Abstract
The cytokine interleukin 1(IL-1) initiates a wide range of proinflammatory cascades and its inhibition has been shown to decrease inflammation in a variety of diseases. IL-1 receptor accessory protein (IL-1RAcP) is an indispensible part of the IL-1R complex that stabilizes IL-1/IL-1R interaction and plays an important role in the signal transduction of the receptor complex. The soluble form of IL-1RAcP (sIL-1RAcP) contains only the extracellular domain and serves as a natural inhibitor of IL-1 signaling. Therefore, increasing sIL-1RAcP levels might be an attractive therapeutic strategy to inhibit IL-1–driven inflammation. To achieve this we designed specific antisense oligonucleotides (AON), to redirect pre-mRNA IL-1RAcP splicing by skipping of the transmembrane domain encoding exon 9. This would give rise to a novel Δ9IL-1RAcP mRNA encoding a soluble, secreted form of IL-1RAcP, which might have similar activity as natural sIL-1RAcP. AON treatment resulted in exon 9 skipping both in vitro and in vivo. A single dose injection of 10 mg AON/kg body weight induced 90% skipping in mouse liver during at least 5 days. The truncated mRNA encoded for a secreted, soluble Δ9IL-1RAcP protein. IL-1RAcP skipping resulted in a substantial inhibition of IL-1 signaling in vitro. These results indicate that skipping of the transmembrane encoding exon 9 of IL-1RAcP using specific AONs might be a promising therapeutic strategy in a variety of chronic inflammatory diseases.
Keywords: antisense oligonucleotide, exon skipping, inflammation, interleukin 1, soluble IL-1R accessory protein
Introduction
Interleukin 1 (IL-1) plays a central role in the generation of inflammatory responses. After binding of IL-1 to the cell surface receptor IL-1RI, IL-1 receptor accessory protein (IL-1RAcP) facilitates stabilization of the ligand–receptor complex. Furthermore, IL-1RAcP is a crucial co-receptor in this complex that enables recruitment and binding of intracellular proteins such as MyD88 and a series of IL-1R–associated kinases, which finally lead to nuclear factor-κB activation. IL-1 is a very effective cytokine and minute amounts are sufficient to induce a cellular response. Activation of IL-1RI is counterbalanced by a large variety of inhibiting molecules and mechanisms. First, there is the other member of the IL-1 receptor family, IL-1RII which also associates with IL-1RAcP upon binding of IL-1. However, this receptor lacks the intracellular domain present in IL-1RI, and therefore is considered as decoy receptor.1,2 Both transmembrane and soluble forms of IL-1RII have been found.3 In addition, IL-1R antagonist (IL-1Ra) is a natural inhibitor of IL-1 signaling which competes with IL-1 for binding to IL-1RI. IL-1Ra binds poorly to IL-1RII and very high amounts of IL-IRa are needed to occupy all IL-1RI.4 Moreover, the soluble form of IL-1RI binds strongly to IL-1Ra than with IL-1, diminishing the inhibiting effect of IL-1Ra.5 A soluble isoform of IL-1RAcP (sIL-1RAcP) is involved in a third mechanism of inhibition of IL-1 signaling. sIL-1RAcP, which lacks the transmembrane and intracellular domain present in IL-1RAcP6,7 and therefore unable to facilitate signal transduction, is mainly produced by the liver as an acute phase protein and circulates systemically.8 It interacts with the cell surface IL-1RI, IL-1RII, and possibly with soluble IL-1RII in the extracellular space, forming a high affinity IL-1 scavenger.9 In this way, sIL-1RAcP provides at least three mechanisms to entrap secreted IL-1.2,10,11 It has been shown that sIL-1RAcP selectively reduces IL-1 activity on cells such as B lymphocytes and chondrocytes, which express more surface type II decoy receptors.2 Low levels of type II receptors on T lymphocytes prevent suppression of the IL-1 responses of these cells in vivo and in vitro by sIL-1RAcP. Since IL-1Ra preferentially binds to the type I receptor, which is present on all nucleated cells, it inhibits IL-1 responses of both B and T lymphocytes. Therefore, increasing sIL-1RAcP levels might be the preferred approach over administration of IL-Ra for long-term treatment of a disease in which IL-1–mediated activation of B cells plays an important role, without affected T lymphocyte function.
Its specific inhibitory role makes sIL-1RAcP an attractive therapeutic target. It has been shown that increased production of sIL-1RAcP encoded by an adenoviral vector ameliorates collagen-induced arthritis in mice.12 Alternatively, a novel form of sIL-1RAcP might be produced by modulating IL-1RAcP splicing using antisense oligonucleotide (AON)-mediated exon skipping. AONs are chemically modified, short nucleic acid sequences, which can modulate pre-mRNA splicing when they bind to the specific regions of the target transcript.13 Some of the chemical modifications protect AONs from endo and exonucleases and some others increase bioavailability by preventing renal clearance (reviewed in ref. 14). The effectiveness of AON-mediated exon skipping has been successfully confirmed on wide range of target genes in various in vitro and in vivo studies, among which restoring the reading frame in Duchenne muscular dystrophy patients to get partially functional dystrophin, changing levels of alternatively splice products of Bcl-x to change the balance from anti- to pro-apoptotic Bcl-x and producing a novel splice variant in tumor necrosis factor receptor-II to switch its transmembrane form to a novel soluble form by deleting the transmembrane domain-encoding exon (reviewed in ref. 15).
