Abstract
Despite growing reports on the biological action of nitric oxide (NO) as a function of NO payload, the validity of such work is often questionable due to the manner in which NO is measured and/or the solution composition in which NO is quantified. To highlight the importance of measurement technique for a given sample type, NO produced from a small molecule NO donor (N-diazeniumdiolated l-proline, PROLI/NO) and a NO-releasing xerogel film were quantified in a number of physiological buffers and fluids, cell culture media, and bacterial broth using the Griess assay, a chemiluminescence analyzer, and an amperometric NO sensor. Despite widespread use, the Griess assay proved to be inaccurate for measuring NO in many of the media tested. In contrast, the chemiluminescence analyzer provided superb kinetic information in most buffers, but was impractical for NO analysis in proteinaceous media. The electrochemical NO sensor enabled greater flexibility across the various media with potential for spatial resolution, albeit at lower than expected NO totals versus either the Griess assay or chemiluminescence. The results of this study highlight the importance of measurement strategy for accurate NO analysis and reporting NO-based biological activity.
Keywords: Nitric oxide, Griess assay, chemiluminescence, electrochemistry, biological media
Introduction
Nitric oxide (NO), an endogenous free radical produced by a collection of enzymes known as NO synthases (NOS), is a physiological mediator of the cardiovascular, immune and nervous systems.1 For example, NO produced in the vasculature by endothelial NOS serves as a vasodilator and blood pressure regulator.2, 3 The immune system produces NO at high concentrations via inducible NOS to serve as a signaling molecule4, 5 and potent antimicrobial agent.6 In the brain, NO produced by neuronal NOS functions as a neurotransmitter and is involved in memory formation.7 The physiological significance of NO has led to increased research on NO and NO-releasing scaffolds as potential therapeutics.8-14 Given that the location and concentration of NO governs its biological activity, the validity of analytical methods for measuring NO is critical.
While understanding NO's behavior in vivo is important, this task is far from trivial. Nitric oxide is reactive and short lived with a lifetime on the order of seconds to minutes.15 Additionally, the diffusion of NO is rapid, with diffusion coefficients approaching 3300 μm2 s-1 in physiological buffer.16, 17 Complicating matters further, the physiological concentration of NO spans six orders of magnitude (pM to μM).18 Sensitive analytical tools for measuring NO over wide concentration ranges are thus a necessity.19 To date, three analytical techniques account for the majority of NO measurements used in the literature to quantify NO.12, 20 First described in 1864,21 the Griess assay (“Griess”) allows for the estimation of total NO concentrations via nitrite analysis. Although inexpensive, readily available commercially, and useful for the determination of NO totals, the limit of detection for Griess is only ~0.5 μM.12, 22, 23 In contrast, chemiluminescence detection via a commercially available NO analyzer is a more costly method primarily due to the instrumentation, but the NO is measured more directly via reaction with ozone.24 Since NO is also redox active, electrochemical sensors have also been developed to quantify NO from sources as small as single cells made possible via sensor miniaturization.23
While each of these techniques has inherent advantages for measuring NO, it is important to consider the environment (i.e., solution or sample) in which the measurement takes place. Since NO readily reacts with free radical species (e.g., superoxide, thiyl radicals, and lipid peroxyls), metal-containing proteins (e.g., hemoglobin), and (to a lesser extent) thiols, the available (i.e., free) NO will vary depending on the sample medium.25-27 While recent reviews have focused on the methodologies for measuring NO in biological samples,23, 24, 28-35 an understanding of the influence of the technique and sample milieu on the validity of such measurements is lacking. Herein, we evaluate the accuracy of NO analysis with respect to measuring NO in physiological buffers and fluids, cell culture media, and bacterial broth using the Griess assay, a chemiluminescence NO analyzer, and an amperometric NO sensor.
Experimental Section
Reagents and Materials
(Heptadecafluoro-1,1,2,2-tetrahydrodecyl)trimethoxysilane (17FTMS), isobutyltrimethoxysilane (BTMOS), and N-(6-aminohexyl)aminopropyltrimethoxysilane (AHAP3) were purchased from Gelest (Morrisville, PA). Methyltrimethoxysilane (MTMOS) was purchased from Fluka (Buchs, Switzerland). Griess assay reagents were purchased from Promega (Madison, WI). Tryptic soy broth (TSB) and brain heart infusion (BHI) broth were purchased from BD Biosciences (San Jose, CA). Dulbecco's Modified Eagle Medium (DMEM), McCoy's Medium 5A Modified, fetal bovine serum (FBS), dipotassium ethylenediaminetetraacetic acid (K2EDTA), nicotinamide adenine dinucleotide phosphate (NADPH), and nitrate reductase (from Aspergillus niger) were purchased from Sigma (St. Louis, MO). Opti-MEM I (a reduced serum medium) was purchased from Life Technologies (Grand Isle, NY). Leibovitz medium (L-15; a carbon dioxide-free cell culture medium) was purchased from Lonza (Basel, Switzerland). Porcine blood was obtained from the Francis Owen Blood Research Laboratory (University of North Carolina; Chapel Hill, NC). Blood serum was obtained by collecting porcine blood without the addition of anticoagulant. After allowing the blood to clot, it was centrifuged at 2500 rpm for 15 min and the supernatant (i.e., serum) was removed. To obtain blood plasma, porcine blood was drawn into a tube with K2EDTA (~1.8 mg mL-1), mixed immediately, and then centrifuged at 2500 rpm for 15 min. After centrifugation, three layers were present (from top to bottom: plasma, leukocytes, and erythrocytes); the top layer was removed. A Millipore Milli-Q UV Gradient A10 System (Bedford, MA) was used to purify distilled water to a final resistivity of 18.2 MΩ·cm and a total organic content of ≤6 ppb. Nitrogen and argon gases were purchased from AirGas National Welders (Raleigh, NC). Nitric oxide gas was purchased from Praxair (Danbury, CT). Other solvents and chemicals were analytical-reagent grade and used as received.
