Abstract
Activated T cells have classically been thought to progress unidirectionally through discrete phenotypic states and differentiate into static lineages. It is increasingly evident, however, that T cells exhibit much more complex and flexible dynamic behaviors than initially appreciated, and that these behaviors influence the efficacy of T cell responses to immunological challenges. In this review, we discuss how new technologies for monitoring the dynamics of T cells are enhancing the resolution of the fine phenotypic and functional heterogeneity within populations of T cells and revealing how individual T cells transition among a continuum of states. Such insights into the dynamic properties of T cells should improve immune monitoring and inform strategies for therapeutic interventions.
Keywords: T cells, dynamics, heterogeneity, multiparameter measurements, single-cell analysis
Assessing dynamic behaviors of T cells
Understanding how T cells generate productive and long-lasting responses is important for the development of effective preventions and treatments for infectious diseases and cancer. Conversely, identifying the mechanisms that result in dysregulated T cells is crucial for the development of treatments for autoimmune and inflammatory diseases. The dynamic processes underlying responses by T cells have been extensively studied in the context of both health and disease. Each process occurs over characteristic timescales (Figure 1a) and in a manner that usually yields robust but controlled immune responses.
Figure 1.
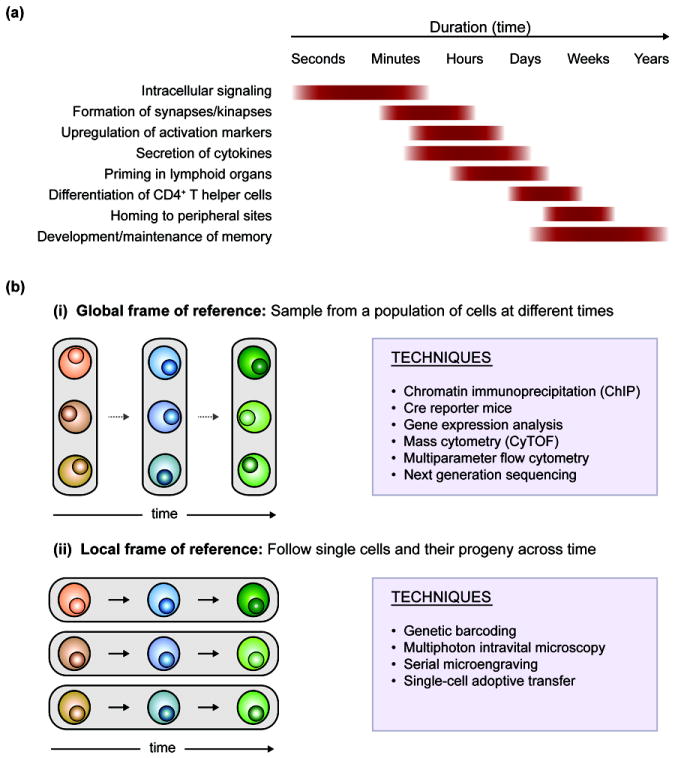
Dynamic T cell processes occur over characteristic time-scales and can be tracked using different experimental frames of reference. (a) Typical durations of dynamic T cell processes. (b) Experimental techniques can use global (Panel i) or local (Panel ii) frames of reference to follow T cells as populations or as individual cells, respectively.
The phenotypic and functional changes that T cells undergo in response to an immunological challenge have conventionally been modeled as unidirectional, stepwise transitions from naïve states to antigen-experienced memory states [1]. These models have been predominantly based on observations collected using standard analytical techniques that rely on a global frame of reference in which an entire population of T cells is tracked by subsampling a set of cells at sequential points in time (Figure 1b, Panel i). A global frame of reference can be used to observe population-wide shifts in behavior, but the properties of an individual cell at two different times cannot be directly linked, even if single-cell measurements are collected at both times. To restore temporal connectivity, new techniques have been developed that use a local frame of reference in which individual cells are marked or isolated so that they and their progeny can be directly tracked through time (Figure 1b, Panel ii). These measurements provide insights into how individual members within a population of T cells behave. Measurements collected using a local frame of reference are beginning to demonstrate that T cells exhibit a much greater diversity of dynamic behaviors than was previously observed by conventional measurements collected using a global frame of reference.
In this review, we focus on three approaches for assessing the dynamic behaviors of T cells that use observations from both global and local frames of reference to generate models of how T cells transition between different states. We discuss (i) how population-level studies are uncovering new features of phenotypic flexibility among T cells (Figure 2a), (ii) how recent advances in high-content single-cell profiling enable inference of the evolution of T cell responses (Figure 2b), and (iii) how methods to directly monitor individual T cells can reveal which specific transitional states are allowed and how dynamic processes in individual cells contribute to diversity within the population (Figure 2c).
Figure 2.
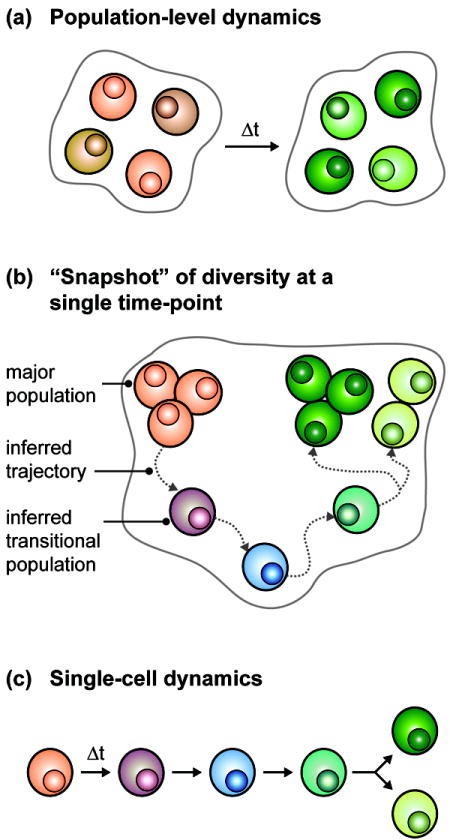
Approaches for assessing dynamic T cell behavior. (a) Global transitions can be analyzed by studying how populations of T cells change over time. Although the cells comprising each population share common features, there is likely cell-to-cell variability within the population that will be masked by bulk analysis. (b) Trajectories of cells as they move through a continuum of states can be inferred from detailed profiles of the T cell population at a single point in time. These profiles do not explicitly track cellular dynamics, but they can identify and characterize putative transitional populations linking more stable major populations. (c) Direct monitoring of an individual cell (and possibly its progeny) over time reveals the dynamic trajectories of each.