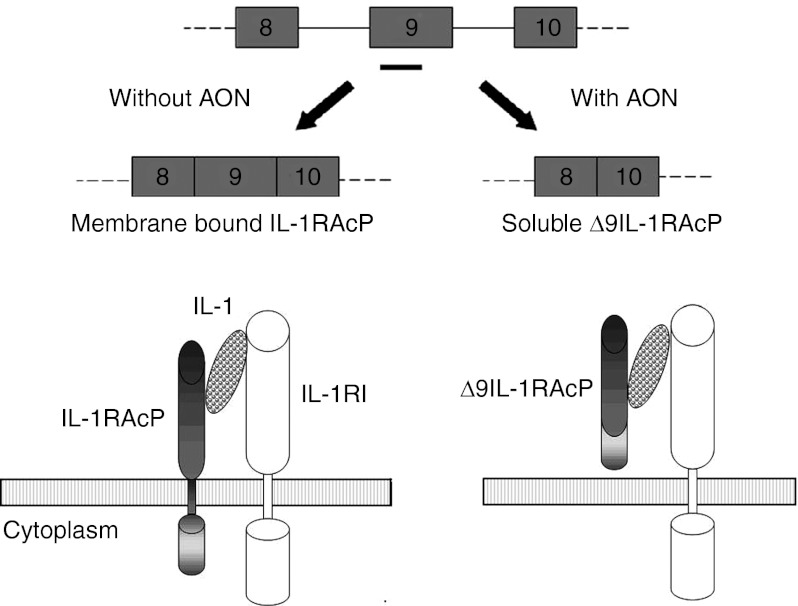
In this study, we employed AONs to mediate skipping of exon 9 to induce the production of a novel form of sIL-1RAcP. We designed AONs consisting of 2′-O-methyl phosphorothioate RNA (2′-O-MePS) or a mix of locked nucleic acid (LNA) and 2′-O-MePS, targeting transmembrane encoding exon 9 IL-1RAcP pre-mRNA. Both in vitro and in vivo treatment of IL-1RAcP-expressing cells with these AONs resulted in a decrease of full-length (FL) IL-1RAcP mRNA up to 90%, which was replaced by a novel shorter transcript lacking exon 9. In the absence of exon 9 the IL-1RAcP mRNA encoded a soluble IL-1RAcP, Δ9IL-1RAcP (Figure 1). Effective skipping of exon 9 resulted in a strong decrease of cellular IL-1 responses of mouse and human cell lines.
Figure 1.
AON-mediated skipping of IL-1RAcP exon 9. Targeting exon 9 in IL-1RAcP pre-mRNA with AON (black bar) induces the expression of an alternatively spiced mRNA encoding a soluble isoform (Δ9IL-1RAcP) that lacks the transmembrane domain. AON, antisense oligonucleotide; IL-1RAcP, interleukin-1 receptor accessory protein.
Results
AONs targeting IL-1RAcP exon 9 induce highly effective exon skipping in vitro
Series of different 2′-O-MePS AONs targeting mouse and human IL-1RAcP exon 9 and the flanking introns were designed (Table 1) based on previously described guidelines.16
Table 1. Overview of the different AONs used to specifically skip exon 9 in IL-1RAcP pre-mRNA.

We evaluated the efficacy of 2′-O-MePS AONs in mouse NIH-3T3 and human HEPG2 cells (Figure 2b,f). The transfection efficiency was tested using 5′-fluorescein (FAM)-labeled control AON. Generally, 70–80% of the cells showed specific nuclear uptake of AON at a concentration of 500 nmol/l (data not shown). Therefore the first series of in vitro tests were performed with an AON concentration of 500 nmol/l. After 24 hours total RNA was isolated and reverse transcription-PCR (RT-PCR) was performed by using primers specific for exons 8 and 10. Almost all AONs specific for mouse IL-1RAcP induced shorter transcript fragments with the size corresponding to the specific skipping of target exon 9 (ENSMUSE00000644552) (Figure 2b). For human IL-1RAcP, only one AON (PS372) was distinctively effective in inducing the skipping of exon 9 (ENSE00002509358) (Figure 2f). Sequence analysis of the shorter PCR products both for mouse and human confirmed the exclusion of exon 9. In addition, a more extensive PCR analysis was performed by using primers specific for exons 5 and 11 showing that the AONs used are specific to exon 9 and do not affect other parts of the IL-1RAcP transcript confirming the prediction from the blast sequence analysis (Supplementary Figure S1).
Figure 2.
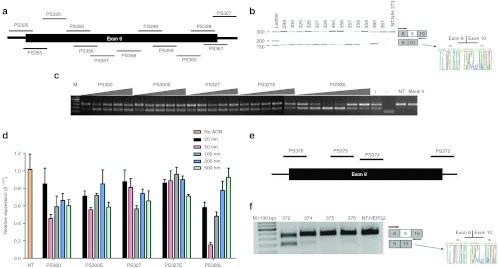
Effect of AONs on mouse and human IL-1RAcP pre-mRNA splicing in vitro. (a) AONs targeted to mouse IL-1RAcP pre-mRNA exon 9 and exon-intron junctions. (b) Test of AONs on NIH-3T3 cells at 500 nmol/l concentration for 24 hours. RT-PCR analysis of samples shows the full-length upper band and the skipped product as a lower band that were amplified with primers specific for exon 8 and 10. Sequence analysis also confirmed exon 9 skipping. (c) RT-PCR results of RNA samples from NIH-3T3 cells transfected with AONs PS300, PS327, 25-mer counterparts of them and PS300L that was designed to increase efficiency of PS300. Triangles show increasing concentrations of 20, 50, 100, 200 and 500 nmol/l for full 2′-O-MePS oligos and additional 10 nmol/l for PS300L (+ = positive control PS300, 100 nmol/l; − = water control, NT = non-transfected cells, Mock tr = only Lipofectamine-2000). (d) Quantification of skipping levels of AON concentration from 20 to 500 nmol/l by qPCR analysis. β-Actin was used as the reference gene and each bar represents the mean value of three different experiments ± SEM. (e) AONs targeted to human IL-1RAcP pre-mRNA exon 9 and exon-intron junctions. (f) Test of AONs on HEPG2 cells at 500 nmol/l for 24 hours. RT-PCR analysis of samples shows the full-length upper band and the skipped product as a lower band that were amplified with exon 8- and 10-specific primers. The sequence analysis also confirmed exon 9 skipping. AON, antisense oligonucleotide; IL-1RAcP, interleukin-1 receptor accessory protein; qPCR, quantitative PCR; RT-PCR, reverse transcription-PCR.
