Abstract
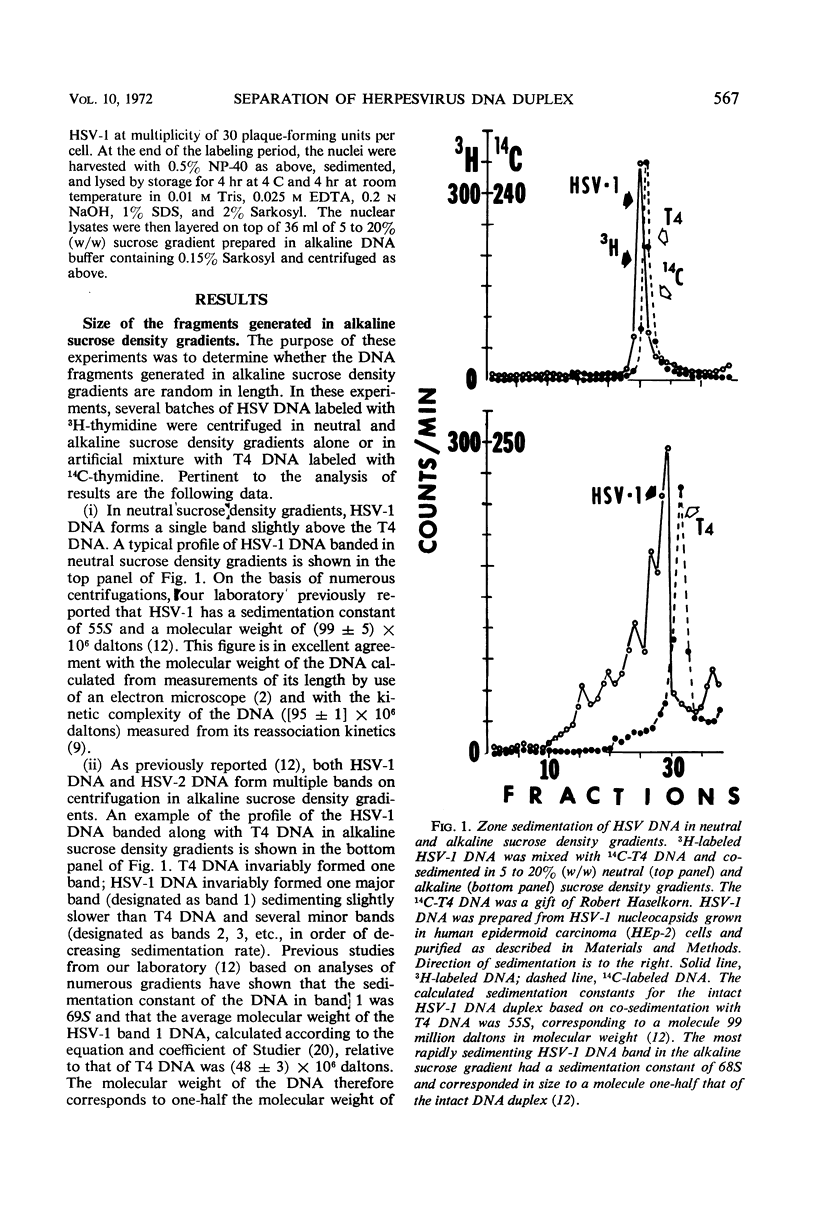
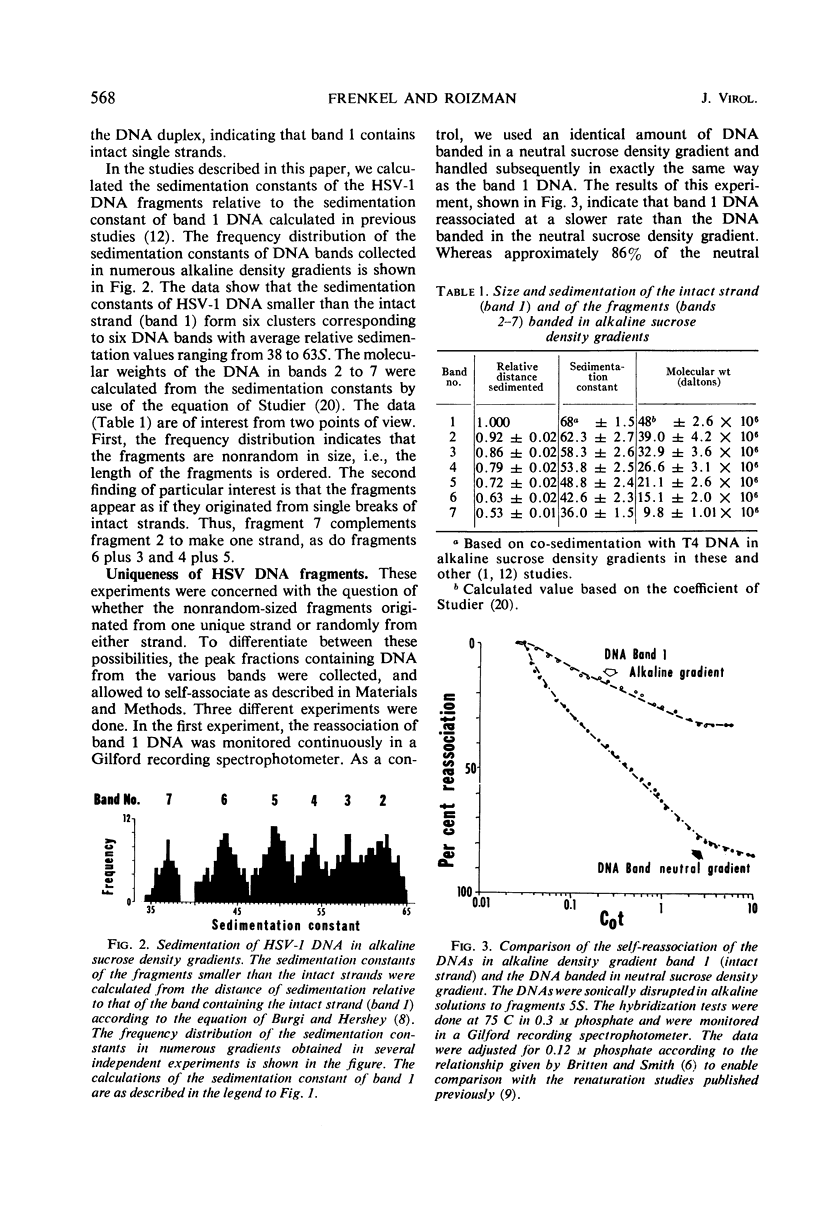
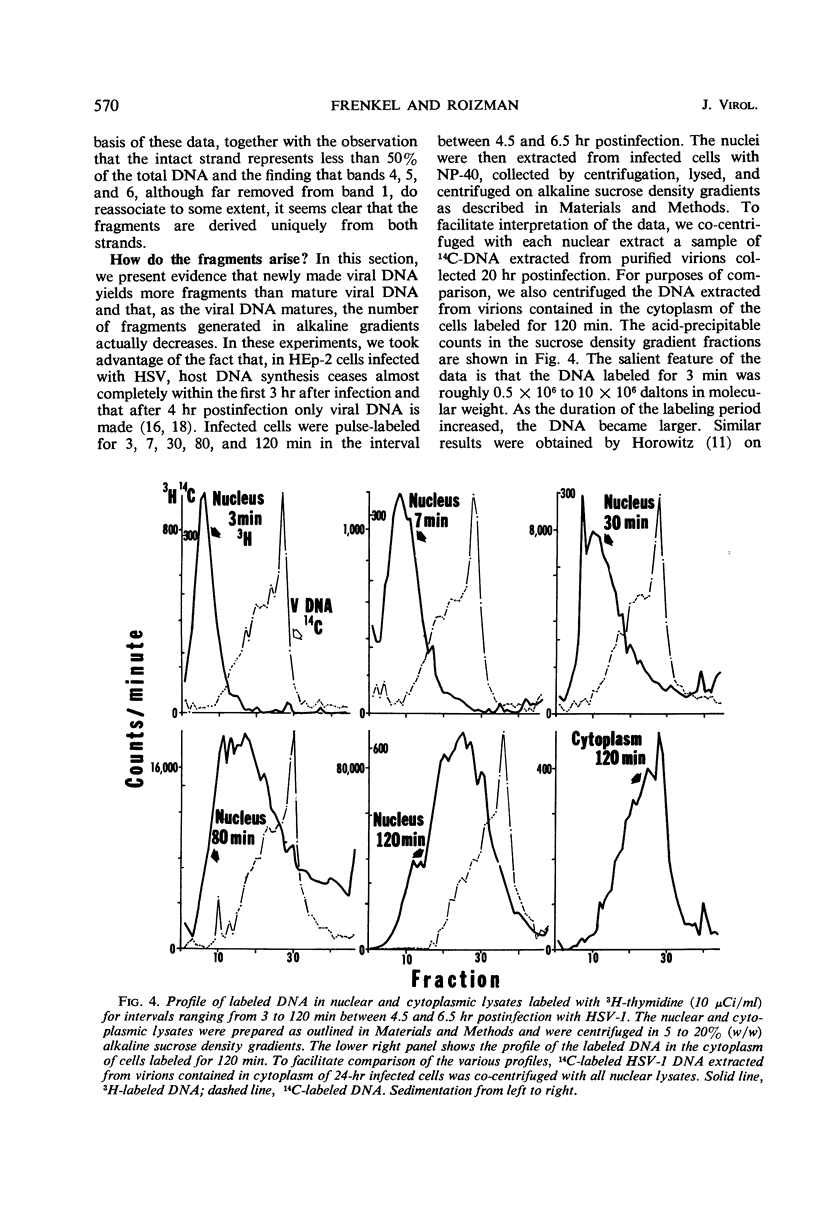
Deoxyribonucleic acid (DNA) extracted from herpes simplex virions forms multiple partially overlapping bands upon denaturation and centrifugation in alkaline sucrose density gradients. The most rapidly sedimenting DNA corresponds to an intact strand 48 × 106 daltons in molecular weight. In this study, we analyzed the DNA fragments generated in alkaline sucrose gradients with respect to size and uniqueness of base sequences. The distribution of sedimentation constants of the various fragments obtained in numerous gradients showed that the fragments smaller than the whole strand fall into six distinct classes ranging in molecular weight from 10 × 106 to 39 × 106 daltons. Four types of DNA strands can be reconstructed from the whole strand and six fragments on the basis of their molecular weights. DNA from each of the bands self-hybridizes to a lower extent than unfractionated viral DNA, indicating that each of the bands preferentially contains sequences from one unique strand. The data permit reconstruction of four possible types of DNA duplexes differing in the positions of the strand interruptions. Analysis of viral DNA extracted from nuclei of cells labeled with 3H-thymidine for intervals from 3 to 120 min showed that nascent DNA is invariably attached to small fragments and that the fragments become elongated only upon prolonged incubation of cells. The experiments suggest that viral DNA replication begins at numerous initiation sites along each strand and that the elongation beyond the size of the replication unit involves repair or ligation, or