Population-level dynamics
Due to the lack of methods for monitoring individual cells over time, the majority of studies addressing T cell dynamics have examined populations of cells (Figure 2a). Initial models generated from these investigations proposed that T cells differentiate to form distinct and relatively static lineages. New experimental observations, however, are beginning to demonstrate that most traditional lineages are metastable states within a continuum of phenotypes, and that T cells move through this continuum following trajectories that are set by their current and past experiences.
Transitions among naïve and memory phenotypes
The development of T cell memory after antigen recognition is a classic example of a dynamic change in populations of T cells. Initial models for this process posited that the primary activation of human CCR7+CD45RA+ naïve T cells proceeds through a linear, unidirectional program of differentiation that first yields CCR7+CD45RA- central memory (TCM) cells and then CCR7-CD45RA- effector memory (TEM) cells (and, in the case of CD8+ T cells, CCR7-CD45RA+ effector memory (TEMRA) cells) [1]. Subsequent experiments focusing on CD8+ T cells demonstrated that this progression was not strictly unidirectional, as TEM cells stimulated in vitro can become phenotypically similar to TCM and naïve cells [2]. In addition, the concept that TEMRA cells are terminally differentiated cells with limited proliferative potential [2] was challenged by studies showing that the majority of antigen-specific CD8+ memory T cells induced by vaccination against yellow fever or vaccinia virus gradually re-express CD45RA over a period of months to years but maintain robust proliferative potential [3,4]. Moreover, it has been shown that >30% of memory cells in some vaccinated subjects regain expression of both CD45RA and CCR7—ostensibly returning them to a naïve phenotype as defined by these canonical markers—yet retain the functional characteristics of memory cells [3]. Surface phenotypes and functional capacities of T cells can also change in a graded—rather than stepwise—manner during the induction of memory [4,5]. These findings demonstrate that populations of T cells can gradually transition through a continuum of memory phenotypes over timescales of months to years and that transit through this continuum is not unidirectional with regard to the surface markers commonly used to define lineages of memory cells.
Plasticity of helper T cells
The differentiation of CD4+ T helper (TH) cells was initially thought to produce distinct, stable lineages as defined by their expression of characteristic transcription factors and cytokines ([6] and reviewed in [7]). It has long been recognized, however, that unambiguous assignment of TH cells to static, well-defined subsets based on the expression of cytokines can be challenging. Early studies of T cell clones demonstrated that the stability of TH subsets depends on the strength and duration of the polarization [8], and that cytokines segregate independently among clones under some conditions [9]. These findings suggested that TH cells can retain functional flexibility and that TH cells can alter stochastically their profiles of expressed cytokines in some instances.
Subsequent studies have added to the dynamic view of TH cells by showing that TH cells can transition between subsets under disease-relevant conditions [10,11] (and reviewed in [7,12]). Transgenic mice have been particularly useful tools for identifying the dynamic transitions that TH cells from a given lineage undergo. For example, mating mice that express Cre recombinase under the control of a lineage-specific promoter with ones that express a Cre-activatable fluorescent reporter gene results in offspring that have cells that heritably and irreversibly express the fluorescent reporter gene upon expression of Cre through the lineage-specific promoter, even if the lineage-specific gene is later turned off [13]. The in vivo plasticity of TH17 cells has been studied using mice in which current TH17 cells and descendants of TH17 cells express enhanced yellow fluorescent protein (EYFP) as a consequence of Cre being expressed from the IL17a [14] or IL17f [15] promoter. These studies have provided genetic evidence of the plasticity of TH17 cells by showing that a considerable fraction of EYFP+CD4+ T cells (i.e., TH cells that presumably expressed interleukin-17 (IL-17) at some time during their history) expressed T-bet and interferon-γ (IFN-γ)—classic markers of TH1 cells. The capacity of CD4+ T cells to transition among alternate states of expression may be explained in part by the observation that some lineage-specific loci can have unstable, stimuli-dependent epigenetic landscapes [16,17]. Because dysregulated TH cells can drive autoimmunity and allergy [18,19], identifying the transitions allowed among different TH subsets—and the conditions required to induce these transitions—could significantly impact the development of treatments for pathological immune responses.
Dynamic patterns of T cell homing
The homing properties of T cells are tightly controlled by the dynamic expression of molecules that target them to specific organs and tissues. Initial models suggested that T cells are “imprinted” with tissue-specific homing properties when they are first primed by antigen-presenting cells (APCs) in the lymph organ draining the affected tissue [20,21]. It has been shown, however, that the homing phenotypes of T cells can still evolve after secondary activation [22,23] or after adoptive transfer into naïve mice [24]. Additional evidence that T cells are not statically imprinted with restricted homing properties has come from the observation that T cells disseminate broadly to organs beyond the site of a challenge [25-28]. Several studies of the dynamics of homing have shown that this promiscuous redistribution is transient. For example, shortly after vaccination, CD8+ T cells transiently express homing markers for both the intestine and skin, but the expression of these markers—and thus the ability to localize to many different sites—is lost after 1–2 weeks [29]. Similarly, acute application of nonspecific irritants to distal skin or vaginal sites induces adoptively transferred, activated CD8+ T cells to traffic to these sites, but if the irritant is applied 15 days after transfer, the enhanced homing abilities are lost [30]. Together, these studies suggest activated CD8+ T cells can go through a transient phase of promiscuous homing before becoming more stable residents of specific sites. This model has significant implications for the design of strategies for vaccination that target the human immunodeficiency virus (HIV) and other pathogens that can enter through and reside in specific mucosal tissues, because it suggests that systemically activated T cells can be directed to home to these sites of localized infection.
