Abstract
Acetate kinases (ACKs) are members of the acetate and sugar kinase/hsp70/actin (ASKHA) superfamily and catalyze the reversible phosphorylation of acetate, with ADP/ATP the most common phosphoryl acceptor/donor. While prokaryotic ACKs have been the subject of extensive biochemical and structural characterization, there is a comparative paucity of information on eukaryotic ACKs, and prior to this report, no structure of an ACK of eukaryotic origin was available. We determined the structures of ACKs from the eukaryotic pathogens Entamoeba histolytica and Cryptococcus neoformans. Each active site is located at an interdomain interface, and the acetate and phosphate binding pockets display sequence and structural conservation with their prokaryotic counterparts. Interestingly, the E. histolytica ACK has previously been shown to be pyrophosphate (PPi)-dependent, and is the first ACK demonstrated to have this property. Examination of its structure demonstrates how subtle amino acid substitutions within the active site have converted cosubstrate specificity from ATP to PPi while retaining a similar backbone conformation. Differences in the angle between domains surrounding the active site suggest that interdomain movement may accompany catalysis. Taken together, these structures are consistent with the eukaryotic ACKs following a similar reaction mechanism as is proposed for the prokaryotic homologs.
Keywords: Acetate kinase, PPi-dependent kinase, ASKHA superfamily
1. Introduction to acetate kinases
Enzyme-catalyzed phosphoryl transfer reactions are important for a range of biological activities. Acetate kinases (ACKs) transfer a phosphoryl group to and from acetate, thus promoting the interconversion of acetate and acetyl phosphate. With this transformation, ACKs play a role in multiple, distinct bioenergetic pathways (Cozzone, 1998; Ingram-Smith et al., 2006; Thauer et al., 2008). For example, in fermentative bacteria, the ACK-catalyzed dephosphorylation of acetyl phosphate has been demonstrated to be essential for the ACK- phosphotransacetylase (ACK-PTA) bioenergetic pathway, which utilizes energy stored in acetyl-CoA (Cozzone, 1998). Similarly, ACK-dependent dephosphorylation of acetyl phosphate facilitates ATP synthesis via the pentose phosphoketolase pathway in fungi (Ingram-Smith et al., 2006). Conversely, in the methanoarchaeon Methanosarcina, ACK activates acetate for its conversion to acetyl-CoA in the first step of acetioclastic methanogenesis (Thauer et al., 2008). While the investigations into ACK enzymes have focused on bacterial and archaeal systems, ACK was identified in the eukaroyote Entamoeba histolytica in the early 1960s (Bragg and Reeves, 1962) and was first biochemically characterized in the 1970s (Reeves and Guthrie, 1975). Genes encoding putative ACKs have since been identified within the genomes of other eukaryotic pathogens, including the basidomycete Cryptococcus neoformans (Ingram-Smith et al., 2006). At least in E. histolytica, the organism does not appear to have homologs for other proteins required for the known bioenergetic pathways that use ACK (Fowler et al., 2012). This suggests that the biological function of the E. histolytica ACK may be different from that demonstrated in prokaryotes, but at present, that function remains unknown.