Each AON was tested at least three times to select the ones that give reproducible results. The most potent mouse IL-1RAcP AONs, PS300 and PS327, were selected for further analyses. PS357 although seems very efficient in skipping, does not give reproducible results so was not selected for further analysis. Different concentrations of AONs PS300 and PS327 were transfected into NIH-3T3 cells and quantitative PCR (qPCR) analysis was performed with the primers targeting exon 9. The maximum skipping efficiency we achieved was 50% with 100 nmol/l of PS300 and 200 nmol/l of PS327 (data not shown). To increase the skipping efficiency of these AONs, their sequences were extended to 25-mer which increases melting temperatures (Tm's). (PS300E and PS327E). However, extensive analysis at different concentrations revealed that the extension did not significantly affect the skipping efficiency (Figure 2d). Therefore, in an alternative approach to enhance efficiency of PS300, we designed a chimeric 2′-O-Me/LNA-PS which contains five LNA nucleotides (PS300L). The ability of this AON to induce skipping was tested in different concentrations (20–500 nmol/l) by qPCR using the primers targeting exon 9, which shows the amount of FL non-skipped IL-1RAcP (Figure 2d). The best skipping was observed with PS300L at a concentration of 50 nmol/l and the analysis of skipping efficiency using qPCR revealed that almost 85–90% skipping could be achieved with this AON (Figure 2d). For PS300L, no direct correlation between the AON concentration and skipping efficiency was observed. This is not surprising because at higher concentrations, especially LNA AONs can hybridize very tightly to other LNA residues so less AON would be available to bind to target RNA for skipping. Moreover, AONs can also become toxic to the cell at higher concentrations. To increase the skipping efficiency of human IL-1RAcP exon 9 also a chimeric 2′-O-Me/LNA-PS version of PS372 was generated (PS372L) and tested in different concentrations. However, this resulted in a more modest increase in skipping efficiency of 5–10% (data not shown).
IL-1RAcP mRNA without exon 9 is translated into secreted sIL-1RAcP
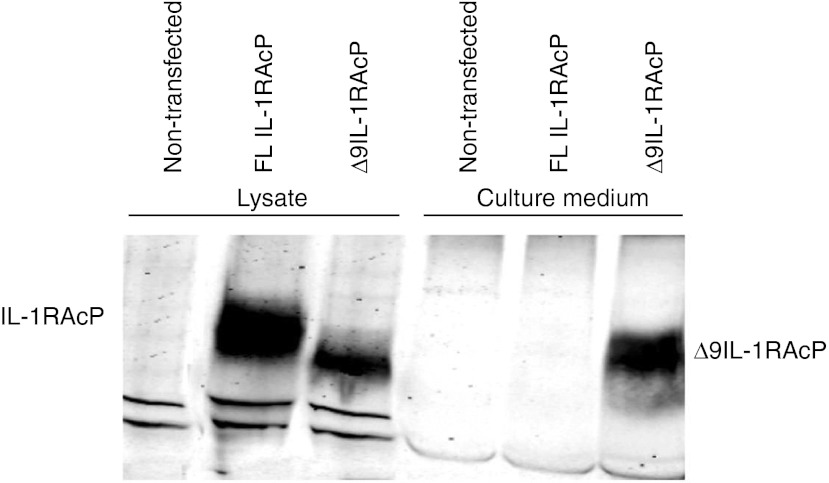
Although IL-1RAcP mRNA could be detected by RT-PCR analysis, it could not be detected on protein level with western blot. Albeit, it cannot be excluded that there is little correlation between IL-1RAcP mRNA and protein levels in the cells used the most likely explanation for this negative result is the much lower sensitivity of the western blot analysis compared with PCR analysis. To prove that removal of exon 9 resulted in a functional mRNA encoding a novel sIL-1RAcP, we transfected HEK293T cells with expression vector pcDNA 3.1B containing FL or truncated (Δ9) IL-1RAcP cDNA to overexpress these proteins. After 48 hours of transfection, cell lysates and culture media were collected for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blot. The complete IL-1RAcP protein could be detected from lysates of HEK293T cells transfected with FL IL-1RAcP cDNA but not in the culture medium of these cells (Figure 3). In contrast, a slightly smaller protein could be detected not only in the lysate of cells transfected with Δ9IL-1RAcP cDNA but also in the culture medium of these cells showing that IL-1RAcP without the exon 9-encoded transmembrane region was secreted.
Figure 3.
Western blot analysis of HEK293 cells and culture medium after transfection with expression vectors encoding either full length (FL) or Δ9 cDNAs of IL-1RAcP. IL-1RAcP, interleukin-1 receptor accessory protein.