Inferring transitional trajectories of T cells
The observation that populations of T cells dynamically transition between different phenotypic and functional states suggests that, when measured at any single point in time, some of the diversity observed within the population as a whole arises from cells at different transitional stages. Single-cell technologies are being developed to map the landscape of diversity within the T cell population with new detail. These profiles enable the identification and characterization of transitional states, and thus facilitate the inference and reconstruction of the transitional trajectories of T cells (Figure 2b).
Snapshots in time: Using heterogeneity to infer dynamic behaviors
To date, flow cytometry has been the method of choice for creating multiparametric, single-cell phenotypic profiles of T cell populations. Spectral overlap between the fluorescent reagents used to label cellular markers, however, has limited the number of analytes that can be simultaneously resolved (typically <20 analytes per cell [31,32]). To increase the depth of profiling, a cytometry technique has been developed that uses mass spectrometry to analyze single cells labeled with antibodies conjugated to elemental (metal) tags instead of fluorophores [33]. This method, called mass cytometry (or, cytometry by time-of-flight (CyTOF)), eliminates the problem of spectral overlap and therefore enables the simultaneous measurement of >36 analytes per cell, with a theoretical range of approximately 100 analytes (reviewed in [34]). New computational approaches have been developed to analyze the large, high-dimensional datasets generated by CyTOF [35]. Profiles of human CD8+ T cells collected using CyTOF have shown that they populate a continuum of intermediary states between major states such as naïve and memory, and that even highly specified subsets of T cells exhibit extensive combinatorial diversity in their expression of cytokines [5]. These findings highlight the complexity and heterogeneity of the CD8+ T cell compartment and demonstrate the utility of CyTOF for identifying cells with transitional phenotypes.
Detailed analysis of the nucleic acid content of single T cells is also uncovering the degree of diversity present among T cells and the dynamic transitions that they undergo. Transcriptional profiling of single murine T cells using a nanofluidic platform has shown that CD8+ memory T cells display distinct patterns of gene expression when responding to the same antigen delivered with different prime-boost strategies [36]. Interestingly, these programmatic variances are not reflected in the frequencies of antigen-specific T cells or the profiles of cytokines produced. These findings illustrate how memory T cells that share common functional properties can still access a variety of unique transcriptional states dictated by their current and past experiences.
Transcriptional profiling has been extended even further by the advancement of singlecell RNA-Seq, which can be used to analyze the entire transcriptome (>10,000 genes) of cells with rare transitional phenotypes [37]. Next-generation sequencing technologies have also been used to study dynamic changes in the repertoires of T cell receptor (TCR) sequences from bulk populations of human T cells, and thus provide another powerful tool for assessing the transitional trajectories of individual clones [38,39]. Although the techniques discussed above are static in the sense that they do not track individual cells over multiple points in time, they nonetheless promise to advance our understanding of dynamic T cell behavior by producing high-dimensional profiles of the states that T cells occupy, which should facilitate the construction of transitional paths by inference.
Dynamic behaviors of individual T cells
Dense static maps of the states that T cells occupy can inform inferences about dynamic patterns. To establish definitive pathways, however, it is beneficial to complement inferential studies with direct observations of individual cells as they transit from one state to another (Figure 2c). Here we discuss approaches for measuring the dynamic properties of individual cells. These approaches are yielding new insights into how the unique history of each T cell influences the transitions that it can make and how these transitions contribute to cell-to-cell variability.
Dynamics of T cell activation
Multiphoton microscopy is a powerful tool for analyzing dynamic interactions among individual cells in intact tissues (reviewed in [40]), and has been used extensively to study the dynamic processes involved in the priming of naïve T cells by antigen-loaded dendritic cells (DCs) in lymph nodes. These experiments have shown that, under certain conditions, T cells go through an initial phase of transient, serial interactions (typically <10 min per interaction) with DCs before transitioning to a phase of long-lived (>1 h) interactions [41,42]. Under other conditions, however, T cells do not go through a prolonged phase of transient interactions, but instead rapidly form stable interactions with DCs [43,44]. One proposed explanation for this variability is that T cells integrate the antigenic, costimulatory, and chemotactic signals they collect from different DCs and their microenvironment and use the cumulative signal to trigger functional responses. According to this model, a T cell may need to serially encounter many DCs presenting weak signals before reaching the same level of activation as a T cell that encounters fewer DCs that each present strong signals [45]. Indeed, several experiments have suggested sted that T cells integrate signals from APCs over a mixture of time, encounters, doses, and potencies, and then use these cumulative signals to induce outcomes such as cell division or the secretion of cytokines [43,45-48]. Additional insights into how T cells collect signals to trigger transitions between states, and into the heterogeneity and dynamics of T cell activation, have come from studies of the interfaces that form between T cells and APCs (Box 1).
Box 1. Role of immunological synapses and kinapses in dynamic T cell behavior.
The properties of immunological synapses (stable interactions) and kinapses (transient, motile interactions) that form between T cells and APCs reflect the prior histories of individual T cells and predict the future paths that they will follow once activated. It is difficult to link measurements of immunological synapses and kinapses to the functional states of the exact same cells after extended periods of time, but several approaches are being used to unravel how the trajectories of T cells are related to the interfaces formed with APCs.