both. Since newly made DNA yields more fragments than viral DNA extracted from mature virions, it is suggested that the fragmentation of mature DNA on denaturation with alkali arises from incomplete processing of specific initiation sites. Comparison of viral DNA extracted from nuclei with that extracted from mature cytoplasmic virions in cells labeled for 120 min indicates that packaged DNA is not randomly selected from among the nuclear DNA population but rather represents DNA molecules which in alkaline gradients yield a minimal number of fragments.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker Y., Dym H., Sarov I. Herpes simplex virus DNA. Virology. 1968 Oct;36(2):184–192. doi: 10.1016/0042-6822(68)90135-9. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Chromatography of nucleic acids on hydroxyapatite. I. Chromatography of native DNA. Biochim Biophys Acta. 1969 Feb 18;174(2):423–434. doi: 10.1016/0005-2787(69)90273-1. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Chromatography of nucleic acids on hydroxyapatite. II. Chromatography of denatured DNA. Biochim Biophys Acta. 1969 Feb 18;174(2):435–448. doi: 10.1016/0005-2787(69)90274-3. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Schekman R., Kornberg A. A possible role for RNA polymerase in the initiation of M13 DNA synthesis. Proc Natl Acad Sci U S A. 1971 Nov;68(11):2826–2829. doi: 10.1073/pnas.68.11.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel N., Roizman B. Herpes vimplex virus: genome size and redundancy studied by renaturation kinetics. J Virol. 1971 Oct;8(4):591–593. doi: 10.1128/jvi.8.4.591-593.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Smith M. G. The chromosome of bacteriophage T5. I. Analysis of the single-stranded DNA fragments by agarose gel electrophoresis. J Mol Biol. 1972 Feb 14;63(3):383–395. doi: 10.1016/0022-2836(72)90435-4. [DOI] [PubMed] [Google Scholar]