Phosphate buffered saline (PBS; 10 mM, pH 7.4), physiosol36 (pH 6.0), artificial urine37 (pH 5.9) and artificial saliva38 (pH 6.7) were prepared as described in Supporting Information. A saturated NO solution (1.9 mM NO) was made by purging ~20 mL of PBS with argon for 30 min to remove oxygen, followed by NO gas for 20 min.
Synthesis of PROLI/NO
N-diazeniumdiolated l-proline (PROLI/NO) was prepared following a previously published protocol.39 Briefly, l-proline (2.05 g) was dissolved in a solution of methanol (25 mL) and sodium methoxide (2.00 g). The solution was then placed in a stainless steel reaction vessel and flushed with Ar six times (three in succession, three for 10 min each), then charged with NO at a pressure of 10 atm for 3 d with constant stirring. Six additional Ar purges were performed after 3 d. The solution was then precipitated by the addition of diethyl ether (150 mL) at –20 °C for 4 h. The white precipitate was isolated by vacuum filtration and dried in vacuo to yield PROLI/NO, which was stored at –20 °C until use. Ultraviolet spectra of a 14.9 μg mL-1 solution of the product (in 1.0 M sodium hydroxide) was acquired using a Thermo Scientific Evolution Array UV-visible spectrophotometer. The molecular weight of pure PROLI/NO was taken to be 251 g/mol.39
Synthesis of NO-releasing xerogels
Nitric oxide-releasing xerogels were fabricated using a procedure similar to one described previously.40 Briefly, 378 μL BTMOS was pre-hydrolyzed in 633 μL ethanol, 190 μL Milli-Q water and 31.7 μL 0.5 M hydrochloric acid for 1 h with agitation. Next, 255 μL AHAP3 was added and the resulting aminosilane/alkylsilane sol was mixed for an additional 1 h. Glass micro slides (Gold Seal, Portsmouth, NH) were cut into 9 × 25 mm sections and sonicated successively in water, ethanol, and acetone. The substrates were dried under a stream of nitrogen and then cleaned in an ultraviolet-ozone cleaner (BioForce Nanosciences; Ames, IA) for 20 min. Following, 40 μL aliquots of the sol were cast onto each substrate, dried for 1 h in ambient conditions and cured at 70 °C for 3 d. To convert the secondary amines in the matrix to N-diazeniumdiolate NO donors, the xerogels were placed in a stainless steel 500 mL Parr bomb, purged copiously with argon gas, and held under 10 atm NO gas for 3 d. After additional argon purging to remove unreacted NO, the films were sealed under nitrogen and stored at –20 °C until use.
Griess assay
To quantify NO via the Griess assay,41 50 μL of a 2 mg mL-1 solution of PROLI/NO in 100 mM sodium hydroxide (NaOH) was added to 15 mL of desired media and incubated at room temperature for at least 24 h. Aliquots (50 μL) of this sample were added to a sulfanilamide solution (50 μL) and incubated in the dark at room temperature for 5 min. Naphthylethylenediamine (50 μL) was added to the mixture to form a colorimetric product with concomitant absorbance measured in each well at 540 nm using a LabSystems MultiSkan RC microplate reader (Helsinki, Finland). Sodium nitrite standards were used to normalize the assay reactivity and associated absorbance.
For analysis of blood constituents (i.e., plasma and serum), NADPH (25 μL) and nitrate reductase (2 μL) were added to the samples and allowed to incubate for at least 30 min prior to the addition of the Griess reagents. Headspace studies using Griess were conducted in the same manner, but the volume of media added to the 20 mL scintillation vial was varied (10, 15, and 20 mL). Concentration dependence studies were conducted by varying the concentration of the stock PROLI/NO solution and injecting aliquots into 15 mL media to yield final PROLI/NO concentrations of 0.67, 6.7, and 67 μg mL-1.
Chemiluminescence detection
Real-time NO release was monitored using a Sievers 280 Chemiluminescent NO Analyzer (Boulder, CO). The instrument was calibrated with a 25.6 ppm gas standard (balance N2) and an atmospheric sample that had been passed through a NO zero filter. Samples were prepared by adding 10 μL of a 2 mg mL-1 solution of PROLI/NO in 100 mM NaOH to 30 mL of desired media that had been degassed in a sample vessel for at least 20 min. Nitric oxide produced in the vessel was carried to the NO analyzer by a stream of nitrogen gas bubbled into the solution (80 mL min-1) and across the headspace of the flask (120 mL min-1), equivalent to 200 mL min-1 flow to the instrument. 1), equivalent to 200 mL min-1 flow to the instrument. Concentration dependence studies were conducted in the same manner, but the concentration of the stock PROLI/NO solutions was varied to yield final concentrations of 0.167, 1.67, and 16.7 μg mL-1 in 30 mL. Release of evolved NO from 40% AHAP (balance BTMOS) xerogels was measured similarly following placement of the substrates directly into deoxygenated PBS using the vessels described above.