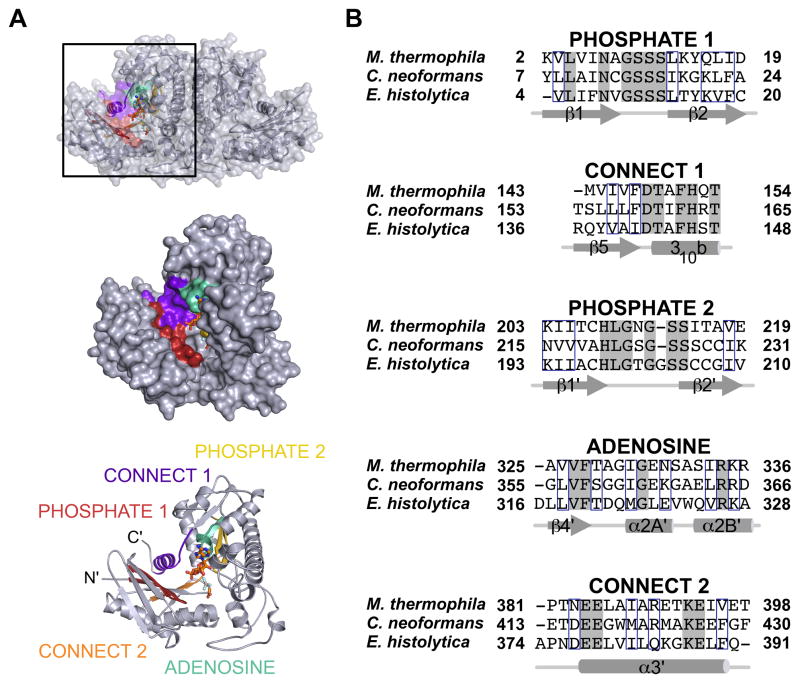
ACKs belong to the acetate and sugar kinase/hsp70/actin (ASKHA) superfamily (Buss et al., 1997). Members of this family use tandem RNase-H like folds as a scaffold for the optimal positioning of five signature sequence motifs, three of which (termed ADENOSINE, PHOSPHATE 1, and PHOSPHATE 2) form the ATP binding pocket (Fig. 1A, B) (Hurley, 1996). While the oligomeric states of ASKHA superfamily members can differ, each protomer fully houses a complete active site.
Figure 1. Conserved motifs of the ASKHA superfamily.
A) Location of the ASKHA superfamily sequence motifs shown in a protomer of C. neoformans ACK. B) A structure-based sequence alignment of each motif shown with secondary structure elements labeled. Fully conserved residues are shaded grey and strongly conserved residues are outlined in blue.
Biochemical and structural investigations of ACK from the methanogenic archaeon Methanosarcina thermophila have been key in developing a mechanistic proposal for ACKs (Buss et al., 2001; Gorrell et al., 2005; Ingram-Smith et al., 2000; Miles et al., 2001; Miles et al., 2002; Singh-Wissmann et al., 2000). It is now generally accepted that phosphoryl transfer between the nucleotide and the substrate occurs directly via an in-line mechanism. This is supported by a crystal structure of the M. thermophila ACK in complex with acetate and the nucleotide transition state analog ADP-AlF3 (Gorrell et al., 2005), which showed the two cosubstrates bound within the active site in a linear array. The structural evidence for the in-line transfer mechanism effectively ended a long-standing debate on the role of ACK phosphoenzyme, which had originally been proposed as a catalytic intermediate.
Domain motions are proposed to facilitate catalysis in M. thermophila ACK by closing around the cosubstrates, which correctly aligns the γ-phosphate of ATP (or pyrophosphate (PPi)) with the phosphoryl acceptor (Gorrell et al., 2005). Fluorescence quenching experiments demonstrated that the differences in interdomain angle observed in the crystal structures of numerous ASKHA enzymes indeed convert to interdomain movement (Gorrell, 2007; Hurley, 1996).
Mirroring the state of knowledge on the biological function of eukaryotic ACKs, few direct studies on the enzymatic mechanism of eukaryotic enzymes have been performed and it is not clear how (or if) the mechanism is modified with respect to the mechanism proposed for the M. thermophila enzyme (Gorrell et al., 2005). Interestingly, the E. histolytica ACK has been shown to use Pi/PPi, rather than ADP/ATP, as the phosphoryl acceptor/donor pair for phosphoryl transfer (Fowler et al., 2012; Reeves and Guthrie, 1975). Further, this enzyme has a strong kinetic advantage for catalyzing the dephosphorylation of acetyl phosphate (Fowler et al., 2012; Reeves and Guthrie, 1975). Although a kinetic characterization has not yet been reported in the literature, C. neoformans ACK has also been shown to kinetically favor acetate formation (Ingram-Smith, C., personal communication). Here, we report the crystal structures of the E. histolytica and C. neoformans ACKs at 2.4 Å and 1.9 Å resolution, respectively. These provide a structural basis for catalysis in the eukaryotic ACKs.