Structure-function relationships: Microscopy-based studies have shown that the prior history of a T cell can affect the structure and composition of the immunological synapse (reviewed in [72]). For example, different types of CD4+ T cells (e.g., Th1 versus Th2 cells [73], or self-reactive versus influenza-specific clones [74]) form morphologically distinct synapses, and anergic CD4+ T cells form synapses that are enriched in negative regulators of TCR signaling [75]. In turn, the patterns of molecular partitioning at the synapse guide the subsequent functional trajectory of the T cell [76,77].
Dynamics of protein localization: In vivo visualization of the dynamics of protein localization at the T cell-APC interface has been achieved by using multiphoton microscopy to monitor enhanced green fluorescent protein (EGFP) fusion proteins in T cells. Using this approach, it was demonstrated that downstream activities associated with antigen recognition can be induced by either transient or stable clustering of TCR molecules at the T cell-APC interface [78], and that migratory and synaptic dynamics are influenced by the history of the T cell (e.g., naïve versus recently activated) and the characteristics of the APC [79].
Dynamics of signaling: T cell signaling during interactions with APCs has been investigated using a technique called dynamic in situ cytometry (DISC), which uses intravital multiphoton microscopy to track cellular interactions while simultaneously monitoring changes in calcium signaling or in the expression of cellular surface markers. DISC analysis of the shedding of CD62L (indicative of TCR signaling) from splenic T cells has shown that signals collected through synapses and those collected through kinapses can both induce the shedding of CD62L, although the shedding is more rapid when stable synapses are formed [80].
Dynamic secretory activity by individual T cells
The cytokines that T cells secrete are important for directing immune responses. Thus, there is considerable interest in understanding how cytokine profiles from individual T cells correlate with outcomes of infection, vaccination, and other immunological processes. Toward this end, intracellular cytokine staining (ICS) combined with multiparameter flow cytometry has been used to show that the breadth and magnitude of cytokines secreted by stimulated T cells shift globally over time [49,50], and that antigen-specific T cells that produce multiple cytokines (“polyfunctional” T cells) are associated with protective immune responses to pathogens such as HIV [51,52], vaccinia virus [53], and Leishmania major [54]. These findings have raised the question of how polyfunctionality arises in individual T cells and how (or if) it is stably maintained. Because the destructive, end-point nature of ICS makes it unsuitable for addressing such questions of single-cell secretory dynamics, our group developed a method, known as microengraving, to longitudinally monitor the secretory activity of individual T cells [55]. In this process, T cells are isolated in an array of subnanoliter wells (nanowells), and then a slide coated with cytokine-specific antibodies is compressed on top of the array to capture and subsequently detect the cytokines secreted by the cell(s) residing in each nanowell [56]. Microengraving is non-destructive and can be repeatedly applied to measure the secretory profiles of the same individual cells at multiple points in time.
Using serial microengraving to analyze IFN-γ, tumor necrosis factor-α (TNF), and IL-2, we observed that individual T cells initiated secretion asynchronously upon polyclonal activation, but that their time-aligned secretory profiles clustered into sets of deterministic trajectories [55]. Secreting T cells were most likely to stably secrete one cytokine or to sequentially transition between single-functional states (e.g., TNF+ → IL-2+). By contrast, the simultaneous secretion of multiple cytokines was primarily a short-lived intermediary state that cells passed through as they transitioned between lower-order secretory states (e.g., TNF+ → TNF+IL-2+ → IL-2+). Although it remains to be seen whether these trajectories are conserved under antigen-specific stimulation, these findings suggest that longitudinal secretory profiles of single T cells may be more informative metrics of immune status than conventional ICS measurements of polyfunctionality. Moreover, these results illustrate how the dynamic and asynchronous secretory behaviors of T cells contribute to the functional heterogeneity observed within populations of T cells any single point in time.
Generation of functional diversity in the progeny of individual T cells
Over longer time-scales (days to years), heterogeneity can also materialize within the T cell compartment as successive generations of T cells take on characteristics distinct from their predecessors (Box 2). Many techniques have been developed to track the fate of T cells across multiple generations (reviewed in [57]). Perhaps the most direct way to determine the potential of a T cell to generate diversity in vivo is to transfer a single, naïve T cell into a host and then examine the phenotypes and functions of its progeny after an immunological challenge. Using this approach, it was demonstrated that a single naïve CD8+ T cell could give rise to diverse subsets of T cells. During the effector phase, donor-derived cells exhibited a range of different secretory and cytolytic characteristics, and after an initial challenge, both effector and central memory cells were produced [58]. The adoptive transfer of genetically barcoded T cells has provided further evidence that single naïve CD8+ T cells can yield both effector and memory cells under a variety of situations (e.g., systemic and local infection, low and high avidity TCR ligands) [59]. Thus, the progeny of individual cells can fill a wide continuum of phenotypes and transition through different phenotypic states as they proliferate. These single-cell observations reinforce population-level observations of gradual phenotypic change and multidirectional transitions during the induction of memory [2-5]. Transgenic strategies to label cells by stochastically expressing different combinations of fluorescent proteins (e.g., using the Cre/lox recombination system of ‘Brainbow’ transgenes) [60] and strategies to label cells from specific microanatomical sites using photoactivatable fluorescent proteins [61] are also promising approaches for tracking cells as they proliferate and migrate.
Box 2. Generating diversity across generations.
Here we highlight two processes by which individual T cells can generate diverse progeny.
Asymmetric cell division: A T cell that receives mitogenic signals from a DC often divides while remaining attached to the DC [81], and ex vivo and in vitro single-cell analyses have demonstrated that fate-determining proteins are unevenly distributed between the proximal and distal daughter T cells [81-83]. This initial asymmetric division can generate daughter cells that consistently differ in their expression of effector- or memory-like properties, lineage-defining transcription factors, receptors for cytokines, signaling proteins, and proteins involved in cell polarity [81-84]. Thus, changes in T cell behavior over generations may be caused in part by the unequal distribution of proteins to each daughter cell formed by APC-induced cell division.
