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 2003;132:161–173. doi: 10.1104/pp.102.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P, Niebel Fde C, Lescure N. Mol Plant Microbe Interact. 1996. pp. 233–242. [DOI] [PubMed]
- El Yahyaoui F, Küster H, Amor BB, Hohnjec N, Pühler A, Becker A, Gouzy J, Vernié T, Gough C, Niebel A, Godiard L, Gamas P. Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 2004;136:3159–3176. doi: 10.1104/pp.104.043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pii Y, Astegno A, Peroni E, Zaccardelli M, Pandolfini T, Crimi M. The Medicago truncatula N5 gene encoding a root-specific lipid transfer protein is required for the symbiotic interaction with Sinorhizobium meliloti. Mol Plant Microbe Interact. 2009;22:1577–1587. doi: 10.1094/MPMI-22-12-1577. [DOI] [PubMed] [Google Scholar]
- Pii Y, Pandolfini T, Crimi M. Signaling LTPs: A new plant LTPs sub-family? Plant Signal Behav. 2010;5:594–597. doi: 10.4161/psb.11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Andriankaja A, Chabaud M, Niebel A, Journet EP, Barker DG, de Carvalho-Niebel F. MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol Plant Microbe Interact. 2005;18:1269–1276. doi: 10.1094/MPMI-18-1269. [DOI] [PubMed] [Google Scholar]
- Vieweg MF, Fruhling M, Quandt HJ, Heim U, Baumlein H, Puhler A, Kuster H, Andreas MP. he promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and non legume plants. Mol Plant Microbe Interact. 2004;17:62–69. doi: 10.1094/MPMI.2004.17.1.62. [DOI] [PubMed] [Google Scholar]
- Fehlberg V, Vieweg MF, Dohmann EM, Hohnjec N, Puhler A, Perlick AM, Kuster H. The promoter of theleghaemoglobingeneVfLb29: functional analysis and identification of modules necessary for its activation in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots. J Exp Bot. 2005;56:799–806. doi: 10.1093/jxb/eri074. [DOI] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GE. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell. 2009;21:545–557. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EJ, Stone JM, Elowsky CG, Arredondo-Peter R, Klucas RV, Sarath G. Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J Exp Bot. 2004;55:1721–1731. doi: 10.1093/jxb/erh211. [DOI] [PubMed] [Google Scholar]
- Ballas N, Wong LM, Theologis A. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum) J Mol Biol. 1993;233:580–596. doi: 10.1006/jmbi.1993.1537. [DOI] [PubMed] [Google Scholar]
- Doherty D, Leigh JA, Glazebrook J, Walker GC. Rhizobium meliloti mutants that overproduce the R. meliloti acidic calcofluor-binding exopolysaccharide. J Bacteriol. 1988;170:4249–4256. doi: 10.1128/jb.170.9.4249-4256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon C, Sousa C, Crespi M, Kondorosi A. Alteration of enod40 expression modifies Medicago truncatula root nodule development induced by Sinorhizobium meliloti. Plant Cell. 1999;11:1953–1966. doi: 10.1105/tpc.11.10.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Goffard N, Nizamidin M, Rolfe BG. Genome-wide transcriptional analysis of super-embryogenic Medicago truncatula explant cultures. BMC Plant Biol. 2008;8:110. doi: 10.1186/1471-2229-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Benedito VA, Wang M, Murray JD, Zhao PX, Tang Y, Udvardi MK. The Medicago truncatula gene expression atlas web server. BMC Bioinformatics. 2009;10:441. doi: 10.1186/1471-2105-10-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Dreyer D, Bonnet D, Howell M, Nony E, VandenBosch K. Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell. 1995;7:43–55. doi: 10.1105/tpc.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V. Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact. 2001;14:737–748. doi: 10.1094/MPMI.2001.14.6.737. [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, Ratet P, Mysore K. Large scale insertional mutagenesis using Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- Wais RJ, Galera C, Oldroyd GE, Catoira R, Penmetsa RV, Cook D, Gough C, Denarié J, Long SR. Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA. 2000;97:13407–13412. doi: 10.1073/pnas.230439797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hartog M, Musgrave A, Munnik T. Nod factor-induced phosphatidic acid and diacylglycerol pyrophosphate formation: a role for phospholipase C and D in root hair deformation. Plant J. 2001;25:55–65. doi: 10.1046/j.1365-313x.2001.00931.x. [DOI] [PubMed] [Google Scholar]