Electrochemical detection
Inlaid 2 mm diameter polycrystalline platinum (Pt) disk electrodes sealed in Kel-F (CH Instruments; Austin, TX) were mechanically polished with successively finer grades of deagglomerated alumina slurries down to 0.05 μm particles (Buehler; Lake Bluff, IL). Residual alumina was removed using an ultrasonic cleaner (in water) and the electrodes were dried with nitrogen. A fluorinated NO-selective xerogel membrane was applied to the electrode as previously described to minimize response to common interferents.42, 43 Briefly, a silane solution was prepared by mixing MTMOS (60 μL) in ethanol (300 μL). To this solution, 17FTMS (15 μL) was added, resulting in a 20% v/v fluoroalkoxysilane (balance MTMOS) mixture. The silane solution was subsequently mixed with water (80 μL) and 0.5 M HCl (5 μL) for 1 h. The resulting sol (1.5 μL) was cast onto Pt working electrodes and allowed to cure for 24 h under ambient conditions. To evaluate the analytical performance of the NO sensors, amperometric measurements were performed using a CH Instruments 730B bipotentiostat (Austin, TX). The electrode assembly (3-electrode configuration) consisted of the xerogel-modified Pt working electrode, a Pt-coiled counter electrode, and a Ag/AgCl reference electrode (3.0 M KCl; CH Instruments). Electrooxidation currents were recorded at an applied potential of +700 mV (vs. Ag/AgCl). When measuring the NO in the various media using PROLI/NO as the NO source, a 2 mg mL-1 solution of the NO donor (i.e., PROLI/NO) was added to a constantly stirring 30 mL solution to achieve a final concentration of 0.1 mg mL-1. Of note, the larger volume of media was necessary to accommodate the working, reference, and counter electrodes in the flask. Concentration dependence studies were conducted in the same manner, but the concentration of the stock PROLI/NO solutions was varied to yield final PROLI/NO concentrations of 0.67, 6.7, and 67 μg mL-1. Xerogel film studies were conducted by placing the film directly below a working electrode and using a micromanipulator to adjust the electrode distance from the NO-releasing surface.
Results and Discussion
Diverse biological media were chosen for these experiments to properly represent environments in which NO measurements are relevant (e.g., in vivo, cell/tissue culture, bacteria culture). Solutions included simple physiological buffers (PBS and physiosol), simulated biological fluids (saliva, urine, and wound fluid), cell culture media (DMEM, McCoy's, L-15, and Opti-MEM), bacterial broth (TSB and BHI), whole blood, plasma, and serum. The salt, amino acid, protein, and vitamin content of these media vary significantly (Tables 1–4 in the Supporting Information) and thus were predicted to impact the results.
NO determination via Griess
The Griess assay allows for indirect measurement of NO by quantifying nitrite, NO's reaction product in oxygenated media (eqs 1–3):
| (1) |
| (2) |
| (3) |
Once formed, the nitrite is reacted with the Griess reagents, sulfanilamide and N-(1-naphyl)ethylenediame, to produce an azo dye with an absorbance maximum at 540 nm. To accurately quantify NO, calibration curves are constructed using a standard nitrite solution in the sample medium.41
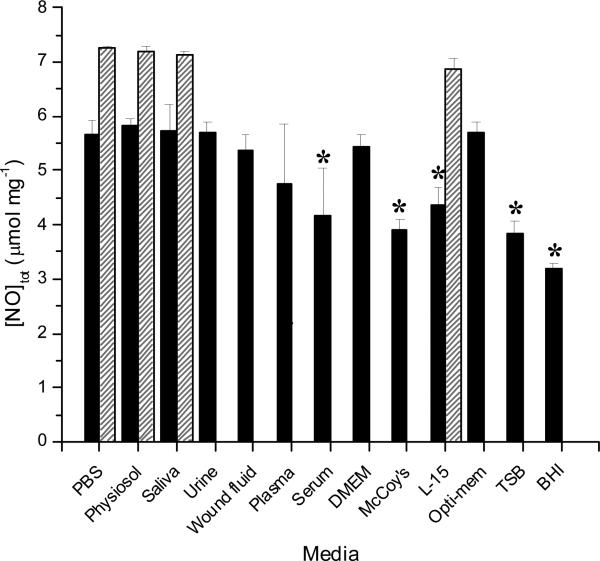
In theory, one mole of PROLI/NO decomposes to release two moles of NO, producing 7.9 μmol NO mg-1 PROLI/NO total.39 Following synthesis, PROLI/NO was characterized using UV/VIS spectroscopy (Supporting Information, Figure S1). An observed λmax at 252 nm confirmed N-diazeniumdiolate formation from the L-proline precursor, consistent with prior reports.39 A 14.9 μg ml-1 solution was prepared in 1.0 M sodium hydroxide and the molar absorptivity coefficient (ε) reported previously by Saavedra et al at 252 nm (8.4 mM-1 cm-1)2 was used to determine a PROLI/NO concentration of 55.7 μM. Assuming total purity, the concentration of this solution would be 59.5 μM, implying a relative purity of 93.5%. This value is also supported by the chemiluminescent NO release totals in buffers lacking scavenging components (i.e., PBS). Estimated NO release totals derived from the total nitrite present in each type of media as determined using the Griess assay are given in Figure 1 along with totals derived from chemiluminescence detection. The total NO measured in PBS via Griess was only 5.67 ± 0.27 μmol mg-1, roughly 30% lower than the theoretical amount and that detected via the chemiluminescence method (7.24 ± 0.04 μmol mg-1). Of note, care was taken to add fresh PROLI/NO solution to the media quickly. In addition, multiple samples were analyzed to obtain standard deviations. Similar depressed NO levels were observed in all of the media types tested using Griess. We attribute the deviation between Griess and theoretical to lost (e.g., escaped) NO gas (from solution) that subsequently was not accounted for in the total nitrite levels. Indeed, when the headspace in a 20 mL scintillation vial was reduced by adding 20 mL of PBS versus 15 mL, the total nitrite detected was increased by 44 ± 12%. Conversely, when the total buffer volume was decreased to 10 mL, the total nitrite recovered was reduced by 32 ± 21% (data not shown). The most significant deviations of measured vs. theoretical NO totals were observed in bacterial broth (i.e., TSB and BHI), McCoy's and L-15 cell culture media, and plasma and serum. These results were somewhat expected as both cell culture and bacterial growth media consist of complex mixtures of amino acids, proteins, sugars, and vitamins of various concentrations (Tables SI21–4 in the Supporting Information). Proteins and other additives have been shown to interfere with the Griess assay previously,44-46 acting as either positive or negative interferents. For example, positive interferents such as NOS and hemoglobin absorb light around 