Epigenetic modifications: At the population level, epigenetic landscapes at the loci of cytokine genes change as cells divide and differentiate; these changes cause the corresponding cytokines to be produced with characteristic dynamic patterns [85-89]. The loci of effector genes that are important in the cytolytic activity of CD8+ T cells, such as perforin (Prf1) and granzyme B (Gzmb), also undergo temporal changes in epigenetic modifications as cells progress from the naïve to the memory state [90,91]. At the single-cell level, stochastic changes in chromatin accessibility are thought to contribute to cell-to-cell variability in cytokine production at any given time [86,92,93]. Multiparameter flow cytometric assays measuring histone modifications, surface markers, and intracellular cytokines have directly demonstrated the relationship between certain patterns of histone acetylation and the phenotypic and functional properties of the same individual T cells [94]. Epigenetic modifications therefore drive global alterations in the functional attributes of T cells while also generating heterogeneity within the T cell population.
In addition, inputs received by T cells early during an immune response—prior to the initiation of proliferation—can contribute to heterogeneity among clones.
T cell-DC interactions: Upon activation by antigen-bearing DCs, antigen-specific CD8+ T cells exhibit considerable cell-to-cell variability in the expression of the IFN-γ gene (as measured by the expression of a fluorescent reporter) at the time of their first cell division [95]. The progenies of the individual cells maintain these variations as they proliferate. Thus, some aspects of functional heterogeneity are initiated prior to the first cell division.
Dynamic modulation of the TCR specificity of individual peripheral T cells
The specificity of the TCR is traditionally regarded as being constant and immutable in peripheral T cells. Several lines of evidence, however, suggest that individual T cells can alter their TCRα and TCRβ chains—even after exiting the thymus. This process, called TCR revision, has been studied in mice bearing transgenic [62] or knock-in [63,64] TCR chains that interact with superantigens. In these mice, a fraction of the peripheral T cells that are exposed to the superantigen revise their TCR by expressing recombination-activating genes (RAG1, RAG2) and replacing the transgenic or knock-in TCR with an endogenous TCR. TCR revision has also been observed in peripheral mouse T cells that recognize self-antigens [64-66], and there is indirect evidence that TCR revision may occur in humans [67]. Some combinations of TCRs and antigens, however, fail to induce TCR revision, so the generality of this process is not yet fully known [63,65]. Several intriguing hypotheses have been generated about how the ability to dynamically transition among different antigen-specific states may relate to the induction of tolerance, autoimmunity, or pathogen-specific immune responses [63,65,66,68,69]. Considering the history-dependent and heterogeneous nature of TCR revision, it is likely that single-cell technologies that combine TCR sequencing with tracking of progeny will help to address unresolved questions about the functional significance of TCR revision and its regulation.
Concluding remarks
Recent research has demonstrated that T cells transition among multiple phenotypic states. These observations must be reconciled with the classic model of T cells differentiating into distinct, static lineages. Probabilistic, non-linear frameworks have previously been used to conceptualize the graded transitions that occur in multi-lineage cellular systems [12,70,71]. Similarly, we propose that it may be useful to conceptualize the phenotype of a T cell not as a static entity but as a probability distribution representing a continuum of possible states (Figure 3). In this model, the phenotype of each T cell is a function both of the universal programs that are hardwired into its genome and of the unique epigenetic and proteomic modifications induced by past and present stimuli. By analogy to chemical reactions and transition state theory, T cells progress toward local “low-energy” phenotypic states in their probability distributions that correspond to classic T cell lineages and phenotypes. If new stimuli provide enough “energy”, however, a T cell becomes capable of transitioning to alternative minima in its distribution, which itself has also likely been reconfigured by the new stimuli. New single-cell technologies enable direct observation of the transitional paths of individual cells and facilitate construction of highly defined profiles of phenotypic probability distributions. Knowledge of these trajectories and distributions should shed light on how dynamic T cell behavior contributes to the inception and maintenance of heterogeneity among T cells. Detailed analysis of phenotypic “transition states” should provide considerable information about the underlying regulatory networks controlling phenotypic distributions and the signals that push T cells out of local phenotypic minima into alternative states. Many questions remain about dynamic T cell behavior and how it can be experimentally analyzed (Box 3). Given the central role of properly timed and executed T cell responses in clinical immunology, an in-depth understanding of dynamic T cell behavior will likely facilitate breakthroughs in the treatment of autoimmune diseases, allergies, and infectious diseases.
Figure 3.
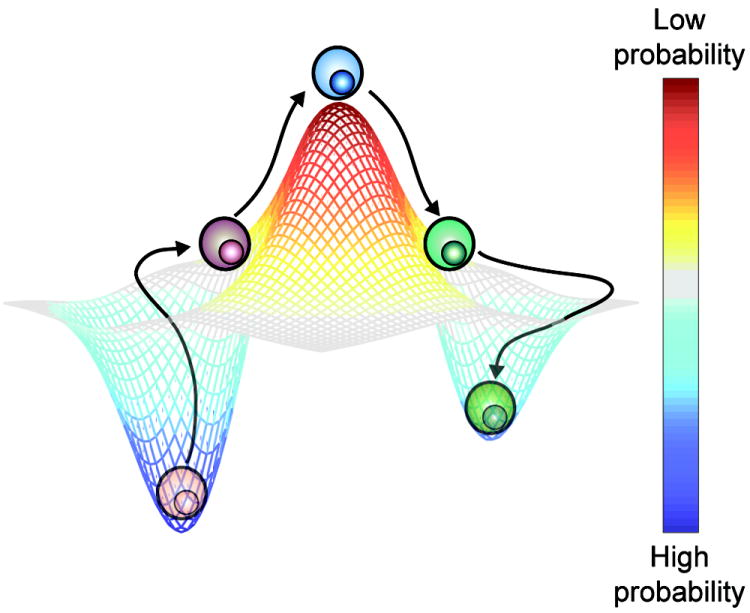
Conceptual model of the phenotypic probability distribution of a T cell. The phenotypic probability distribution of each T cell is shaped by the unique properties and history of that cell. The T cell will most likely be found in a high-probability state (analogous to an energy minimum in a chemical reaction) that corresponds to a relatively stable phenotype. External stimuli or cell-intrinsic programs may induce the T cell to transition to a different high-probability state. During this transition, the T cell may be observed in a low-probability state corresponding to a short-lived, unstable transitional phenotype.