2. Protein expression and purification
The E. histolytica ACK gene cloned into the pQE30 plasmid was co-transformed with the lacI-containing plasmid pREP-4 into Escherichia coli YBS121 (a generous gift of George Bennett, Rice University). The C. neoformans ACK gene was cloned into the pET21b plasmid and transformed into E. coli Rosetta2 (DE3). Both were expressed and purified using similar methods to those described for E. histolytica ACK (Fowler et al., 2012). Briefly, expression cultures were grown in LB broth containing the appropriate antibiotic at 37°C with shaking until the OD600 reached 0.9. Expression was induced with the addition of IPTG to a final concentration of 1 mM. Cultures were shaken overnight at ambient temperature.
Cells were harvested by centrifugation and resuspended in purification buffer (25 mM Tris, 150 mM NaCl, 20 mM imidazole, and 10% glycerol, pH 7.4). Cells were lysed using a French pressure cell and the cellular debris removed by centrifugation at 100,000 × g for 1 hour. ACK was purified from clarified lysate using a 5 mL HisTrap Ni-affinity column and eluted with a linear gradient from 20 mM to 500 mM imidazole in purification buffer. Each protein was pooled and dialyzed against buffer containing 25 mM Tris, 150 mM NaCl, and 10% glycerol, pH 7.4 and further purified by size exclusion chromatography using a Superdex S200 10/300GL column.
3. Crystallization, data collection, structure determination and refinement
E. histolytica ACK was crystallized using the hanging drop vapor diffusion method by mixing 1 μL protein (8 mg/mL in 25 mM HEPES, pH 7.5) with 1 μL reservoir solution (50 mM ADA, 0.6 M potassium-sodium tartrate, 10 mM FeCl3, pH 6.6) and equilibrating against 1 mL reservoir solution at 20°C. Crystals formed within 3 days and were cryo-protected in a solution containing all of the crystallization components and 30% ethylene glycol prior to flash cooling in liquid nitrogen.
C. neoformans ACK was crystallized by the hanging drop vapor diffusion method by mixing 1 μL protein (3 mg/mL in 25 mM Tris, pH 7.4) with 1 μL reservoir solution (50 mM ADA, 100 mM sodium tartrate, 18.5% PEG 2000, pH 6.2) and equilibrated against 1 mL reservoir solution at 4°C for 4 days. Crystals were cryo-protected by soaking in a solution containing all of the components of the crystallization reaction, but with the PEG 2000 concentration increased to 30% and then flash cooled in liquid nitrogen.
Crystallographic data were collected at the Advanced Photon Source LS-CAT beamlines (Table 1) and processed using HKL2000 (Otwinowski and Minor, 1997). The structures of both eukaryotic ACKs were determined by molecular replacement using the program PHASER (McCoy et al., 2007) and a polyalanine model of the M. thermophila ACK structure (PDB entry 1G99; (Buss et al., 2001)) as the search model. Preliminary phases for the C. neoformans ACK model were calculated in DM (Cowtan, 1994) and improved by solvent-flattening and two-fold non-crystallographic symmetry (NCS) averaging. Manual model building was performed in COOT (Emsley and Cowtan, 2004) and refinement was performed in CNS (Brunger et al., 1998) and REFMAC (Murshudov et al., 1997). Tight NCS restraints applied to individual domains of the C. neoformans ACK model were reduced as the model quality improved. Final model quality was assessed with PROCHECK (Laskowski et al., 1993). Figures were prepared with PyMOL (Schrodinger, 2010).
Table 1.