540 nm. Other interferents including cysteine, tyrosine, ascorbate, and NADPH react with nitrite to negatively skew NO totals. Although interfering proteins could be removed by chemical precipitation or ultrafiltration,28 such solution conditioning is tedious and would be at the expense of biological relevance. As anticipated, NO was not quantifiable in whole blood via Griess due to the opacity of blood and presence of interfering proteins.45 Additionally, NO and nitrite are readily oxidized to nitrate in whole blood by oxyhemoglobin.47, 48 Nevertheless, nitrite levels were measureable in plasma and serum samples extracted from the blood with the caveat that nitrate reductase and NADPH were used to revert nitrate that formed back to nitrite. Despite this, NO lost by its conversion to nitrosylheme via the reaction with deoxyhemoglobin could not be recovered using this assay. In both plasma and serum, the NO totals were >15% lower (4.75 ± 1.1 and 4.18 ± 0.85 μmol mg-1, respectively) than that in PBS, further indicating measurement inaccuracies in protein-rich samples. Of note, nitrite recovery is highly variable and dependent on enzyme activity.28, 33, 49
Figure 1.
Total NO released from PROLI/NO in several types of biological media determined via Griess assay (solid) and chemiluminescence (striped). Theoretical NO release from PROLI/NO is 7.9 4.82 ± 0.21 μmol mg-1. *Denotes a significant difference (p <0.05) relative to PBS.
The accurate detection of NO via the Griess assay is also concentration dependent in certain types of media. While no significant differences were observed for non-proteinaceous media, detection in the more complex media (i.e., cell culture media and bacterial broth) varied significantly depending on the amount of PROLI/NO added to solution (Figure S2 in Supporting Information). For example, while the addition of 0.67 μg mL-1 PROLI/NO in DMEM yielded nitrite totals that were only 48 ± 9% of those achieved at the highest concentration of PROLI/NO, a 10X more concentrated sample yielded nitrite amounts that nearly matched those of the highest concentration (97 ± 1%). The same trend held true for the bacterial broth TSB. This can be attributed to the effect of scavenging, as the concentration of such scavengers in a given media is constant, so adding more NO allows this effect to be overcome to some extent.
Given these results, the use of Griess for quantifying NO in most biological media leads to questionable results. Nevertheless, the potential for high throughput analysis via microtiter plates and readers makes Griess useful for initial screening of NO-release materials in less complex media (e.g., PBS) so long as such data is supplemented with more rigorous analysis in relevant milieu prior to drawing conclusions regarding clinical utility. In addition, some kinetic data is obtainable using Griess by taking aliquots from a sample solution at set periods or moving bulk (i.e., larger) substrates (e.g., polymer-coated slides) in and out of a soak solution, and subsequently sampling those solutions. For example, AHAP films soaked in select media yielded totals similar to those obtained under the same conditions using chemiluminescent detection (Table S5 in Supporting Information) after one week. Although the low levels of NO release were undetectable with chemiluminescence after 2 weeks, the accumulation of nitrite was still measureable using the Griess assay and indicated a continued release of NO from the substrates.
Chemiluminescence NO analyzer
To facilitate real-time measurement of NO with greater time resolution and lower limits of detection relative to the Griess assay, chemiluminescence-based instrumentation is a standard tool for solution-based analysis.20 Using measurement principles from its original use (i.e., respiratory applications), NO is carried from the sample (e.g., exhaled breath, solution vessel, etc.) by an inert gas (e.g., nitrogen, argon) to an instrument where the NO is reacted with ozone to form an excited-state nitrogen dioxide species that emits a photon upon relaxation back to the ground state. This light, ranging from 600–875 nm, is detected by a photomultiplier tube and is proportional to the NO present in the sample. The reaction of NO with ozone is both specific for and sensitive to NO with a detection limit approaching 0.5 ppb (0.66 pM in 100 mL) in PBS.12 Detection limits in solutions other than PBS have not been reported previously.
Analogous to the Griess experiments above, samples of PROLI/NO (0.67 μg mL-1) were introduced into a solution, but analyzed using chemiluminescence detection to quantify NO. Unexpectedly, the media compatible with this method was limited to solutions that did not foam as a result of purging with nitrogen gas (necessary to carry the NO to the instrument and eliminate auto-oxidation to nitrite by removing oxygen). Foaming due to nitrogen bubbling was most prominent for culture media, bacterial broth solutions, and blood (i.e., whole, plasma, serum) due to their high protein content.50 Nitric oxide release totals via chemiluminescence for the compatible media are also shown in Figure 1, allowing for direct comparison of total NO detected with Griess as a function of media type. Of note, the 7.24 ± 0.04 μmol NO mg-1 measured in PBS via chemiluminescence was near the predicted NO payload for PROLI/NO based on the theoretical total and calculated relative purity. The NO totals measured in low protein content media using chemiluminescence were not significantly different from theoretical NO payloads, indicating high accuracy of this technique for measuring total NO. While L-15 cell culture media contains biomolecular components that may scavenge NO, such reactivity was limited as the NO was rapidly moved out of the sample vessel to the instrument by the carrier gas after formation.
