Abstract
The synapsin family of neuronal phosphoproteins is composed of three genes (SYN1, SYN2 and SYN3) with alternative splicing resulting in a number of variants with various levels of homology. These genes have been postulated to play significant roles in several neuropsychiatric disorders, including bipolar disorder, schizophrenia and epilepsy. Epigenetic regulatory mechanisms, such as histone modifications in gene regulatory regions, have also been proposed to play a role in a number of psychiatric disorders, including bipolar disorder and major depressive disorder. One of the best characterized histone modifications is histone 3 lysine 4 tri-methylation (H3K4me3), an epigenetic mark shown to be highly enriched at transcriptional start sites and associated with active transcription. In the present study we have quantified the expression of transcript variants of the three synapsin genes and investigated their relationship to H3K4me3 promoter enrichment in post-mortem brain samples. We found that histone modification marks were significantly increased in bipolar disorder and major depression and this effect was correlated with significant increases in gene expression. Our findings suggest that synapsin dysregulation in mood disorders is mediated in part by epigenetic regulatory mechanisms.
Keywords: Bipolar disorder, epigenetics, gene expression, H3K4me3, synapsin
Introduction
The synapsin family of neuronal phosphoproteins is composed of three genes (SYN1, SYN2, SYN3) with alternative splicing giving rise to 10 reported variants expressed at various developmental time-points and in various cell types (Porton et al. 1999; Sudhof et al. 1989). The genes are involved in synaptogenesis, synaptic transmission and synaptic plasticity (Corradi et al. 2008). Of the three synapsin genes, SYN1 and SYN2 are predominantly expressed by mature neurons, where they have been shown to associate with the cytoplasmic surface of synaptic vesicles and to represent >6% of their protein content (Cesca et al. 2010; Ferreira et al. 2000; Grafodatskaya et al. 2010; Huttner et al. 1983; Takamori et al. 2006). SYN1 maps to chromosome Xp11.23 and has two known variants, SYN1a and SYN1b (Cesca et al. 2010; Kile et al. 2010) and SYN2 maps to chromosome 3p25 and has two known variants, SYN2a and SYN2b (Cesca et al. 2010). Both SYN1 and SYN2 are differentially expressed in nerve terminals in the majority of the adult brain with demonstrated homology across numerous vertebrate and invertebrate organisms (Cesca et al. 2010; Grafodatskaya et al. 2010; Kile et al. 2010). SYN3 maps to chromosome 22q12.3 and has been shown to produce up to six variants, although not all are expressed in the adult brain (Grafodatskaya et al. 2010; Porton et al. 1999). Its expression is much lower than that of SYN1 or SYN2 (Kao et al. 1998). The full-length SYN1a and SYN1b protein (isoform 3a) exhibits protein homology with the other two synapsins and consequently possible functional homology as well, while the other variants have been shown to have developmentally specific-expression and the majority to be limited to foetal neuron expression (Grafodatskaya et al. 2010; Porton et al. 1999). The only other SYN3 variant that shows adult expression in the human brain is SYN3g. The function of the SYN3 variants is not as well understood as that of SYN1 or SYN2, but it has been suggested to be mainly localized to regions outside of the synapse in the adult brain and function in neurogenesis and synaptic plasticity (Corradi et al. 2008). The majority of brain regions jointly express synapsin variants at similar levels, suggesting that they are functionally complementary (Ullrich & Sudhof, 1995). However, deleting each of the three synapsin genes produces different phenotypes, indicating that the various gene products must differ in their function to some degree (Feng et al. 2002; Gitler et al. 2004; Li et al. 1995; Rosahl et al. 1995).
Synapsin genes have been proposed to play roles in several psychiatric disorders such as schizophrenia, bipolar disorder (BD) and epilepsy (Cesca et al. 2010; Fassio et al. 2011) in both genetic (Chen et al. 2004; Lee et al. 2005; Saviouk et al. 2007) and functional studies (Browning et al. 1993; Grebb & Greengard, 1990; Lopez de Lara et al. 2010; Mirnics et al. 2000; Nowakowski et al. 2002; Schroeder et al. 2010; Vawter et al. 2002). Given the evidence suggesting differential expression of synapsin genes in association with psychiatric phenotypes, it is interesting to study potential regulatory mechanisms that may underlie these changes. In this study, we set out to investigate epigenetic mechanisms, specifically the role of histone modifications, in explaining differential synapsin expression in BD.
Epigenetic modifications have been investigated in various psychiatric phenotypes, including schizophrenia (Akbarian, 2010a, b), autism (Grafodatskaya et al. 2010), major depression (Schroeder et al. 2010) and suicide (Akbarian, 2010b). Interestingly, valproate, one of the most commonly used mood stabilizers in BD, is an inhibitor of histone deacetylases (Arent et al. 2011; Kielland et al. 2006; Machado-Vieira et al. 2010; Tsankova et al. 2007), and thus it is possible that its stabilizing role in the disorder is mediated through inhibition of histone deacetylases. One of the best understood epigenetic mechanisms is histone methylation, particularly the tri-methylation of the fourth lysine tail on histone 3 (H3K4me3; Berger, 2007). This modification has been shown to be most abundant at transcriptional start sites (TSS) of genes and has been associated with increased transcription (Bernstein et al. 2005; Santos-Rosa et al. 2002; Schneider et al. 2004). H3K4me3 functions by opening up the chromatin and allowing transcriptional machinery to bind to the promoter region of genes, thus leading to the initiation of transcription. Enrichment of this mark typically leads to an increase in expression levels (Bannister & Kouzarides, 2011; Kouzarides, 2007; Santos-Rosa et al. 2002).