Treatment with IL-1RAcP–specific AON decreased the expression of IL-1 responsive genes in vitro
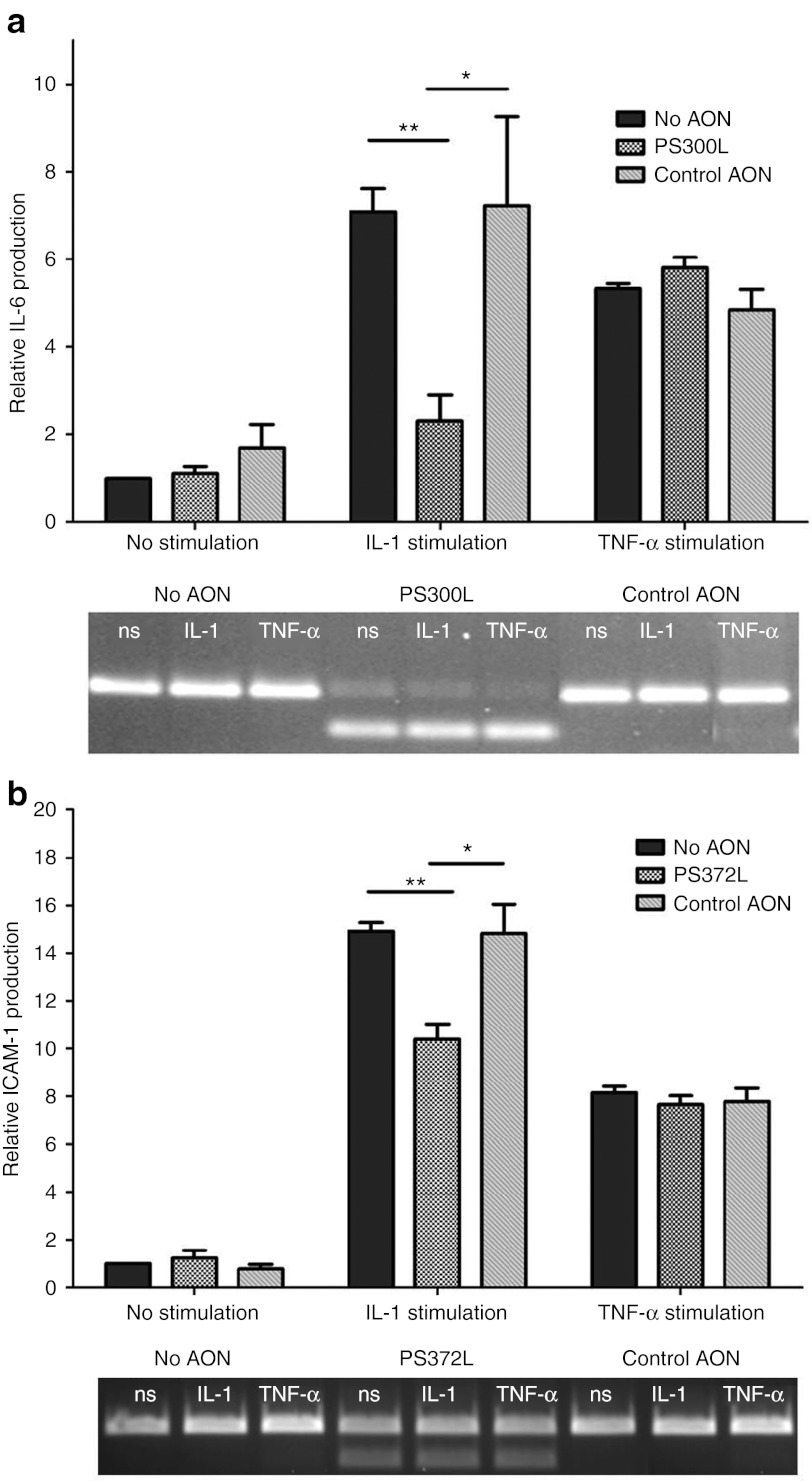
The direct biological effect of AON-mediated exon skipping of IL-1RAcP was determined by measuring mRNA expression levels of the IL-1–responsive genes IL-6 or intercellular adhesion molecule-1 (ICAM-1) in the mouse cell line NIH-3T3 and the human cell line HEPG2, respectively using qPCR. The adhesion molecule ICAM-1 and the cytokine IL-6 are normally expressed at low levels in resting cells but their expression increases upon response to IL-1. Effective exon skipping induced by 300L in NIH-3T3 cells (Figure 4a, lower panel) resulted in reduced IL-1β–dependent upregulation of IL-6 compared with non-transfected NIH-3T3 cells, whereas no difference in IL-6 mRNA expression was seen between transfected and non-transfected cells after tumor necrosis factor-α stimulation (Figure 4a). Comparable results were found with AON PS372L-mediated skipping of exon 9 of human IL-1RAcP in HEPG2 cells (Figure 4b). Upon treatment with IL-1 cells transfected with the AON PS372L showed a decreased mRNA expression of the IL-1–responsive ICAM-1 expression compared with untransfected cells.
Figure 4.
Inhibition of IL-1 signaling with AON targeting IL-1RAcP. (a) IL-1– and TNF-α–induced IL-6 production levels were measured by qPCR on the NIH-3T3 cells transfected with PS300L and control AON for 48 hours and stimulated for 5 hours. **P < 0.002, *P < 0.05. (b) IL-1– and TNF-α–induced ICAM expression levels were measured by qPCR on the HEPG2 cells transfected with PS372L and control AON for 48 hours and stimulated for 5 hours. **P = 0.002, *P < 0.05, β-actin was used as the reference gene and each bar represents the mean value of three different experiments ± SEM. AON, antisense oligonucleotide; ICAM, intercellular adhesion molecule; IL-1RAcP, interleukin-1 receptor accessory protein; ns, no stimulation; qPCR, quantitative PCR; TNF, tumor necrosis factor.
PS300L effectively induced IL-1RAcP exon 9 skipping in mouse liver
To determine whether the effective skipping observed in vitro could be confirmed in vivo, exon skipping was analyzed in liver from mice injected intravenously with the most effective AONs. Administration (intravenously in tail vein, 100 mg/kg of mouse, for 4 consecutive days) of PS300 resulted in skipping efficiency <50% in liver, as determined 1 day after the last AON injection, with no detectable skipping in spleen and kidney (data not shown). Moreover, no skipped product was detected in either saline or control AON-treated mice.