Crystallographic data collection and refinement statistics
| E. histolytica ACK | C. neoformans ACK | |
|---|---|---|
| Data collection | ||
| APS Beamline | 21-ID-G | 21-ID-D |
| Wavelength | 0.979 Å | 1.127 Å |
| Space group | I222 | P21 |
| Unit cell dimensions | a=98.8 Å | a=51.4 Å |
| b=126.9 Å | b=107.6 Å | |
| c=145.6 Å | c=79.1 Å | |
| β=90° | β=99.8° | |
| Resolution Range | 44 - 2.4 Å (2.46 - 2.4 Å)a | 39 - 1.9 Å (1.97 - 1.9 Å) |
| Number of reflections | 273,804 | 224,881 |
| Unique reflections | 35,682 | 64,297 |
| Rsymb | 13.8% (44.0%) | 5.8% (30.9%) |
| <I>/<σ>c | 15.6 (3.0) | 27.0 (5.2) |
| Redundancy | 7.7 (5.8) | 3.5 (3.0) |
| Completeness | 99.3% (93.7%) | 95.7% (89.8%) |
| Refinement | ||
| Rcrystd | 21.0% | 17.8% |
| Rfreee | 23.1% | 21.9% |
| Rms deviation | ||
| Bond Length | 0.004 | 0.01 |
| Bond Angle | 0.93 | 1.2 |
Values in parentheses are for the highest resolution shell.
Rsym = Σhkl Σj |Ij − 〈I〉|/Σhki ΣIj where j is the jth measurement and <I> is the weighted mean of I.
<I>/<σ> is the mean intensity divided by the mean error.
Rcryst=Σhkl ||Fo| −K|Fc||/Σhki |Fo| where Fo and Fc are the observed and calculated structure factor amplitudes, and k is a weighting factor.
Rfree is the same as Rcryst calculated on 5% of the reflections in E. histolytica ACK (1999 reflections) and C. neoformans ACK (3188 reflections).
4. Overall structures
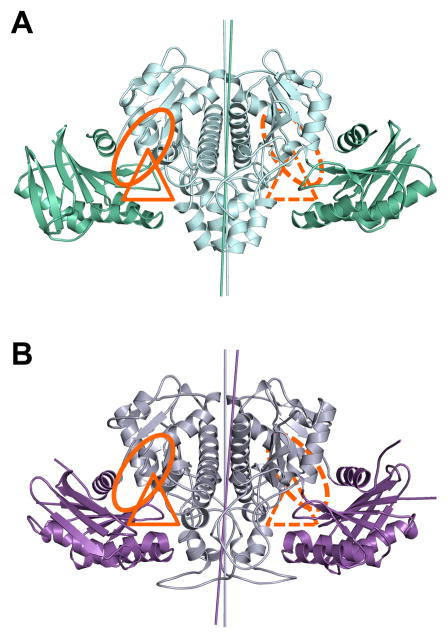
On a global level, the structures of both eukaryotic ACKs (Fig. 2A, B) are similar to that of the previously reported M. thermophila ACK. This dimer has been described as resembling a bird with wings spread (Buss et al., 2001). The ‘body’ of the bird contains the C-terminal RNase-H like domain and mediates dimerization, while the ‘wing’ is organized around the N-terminal RNase-H like domain.
Figure 2. Structures of the eukaryotic ACKs.
A) E. histolytica ACK with the N-terminal wing domain colored green and the C-terminal body domain colored cyan. B) C. neoformans ACK with the wing domain colored purple and the body domain colored grey. The putative acetate and nucleotide binding sites are highlighted with a triangle and a circle, respectively. The rotation axes relating each domain of the dimer are highlighted with a line colored similarly to the corresponding domain.
Superposition of each protomer of the E. histolytica and C. neoformans ACKs revealed that while each domain is folded similarly, there is a difference in the interdomain angle between the body and wing domains. This gives rise to unique rotation axes that superimpose the body and wing domains in both structures (Fig 2A, B). The difference in angle was calculated using DynDom (Hayward and Lee, 2002) and revealed a difference in interdomain angle of 9° between the two protomers of E. histolytica ACK and 14° between the two protomers of the C. neoformans ACK.