In this study, we analysed expression of SYN1a, SYN1b, SYN2a, SYN2b, SYN3a, and SYN3g (Supplementary Fig. 1) in post-mortem brains from BD patients, focusing on Brodmann Area 10 (BA 10) of the prefrontal cortex (PFC). Our choice to focus on the PFC was based on studies showing its importance in mood regulation as well as documented deficits in PFC-mediated working memory and executive function in BD patients (Blumberg et al. 2003; Quraishi & Frangou, 2002; Robinson et al. 2008). In addition, imaging studies have shown abnormalities in PFC biochemistry and function in BD patients during manic and depressive episodes, as well as during euthymia, suggesting the possibility of persistent neuropsychological deficits in BD (Malhi et al. 2007; Thompson et al. 2005). Furthermore, the mediofrontal cortex has been previously linked to mood regulation in BD. A study comparing BD patients with their at-risk but healthy siblings showed regional cerebral blood flow decreases in this region (BA 9/10) in patients but an increase in their siblings, suggesting that this brain region may be involved in BD (Kruger et al. 2006).
Since BD is characterized by alternating episodes of depression and mania, and the BD subjects investigated in this study died by suicide during a depressive episode, we included a comparison group of subjects with major depressive disorder (MDD) in order to control for possible effects that may be associated with depressive symptomatology. We compared both groups with a group of matched psychiatrically healthy controls.
Method and materials
Subjects
Post-mortem PFC brain tissue from BA 10 used in this study was obtained from the Quebec Suicide Brain Bank (www.douglasrecherche.qc.ca/suicide) as described elsewhere (Klempan et al. 2009; Lopez de Lara et al. 2010). Clinical information, toxicology and history of psychoactive prescription drugs were collected for both cases and controls. These data were found to have no influence on our results ; a detailed discussion is presented in Supplementary Methods, Supplementary Tables 1 and 2. All procedures in this study were approved by the ethics review board of our institution. Cases in this study were individuals who had a diagnosis of BD type I or type II (n=13) or MDD (n=18) and died by suicide. Controls were individuals who died suddenly and could not have undergone any resuscitation procedures or other type of medical intervention (n=14). Controls had neither current nor past psychiatric diagnoses. There were no significant group differences in gender, age, post-mortem delay, pH and RNA integrity numbers (Table 1). We chose to focus on BA 10 as a representative PFC region and extracted total RNA from post-mortem brains.
Table 1.
Brain sample group demographics (presented as mean±S.E.M.) for BD, MDD and controls
| Status | Gender Male/female | Age | Brain pH | Post-mortem delay | RNA integrity no. |
|---|---|---|---|---|---|
| BD | 9/4 | 44.00±4.05 | 6.63±0.07 | 30.38±6.31 | 6.63±0.30 |
| MDD | 11/7 | 52.00±3.81 | 6.72±0.06 | 20.28±4.32 | 6.34±0.21 |
| Con | 12/3 | 41.73±6.04 | 6.56±0.05 | 24.03±4.62 | 6.48±0.18 |
| Group differences | n.s. | 0.076 | 0.161 | 0.458 | 0.563 |
BD, Bipolar disorder; MDD, major depressive disorder; Con, controls.
Group differences were computed using one-way analysis of variance.
Gene expression
Total messenger RNA (mRNA) was extracted from frozen brain tissue using the RNeasy Lipid Tissue Mini Kit (Qiagen, The Netherlands). For synthesis of complementary DNA, M-MLV reverse transcriptase (Gibco, USA) and oligo(dT)16 primers (Invitrogen, USA) were used.
Chromatin immunoprecipitation
DNA for chromatin immunoprecipitation (ChIP) was prepared from BA 10 of post-mortem brain tissues (regions adjacent to those selected for mRNA experiments) as described by Matevossian & Akbarian (2008). Briefly, 80 mg tissue was cleaved between adjacent nucleosomes with micrococcal nuclease (Sigma Aldrich, USA). A portion of selected intact nucleosomes was treated with anti-H3K4me3 antibody (Millipore, USA) and purified with protein G agarose beads (Millipore). The remainder was used as input control. Both input and bound fractions were digested with proteinase K before purifying DNA by phenol/chloroform extraction (Fiori & Turecki, 2011; Matevossian & Akbarian, 2008).
Quantitative real-time polymerase chain reaction
Samples were run on the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, USA) in quadruplicate using standard quantitative real-time polymerase chain reaction (qRT-PCR) conditions and the TaqMan Fast Master Mix or the Power SYBR1 Green PCR Master Mix (Applied Biosystems) as applicable. Relative expression for both mRNA and ChIP was calculated using the relative quantitation method (ΔΔ-Ct) with GAPDH as an endogenous control in the RQ Manager 1.2 software. TaqMan assays were used for gene expression (Applied Biosystems). Expression values are presented as relative quantification (RQ) values throughout and they represent 2−ΔΔ-Ct metrics in reference to a pooled calibrator sample. For ChIP quantification, ratios of bound:input fractions were calculated for each sample by using custom SYBR Green primers designed (IDT, USA) in the promoter region ~500 bp upstream of the transcription start site. Primer sequences are available upon request.
Data analysis
Test coefficients and probability distributions were calculated using statistical software GraphPad Prism 5 and SPSS. Before any other statistical computation or graphical representation of results, outlier analyses were performed for each dataset. For this reason, select subjects may be missing from analyses on a case-by-case basis. For qRT–PCR experiments, relative quantitation was performed with GAPDH as an endogenous control in the RQ Manager 1.2 software (Applied Biosystems).