A key advantage of chemiluminescence NO detection is the ability to obtain information about NO release from NO-donor systems. In addition to total NO amount, the maximum NO flux, time to the maximum release (tmax) and NO-release half-life (t1/2) may be determined for a particular NO source (e.g., small molecule, macromolecular scaffold). These parameters were extracted from the NO-release profile of PROLI/NO in PBS, physiosol, artificial saliva, and L-15 (Table 1). Little variation in the NO-release kinetics for PROLI/NO in these media was noted using chemiluminescence, with two exceptions. In artificial saliva, the maximum NO release was significantly lower than that in PBS (73,360 ± 1,100 versus 86,200 ± 3,700 pmol s-1 mg-1, respectively). This behavior may be attributed to greater scavenging of NO by components in saliva relative to PBS. Likewise, the half2life of PROLI/NO in L-15 cell culture media was smaller (reduced by ~8 s) than that measured in PBS. These variances are not surprising given the complex and varying nature of biological media (Supporting Information).
Table 1.
Kinetic parameters of NO release from PROLI/NO in PBS, physiosol, L-15, and artificial saliva.
| tmax (s) | max NO release (pmol s-1 mg-1) | t1/2 (s) | |
|---|---|---|---|
| PBS | 42 ± 1.9 | 86,200 ± 3,700 | 65 ± 3.0 |
| Physiosol | 40 ± 1.7 | 81,200 ± 36 | 64 ± 1.1 |
| L-15 | 40 ± 1.2 | 96,600 ± 3,100 | 57 ± 0.7* |
| Saliva | 47 ± 5.2 | 73,400 ± 1,100* | 74 ± 3.5 |
Denotes a significant difference (p <0.05) relative to PBS.
Analogous to the Griess assay, the accurate measurement of NO totals from PROLI/NO was dependent on the concentration of the NO donor introduced into the sample flask (Figure S1 in Supporting Information). As expected, no difference was observed between concentrations in the buffer solution (PBS) or the simulated biological solution tested (saliva), as these are simply composed of various salts. However, a slight decrease of 9 ± 1% was observed for the lowest concentration relative to the 100X more concentrated sample for the cell culture media tested (L-15 in this case, as DMEM foams too significantly to be used with this technique).
Despite the advantage of kinetic NO-release parameters, the required purging of the sample vessel remains problematic for measuring NO via chemiluminescence in biological samples due to undesirable frothing. Furthermore, the reaction vessel should be free of oxygen to minimize any side reactions that would decrease NO entering the instrument.12, 24 Unfortunately, mammalian cells and tissues that produce NO require an aerobic environment for survival, thus excluding chemiluminescence for measuring NO production from cells and/or tissue in their native environment for extended periods.
Amperometric NO sensor
In contrast to Griess and chemiluminescence, electrochemical sensors allow for the measurement of NO in almost any biological setting down to the single cell level.51-53 The advantages of electrochemical detection of NO include superior spatial and temporal resolutions, excellent limits of detection and dynamic ranges, and the ability to miniaturize the sensor. Both the selectivity and sensitivity of the sensor are tunable by changing the applied potential to the working electrode and/or modifying the working electrode with a catalyst and/or permselective membrane.12, 54 Due to these inherent advantages, amperometric NO sensors have been used extensively to examine both endogenous11, 52, 53, 55-58 and exogenous59-61 NO production. Nitric oxide measured using an amperometric sensor results in current proportional to the NO concentration upon the oxidation of NO via a three-electron process (eqs 4–6):62, 63
| (4) |
| (5) |
| (6) |
Depending on the thickness of the permselective membrane, the response to NO can be fast (subsecond) with limits of detection approaching 83 pM.51 For this study, a NO-selective fluorosilane-based xerogel-coated platinum working electrode42 was used to measure the electrooxidation of NO as current at an applied potential of +700 mV vs. Ag/AgCl reference electrode in a well-stirred solution. Total NO release was determined by integrating the current vs. time response for each sample, since the charge at the electrode surface is proportional to the moles of analyte oxidized.64 More accurate predictions of NO totals in the bulk solution were then obtained by relating these values to a calibration curve created using aliquots of a saturated NO solution (1.9 mM) in PBS. As shown in Figure 2, the integrated totals yielded NO concentrations significantly lower than the theoretical amount of NO produced from PROLI/NO regardless of sample medium. These results were somewhat expected since the electrode is only able to oxidize (i.e., measure) a portion of the NO in its vicinity due to probe geometry despite vigorous stirring. While electrochemical sensors are clearly less suitable for characterization of NO donors or drugs that produce/consume NO, useful information may be deduced by comparing the relative NO totals in different media and when quantifying more localized NO generation (e.g., from a surface or single cell). The solutions resulting in the greatest total NO detected when oxygenated were PBS and physiosol (~1.07 ± 0.09 μmol mg-1), not surprising due to the similarities of these solutions; only their salt concentration and pH differ slightly. The NO totals in oxygenated L-15 and Opti-MEM cell culture media were also in line with that in PBS despite significant protein content. In assessing trends, we noted that L-15 and Opti-MEM solutions do not contain fetal bovine serum (FBS) as do the DMEM and McCoy's solutions, the latter resulting in NO scavenging via sulfhydryl-containing proteins (e.g., albumin, fibrinogen, macroglobulins, glycoproteins).65, 66 The amount of NO measured in simulated wound fluid (10% v/v FBS in water) with the electrochemical sensor was also significantly lower (0.40 ± 0.05 μmol mg-1). In whole blood, undoubtedly the most complex of all the media tested, the measured NO total was the lowest (12.0 ± 3.2 × 10-4 μmol mg-1) due to the anticipated reaction of NO with blood proteins including oxyhemoglobin.67 Despite the lower totals in whole blood, NO concentrations were still measurable using the amperometric sensor, as was construction of a calibration curve (in whole blood). Nitric oxide totals in plasma and serum measured greater than in blood (0.27 ± 0.09 and 0.11 ± 0.04 μmol mg-1, respectively) as would be predicted due to the reduced hemoglobin concentration in these samples.