5. Active site architecture
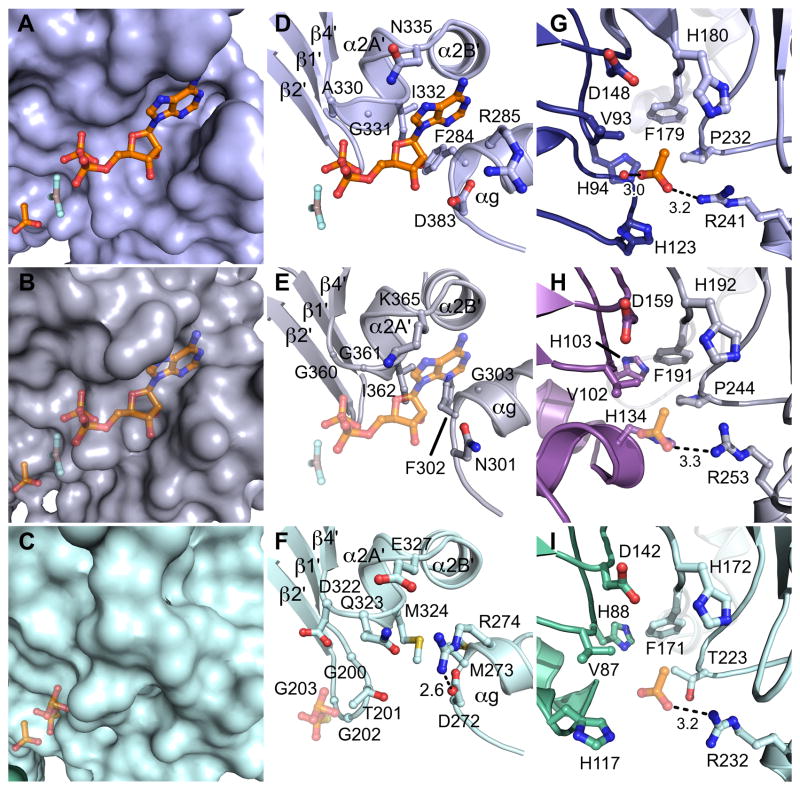
The M. thermophila ACK is the closest structurally characterized homolog of the eukaryotic ACKs, and will be used for comparisons in this report. Comparison of each eukaryotic ACK to M. thermophila ACK determined in the presence of acetate, ADP-AlF3, and thiopyrophosphate (PPS) (Gorrell et al., 2005) supports the assignment of the active site at the interface of the body and wing domains and suggests a binding site for the phosphoryl donor (ATP or PPi) and acetate. The three signature motifs that mediate ATP binding in the ASKHA superfamily, termed ADENOSINE, PHOSPHATE 1, and PHOSPHATE 2 (Bork et al., 1992), surround the putative phosphoryl donor binding pocket in both of the eukaryotic ACKs (Fig. 1A). Co-crystallization of the E. histolytica and C. neoformans ACKs with either PPi or nucleotide analogs, respectively, did not result in the appearance of new electron density corresponding to a bound phosphoryl donor in this site. Instead, superpositions with the structure of acetate and ADP-AlF3 bound M. thermophila ACK (Fig. 3A, D) were used to evaluate whether the phosphoryl donor could reasonably be accommodated in a similar location within the eukaryotic ACKs. Manual modeling of ADP-AlF3 into the structure of the C. neoformans ACK (Fig. 3B, E) and PPS into the structure of the E. histolytica ACK (Fig. 3C, F) resulted in reasonable contacts between protein and the respective nucleotide analogs.
Figure 3. Active site architecture.
A) A surface representation of the M. thermophila ACK (PDB ID 1TUY) with the N-terminal wing domain colored dark blue and the C-terminal body domain colored light blue. B) A surface representation of the C. neoformans ACK colored as in Figure 2. ADP-AlF3 and acetate are modeled into putative binding sites. C) A surface representation of the E. histolytica ACK colored as in Figure 2 with PPS and acetate modeled into putative binding sites. D–F) Close up views of the nucleotide or PPi binding sites in ACKs. D) M. thermophila ACK, E) C. neoformans ACK, and F) E. histolytica ACK. In panels E) and F), the position is modeled according to methods listed in the text. G–I) Close up views of the substrate binding site. G) M. thermophila ACK H) C. neoformans ACK, and I) E. histolytica ACK. The acetate is modeled in panels H) and I).