- Kelly MN, Irving HR. Nod factors activate both heterotrimeric and monomeric G-proteins in Vigna unguiculata (L.) Walp. Planta. 2003;216:674–685. doi: 10.1007/s00425-002-0900-8. [DOI] [PubMed] [Google Scholar]
- Pingret JL, Journet EP, Barker DG. Rhizobium Nod factor signalling: evidence for a G protein-mediated transduction mechanism. Plant Cell. 1998;10:659–671. doi: 10.1105/tpc.10.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke D, Fang Q, Chen C, Zhu H, Chen T, Chang X, Yuan S, Kang H, Ma L, Hong Z, Zhang Z. The small GTPase ROP6 interacts with NFR5 and is involved in nodule formation in Lotus japonicus. Plant Physiol. 2012;159:131–143. doi: 10.1104/pp.112.197269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving HR, Boukli NM, Kelly MN, Broughton WJ. In: Root Hairs: Cell and Molecular Biology. Ridge RW, Emons AM, editor. Tokyo: Springer-Verlag; 2000. Nod-factors in symbiotic development of root hairs; pp. 241–265. [Google Scholar]
- Kelly MN, Irving HR. Nod factors stimulate plasma membrane delimited phospholipase C activity in vitro. Physiol Plant. 2001;113:461–468. doi: 10.1034/j.1399-3054.2001.1130404.x. [DOI] [Google Scholar]
- Kelly-Skupek MN, Irving HR. Pharmacological evidence for activation of phospholipid and small GTP binding protein signalling cascades by Nod factors. Plant Physiol Biochem. 2006;44:132–142. doi: 10.1016/j.plaphy.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Laxalt AM, Goedhart J, Gadella TW, Munnik T. Phospholipase D activation correlates with microtubule reorganization in living plant cells. Plant Cell. 2003;15:2666–2679. doi: 10.1105/tpc.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Arisz SA, De Vrije T, Musgrave A. G Protein activation stimulates Phospholipase D signaling in plants. Plant Cell. 1995;7:2197–2210. doi: 10.1105/tpc.7.12.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, Parniske M. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Blanco FA, Beker MP, Battaglia M, Aguilar OM. A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean–Rhizobium etli symbiosis. Plant Cell. 2010;22:4142–4157. doi: 10.1105/tpc.110.079137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto MJ, Domínguez-Ferreras A, Pérez-Mendoza D, Sanjuán J, Olivares J. Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell Microbiol. 2009;11:381–388. doi: 10.1111/j.1462-5822.2009.01282.x. [DOI] [PubMed] [Google Scholar]
- Mortier V, Holsters M, Goormachtig S. Never too many? How legumes control nodule numbers. Plant Cell Environ. 2012;35:245–258. doi: 10.1111/j.1365-3040.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- Brechenmacher L, Kim MY, Benitez M, Li M, Joshi T, Calla B, Lee MP, Libault M, Vodkin LO, Xu D, Lee SH, Clough SJ, Stacey G. Transcription profiling of soybean nodulation by Bradyrhizobium japonicum. Mol Plant Microbe Interact. 2008;21:631–645. doi: 10.1094/MPMI-21-5-0631. [DOI] [PubMed] [Google Scholar]
- Vasse J, de Billy F, Truchet G. Abortion of infection during the Rhizobium meliloti-alfalfa symbiotic interactions is accompanied by a hypersensitive reaction. Plant J. 1993;4:555–566. doi: 10.1046/j.1365-313X.1993.04030555.x. [DOI] [Google Scholar]
- Tellstrom V, Usadel B, Thimm O, Stitt M, Kuster H, Niehaus K. The lipopolysaccharide of Sinorhizobium meliloti suppresses defense-associated gene expression in cell cultures of the host plant Medicago truncatula. Plant Physiol. 2007;143:825–837. doi: 10.1104/pp.106.090985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Ott T. Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr Opin Plant Biol. 2011;14:458–467. doi: 10.1016/j.pbi.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Hirsch AM, Bhuvaneswari TV, Torrey JG, Bisseling T. Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc Natl Acad Sci USA. 1989;86:1244–1248. doi: 10.1073/pnas.86.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraioli S, Tate R, Rogato A, Chiurazzi M, Patriarca EJ. Development of ectopic roots from abortive nodule primordia. Mol Plant Microbe Interact. 2004;17:1043–1050. doi: 10.1094/MPMI.2004.17.10.1043. [DOI] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs) Protein Sci. 2008;17:191–198. doi: 10.1110/ps.073300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Lord EM. Pollen tube growth and guidance: roles of small, secreted proteins. Ann Bot. 2011;108:627–636. doi: 10.1093/aob/mcr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado AM, Dixon PA, Lamb CJ, Cameron RK. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature. 2002;419:399–403. doi: 10.1038/nature00962. [DOI] [PubMed] [Google Scholar]