Since the efficacy of in vivo skipping needs to be increased to expect a biological effect, the AONs were complexed with Invivofectamine, lipid-based nanoparticles that allows highly efficient delivery of synthetic oligonucleotides used in RNA interference to the mouse liver i.e., hepatocytes, the cell type within the liver IL-1RAcP is mainly expressed. Mice were injected intravenously in the tail vein on day 0, once, with the maximum dose of 10 mg/kg of AON PS300. On day 1 mice were killed and livers were harvested for RNA isolation. By RT-PCR, presence of skipped product could be detected in two out of three mice administered with Invivofectamine-PS300 (Supplementary Figure S2), confirmed by sequence analysis (Supplementary Figure S2). The skipped band was not detected in the liver samples of mice injected with noncomplexed PS300 (n = 3) which confirms that without Invivofectamine a single dose of 10 mg/kg is too low to induce detectable skipping. No skipped product was detected in the liver samples of mice injected with control non-target–specific oligonucleotide Invivofectamine (n = 2) which eliminates the effect of Invivofectamine in formation of the shorter product.
Based on these findings, PS300 and PS300L, both complexed to Invivofectamine, were injected 10 mg/kg intravenously (n = 4/group) on day 0 in tail vein. On day 2, partial hepatectomy was performed to isolate RNA from the liver. RT-PCR analysis showed that both PS300 and PS300L were able to induce the production of skipped product (Figure 5) with PS300L skipping efficiency was 90%. The skipping persisted for at least 5 days as demonstrated by the analysis of liver RNA from mice killed 5 days after the administration of the AONs. In both timepoints, the skipping efficiency of PS300L was larger than that of PS300. Slight variations of skipping efficiencies were observed between individual mice in the same group.
Figure 5.
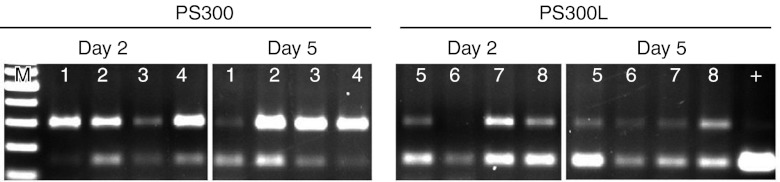
RT-PCR analysis of liver samples of the mice administered intravenously with Invivofectamine-complexed PS300 or PS300L. Dosage of 10 mg/kg was injected on day 0, partial hepatectomy was performed on day 2, and the mice were killed on day 5 and the livers were harvested for RNA isolation. 1–4 = mice injected with PS300; 5–8 = mice injected with PS300L; M = 100 bp marker, + = PS300L in vitro, positive control. RT-PCR, reverse transcription-PCR.
Discussion
Strategies to manipulate splicing can be used to correct aberrant splicing caused by mutations, to inhibit protein expression by generating out-of-frame transcripts and to alter the function of a protein by skipping or including exons.17 Here, we show a convincing example of the third concept by skipping the transmembrane domain encoding exon 9 of IL-1RAcP, resulting in not only the decrease of the expression of membrane bound IL-1RAcP mRNA but also increase in Δ9IL-1RAcP mRNA which is supposed to express a novel soluble inhibiting protein. The inhibiting effect was demonstrated by the decrease in expression of the IL-1–responsive cytokine IL-6 and chemokine ICAM-1.
sIL-1RAcP downregulates IL-1 signaling in two different mechanisms; by competing with membrane bound IL-1RAcP for association with IL-1RI and by forming high affinity IL-1 scavenger with sIL-1RII in the extracellular space.7,9 In our in vitro experiments, the inhibitory effect on IL-1 signaling was only determined for the first mechanism, decrease of membrane bound IL-1RAcP, because the second mechanism, the formation of the soluble scavenger, requires soluble IL-1RII that we think is not present in the culture media. Therefore, on the basis of the in vitro results, we most likely underestimate the inhibiting effect of Δ9IL-1RAcP in vivo, where sIL-1RII is present to form the scavenger for an additional systemic inhibitory effect.
Most of the AONs targeting mouse IL-1RAcP exon 9 induced quite efficient skipping. Each AON was tested at least three times in different experiments to select the ones that gave best skipping efficiency reproducibly. Although some AONs cover overlapping regions (e.g., PS327 and PS361, PS356 and PS300, PS325 and PS355) on the target pre-mRNA, there might be slight differences in terms of targeting different splice enhancer sites, which makes them more or less effective. The length of the AON also affects the binding properties; increasing the length causes increase in Tm but it may also change secondary structure that would increase or decrease AON's skipping efficiency. Moreover, presence of CCC and GGG repeats and G/C contents of targeted region change the effect.
The human and mouse IL-1RAcP exon 9 sequences are around 20% different. Therefore, they have different secondary structures which makes some target sites less or more accessible for the AONs. For human IL-1RAcP exon 9, we have shown proof-of-concept of the exon skipping approach. The efficacy of the only tested chimeric AON PS372L specific for human IL-1RAcP was much lower compared with the efficacy of the mouse-specific AON PS300L. However, it is still possible to design more chimeric AONs with varying the position and the number of LNAs to achieve better skipping and biological effect in the human system.