- Horwitz M. S. Intermediates in the synthesis of type 2 adenovirus deoxyribonucleic acid. J Virol. 1971 Nov;8(5):675–683. doi: 10.1128/jvi.8.5.675-683.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E., Hoyer B., Bachenheimer S., Roizman B. Genetic relatedness of type 1 and type 2 herpes simplex viruses. J Virol. 1972 May;9(5):738–745. doi: 10.1128/jvi.9.5.738-745.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. F., Kieff E. D., Bachenheimer S. L., Roizman B., Spear P. G., Burmester B. R., Nazerian K. Size and composition of Marek's disease virus deoxyribonucleic acid. J Virol. 1971 Mar;7(3):289–294. doi: 10.1128/jvi.7.3.289-294.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonoyama M., Pagano J. S. Detection of Epstein-Barr viral genome in nonproductive cells. Nat New Biol. 1971 Sep 22;233(38):103–106. doi: 10.1038/newbio233103a0. [DOI] [PubMed] [Google Scholar]
- ROIZMAN B., ROANE P. R., Jr THE MULTIPLICATION OF HERPES SIMPLEX VIRUS. II. THE RELATION BETWEEN PROTEIN SYNTHESIS AND THE DUPLICATION OF VIRAL DNA IN INFECTED HEP-2 CELLS. Virology. 1964 Feb;22:262–269. doi: 10.1016/0042-6822(64)90011-x. [DOI] [PubMed] [Google Scholar]
- Roizman B., Spear P. G. Preparation of herpes simplex virus of high titer. J Virol. 1968 Jan;2(1):83–84. doi: 10.1128/jvi.2.1.83-84.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The herpesviruses--a biochemical definition of the group. Curr Top Microbiol Immunol. 1969;49:3–79. [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]