Results
SYN1 and SYN2 have different expression profiles in BD and MDD
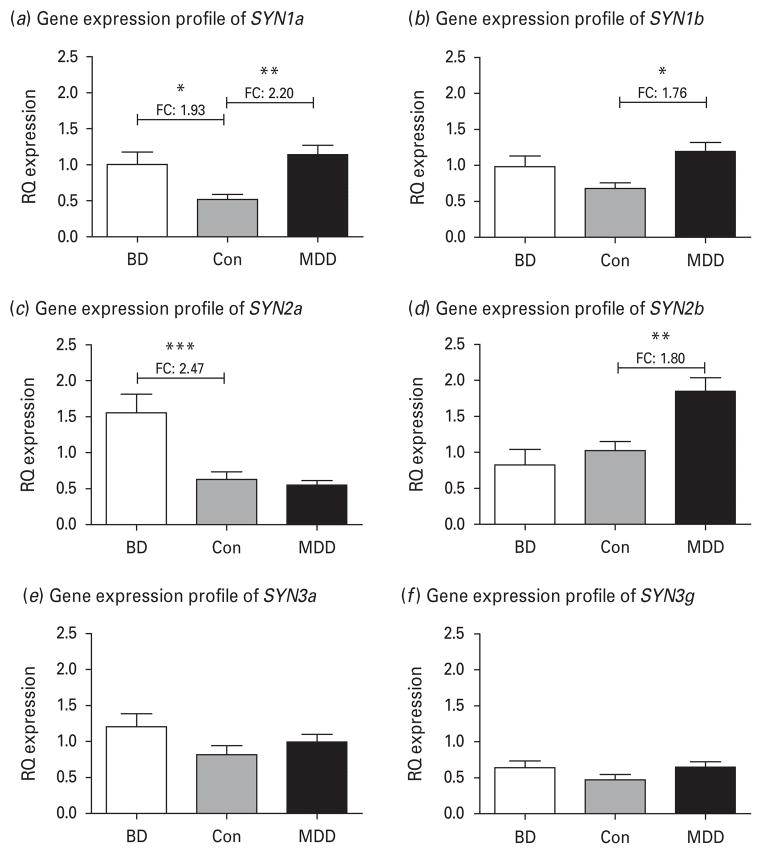
Demographic and post-mortem characteristics of the subjects included in the post-mortem expression study are reported in Table 1. As there were no significant differences between groups in these variables, we performed one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc tests to assess the differences in expression between subjects with BD and controls, as well as MDD and controls for the six synapsin variants (SYN1a, SYN1b, SYN2a, SYN2b, SYN3a and SYN3g) that are expressed in the adult human brain and that are structurally and functionally similar (Cesca et al. 2010). As shown in Fig. 1a, b, the SYN1a variant was differentially up-regulated in both BD and MDD (ANOVA p=0.0045), while the SYN1b variant was only significantly up-regulated in MDD (ANOVA p=0.0172). These results suggest distinct patterns between the two SYN1 variants.
Fig. 1.
Brain expression relative quantification (RQ) values from quantitative real-time polymerase chain reaction relative to GAPDH as an endogenous control. Data are presented as RQ expression values, which represent 2−ΔΔ-Ct metrics. The groups compared are bipolar disorder (BD), major depressive disorder (MDD) and non-psychiatric controls (Con) using analysis of variance (ANOVA) analyses followed by Tukey’s post tests. (a) Relative quantitative expression for variant SYN1a (ANOVA p=0.0045 ; after outlier analysis BD n=13, MDD n=15, Con n=11). (b) Relative quantitative expression for variant SYN1b (ANOVA p=0.0172 ; after outlier analysis BD n=13, MDD n=15, Con n=11). (c) Relative quantitative expression for variant SYN2a (ANOVA p=0.0001 ; after outlier analysis BD n=13, MDD n=12, Con n=13). (d) Relative quantitative expression for variant SYN2b (ANOVA p=0.0005 ; after outlier analysis BD n=13, MDD n=15, Con n=11). (e) Relative quantitative expression for variant SYN3a (ANOVA p=0.2121 ; after outlier analysis BD n=12, MDD n=12, Con n=12). (f) Relative quantitative expression for variant SYN3g (ANOVA p=0.1551 ; after outlier analysis BD n=12, MDD n=14, Con n=12).
The result for SYN2 showed opposing expression patterns for the two variants. As shown in Fig. 1c, d, SYN2a was significantly up-regulated in BD with no effect in MDD (ANOVA p=0.0001), while the converse was true for SYN2b (ANOVA p=0.0005). Considering that gene expression changes in the brain are usually subtle, we note that these significant results were accompanied by fairly high fold changes of 2.47 and 1.80 respectively. Furthermore, these differences between BD and MDD are highly significant when comparing the groups to one another. For SYN2a, the BD group has an average RQ expression value 2.81 times higher than the MDD group, while for SYN2b the MDD groups has an average RQ expression value 2.23 times higher than the BD group (see Table 2 for significance coefficients).
Table 2.