We have designed more AONs for mouse IL-1RAcP exon 9, covering most of the possible target regions. Therefore, it was more likely to find AONs with better skipping efficacy for mouse compared with human. The first selection was followed by further optimization to maximize the outcome both in vitro and in vivo. We added LNA bases to the most promising mouse 2′-O-MePS AON (PS300), making a chimeric (PS300L), to improve its efficiency, as LNA bases display a remarkably increased thermodynamic stability and enhanced nucleic acid recognition.18 This resulted in an increase in efficiency up to 90%. PS300L contains five LNA bases, mainly placed on G and Cs in order to provide higher increase in Tm (>15 °C increase in Tm compared with PS300). Although it was already partially prevented by 2′-O-Me modification, an unmethylated C of an internal CpG was LNA modified to prevent its recognition as non-self causing immunostimulation.19
The main challenge with the in vivo experiments is the delivery of the AON to the target organ or cell type. Previous studies have shown that after systemic administration, AONs mainly accumulate in the liver and are taken up by both hepatocytes and Kupffer cells. In order to direct the cellular uptake more in favor of hepatocytes, we used a lipid-based delivery method20 and complexed the AON to Invivofectamine. The increased uptake of AON by hepatocyes resulted in dramatic increase in IL-1RAcP exon 9 skipping up to 90% even with low dose. However, not only liver cells express IL-1RAcP. IL-1RAcP mRNAs were observed to vary in a tissue or cell type-specific manner, making certain organs more responsive and susceptible to IL-1–induced inflammation.21 Therapeutic control of IL-1 responsiveness might be improved by targeting effector cells, especially macrophages, directly, providing both a reduction of membrane bound IL-1RAcP on these cells and an increase of Δ9IL-1RAcP that forms high affinity IL-1 scavenger. Nevertheless, targeting of AONs to immune cells is not very effective. One approach to achieve transit across the cell membrane is conjugation of AONs to arginine-rich or cell-specific peptides. A variety of different arginine-rich peptides have been used for the delivery of AONs to T-cells and dendritic cells.17 New delivery methodologies will boost the development of new therapeutic interventions in the immune system based on AON-mediated exon skipping.
In conclusion, we have shown that AONs specific for exon 9 can modify very effectively the pre-mRNA splicing of IL-1RAcP in vitro and in vivo, in mouse liver. In vitro we demonstrated that this resulted in decrease in IL-1β–mediated signaling. Thus, AONs have the capacity to induce a novel splice variant with therapeutic potential and this might be an attractive therapeutic approach to cope with IL-1–induced inflammatory conditions. The development of specific carrier molecules that direct the efficient uptake of the AONs to certain cell types, e.g., immune cells would considerably enhance the applicability and will improve the efficacy of exon skipping-based therapies in the near future.
Materials and Methods
Design of AON. Based on the criteria published in Aartsma-Rus et.al.16 a series of AONs were designed targeting exon-internal sequences or exon-intron junctions (Table 1). AONs with phosphorothioate backbones and 2′-O-methyl ribose modifications (2′-O-MePS) were synthesized by Prosensa (Leiden, Netherlands) and AONs with 2′-O-MePS with additional LNA modifications were synthesized by Eurogentec (Seraing, Belgium). Transfection efficiency was assessed using a control AON with 5′-fluorescein group (6-FAM). All AONs were high-pressure liquid chromatography purified.
Cell culture and AON transfection. NIH-3T3 (mouse embryonic fibroblast cell line), HEPG2 (human hepatocellular liver carcinoma cell line), and HEK293T (human embryonic kidney) cells were cultured in Dulbecco's modified Eagle's medium + 10% fetal bovine serum + penicillin/streptomycin at 37 °C with 5% CO2 and passaged when confluent using 0.25% trypsin diluted with phosphate-buffered saline. Cells were plated in 6-well plates (1 × 106 cells per well) and transfected with 10–500 nmol/l AON in 2 ml Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instruction. The nuclear localization and transfection efficiency of the 6-FAM–labeled AON was assessed using a fluorescent microscope starting from 6–8 hours after transfection. Medium was changed 6–8 hours after transfection and the cells were cultured further to confluency for 24 or 48 hours for RNA or protein isolation, respectively.
RNA isolation, RT-PCR, and qPCR. Total RNA was isolated by using Trizol (Invitrogen) reagent according to the manufacturer's protocol from minimally 1 × 106 cells or 10 mg of tissue. For isolation of RNA from mouse liver samples, 20–30 mg of tissue were homogenized in Magna Lyser Green Beads (Roche Applied Science, Nijmegen, Netherlands) containing 500 µl phosphate-buffered saline with 5 µl 2-mercaptoehtanol for 20 seconds at 7,000 rpm, followed by another 10 seconds at 7,000 rpm in the Magna Lyser (Roche Applied Science). Homogenized tissue of 250 µl was added to 750 µl Trizol and RNA was isolated according to the manufacturer's protocol. About 1 µg of total RNA was used for first strand cDNA synthesis with random hexamer primers (Roche Applied Science). For the analysis of IL-1RAcP exon skipping, 1 µl of 20 µl cDNA was then amplified using the following primer sets, mouse IL-1RAcP forward (exon 8) primer GAGGATCTCAGGCGCAACTA, reverse (exon 10) primer TCAGCAGCACAAATTCCTCTT and forward (exon 5) primer CGTTTCATCTCACCAGGACTC, reverse (exon 11) primer GTTGGGGCTTAGAACAACCA; human IL-1RAcP forward (exon 8) primer CAAGCGCAGCTATGTCTGTC, reverse (exon 10) primer TCTCGGTCAAAGATGCACAG and forward (exon 5) primer CGTTTCATCTCACCAGGACTC, reverse (exon 11) primer GTTGGGGCTTAGAACAACCA and the following PCR conditions: 25 cycles at 94 °C, 30 seconds; 60 °C, 30 seconds; 72 °C, 1 minute (30 cycles for in vivo samples). PCR products were analyzed on 1.5% agarose gel stained with ethidium bromide To confirm proper exon skipping, PCR products were purified from gel with NucleoSpin Extract II Kit (Macherey-Nagel, Düren, Germany) following the manufacturer's instructions and analyzed by Sanger sequencing. PCR products were also run on a DNA1000 LabChip on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) which allows semi-quantitative assessment of exon skipping levels. qPCR reactions containing FastStart Universal SYBR Green Master mix (Roche Applied Science) and 10 pmol of each qPCR primer and 5 µl of 10 times diluted cDNA in a 20 µl total volume for each sample were performed on the Roche LightCycler 48. Data were normalized to values of β-actin gene. The primer sets used in qPCR reactions were: mouse IL-1RAcP forward primer: AGAACTCGCCTGTGGTTTTG and reverse primer: TTCCAAAGTGAGCTCGGTAAA, mouse β-actin forward primer: TGCGTGACATCAAAGAGAAG and reverse primer: GATGCCACAGGATTCCATA. For qPCR analysis of IL-1–induced IL-6 and ICAM-1 mRNA expression, the following primers were used: Mouse IL-6 forward primer: AGTTGCCTTCTTGGGACTGA, reverse primer: GTCTCCTCT CCGGACTTGTG; human ICAM-1 forward primer: CATAGAGA CCCCGTTGCCTAAA, reverse primer: TGGCTATCTTCTT GCACATTGC; human β-actin forward primer: AATGTCGC GGAGGACTTTGATTGC, reverse primer: AGGATGGCAA GGGACTTCCTGTAA.