The ADENOSINE motif normally positions the protein side chains into conformations that promote the interaction between protein and the adenosine base of ATP in ASKHA superfamily enzymes (Bork et al., 1992). Interestingly, both the sequence and the backbone structure of the ADENOSINE motif are conserved in the E. histolytica ACK (Fig. 1A, 3F), which has been demonstrated to use PPi, and not ATP, as a phosphoryl donor (Fowler et al., 2012). Inspection of the E. histolytica ACK structure shows that substitution of a conserved glycine to glutamine and isoleucine to methionine (Gln-323 and Met-324 on α2A′) sterically occludes the ATP-binding cleft (Fig. 3C, F). Additionally, a salt bridge between Asp-272 and Arg-274 on αg stabilizes an alternate conformation of Arg-274 which positions its guanidino group into the ATP-binding cleft further contributing to the occlusion. These features may be important in the conversion of phosphoryl donor selectivity from ATP to PPi.
The structure of the M. thermophila ACK in complex with its substrate acetate (Fig. 3A, G) revealed a hydrophobic substrate-binding pocket between the wing and body domains (Gorrell et al., 2005). Manual modeling of acetate into both the C. neoformans (Fig. 3B, H) and E. histolytica ACKs (Fig. 3C, I) again resulted in reasonable contacts between protein and substrate. Indeed, the residues surrounding the acetate-binding pocket are almost completely conserved in the eukaryotic ACKs with the exception of a proline to threonine substitution at position 223 in the E. histolytica enzyme (Fig. 3I). However, site-directed mutagenesis studies of Thr-223 in E. histolytica ACK did not reveal a specific role for this side chain (Fowler et al., 2012).
6. Mechanistic implications
Given the similarities observed within the active sites of the M. thermophila, C. neoformans, and E. histolytica ACKs, it is reasonable to use the mechanism proposed for the M. thermophila enzyme as a starting proposal for eukaryotic ACKs. Each of the enzymes has an active site that would support binding of the acetate and the phosphoryl donor (either ATP or PPi) in a linear arrangement (Fig. 3A–C). This binding mode is consistent with the in-line transfer mechanism proposed for M. thermophila ACK, where the phosphoryl transfer occurs directly between the two properly aligned cosubstrates (Gorrell et al., 2005).
The biological role of ACK in M. thermophila is to produce acetyl phosphate (and ADP) from acetate and ATP during methanogenesis (Buss et al., 2001). The reaction is therefore commonly discussed in the acetyl phosphate forming direction, although in vitro, the M. thermophila enzyme catalyzes the reverse reaction at a similar rate (Miles et al., 2001; Fowler et al., 2012). In contrast, kinetic characterization of both the E. histolytica ACK (Fowler et al., 2012; Reeves and Guthrie, 1975) and the C. neoformans ACK (Ingram-Smith, C., personal communication) revealed faster turnover in the acetate-forming direction. It is unclear from the structures why the reaction is favored in one direction while the M. thermophila enzyme appears to catalyze the same reaction bidirectionally with comparable efficiency (Miles et al., 2001). Nevertheless, it is conceivable that the in-line transfer could work in reverse. In this scenario, acetyl phosphate and Pi/ADP would bind in a linear array within the active site, and the phosphoryl group would be transferred from the acetyl phosphate to the Pi/ADP.
The difference in interdomain angle observed in both the E. histolytica and C. neoformans ACKs mirrors that observed in other ASKHA superfamily members (Hurley, 1996). Enzymes with a global architecture similar to ACKs commonly employ domain closure to facilitate catalysis (for example, see (Hayward, 2004)). The ability to adopt multiple interdomain angles in the eukaryotic ACKs suggests that interdomain motions could similarly contribute to catalysis, as has been shown for the M. thermophila enzyme (Gorrell and Ferry, 2007).
8. Accession numbers
Coordinates and structure factors have been deposited with the RCSB Protein Data Bank with accession numbers 4H0O (E. histolytica ACK) and 4H0P (C. neoformans ACK).