Gene expression results
| BD vs. Con | MDD vs. Con | BD vs. MDD | |
|---|---|---|---|
| SYN1a | |||
| ANOVA p value | 0.0045** | ||
| Tukey’s multiple comparison test (q) | 3.575 | 4.844 | 1.089 |
| Significance | * | ** | n.s. |
| SYN1b | |||
| ANOVA p value | 0.0172* | ||
| Tukey’s multiple comparison test (q) | 2.356 | 4.244 | 1.736 |
| Significance | n.s. | * | n.s. |
| SYN2a | |||
| ANOVA p value | 0.0001*** | ||
| Tukey’s multiple comparison test (q) | 5.661 | 0.487 | 6.037 |
| Significance | *** | n.s. | *** |
| SYN2b | |||
| ANOVA p value | 0.0005*** | ||
| Tukey’s multiple comparison test (q) | 1.049 | 4.624 | 5.626 |
| Significance | n.s. | ** | *** |
| SYN3a | |||
| ANOVA p value | 0.2121 | ||
| Tukey’s multiple comparison test (q) | 2.098 | 2.278 | 0.054 |
| Significance | n.s. | n.s. | n.s. |
| SYN3g | |||
| ANOVA p value | 0.1551 | ||
| Tukey’s multiple comparison test (q) | 2.798 | 1.329 | 1.567 |
| Significance | n.s. | n.s. | n.s. |
For each of the six synapsin variants, gene expression was quantified using quantitative real-time polymerase chain reaction. Data are presented as relative quantification expression values, which represent 2−ΔΔ-Ct metrics one-way analyses of variance (ANOVA) and Tukey’s post-hoc tests were computed for the three diagnostic groups: bipolar disorder (BD), control (Con) and major depressive disorder (MDD).
p Values are presented along with significance levels (* p≤0.05; ** p≤0.001; ** p≤0.0001).
For SYN3 we only detected SYN3a and SYN3g at quantifiable levels in our brain samples. The two variants have perfect homology in regard to their coding exons, although at the mRNA level SYN3g expresses an additional exon at the 5′ end. However, we did not detect differential expression in either the SYN3a variant (Fig. 2e) or the SYN3g variant (Fig. 2f ) (ANOVA p=0.2121 and 0.1551 respectively).
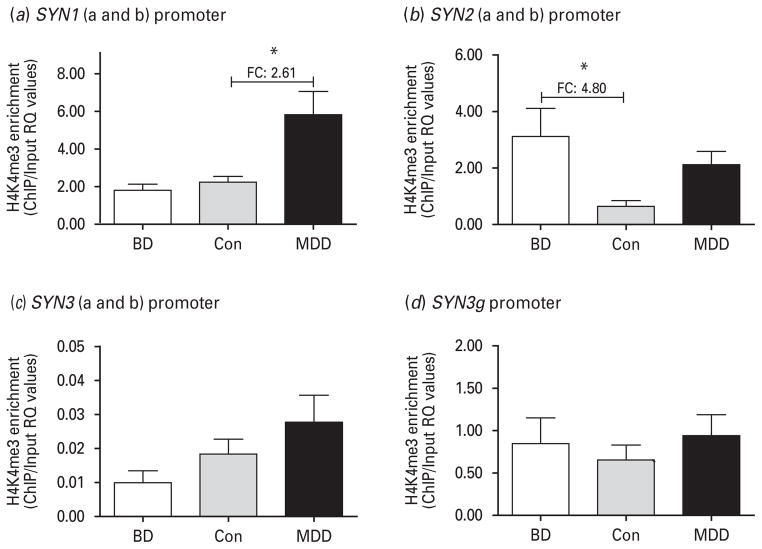
Fig. 2.
Histone 3 lysine 4 tri-methylation (H3K4me3) enrichment results for four different promoter regions representing the specific synapsin variants. Data are presented as relative quantification (RQ) expression values, which represent 2−ΔΔ-Ct metrics. The groups compared are bipolar disorder (BD), major depressive disorder (MDD) and non-psychiatric controls (Con) using analyses of variance (ANOVA) followed by Tukey’s post tests. (a) H3K4me3 enrichment for the shared promoter of variants SYN1a and SYN1b (ANOVA p=0.005 ; after outlier analysis BD n=12, MDD n=15, Con n=12). (b) H3K4me3 enrichment for the shared promoter of variants SYN2a and SYN2b (ANOVA p=0.0187 ; after outlier analysis BD n=9, MDD n=8, Con n=10).
SYN2 expression is modulated by H3K4me3 enrichment at the promoter region distinctly for BD and MDD
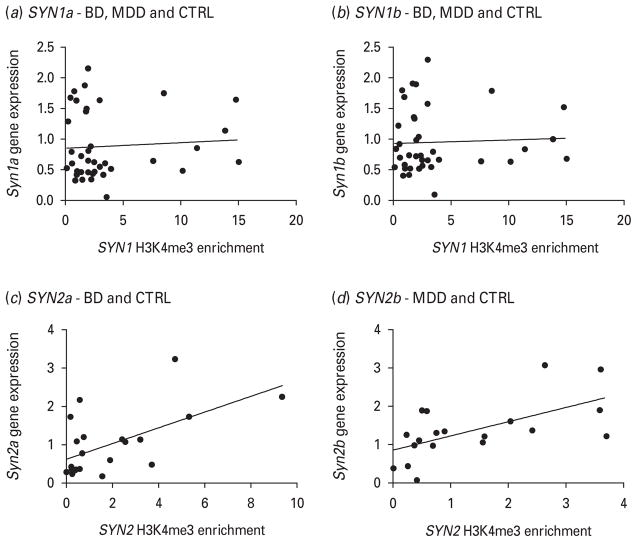
Given that expression of synapsin variants was increased in BD cases, we chose to investigate whether these changes were epigenetically regulated. We investigated levels of tri-methylation of the fourth lysine tail of histone 3 (H3K4me3) using ChIP and designed primers for each independent promoter in the first 500 bp upstream of the TSS, since H3K4me3 has been shown to be enriched in this region. The SYN1a and SYN1b variants share a promoter (Supplementary Fig. 1). As shown in Fig. 2a, this promoter was highly enriched in the MDD group with no change in the BD group (ANOVA p=0.005). There was also a significant difference when comparing the BD and MDD groups to one another, with a fold change of 3.22 (see Table 3 for significance coefficients). However, when following up this analysis with a Pearson’s correlation between expression and H3K4me3 enrichment RQ values (Fig. 3a, b), we found no significant effect. For simplicity, all three diagnostic groups were included in this analysis since the expression patterns were very similar for the SYN1a and SYN1b variants. However, we found that separate analyses by diagnostic status (as for SYN2 below) yield the same non-significant correlation results (data not shown).