Figure 2.
Nitric oxide totals measured using an amperometric NO sensor and PROLI/NO as the NO source as a function of biological media that with (black) and without (gray) oxygen present. Theoretical NO release from PROLI/NO is 7.9 μmol mg-1. *Significant difference (p <0.05) relative to PBS. #Significant difference (p <0.05) relative to deoxygenated solution.
While testing of deoxygenated media was limited to biological solutions that did not foam upon purging with nitrogen, the measured NO levels were greater as would expected upon eliminating NO's propensity to react with oxygen.68, 69 In blood and other media containing high protein content and cells, fouling of the electrode resulting from protein/cell adsorption may also impact the analytical accuracy of the measurement for electrochemical sensors.70-73 In the short experiments described herein, such fouling only accounted for a 3–5% decrease in sensitivity (data not shown), and therefore did not contribute significantly to the differences observed in the NO totals.
Analogous to the Griess assay and chemiluminescent detection, the accurate electrochemical detection of NO was also dependent on the concentration of the NO donor. However, a concentration-dependent effect was observed in all media (Figure S1 in Supporting Information). We attribute this effect to the finite surface area of the working electrode whereby greater concentrations of the NO donor readily alter the local NO concentrations.
As the amount of NO donor near the electrode surface influences the concomitant NO measured, the electrode distance from an NO source should have a similar effect on NO measurement. To examine this methodically, a NO-selective electrode was placed 25, 50, and 100 μm above a NO-releasing surface (xerogel polymer cast on glass) under ambient conditions in PBS. The amperometric signal (current) obtained when the working electrode was placed 50 μm above the surface at 95 min was ~45% that recorded at 25 μm (Figure S3 in Supporting Information). Similarly, the signal was reduced by ~73% at 100 μm above the surface (relative to the 25 μm working electrode placement). While such NO source/electrode distance-dependence will clearly impact the analytical accuracy of a measurement, it may be useful for measuring the diffusion of NO from surfaces or cells as a function of biological medium.
Despite the lower NO totals quantified electrochemically, the use of amperometric sensors remains advantageous due to unparalleled spatial and temporal resolutions.74 As such, electrochemistry remains the method of choice for the characterization of NO generation from cells11, 52, 53, 55-58 and confined surface locations.75 Unfortunately, both commercial and home-built sensors are generally not robust, requiring frequent (e.g., after analysis of 1 or 2 samples) calibration and performance testing to maintain data integrity.
Conclusions
The analytical measurement of NO is clearly complex,12 requiring careful scrutiny of the analysis method with respect to performance. While prior work has described the utility of the Griess assay, chemiluminescence analyzers, and electrochemical sensors for measuring NO in solution, performance discrepancies as a function of sample type have been disregarded. The data reported here clearly demonstrate the significant variations between analysis technique and sample composition. As such, the caveats of the analytical method employed must be carefully considered with respect to sample, desired data (e.g., NO totals, flux, kinetics and bioavailability), and result integrity.
Supplementary Material
ACKNOWLEDGMENT
The authors acknowledge research support from the National Institutes of Health (NIH Grant AI097539) and the Francis Owen Blood Lab for porcine blood.
Footnotes
Supporting Information Available: Methods for preparing biological solutions (artificial saliva, artificial urine, and physiosol), and concentrations of key components in the solutions. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Epstein FH, Moncada S, Higgs A. N. Engl. J. Med. 1993;329:2002–2012. doi: 10.1056/NEJM199312303292706. [DOI] [PubMed] [Google Scholar]
- 2.Ignarro LJ, Buga GM, Wood KS, Byrns RE, Chaudhuri G. Proc. Natl. Acad. Sci. U. S. A. 1987;84:9265–9269. doi: 10.1073/pnas.84.24.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moncada S, Radomski MW, Palmer RMJ. Biochem. Pharmacol. 1988;37:2495–2501. doi: 10.1016/0006-2952(88)90236-5. [DOI] [PubMed] [Google Scholar]
- 4.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. Free Radical Biol. Med. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snyder SH, Bredt DS. Sci. Am. 1992;266:74–77. doi: 10.1038/scientificamerican0592-68. [DOI] [PubMed] [Google Scholar]