In vitro human IL-1RAcP expression system. Human FL IL-RAcP and IL-1RAcP without exon 9 (Δ9IL-1RAcP) cDNA were synthesized according to the method described by Ko et al.22 cDNA was made with total random hexamers using RNA isolated from the human liver cell line HEPG2. From this cDNA FL IL-1RAcP cDNA was cloned into the pcDNA3.1B (−) expression vector using primers with restriction endonuclease sites. For the exon 9 deletion construct, exons 1 to 8 and exons 10 and 11 fragments were generated by PCR using primers with restriction enzyme sites, which allowed cloning into pcDNA3.1B (−). The restriction enzyme used to fuse exon 8 to exon 10 is type IIs restriction enzyme. Type IIs restriction enzymes cut outside their recognition site, the actual cut is sequence independent. The resulting sticky ends allow a seamless joining of the coding sequences of exon 8 and exon 10.
Analysis of IL-1RAcP protein. HEK293T cells (1 × 106 per well of a 6-well plate) were transfected with 8 µg plasmids using Lipofectamine 2000. Whole cell extracts were prepared after 48 hours after transfection by lysing the cells with RIPA buffer (Therma Scientific, Waltham, MA). Cells were incubated on ice for 15 minutes with gentle shaking and then disrupted by multiple passages through a syringe with a 19-G needle. Cell debris was removed by centrifugation at 4 °C at 12,000g for 10 minutes. The protein concentration was determined by the Quant-it Protein assay kit (Invitrogen). In addition, cell culture supernatant was collected for protein analysis.
For western blot 15 µl of each sample was mixed with 5 µl loading buffer, boiled for 5 minutes and loaded and run first at 70 V for 30 minutes then at 100 V on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by transfer onto a nitrocellulose membrane. The membrane was blocked in Odyssey Blocking Buffer (Li-cor, Lincoln, NE) for 1 hour at room temperature and subsequently incubated overnight at 4 °C with a 1:1,000 dilution of goat anti-human IL-1RAcP antibody (R&D Systems, Minneapolis, MN). After five washes in 0.1% Tween 20 in phosphate-buffered saline, membranes were incubated for 1 hour at room temperature with the fluorescently labeled donkey anti-goat secondary antibody (Li-cor) diluted 1: 5,000 in Li-cor Odyssey blocking buffer and scanned in Odyssey Imaging System (Li-cor).
IL-1–induced cytokine production assay. 1 × 106 NIH-3T3 and HEPG2 cells were seeded in each well of a 6-well plate in 2 ml Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum on day 0. On day 1, cells were transfected with PS300L (50 nmol/l) and PS372L (100 nmol/l), respectively. On day 2, the medium was refreshed and transfection was repeated only for HEPG2 with PS372L. On day 3 NIH-3T3 and on day 4 HEPG2 cells were stimulated with 10 ng/ml mouse IL-1β (Miltenyi Biotec, Bergisch Gladbach, Germany) and 50 ng/ml TNF-α (Miltenyi Biotec) for 5 hours. After IL-1 stimulation expression of IL-6 and ICAM-1, from mouse and human cell line, respectively was determined by qPCR.
IL-1RAcP skipping in mouse liver. AON PS300, PS 300L, and control small-interfering RNA were complexed with Invivofectamine 2.0 Reagent (Invitrogen, Bleiswijk, Netherlands) according to the manufacturer's protocol with the amount of AON being 2 mg/ml of Invivofectamine 2.0 Reagent. Subsequently, 6 weeks old female C57BL/6J mice were intravenously injected via the tail vein with 200 µl complexed AONs at a dose of ~10 mg AON per kg body weight. At 2 days after AON injection, all mice were subjected to liver biopsy as described previously23 and they were killed on day 5.
SUPPLEMENTARY MATERIAL Figure S1. RT-PCR analysis by using primers targeting exon 5 and 11. Figure S2. RT-PCR analysis of liver samples of mice treated with Invivofectamine-complexed PS300.
Acknowledgments
This work was supported by the Netherlands Ministry of Economic Affairs, IOP grant IGE07001 (Innovatiegerichte onderzoeksprogramma). G.-J. van O. is an unpaid member of the Scientific Advisory Board of Prosensa. A patent has been filed (application no. PCT/NL2010/050882) “Molecule for treating an inflammatory disorder”. The listed inventors are G.-J. van O., A.A.-R., J. van D., S. de K., A.S.Y.-E., and J.S.V. We thank Ka Lei Cheung (Department of Thrombosis and Hemostasis, Leiden University Medical Center, Leiden, the Netherlands) for technical assistance with liver biopsies and all the staff of the animal facilities of the Leiden University Medical Center (PDC) for excellent animal care. The research of H.S. is financially supported by the Dutch Organization for Scientific Research (NWO-TOP grant no. 40-00812-98-07-045). G.-J. van O. is an unpaid member of the Scientific Advisory Board of Prosensa Therapeutics BV, J. van D. and S. de K. are employees of Prosensa Therapeutics BV. The other authors declared no conflict of interest.
Supplementary Material
RT-PCR analysis by using primers targeting exon 5 and 11.