Table 3.
Histone 3 lysine 4 tri-methylation (H3K4me3) enrichment results
| BD vs. Con | MDD vs. Con | BD vs. MDD | |
|---|---|---|---|
| SYN1 a+b | |||
| ANOVA p value | 0.005** | ||
| Tukey’s multiple comparison test (q) | 0.4217 | 4.046 | 4.211 |
| Significance | n.s. | * | * |
| SYN2 a+b | |||
| ANOVA p value | 0.0187* | ||
| Tukey’s multiple comparison test (q) | 4.267 | 2.366 | 1.501 |
| Significance | * | n.s. | n.s. |
For each of the four different synapsin promoter regions, chromatin immunoprecipitation (ChIP) or input enrichment was quantified using quantitative real-time polymerase chain reaction.
We report ChIP: input ratios of relative quantification expression values, which represent 2−ΔΔ-Ct metrics.
One-way analyses of variance (ANOVA) and Tukey’s post-hoc tests were computed for the three diagnostic groups: bipolar disorder (BD), control (Con) and major depressive disorder (MDD).
p Values are presented along with significance levels (* p≤0.05 ; ** p≤0.001 ; ** p≤0.0001).
Fig. 3.
Pearson’s correlations of gene expression relative quantification (RQ) values vs. histone 3 lysine 4 tri-methylation (H3K4me3) enrichment RQ values at the promoter region of the various synapsin variants. (a) For the correlation between the SYN1a variant expression and the SYN1 promoter H3K4me3 enrichment, the two-tailed p value is 0.6833 (not significant). (b) For the SYN1b variant the same correlation is also not significant, with a p value of 0.7825. For the SYN2 variants, since gene expression was so discrepant across diagnostic groups, with each variant showing an effect in a different disorder, correlations were computed accordingly. (c) For the SYN2a variant, the correlation for the bipolar disorder and control (Con) groups had a two-tailed p value of 0.0052. (d) For the SYN2b variant the correlation for the major depressive disorder and Con groups had a two-tailed p value of 0.0054.
The SYN2a and the SYN2b variants also share a promoter (Supplementary Fig. 1), which was significantly highly enriched in the H3K4me3 modification (ANOVA p=0.0187), as shown in Fig. 2b. Only the BD group, however, was significantly different from controls in Tukey’s post-hoc test. Given the divergent expression of variants in the two disorders, Pearson’s correlations were computed on the groups that had significantly different gene expression effects – BD–control (Con) for SYN2a and MDD–Con for SYN2b – and these correlations were highly significant (Fig. 3c, d).
Discussion
In this study we investigated expression patterns of synapsin variants and possible epigenetic regulatory mechanisms in the PFC (BA 10) of post-mortem brains from patients with BD, as well as MDD and controls with no psychiatric history. We focused on the PFC because of its involvement in mood regulation, working memory and executive function (Blumberg et al. 2003; Quraishi & Frangou, 2002; Robinson et al. 2008). Overall, we found that SYN1a and SYN2a were upregulated in the BD brain samples. The most striking gene expression finding was for SYN2, where the gene was over-expressed in BD compared to controls and this effect was accounted for by the longer variant, SYN2a. The converse was found in post-mortem brains from patients with MDD, where we saw a significant up-regulation of the SYN2b variant, but no change for SYN2a. This expression difference between the two disorders may not be aetiologically relevant, considering evidence that synapsin variants have overlapping function in the brain (Cesca et al. 2010). However, when looking to identify functional individualities in various synapsin isoforms, Gitler et al. found a unique role for SYN2a during synaptic activity at glutamatergic synapses (Gitler et al. 2008). This is of potential relevance, as alterations in glutamatergic transmission and plasticity have been indicated in BD (Chen et al. 2010; Eastwood & Harrison, 2010; Ongur et al. 2008; Sanacora et al. 2008). Furthermore, in a separate investigation of the effect of lithium treatment on synapsin expression in neuronal cell lines we found that this mood stabilizer classically used in BD treatment affected SYN2a but not SYN2b expression (Cruceanu et al. 2012). Based on this evidence, our findings could reflect a subtle but distinct mechanism of regulation of the SYN2 gene in the brains of patients with different mood disorders.
The second part of our study was to determine whether the observed up-regulation in gene expression was mediated through epigenetic modifications. To our knowledge, no previous studies have tried to identify histone modifications in the synapsin genes in relation to mood disorders, so we quantified H3K4me3 levels in the promoter regions of synapsin variants. Overall, we showed an increase in H3K4me3 levels at synapsin promoters in mood disorders, with patterns that are disease-specific. For the SYN1 variants, there was no significant correlation between mRNA expression and H3K4me3 enrichment. Although both the gene expression and the epigenetic findings for SYN1a and SYN1b are interesting, the two appear to be independent phenomena or part of a much more complex mechanism.