Box 3. Outstanding questions.
How stable (or, rather, unstable) are conventionally defined states of memory and differentiation?
What epigenetic mechanisms are responsible for maintaining differentiated states and what cell-intrinsic or cell-extrinsic signals induce transitions between states?
How do stochastic factors shape the dynamic behaviors of T cells?
What technological developments are needed to measure >100 protein analytes in single cells in a high-throughput manner, and how can these measurements be acquired dynamically from viable single cells?
What are the best mathematical frameworks for analyzing large, multiparameter, temporal datasets?
How do dynamic responses observed in vitro reflect the in vivo behavior of T cells?
How can the dynamic patterns of T cell homing be used to improve strategies for vaccination?
How can dynamic profiles of T cell behavior supplement static profiles to improve immune monitoring, diagnosis, and treatment?
What is the in vivo frequency of TCR revision in peripheral human T cells, and what is the functional significance of this process?
Acknowledgments
We thank Qing Han, Rita Lucia Contento, and Alexis Torres for helpful discussions and comments on the manuscript. This work was supported in part by funding from the Charles A. Dana Foundation, the W.M. Keck Foundation, and NIH/NIAID (4U19AI089992, 5P01AI045757, UMI AI068618). Y.J.Y is supported in part by a fellowship from the National Science Foundation and the Collamore-Rogers Fellowship. T.M.G. was supported in part by a post-doctoral fellowship from the Ragon Institute of MGH, MIT and Harvard. J.C.L. is a Latham Family Career Development Professor and Camille Dreyfus Teacher-Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sallusto F, et al. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 2.Champagne P, et al. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410(6824):106–111. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- 3.Akondy RS, et al. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol. 2009;183(12):7919–7930. doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller JD, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28(5):710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Newell EW, et al. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36(1):142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosmann TR, et al. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136(7):2348–2357. [PubMed] [Google Scholar]
- 7.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327(5969):1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy E, et al. Reversibility of T helper 1 and 2 populations is lost after long-term stimulation. J Exp Med. 1996;183(3):901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelso A, et al. Evidence for the stochastic acquisition of cytokine profile by CD4+ T cells activated in a T helper type 2-like response in vivo. Eur J Immunol. 1995;25(5):1168–1175. doi: 10.1002/eji.1830250506. [DOI] [PubMed] [Google Scholar]
- 10.Amarnath S, et al. The PDL1-PD1 axis converts human TH1 cells into regulatory T cells. Sci Transl Med. 2011;3(111):111ra120. doi: 10.1126/scitranslmed.3003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panzer M, et al. Rapid in vivo conversion of effector T cells into Th2 cells during helminth infection. J Immunol. 2012;188(2):615–623. doi: 10.4049/jimmunol.1101164. [DOI] [PubMed] [Google Scholar]
- 12.Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat Immunol. 2010;11(8):674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croxford AL, et al. Cutting edge: an IL-17F-CreEYFP reporter mouse allows fate mapping of Th17 cells. J Immunol. 2009;182(3):1237–1241. doi: 10.4049/jimmunol.182.3.1237. [DOI] [PubMed] [Google Scholar]
- 14.Hirota K, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12(3):255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurschus FC, et al. Genetic proof for the transient nature of the Th17 phenotype. Eur J Immunol. 2010;40(12):3336–3346. doi: 10.1002/eji.201040755. [DOI] [PubMed] [Google Scholar]
- 16.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30(1):155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukasa R, et al. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32(5):616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umetsu DT, DeKruyff RH. The regulation of allergy and asthma. Immunol Rev. 2006;212:238–255. doi: 10.1111/j.0105-2896.2006.00413.x. [DOI] [PubMed] [Google Scholar]
- 19.Wilke CM, et al. Deciphering the role of Th17 cells in human disease. Trends Immunol. 2011;32(12):603–611. doi: 10.1016/j.it.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudda JC, et al. Dendritic cell immunization route determines CD8+ T cell trafficking to inflamed skin: role for tissue microenvironment and dendritic cells in establishment of T cell-homing subsets. J Immunol. 2004;172(2):857–863. doi: 10.4049/jimmunol.172.2.857. [DOI] [PubMed] [Google Scholar]
- 21.Mora JR, et al. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 2003;424(6944):88–93. doi: 10.1038/nature01726. [DOI] [PubMed] [Google Scholar]
- 22.Dudda JC, et al. Dendritic cells govern induction and reprogramming of polarized tissue-selective homing receptor patterns of T cells: important roles for soluble factors and tissue microenvironments. Eur J Immunol. 2005;35(4):1056–1065. doi: 10.1002/eji.200425817. [DOI] [PubMed] [Google Scholar]
- 23.Mora JR, et al. Reciprocal and dynamic control of CD8 T cell homing by dendritic cells from skin- and gut-associated lymphoid tissues. J Exp Med. 2005;201(2):303–316. doi: 10.1084/jem.20041645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jennrich S, et al. Long-term commitment to inflammation-seeking homing in CD4+ effector cells. J Immunol. 2007;178(12):8073–8080. doi: 10.4049/jimmunol.178.12.8073. [DOI] [PubMed] [Google Scholar]
- 25.Liu L, et al. Dynamic programming of CD8+ T cell trafficking after live viral immunization. Immunity. 2006;25(3):511–520. doi: 10.1016/j.immuni.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Masopust D, et al. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291(5512):2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 27.Masopust D, et al. Activated primary and memory CD8 T cells migrate to nonlymphoid tissues regardless of site of activation or tissue of origin. J Immunol. 2004;172(8):4875–4882. doi: 10.4049/jimmunol.172.8.4875. [DOI] [PubMed] [Google Scholar]