- 6.Fang FC. J. Clin. Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyder SH. Curr. Opin. Neurobiol. 1992;2:323–327. doi: 10.1016/0959-4388(92)90123-3. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter AW, Schoenfisch MH. Chem. Soc. Rev. 2012;41:3742–3752. doi: 10.1039/c2cs15273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riccio DA, Schoenfisch MH. Chem. Soc. Rev. 2012;41:3731–3741. doi: 10.1039/c2cs15272j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nichols SP, Storm WL, Koh A, Schoenfisch MH. Adv. Drug Del. Rev. 2012 doi: 10.1016/j.addr.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel BA, Galligan JJ, Swain GM, Bian X. Neurogastroenterol. Motil. 2008;20:1243–1250. doi: 10.1111/j.1365-2982.2008.01177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hetrick EM, Schoenfisch MH. Annu. Rev. Anal. Chem. 2009;2:409–433. doi: 10.1146/annurev-anchem-060908-155146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damodaran VB, Joslin JM, Wold KA, Lantvit SM, Reynolds MM. J. Mater. Chem. 2012;22:5990–6001. [Google Scholar]
- 14.Seabra AB, Martins D, Simões MMSG, Da Silva R, Brocchi M, De Oliveira MG. Artif. Organs. 2010;34:E204–E214. doi: 10.1111/j.1525-1594.2010.00998.x. [DOI] [PubMed] [Google Scholar]
- 15.Lowenstein CJ, Dinerman JL, Snyder SH. Ann. Intern. Med. 1994;120:227–237. doi: 10.7326/0003-4819-120-3-199402010-00009. [DOI] [PubMed] [Google Scholar]
- 16.Lancaster JR. Nitric Oxide. 1997;1:18–30. doi: 10.1006/niox.1996.0112. [DOI] [PubMed] [Google Scholar]
- 17.Malinski T, Taha Z, Grunfeld S, Patton S, Kapturczak M, Tomboulian P. Biochem. Biophys. Res. Commun. 1993;193:1076–1082. doi: 10.1006/bbrc.1993.1735. [DOI] [PubMed] [Google Scholar]
- 18.Wink DA, Mitchell JB. Free Radical Biol. Med. 1998;25:434–456. doi: 10.1016/s0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 19.Moncada S, Palmer RM, Higgs EA. Pharmacol. Rev. 1991;43:109–142. [PubMed] [Google Scholar]
- 20.Coneski PN, Schoenfisch MH. Chem. Soc. Rev. 2012;41:3753–3758. doi: 10.1039/c2cs15271a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griess P. Philos. Trans. R. Soc. London. 1864;154:667–731. [Google Scholar]
- 22.Sun J, Zhang XJ, Broderick M, Fein H. Sensors. 2003;3:276–284. [Google Scholar]
- 23.Bryan NS, Grisham MB. Free Radical Biol. Med. 2007;43:645–657. doi: 10.1016/j.freeradbiomed.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bates JN. Neuroprotocols. 1992;1:141–149. [Google Scholar]
- 25.Williams RJP. Chem. Soc. Rev. 1996;25:77–83. [Google Scholar]
- 26.Möller MN, Li Q, Lancaster JR, Denicola A. IUBMB Life. 2007;59:243–248. doi: 10.1080/15216540701311147. [DOI] [PubMed] [Google Scholar]
- 27.Hall CN, Garthwaite J. Nitric Oxide. 2009;21:92–103. doi: 10.1016/j.niox.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsikas D. J. Chromatogr. B. 2007;851:51–70. doi: 10.1016/j.jchromb.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 29.Taha ZH. Talanta. 2003;61:3–10. doi: 10.1016/S0039-9140(03)00354-0. [DOI] [PubMed] [Google Scholar]
- 30.Allen BW, Liu J, Piantadosi CA, Lester P, Enrique C. Methods Enzymol. Vol. 396. Academic Press; 2005. pp. 68–77. [DOI] [PubMed] [Google Scholar]
- 31.Bedioui F, Villeneuve N. Electroanalysis. 2003;15:5–18. [Google Scholar]
- 32.Yao D, Vlessidis AG, Evmiridis NP. Microchim. Acta. 2004;147:1–20. [Google Scholar]
- 33.Giustarini D, Rossi R, Milzani A, Dalle-Donne I. Nitric Oxide. 2008;440:361–380. doi: 10.1016/S0076-6879(07)00823-3. [DOI] [PubMed] [Google Scholar]
- 34.Tsikas D. Free Radical Res. 2005;39:797–815. doi: 10.1080/10715760500053651. [DOI] [PubMed] [Google Scholar]
- 35.Guevara I, Iwanejko J, Dembinska-Kiec A, Pankiewicz J, Wanat A, Anna P, Golabek I, Bartus S, Malczewska-Malec M, Szczudlik A. Clin. Chim. Acta. 1998;274:177–188. doi: 10.1016/s0009-8981(98)00060-6. [DOI] [PubMed] [Google Scholar]
- 36.NIH, editor. DailyMed. U.S. National Library of Medicine; Bethesda: 2006. [Google Scholar]
- 37.Kark RM, Lawrence JR, Pollack VE, Pirani CL, Muehrcke RC, Silva H. A Primer of Urinalysis. 2nd ed. Hoeber Medical Division, Harper & Row; New York: 1964. [Google Scholar]
- 38.Arvidson K, Johansson EG. Eur. J. Oral Sci. 1985;93:467–473. [Google Scholar]
- 39.Saavedra JE, Southan GJ, Davies KM, Lundell A, Markou C, Hanson SR, Adrie C, Hurford WE, Zapol WM, Keefer LK. J. Med. Chem. 1996;39:4361–4365. doi: 10.1021/jm960616s. [DOI] [PubMed] [Google Scholar]
- 40.Marxer SM, Rothrock AR, Nablo BJ, Robbins ME, Schoenfisch MH. Chem. Mater. 2003;15:4193–4199. [Google Scholar]
- 41.Schmidt H, Kelm M. Feelisch M, Stamler JS, editors. Methods in nitric oxide research. 1996. pp. 491–497.