The most interesting epigenetic finding was the enrichment of H3K4me3 at the SYN2 promoter. Unlike the SYN1 data, the H3K4me3 enrichment in the SYN2 promoter correlated with the expression up-regulation shown for the individual variants on disease-specific lines. This finding suggests that gene expression of SYN2a in BD, and SYN2b in MDD, are regulated, at least in part, by changes in H3K4me3 levels at the SYN2 promoter. H3K4me3 is a marker for open chromatin and subsequent enhanced expression, so once the chromatin has been opened, transcription levels are dependent on transcription factors binding. The promoter region where we detected H3K4me3 enrichment is between 176 bp and 395 bp upstream of the transcription start site. Our attempt to design primers in regions closer to the TSS did not yield quantifiable H3K4me3 levels. Interestingly, within this region there are two binding sites for the transcription factor adaptor-related protein complex 2, α-1 subunit (AP-2α). These sites were first identified by Petersohn et al. through DNA-protein binding assays in vitro (Petersohn et al. 1995) and the direct role of AP-2α in regulating SYN2 expression was validated through knock-down experiments in primary mid-brain embryonic mouse neurons by Skoblenick et al. (2010). The latter showed an increase in neuronal SYN2 expression mediated through AP-2α following dopamine D1 receptor stimulation or dopamine D2 receptor inhibition (Skoblenick et al. 2010). As dopamine dysfunction has been well characterized in both BD and MDD (Hashimoto et al. 2007), AP-2α is a likely candidate for mediating the role of SYN2 in these disorders. Furthermore, AP-2α has been shown to be regulated by lithium and carbamazepine (Akbarian, 2010a ; Rao et al. 2007), two common mood stabilizer treatments used for BD, as well as by antidepressants such as citalopram and imipramine (Damberg et al. 2000).
Although the H3K4me3 findings are of interest, considering that the two SYN2 variants share a promoter, the disease-specific expression cannot alone be explained by this epigenetic mechanism. Since the SYN2 variants are only dissimilar at the 3′ end, other regulatory mechanisms could explain the differential expression of these two SYN2 transcripts in BD and MDD. One such mechanism could be microRNA regulation, a class of regulatory molecules that frequently act at 3′ sites and have been shown to be dysregulated in BD post-mortem brains (Kim et al. 2010; Miller & Wahlestedt, 2010; Moreau et al. 2011).
As with all post-mortem brain studies, there are technical limitations to take into account, such as the relatively small sample size and the possible confounders associated with using frozen tissue for expression studies. To account for this, we ensured that the three diagnostic groups had no significant differences in brain pH, post-mortem delay, as well as RNA integrity for expression studies (Table 1). Furthermore, as explained in greater detail in Supplementary Methods, we performed thorough post-mortem investigations on all subjects in an attempt to gather all the relevant medical history information as well as toxicology analyses at time of death. No significant effect of these potential covariates was identified in this study in regard to gene expression or epigenetic modifications (Supplementary Methods, Supplementary Tables 1 and 2).
Another limitation of this study is that we only investigated one epigenetic modification to try to explain our gene expression findings. It has been noted in the literature that epigenetic mechanisms seem to work in concert (Kouzarides, 2007) and, accordingly, it is entirely possible that H3K4me3 enrichment is only one piece of the puzzle, particularly concerning the results for SYN1 variants. The present study serves to demonstrate the involvement of epigenetic mechanisms in synapsin gene regulation in mood disorders, but it would be interesting to follow up our findings with a more in-depth look at various levels of epigenetic regulation, not just in terms of histone modifications but also DNA methylation.
The main findings of this study are two-fold. First, we showed distinct synapsin profiles for BD and MDD post-mortem brain mRNA expression. These findings are interesting because they potentially indicate a molecular marker for distinguishing the two clinically similar disorders. Second, we showed that for SYN2 the changes in expression are correlated with enrichment of H3K4me3, an epigenetic mark associated with transcriptional activation. To our knowledge, this is the first study to identify an epigenetic mechanism to be involved in the regulation of this gene. As with any molecular studies of disease, independent replication in additional post-mortem sample sets is extremely important to validate that the findings are truly relevant for the disorder and do not merely characterize the studied population. Future studies are warranted to understand the extent of epigenetic regulation of the SYN2 gene in BD, as well as the processes by which the SYN2a and SYN2b variants are distinctly expressed in the PFC.
Supplementary Material
Acknowledgments
This work was supported by the Canadian Institute of Health Research (CIHR) grant 64410.
Footnotes
For supplementary material accompanying this paper, visit http://dx.doi.org/10.1017/S1461145712000363.
Statement of Interest
None.