Acknowledgments
This work was supported by NSF grant 0920274 (KSS), NIH grants GM084417 (KSS), GM079419 (TMI), EY018435 (TMI) and AI079558 (TMI), and Bundesministerium für Bildung und Forschung (BMBF) ZIK program grant FKZ 03Z2HN21 (MT). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the Life Sciences Collaborative Access Team (LS-CAT) ID21-D, and -G at Advanced Photon Source (APS) was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). A portion of this work used facilities that were supported by the Vanderbilt Core Grant in Vision Research (P30EY008126). This paper is Technical Contribution No. 6077 of the Clemson University Experiment Station. We thank Kathryn McCulloch, Prashant Singh, and Kendra Vann for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg PD, Reeves RE. Pathways of glucose dissimilation in Laredo strain of Entamoeba histolytica. Exp Parasitol. 1962;12:393–400. doi: 10.1016/0014-4894(62)90050-4. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D: Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Buss KA, Ingram-Smith C, Ferry JG, Sanders DA, Hasson MS. Crystallization of acetate kinase from Methanosarcina thermophila and prediction of its fold. Protein Sci. 1997;6:2659–2662. doi: 10.1002/pro.5560061222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss KA, Cooper DR, Ingram-Smith C, Ferry JG, Sanders DA, et al. Urikinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J Bacteriol. 2001;183:680–686. doi: 10.1128/JB.183.2.680-686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. Joint CCP4 and ESF-EACBM newsletter on protein crystallography. 1994;31:34–38. [Google Scholar]
- Cozzone AJ. Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annu Rev Microbiol. 1998;52:127–164. doi: 10.1146/annurev.micro.52.1.127. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fowler ML, Ingram-Smith C, Smith KS. A novel pyrophosphate-forming acetate kinase from the protist Entamoeba histolytica. Eukaryot Cell. 2012 doi: 10.1128/EC.00169-12. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrell A, Ferry JG. Investigations of the acetate kinase mechanism by fluorescence quenching. Biochemistry. 2007;46:14170–14176. doi: 10.1021/bi701292a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrell A, Lawrence SH, Ferry JG. Structural and kinetic analyses of arginine residues in the active site of the acetate kinase from Methanosarcina thermophila. J Biol Chem. 2005;280:10731–10742. doi: 10.1074/jbc.M412118200. [DOI] [PubMed] [Google Scholar]
- Hayward S. Identification of specific interactions that drive ligand-induced closure in five enzymes with classic domain movements. J Mol Biol. 2004;339:1001–1021. doi: 10.1016/j.jmb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model. 2002;21:181–183. doi: 10.1016/s1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]
- Hurley JH. The sugar kinase/heat shock protein 70/actin Superfamily: implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- Ingram-Smith C, Barber RD, Ferry JG. The role of histidines in the acetate kinase from Methanosarcina thermophila. J Biol Chem. 2000;275:33765–33770. doi: 10.1074/jbc.M005303200. [DOI] [PubMed] [Google Scholar]
- Ingram-Smith C, Martin SR, Smith KS. Acetate kinase: not just a bacterial enzyme. Trends Microbiol. 2006;14:249–253. doi: 10.1016/j.tim.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles RD, Iyer PP, Ferry JG. Site-directed mutational analysis of active site residues in the acetate kinase from Methanosarcina thermophila. J Biol Chem. 2001;276:45059–45064. doi: 10.1074/jbc.M108355200. [DOI] [PubMed] [Google Scholar]
- Miles RD, Gorrell A, Ferry JG. Evidence for a transition state analog, MgADP-aluminum fluoride-acetate, in acetate kinase from Methanosarcina thermophila. J Biol Chem. 2002;277:22547–22552. doi: 10.1074/jbc.M105921200. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D: Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Reeves RE, Guthrie JD. Acetate kinase (pyrophosphate). A fourth pyrophosphate-dependent kinase from Entamoeba histolytica. Biochem Biophys Res Commun. 1975;66:1389–1395. doi: 10.1016/0006-291x(75)90513-6. [DOI] [PubMed] [Google Scholar]
- Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- Singh-Wissmann K, Ingram-Smith C, Miles RD, Ferry JG. Identification of essential glutamates in the acetate kinase from Methanosarcina thermophila. J Bacteriol. 1998;180:1129–1134. doi: 10.1128/jb.180.5.1129-1134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]