- 28.Stevceva L, et al. Both mucosal and systemic routes of immunization with the live, attenuated NYVAC/simian immunodeficiency virus SIV(gpe) recombinant vaccine result in gag-specific CD8(+) T-cell responses in mucosal tissues of macaques. J Virol. 2002;76(22):11659–11676. doi: 10.1128/JVI.76.22.11659-11676.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masopust D, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207(3):553–564. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackay LK, et al. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci U S A. 2012;109(18):7037–7042. doi: 10.1073/pnas.1202288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chattopadhyay PK, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12(8):972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 32.Perfetto SP, et al. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4(8):648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 33.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81(16):6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 34.Bendall SC, et al. A deep profiler’s guide to cytometry. Trends Immunol. 2012 doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29(10):886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flatz L, et al. Single-cell gene-expression profiling reveals qualitatively distinct CD8 T cells elicited by different gene-based vaccines. Proc Natl Acad Sci U S A. 2011;108(14):5724–5729. doi: 10.1073/pnas.1013084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci U S A. 2010;107(4):1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warren RL, et al. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 2011;21(5):790–797. doi: 10.1101/gr.115428.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Germain RN, et al. Dynamic imaging of the immune system: progress, pitfalls and promise. Nat Rev Immunol. 2006;6(7):497–507. doi: 10.1038/nri1884. [DOI] [PubMed] [Google Scholar]
- 41.Miller MJ, et al. Imaging the single cell dynamics of CD4+ T cell activation by dendritic cells in lymph nodes. J Exp Med. 2004;200(7):847–856. doi: 10.1084/jem.20041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mempel TR, et al. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427(6970):154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 43.Celli S, et al. Real-time manipulation of T cell-dendritic cell interactions in vivo reveals the importance of prolonged contacts for CD4+ T cell activation. Immunity. 2007;27(4):625–634. doi: 10.1016/j.immuni.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Shakhar G, et al. Stable T cell-dendritic cell interactions precede the development of both tolerance and immunity in vivo. Nat Immunol. 2005;6(7):707–714. doi: 10.1038/ni1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henrickson SE, et al. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat Immunol. 2008;9(3):282–291. doi: 10.1038/ni1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Celli S, et al. CD4 T cells integrate signals delivered during successive DC encounters in vivo. J Exp Med. 2005;202(9):1271–1278. doi: 10.1084/jem.20051018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark CE, et al. A role for the immediate early gene product c-fos in imprinting T cells with short-term memory for signal summation. PLoS One. 2011;6(4):e18916. doi: 10.1371/journal.pone.0018916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skokos D, et al. Peptide-MHC potency governs dynamic interactions between T cells and dendritic cells in lymph nodes. Nat Immunol. 2007;8(8):835–844. doi: 10.1038/ni1490. [DOI] [PubMed] [Google Scholar]
- 49.Mascher B, et al. Expression and kinetics of cytokines determined by intracellular staining using flow cytometry. J Immunol Methods. 1999;223(1):115–121. doi: 10.1016/s0022-1759(98)00200-2. [DOI] [PubMed] [Google Scholar]
- 50.Sandberg JK, et al. Functional heterogeneity of cytokines and cytolytic effector molecules in human CD8+ T lymphocytes. J Immunol. 2001;167(1):181–187. doi: 10.4049/jimmunol.167.1.181. [DOI] [PubMed] [Google Scholar]
- 51.Almeida JR, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204(10):2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Precopio ML, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med. 2007;204(6):1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darrah PA, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13(7):843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 55.Han Q, et al. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc Natl Acad Sci U S A. 2011;109(5):1607–1612. doi: 10.1073/pnas.1117194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han Q, et al. Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab Chip. 2010;10(11):1391–1400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schumacher TN, et al. Mapping the life histories of T cells. Nat Rev Immunol. 2010;10(9):621–631. doi: 10.1038/nri2822. [DOI] [PubMed] [Google Scholar]
- 58.Stemberger C, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27(6):985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 59.Gerlach C, et al. One naive T cell, multiple fates in CD8+ T cell differentiation. J Exp Med. 2010;207(6):1235–1246. doi: 10.1084/jem.20091175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 61.Victora GD, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143(4):592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McMahan CJ, Fink PJ. RAG reexpression and DNA recombination at T cell receptor loci in peripheral CD4+ T cells. Immunity. 1998;9(5):637–647. doi: 10.1016/s1074-7613(00)80661-5. [DOI] [PubMed] [Google Scholar]
- 63.Huang CY, et al. Superantigen-induced TCR alpha locus secondary rearrangement: role in tolerance induction. J Immunol. 2002;168(7):3259–3265. doi: 10.4049/jimmunol.168.7.3259. [DOI] [PubMed] [Google Scholar]
- 64.Takase M, et al. Age-dependent TCR revision mediated by interaction between alphabeta TCR and self-antigens. J Immunol. 2007;179(4):2163–2169. doi: 10.4049/jimmunol.179.4.2163. [DOI] [PubMed] [Google Scholar]