- 42.Shin JH, Privett BJ, Kita JM, Wightman RM, Schoenfisch MH. Anal. Chem. 2008;80:6850–6859. doi: 10.1021/ac800185x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin JH, Weinman SW, Schoenfisch MH. Anal. Chem. 2005;77:3494–3501. doi: 10.1021/ac048153i. [DOI] [PubMed] [Google Scholar]
- 44.Indika PN, Bayachou M. Anal. Bioanal. Chem. 2004;379:1055–1061. doi: 10.1007/s00216-004-2674-2. [DOI] [PubMed] [Google Scholar]
- 45.Tsikas D, Caidahl K. J. Chromatogr. B. 2005;814:1–9. doi: 10.1016/j.jchromb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Fox JB. Anal. Chem. 1979;51:1493–1502. [Google Scholar]
- 47.Moshage H, Kok B, Huizenga JR, Jansen PL. Clin. Chem. 1995;41:892–896. [PubMed] [Google Scholar]
- 48.Miranda KM, Espey MG, Wink DA. Nitric Oxide. 2001;5:62–71. doi: 10.1006/niox.2000.0319. [DOI] [PubMed] [Google Scholar]
- 49.Tsikas D, Gutzki F-M, Rossa S, Bauer H, Neumann C, Dockendorff K, Sandmann J, Frolich JC. Anal. Biochem. 1997;244:208–220. doi: 10.1006/abio.1996.9880. [DOI] [PubMed] [Google Scholar]
- 50.Kitabatake N, Doi E. J. Food Sci. 1982;47:1218–1221. [Google Scholar]
- 51.Privett BJ, Shin JH, Schoenfisch MH. Chem. Soc. Rev. 2010;39:1925–1935. doi: 10.1039/b701906h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amatore C, Arbault S, Bouton C, Drapier J-C, Ghandour H, Koh ACW. ChemBioChem. 2008;9:1472–1480. doi: 10.1002/cbic.200700746. [DOI] [PubMed] [Google Scholar]
- 53.Cha W, Tung Y-C, Meyerhoff ME, Takayama S. Anal. Chem. 2010;82:3300–3305. doi: 10.1021/ac100085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davies IR, Zhang X, Robert KP. Methods Enzymol. Vol. 436. Academic Press; 2008. pp. 63–95. [DOI] [PubMed] [Google Scholar]
- 55.Lee Y, Yang J, Rudich SM, Schreiner RJ, Meyerhoff ME. Anal. Chem. 2004;76:545–551. doi: 10.1021/ac035065+. [DOI] [PubMed] [Google Scholar]
- 56.Patel BA, Arundell M, Parker KH, Yeoman MS, O'Hare D. Anal. Chem. 2006;78:7643–7648. doi: 10.1021/ac060863w. [DOI] [PubMed] [Google Scholar]
- 57.Amatore C, Arbault S, Bouton C, Coffi K, Drapier J-C, Ghandour H, Tong Y. ChemBioChem. 2006;7:653–661. doi: 10.1002/cbic.200500359. [DOI] [PubMed] [Google Scholar]
- 58.Amatore C, Arbault S. p., Koh ACW. Anal. Chem. 2010;82:1411–1419. doi: 10.1021/ac902486x. [DOI] [PubMed] [Google Scholar]
- 59.Riccio DA, Nutz ST, Schoenfisch MH. Anal. Chem. 2012;84:851–856. doi: 10.1021/ac2031805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X, Cardosa L, Broderick M, Fein H, Davies IR. Electroanalysis. 2000;12:425–428. [Google Scholar]
- 61.Hou Y, Wu X, Xie W, Braunschweiger PG, Wang PG. Tetrahedron Lett. 2001;42:825–829. [Google Scholar]
- 62.Malinski T, Bailey F, Zhang ZG, Chopp M. J. Cereb. Blood Flow Metab. 1993;13:355–358. doi: 10.1038/jcbfm.1993.48. [DOI] [PubMed] [Google Scholar]
- 63.Trevin S, Bedioui F, Devynck F. Talanta. 1996;43:303–311. doi: 10.1016/0039-9140(95)01752-6. [DOI] [PubMed] [Google Scholar]
- 64.Bard AJ, Faulkner LR. Electrochemical Methods: Fundamentals and Applications. 2nd ed. John Wiley and Sons, Inc.; Hoboken: 2001. [Google Scholar]
- 65.Elsadek B, Kratz F. J. Controlled Release. 2012;157:4–28. doi: 10.1016/j.jconrel.2011.09.069. [DOI] [PubMed] [Google Scholar]
- 66.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. Proc. Natl. Acad. Sci. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim-Shapiro DB, Schechter AN, Gladwin MT. Arterioscler. Thromb. Vasc. Biol. 2006;26:697–705. doi: 10.1161/01.ATV.0000204350.44226.9a. [DOI] [PubMed] [Google Scholar]
- 68.Solc M. Nature. 1966;209:706–706. [Google Scholar]
- 69.Lewis RS, Deen WM. Chem. Res. Toxicol. 1994;7:568–574. doi: 10.1021/tx00040a013. [DOI] [PubMed] [Google Scholar]
- 70.Wisniewski N, Moussy F, Reichert WM. Fresenius J. Anal. Chem. 2000;366:611–621. doi: 10.1007/s002160051556. [DOI] [PubMed] [Google Scholar]
- 71.Wisniewski N, Klitzman B, Miller B, Reichert WM. J. Biomed. Mater. Res. 2001;57:513–521. doi: 10.1002/1097-4636(20011215)57:4<513::aid-jbm1197>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 72.Frost M, Meyerhoff ME. Anal. Chem. 2006;78:7370–7377. doi: 10.1021/ac069475k. [DOI] [PubMed] [Google Scholar]
- 73.Voskerician G, Anderson J. Wiley Encyclopedia of Biomedical Engineering. John Wiley & Sons, Inc.; 2006. [Google Scholar]
- 74.Wightman RM. Science. 2006;311:1570–1574. doi: 10.1126/science.1120027. [DOI] [PubMed] [Google Scholar]
- 75.Oh BK, Robbins ME, Schoenfisch MH. Analyst. 2006;131:48–54. doi: 10.1039/b507981k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.