RT-PCR analysis of liver samples of mice treated with Invivofectamine-complexed PS300.
References
- Huang J, Gao X, Li S., and, Cao Z. Recruitment of IRAK to the interleukin 1 receptor complex requires interleukin 1 receptor accessory protein. Proc Natl Acad Sci USA. 1997;94:12829–12832. doi: 10.1073/pnas.94.24.12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Knop J, Wesche H, Raffetseder U, Kurrle R, Boraschi D.et al. (1998The type II IL-1 receptor interacts with the IL-1 receptor accessory protein: a novel mechanism of regulation of IL-1 responsiveness J Immunol 1616871–6877. [PubMed] [Google Scholar]
- Symons JA, Young PR., and, Duff GW. Soluble type II interleukin 1 (IL-1) receptor binds and blocks processing of IL-1 beta precursor and loses affinity for IL-1 receptor antagonist. Proc Natl Acad Sci USA. 1995;92:1714–1718. doi: 10.1073/pnas.92.5.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets RL, Joosten LA, Arntz OJ, Bennink MB, Takahashi N, Carlsen H.et al. (2005Soluble interleukin-1 receptor accessory protein ameliorates collagen-induced arthritis by a different mode of action from that of interleukin-1 receptor antagonist Arthritis Rheum 522202–2211. [DOI] [PubMed] [Google Scholar]
- Burger D, Chicheportiche R, Giri JG., and, Dayer JM. The inhibitory activity of human interleukin-1 receptor antagonist is enhanced by type II interleukin-1 soluble receptor and hindered by type I interleukin-1 soluble receptor. J Clin Invest. 1995;96:38–41. doi: 10.1172/JCI118045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LE, Muzio M, Mantovani A., and, Whitehead AS. IL-1 signaling cascade in liver cells and the involvement of a soluble form of the IL-1 receptor accessory protein. J Immunol. 2000;164:5277–5286. doi: 10.4049/jimmunol.164.10.5277. [DOI] [PubMed] [Google Scholar]
- Jensen LE., and, Whitehead AS. Expression of alternatively spliced interleukin-1 receptor accessory protein mRNAs is differentially regulated during inflammation and apoptosis. Cell Signal. 2003;15:793–802. doi: 10.1016/s0898-6568(03)00039-1. [DOI] [PubMed] [Google Scholar]
- Gabay C, Smith MF, Eidlen D., and, Arend WP. Interleukin 1 receptor antagonist (IL-1Ra) is an acute-phase protein. J Clin Invest. 1997;99:2930–2940. doi: 10.1172/JCI119488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Hanna R, Della Friend, Moore H, Chen H, Farese AM.et al. (2003The soluble form of IL-1 receptor accessory protein enhances the ability of soluble type II IL-1 receptor to inhibit IL-1 action Immunity 1887–96. [DOI] [PubMed] [Google Scholar]
- Dinarello CA. The many worlds of reducing interleukin-1. Arthritis Rheum. 2005;52:1960–1967. doi: 10.1002/art.21107. [DOI] [PubMed] [Google Scholar]
- Malinowsky D, Lundkvist J, Layé S., and, Bartfai T. Interleukin-1 receptor accessory protein interacts with the type II interleukin-1 receptor. FEBS Lett. 1998;429:299–302. doi: 10.1016/s0014-5793(98)00467-0. [DOI] [PubMed] [Google Scholar]
- Smeets RL, van de Loo FA, Joosten LA, Arntz OJ, Bennink MB, Loesberg WA.et al. (2003Effectiveness of the soluble form of the interleukin-1 receptor accessory protein as an inhibitor of interleukin-1 in collagen-induced arthritis Arthritis Rheum 482949–2958. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A., and, van Ommen GJ. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007;13:1609–1624. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurreck J. Antisense technologies Improvement through novel chemical modifications. Eur J Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- Kole R, Krainer AR., and, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, van Vliet L, Hirschi M, Janson AA, Heemskerk H, de Winter CL.et al. (2009Guidelines for antisense oligonucleotide design and insight into splice-modulating mechanisms Mol Ther 17548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourich DV., and, Iversen PL. Splicing in the immune system: potential targets for therapeutic intervention by antisense-mediated alternative splicing. Curr Opin Mol Ther. 2009;11:124–132. [PubMed] [Google Scholar]
- Braasch DA, Liu Y., and, Corey DR. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 2002;30:5160–5167. doi: 10.1093/nar/gkf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer J, Jepsen JS, Uhlmann E, Schetter C, Jurk M, Wader T.et al. (2004Modulation of CpG oligodeoxynucleotide-mediated immune stimulation by locked nucleic acid (LNA) Oligonucleotides 1423–31. [DOI] [PubMed] [Google Scholar]
- Raouane M, Desmaele D, Urbinati G., and, CouvVeur P. Lipid conjugated oligonucleotides: a useful strategy for delivery. Bioconjugate Chem. 2012;23:1091−1104. doi: 10.1021/bc200422w. [DOI] [PubMed] [Google Scholar]
- Jensen LE., and, Whitehead AS. The 3' untranslated region of the membrane-bound IL-1R accessory protein mRNA confers tissue-specific destabilization. J Immunol. 2004;173:6248–6258. doi: 10.4049/jimmunol.173.10.6248. [DOI] [PubMed] [Google Scholar]
- Ko JK., and, Ma J. A rapid and efficient PCR-based mutagenesis method applicable to cell physiology study. Am J Physiol, Cell Physiol. 2005;288:C1273–C1278. doi: 10.1152/ajpcell.00517.2004. [DOI] [PubMed] [Google Scholar]
- Greene AK., and, Puder M. Partial hepatectomy in the mouse: technique and perioperative management. J Invest Surg. 2003;16:99–102. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT-PCR analysis by using primers targeting exon 5 and 11.
RT-PCR analysis of liver samples of mice treated with Invivofectamine-complexed PS300.