- Chae K, Kieslich CA, Morikis D, Kim SC, Lord EM. A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell. 2009;21:3902–3914. doi: 10.1105/tpc.109.070854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt HJ, Pühler A, Broer I. Transgenic root nodules ofVicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact. 1993;6:699–706. doi: 10.1094/MPMI-6-699. [DOI] [Google Scholar]
- Pii Y, Crimi M, Cremonese G, Spena A, Pandolfini T. Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol. 2007;7:21. doi: 10.1186/1471-2229-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom EM, Ehrhardt DW, Mitra RM, Long SR. Pharmacological analysis of Nod factor-induced calcium spiking in Medicago truncatula. Evidence for the requirement of type IIA calcium pumps and phosphoinositide signaling. Plant Physiol. 2002;128:1390–1400. doi: 10.1104/pp.010691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molesini B, Pandolfini T, Pii Y, Korte A, Spena A. Arabidopsis thaliana AUCSIA-1 regulates auxin biology and physically interacts with a kinesin-related protein. PLoS One. 2012;7:e41327. doi: 10.1371/journal.pone.0041327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Long SR. Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol. 2003;131:1027–1032. doi: 10.1104/pp.102.010710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veereshlingam H, Haynes JG, Penmetsa RV, Cook DR, Sherrier DJ, Dickstein R. nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol. 2004;136:3692–3702. doi: 10.1104/pp.104.049064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker DG. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact. 2001;14:695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- Molesini B, Pandolfini T, Rotino GL, Dani V, Spena A. Aucsia gene silencing causes parthenocarpic fruit development in tomato. Plant Physiol. 2009;149:534–548. doi: 10.1104/pp.108.131367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide sequence of the putative MtN5 promoter and MtN5 open reading frame. The beginning of the ORF is identified by +1. The region analysed as the putative promoter encompasses 1.54 kb upstream the initiation codon. TATA box and CAAT box are double underlined and underlined with dashed line, respectively. The other motifs highlighted are listed as follow: OSE1ROOTNODULE (AAAGAT occurring in antisense orientation), OSE2ROOTNODULE (AAGAG occurring in both sense and antisense orientations), NRE (AATTT) , ARR1AT (AGATT) and AUXREPSIAA4 (GTCCCAT occurring in antisense orientation).
MtN5 promoter activity in non-inoculated M. truncatula root tissue. A. Representative GUS staining pattern in M. truncatula transgenic adventitious roots harbouring the MtN5::GUS construct. Insets show the representative GUS staining pattern at the lateral root apex (B) and at the lateral root emergence (C), respectively.
Description of NF4257 insertional mutant and determination of DMI1 expression in the mutated background. A. Schematic drawing of the Tnt1 insertion and representation of the oligonucleotides used for the genetic characterization of the insertional line. B. qRT-PCR analysis of DMI1 expression in M. truncatula R108 wild-type and NF4257 roots inoculated with S. meliloti. The expression data were normalized to an internal actin control. The relative expression ratio was calculated using inoculated wild type roots as calibrator sample. The values reported are means ± SE (n=3). Student’s t test was applied. **, P < 0.01.
Root hair deformations and nodule primordia in MtN5-silenced and control roots. A-B. Representative micrographs of root hair curling in M. truncatula roots inoculated with S. meliloti. A. Transgenic adventitious root bearing the MtN5hp gene construct. B. Transgenic adventitious root bearing an empty T-DNA (Control sample). C-D. Representative micrographs of root nodule primordia in M. truncatula roots inoculated with S. meliloti carrying the pXLGD4 plasmid. The presence of rhizobia within the primordia was highlighted through the staining for the β-galactosidase activity. C. Nodule primordium generated on transgenic adventitious root bearing the MtN5hp gene construct. D. Nodule primordium generated on transgenic adventitious root bearing an empty T-DNA (Control sample).
Primers used for RT-PCR. List of the oligonucleotides used as primers in the qRT-PCR experiments.