References
- Akbarian S. Epigenetics of schizophrenia. Current Top Behavioral Neuroscience. 2010a;4:611–628. doi: 10.1007/7854_2010_38. [DOI] [PubMed] [Google Scholar]
- Akbarian S. The molecular pathology of schizophrenia – focus on histone and DNA modifications. Brain Research Bulletins. 2010b;83:103–107. doi: 10.1016/j.brainresbull.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arent CO, Valvassori SS, Fries GR, Stertz L, et al. Neuroanatomical profile of antimaniac effects of histone deacetylases inhibitors. Molecular Neurobiology. 2011;43:207–214. doi: 10.1007/s12035-011-8178-0. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Research. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Blumberg HP, Leung HC, Skudlarski P, Lacadie CM, et al. A functional magnetic resonance imaging study of bipolar disorder, state- and trait-related dysfunction in ventral prefrontal cortices. Archives of General Psychiatry. 2003;60:601–609. doi: 10.1001/archpsyc.60.6.601. [DOI] [PubMed] [Google Scholar]
- Browning MD, Dudek EM, Rapier JL, Leonard S, et al. Significant reductions in synapsin but not synaptophysin specific activity in the brains of some schizophrenics. Biological Psychiatry. 1993;34:529–535. doi: 10.1016/0006-3223(93)90195-j. [DOI] [PubMed] [Google Scholar]
- Cesca F, Baldelli P, Valtorta F, Benfenati F. The synapsins, key actors of synapse function and plasticity. Progress in Neurobiology. 2010;91:313–348. doi: 10.1016/j.pneurobio.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Chen Q, He G, Qin W, Chen QY, et al. Family-based association study of synapsin II and schizophrenia. American Journal of Human Genetics. 2004;75:873–877. doi: 10.1086/425588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Henter ID, Manji HK. Presynaptic glutamatergic dysfunction in bipolar disorder. Biological Psychiatry. 2010;67:1007–1009. doi: 10.1016/j.biopsych.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi A, Zanardi A, Giacomini C, Onofri F, et al. Synapsin-I- and synapsin-II-null mice display an increased age-dependent cognitive impairment. Journal of Cell Science. 2008;121:3042–3051. doi: 10.1242/jcs.035063. [DOI] [PubMed] [Google Scholar]
- Cruceanu C, Alda M, Grof P, Rouleau GA, et al. Synapsin II is involved in the molecular pathway of lithium treatment in bipolar disorder. PLoS One. 2012;7:e32680. doi: 10.1371/journal.pone.0032680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damberg M, Ekblom J, Oreland L. Chronic pharmacological treatment with certain antidepressants alters the expression and DNA-binding activity of transcription factor AP-2. Life Sciences. 2000;68:669–678. doi: 10.1016/s0024-3205(00)00969-3. [DOI] [PubMed] [Google Scholar]
- Eastwood SL, Harrison PJ. Markers of glutamate synaptic transmission and plasticity are increased in the anterior cingulate cortex in bipolar disorder. Biological Psychiatry. 2010;67:1010–1016. doi: 10.1016/j.biopsych.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassio A, Raimondi A, Lignani G, Benfenati F, et al. Synapsins: from synapse to network hyperexcitability and epilepsy. Seminars in Cell Developmental Biology. 2011;22:408–415. doi: 10.1016/j.semcdb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Feng J, Chi P, Blanpied TA, Xu Y, et al. Regulation of neurotransmitter release by synapsin III. Journal of Neuroscience. 2002;22:4372–4380. doi: 10.1523/JNEUROSCI.22-11-04372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A, Kao HT, Feng J, Rapoport M, et al. Synapsin III : developmental expression, subcellular localization, and role in axon formation. Journal of Neuroscience. 2000;20:3736–3744. doi: 10.1523/JNEUROSCI.20-10-03736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori LM, Turecki G. Epigenetic regulation of spermidine/spermine N(1)-acetyltransferase (SAT1) in suicide. Journal of Psychiatry Research. 2011;45:1229–1235. doi: 10.1016/j.jpsychires.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Gitler D, Cheng Q, Greengard P, Augustine GJ. Synapsin IIa controls the reserve pool of glutamatergic synaptic vesicles. Journal of Neuroscience. 2008;28:10835–10843. doi: 10.1523/JNEUROSCI.0924-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler D, Takagishi Y, Feng J, Ren Y, et al. Different presynaptic roles of synapsins at excitatory and inhibitory synapses. Journal of Neuroscience. 2004;24:11368–11380. doi: 10.1523/JNEUROSCI.3795-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafodatskaya D, Chung B, Szatmari P, Weksberg R. Autism spectrum disorders and epigenetics. Journal of the American Academy of Child and Adolescent Psychiatry. 2010;49:794–809. doi: 10.1016/j.jaac.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Grebb JA, Greengard P. An analysis of synapsin II, a neuronal phosphoprotein, in postmortem brain tissue from alcoholic and neuropsychiatrically ill adults and medically ill children and young adults. Archives of General Psychiatry. 1990;47:1149–1156. doi: 10.1001/archpsyc.1990.01810240069011. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Sawa A, Iyo M. Increased levels of glutamate in brains from patients with mood disorders. Biological Psychiatry. 2007;62:1310–1316. doi: 10.1016/j.biopsych.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Schiebler W, Greengard P, De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. Journal of Cell Biology. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HT, Porton B, Czernik AJ, Feng J, et al. A third member of the synapsin gene family. Proceedings of the National Academy of Sciences USA. 1998;95:4667–4672. doi: 10.1073/pnas.95.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielland A, Erisir A, Walaas SI, Heggelund P. Synapsin utilization differs among functional classes of synapses on thalamocortical cells. Journal of Neuroscience. 2006;26:5786–5793. doi: 10.1523/JNEUROSCI.4631-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile BM, Guillot TS, Venton BJ, Wetsel WC, et al. Synapsins differentially control dopamine and serotonin release. Journal of Neuroscience. 2010;30:9762–9770. doi: 10.1523/JNEUROSCI.2071-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Reimers M, Maher B, Williamson V, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophrenia Research. 2010;124:183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempan TA, Rujescu D, Merette C, Himmelman C, et al. Profiling brain expression of the spermidine/spermine N1-acetyltransferase 1 (SAT1) gene in suicide. American Journal of Medical Genetics B Neuropsychiatric Genetics. 2009;150B:934–943. doi: 10.1002/ajmg.b.30920. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kruger S, Alda M, Young LT, Goldapple K, et al. Risk and resilience markers in bipolar disorder : brain responses to emotional challenge in bipolar patients and their healthy siblings. American Journal of Psychiatry. 2006;163:257–264. doi: 10.1176/appi.ajp.163.2.257. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Song JY, Kim JW, Jin SY, et al. Association study of polymorphisms in synaptic vesicle-associated genes, SYN2 and CPLX2, with schizophrenia. Behavioral and Brain Functions. 2005;1:15. doi: 10.1186/1744-9081-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chin LS, Shupliakov O, Brodin L, et al. Impairment of synaptic vesicle clustering and of synaptic transmission, and increased seizure propensity, in synapsin I-deficient mice. Proceedings of the National Academy of Sciences USA. 1995;92:9235–9239. doi: 10.1073/pnas.92.20.9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Lara C, Jaitovich-Groisman I, Cruceanu C, Mamdani F, et al. Implication of synapse-related genes in bipolar disorder by linkage and gene expression analyses. International Journal of Neuropsychopharmacology. 2010;13:1397–1410. doi: 10.1017/S1461145710000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Vieira R, Ibrahim L, Zarate CA., Jr Histone deacetylases and mood disorders : epigenetic programming in gene–environment interactions. CNS Neuroscience and Therapeutics. 2010;17:699–704. doi: 10.1111/j.1755-5949.2010.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi GS, Ivanovski B, Hadzi-Pavlovic D, Mitchell PB, et al. Neuropsychological deficits and functional impairment in bipolar depression, hypomania and euthymia. Bipolar Disorders. 2007;9:114–125. doi: 10.1111/j.1399-5618.2007.00324.x. [DOI] [PubMed] [Google Scholar]
- Matevossian A, Akbarian S. A chromatin assay for human brain tissue. Journal of Visual Experiments. 2008;13:717. doi: 10.3791/717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Wahlestedt C. MicroRNA dysregulation in psychiatric disease. Brain Research. 2010;1338:89–99. doi: 10.1016/j.brainres.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, et al. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, David-Rus R, Buyske S, et al. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biological Psychiatry. 2011;69:188–193. doi: 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski C, Kaufmann WA, Adlassnig C, Maier H, et al. Reduction of chromogranin B-like immunoreactivity in distinct subregions of the hippocampus from individuals with schizophrenia. Schizophrenia Research. 2002;58:43–53. doi: 10.1016/s0920-9964(01)00389-9. [DOI] [PubMed] [Google Scholar]
- Ongur D, Jensen JE, Prescot AP, Stork C, et al. Abnormal glutamatergic neurotransmission and neuronal–glial interactions in acute mania. Biological Psychiatry. 2008;64:718–726. doi: 10.1016/j.biopsych.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersohn D, Schoch S, Brinkmann DR, Thiel G. The human synapsin II gene promoter. Possible role for the transcription factor zif268/egr-1, polyoma enhancer activator 3, and AP2. Journal of Biological Chemistry. 1995;270:24361–24369. doi: 10.1074/jbc.270.41.24361. [DOI] [PubMed] [Google Scholar]
- Porton B, Kao HT, Greengard P. Characterization of transcripts from the synapsin III gene locus. Journal of Neurochemistry. 1999;73:2266–2271. doi: 10.1046/j.1471-4159.1999.0732266.x. [DOI] [PubMed] [Google Scholar]
- Quraishi S, Frangou S. Neuropsychology of bipolar disorder : a review. Journal of Affective Disorders. 2002;72:209–226. doi: 10.1016/s0165-0327(02)00091-5. [DOI] [PubMed] [Google Scholar]
- Rao JS, Bazinet RP, Rapoport SI, Lee HJ. Chronic administration of carbamazepine down-regulates AP-2 DNA-binding activity and AP-2alpha protein expression in rat frontal cortex. Biological Psychiatry. 2007;61:154–161. doi: 10.1016/j.biopsych.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Robinson JL, Monkul ES, Tordesillas-Gutierrez D, Franklin C, et al. Fronto-limbic circuitry in euthymic bipolar disorder : evidence for prefrontal hyperactivation. Psychiatry Research. 2008;164:106–113. doi: 10.1016/j.pscychresns.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Rosahl TW, Spillane D, Missler M, Herz J, et al. Essential functions of synapsins I and II in synaptic vesicle regulation. Nature. 1995;375:488–493. doi: 10.1038/375488a0. [DOI] [PubMed] [Google Scholar]
- Sanacora G, Zarate CA, Krystal JH, Manji HK. Targeting the glutamatergic system to develop novel, improved therapeutics for mood disorders. Nature Reviews Drug Discovery. 2008;7:426–437. doi: 10.1038/nrd2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Saviouk V, Moreau MP, Tereshchenko IV, Brzustowicz LM. Association of synapsin 2 with schizophrenia in families of Northern European ancestry. Schizophrenia Research. 2007;96:100–111. doi: 10.1016/j.schres.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nature Cell Biology. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Schroeder M, Krebs MO, Bleich S, Frieling H. Epigenetics and depression: current challenges and new therapeutic options. Current Opinions in Psychiatry. 2010;23:588–592. doi: 10.1097/YCO.0b013e32833d16c1. [DOI] [PubMed] [Google Scholar]
- Skoblenick KJ, Argintaru N, Xu Y, Dyck BA, et al. Role of AP-2alpha transcription factor in the regulation of synapsin II gene expression by dopamine D1 and D2 receptors. Journal of Molecular Neuroscience. 2010;41:267–277. doi: 10.1007/s12031-009-9299-z. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Czernik AJ, Kao HT, Takei K, et al. Synapsins: mosaics of shared and individual domains in a family of synaptic vesicle phosphoproteins. Science. 1989;245:1474–1480. doi: 10.1126/science.2506642. [DOI] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Thompson JM, Gallagher P, Hughes JH, Watson S, et al. Neurocognitive impairment in euthymic patients with bipolar affective disorder. British Journal of Psychiatry. 2005;186:32–40. doi: 10.1192/bjp.186.1.32. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nature Reviews Neuroscience. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Ullrich B, Sudhof TC. Differential distributions of novel synaptotagmins: comparison to synapsins. Neuropharmacology. 1995;34:1371–1377. doi: 10.1016/0028-3908(95)00132-p. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Thatcher L, Usen N, Hyde TM, et al. Reduction of synapsin in the hippocampus of patients with bipolar disorder and schizophrenia. Molecular Psychiatry. 2002;7:571–578. doi: 10.1038/sj.mp.4001158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.