- 65.Serra P, et al. RAG-dependent peripheral T cell receptor diversification in CD8+ T lymphocytes. Proc Natl Acad Sci U S A. 2002;99(24):15566–15571. doi: 10.1073/pnas.242321099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bynoe MS, et al. T cells from epicutaneously immunized mice are prone to T cell receptor revision. Proc Natl Acad Sci U S A. 2005;102(8):2898–2903. doi: 10.1073/pnas.0409880102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lantelme E, et al. An in vitro model of T cell receptor revision in mature human CD8+ T cells. Mol Immunol. 2008;45(2):328–337. doi: 10.1016/j.molimm.2007.06.153. [DOI] [PubMed] [Google Scholar]
- 68.Vaitaitis GM, et al. Cutting edge: CD40-induced expression of recombination activating gene (RAG) 1 and RAG2: a mechanism for the generation of autoaggressive T cells in the periphery. J Immunol. 2003;170(7):3455–3459. doi: 10.4049/jimmunol.170.7.3455. [DOI] [PubMed] [Google Scholar]
- 69.Zehn D, et al. Cutting edge: TCR revision affects predominantly Foxp3 cells and skews them toward the Th17 lineage. J Immunol. 2007;179(9):5653–5657. doi: 10.4049/jimmunol.179.9.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ceredig R, et al. Models of haematopoiesis: seeing the wood for the trees. Nat Rev Immunol. 2009;9(4):293–300. doi: 10.1038/nri2525. [DOI] [PubMed] [Google Scholar]
- 71.O’Garra A, et al. Quantitative events determine the differentiation and function of helper T cells. Nat Immunol. 2011;12(4):288–294. doi: 10.1038/ni.2003. [DOI] [PubMed] [Google Scholar]
- 72.Thauland TJ, Parker DC. Diversity in immunological synapse structure. Immunology. 2010;131(4):466–472. doi: 10.1111/j.1365-2567.2010.03366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thauland TJ, et al. Th1 and Th2 cells form morphologically distinct immunological synapses. J Immunol. 2008;181(1):393–399. doi: 10.4049/jimmunol.181.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schubert DA, et al. Self-reactive human CD4 T cell clones form unusual immunological synapses. J Exp Med. 2012;209(2):335–352. doi: 10.1084/jem.20111485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doherty M, et al. Anergic CD4+ T cells form mature immunological synapses with enhanced accumulation of c-Cbl and Cbl-b. J Immunol. 2010;184(7):3598–3608. doi: 10.4049/jimmunol.0902285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maldonado RA, et al. Control of T helper cell differentiation through cytokine receptor inclusion in the immunological synapse. J Exp Med. 2009;206(4):877–892. doi: 10.1084/jem.20082900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maldonado RA, et al. A role for the immunological synapse in lineage commitment of CD4 lymphocytes. Nature. 2004;431(7008):527–532. doi: 10.1038/nature02916. [DOI] [PubMed] [Google Scholar]
- 78.Friedman RS, et al. Real-time analysis of T cell receptors in naive cells in vitro and in vivo reveals flexibility in synapse and signaling dynamics. J Exp Med. 2010;207(12):2733–2749. doi: 10.1084/jem.20091201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Azar GA, et al. Subcellular dynamics of T cell immunological synapses and kinapses in lymph nodes. Proc Natl Acad Sci U S A. 2010;107(8):3675–3680. doi: 10.1073/pnas.0905901107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moreau HD, et al. Dynamic In Situ Cytometry Uncovers T Cell Receptor Signaling during Immunological Synapses and Kinapses In Vivo. Immunity. 2012;37 doi: 10.1016/j.immuni.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 81.Oliaro J, et al. Asymmetric cell division of T cells upon antigen presentation uses multiple conserved mechanisms. J Immunol. 2010;185(1):367–375. doi: 10.4049/jimmunol.0903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang JT, et al. Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division. Immunity. 2011;34(4):492–504. doi: 10.1016/j.immuni.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang JT, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315(5819):1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 84.Ciocca ML, et al. Cutting edge: Asymmetric memory T cell division in response to rechallenge. J Immunol. 2012;188(9):4145–4148. doi: 10.4049/jimmunol.1200176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Avni O, et al. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3(7):643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 86.Mariani L, et al. Short-term memory in gene induction reveals the regulatory principle behind stochastic IL-4 expression. Mol Syst Biol. 2010;6:359. doi: 10.1038/msb.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McKarns SC, Schwartz RH. Biphasic regulation of Il2 transcription in CD4+ T cells: roles for TNF-alpha receptor signaling and chromatin structure. J Immunol. 2008;181(2):1272–1281. doi: 10.4049/jimmunol.181.2.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Denton AE, et al. Differentiation-dependent functional and epigenetic landscapes for cytokine genes in virus-specific CD8+ T cells. Proc Natl Acad Sci U S A. 2011;108(37):15306–15311. doi: 10.1073/pnas.1112520108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Northrop JK, et al. Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol. 2006;177(2):1062–1069. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- 90.Zediak VP, et al. Cutting edge: persistently open chromatin at effector gene loci in resting memory CD8+ T cells independent of transcriptional status. J Immunol. 2011;186(5):2705–2709. doi: 10.4049/jimmunol.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Araki Y, et al. Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B) J Immunol. 2008;180(12):8102–8108. doi: 10.4049/jimmunol.180.12.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo L, et al. Probabilistic regulation of IL-4 production in Th2 cells: accessibility at the Il4 locus. Immunity. 2004;20(2):193–203. doi: 10.1016/s1074-7613(04)00025-1. [DOI] [PubMed] [Google Scholar]
- 93.Calado DP, et al. Stochastic monoallelic expression of IL-10 in T cells. J Immunol. 2006;177(8):5358–5364. doi: 10.4049/jimmunol.177.8.5358. [DOI] [PubMed] [Google Scholar]
- 94.Dispirito JR, Shen H. Histone acetylation at the single-cell level: a marker of memory CD8+ T cell differentiation and functionality. J Immunol. 2010;184(9):4631–4636. doi: 10.4049/jimmunol.0903830. [DOI] [PubMed] [Google Scholar]
- 95.Beuneu H, et al. Visualizing the functional diversification of CD8+ T cell responses in lymph nodes. Immunity. 2010;33(3):412–423. doi: 10.1016/j.immuni.2010.08.016. [DOI] [PubMed] [Google Scholar]


