Introduction—counting molecules is as good as it gets
As analytical chemists, the highest resolution measurement one can make is at the single molecule level—it just doesn’t get any better than that. To determine the concentration of a molecule in solution, the best way is to count the number of molecules in a given volume. As long as the volume contains a statistically large enough number of molecules and is above the Poisson noise limit, molecular counting is the absolute best way to make an accurate measurement. Molecular counting is the method of the future and is beginning to be performed today.
In addition to counting, single molecule analysis provides a resolution that cannot be obtained with ensemble measurements. Ensemble measurements are averages—small or rare differences between ostensibly identical molecules cannot be detected. Let’s do a simple thought experiment. If we have a 3000 amino acid protein with a known sequence and we analyze it using any analytical method—x-ray diffraction, NMR spectroscopy, or mass spectrometry, it will give the predicted structure or spectrum. We can predict the protein sequence from the DNA sequence so it should be no surprise that the protein sequence is identical to our prediction—after all, the genetic and protein codes have been known for nearly 50 years. But wait! There are errors that occur in transcription (DNA to RNA) and errors in translation (RNA to protein). Cumulatively, these error rates are between one in a thousand to a few per ten thousand1. With a 3000 amino acid protein, this means that there will be between 0 and 3 errors per molecule!!! In other words, virtually no two of the molecules will be identical because the errors are random. So, while the ensemble measurement gives exactly what we would predict because the average spectrum or crystal structure converges on the predicted structure, in reality we have a heterogeneous mixture of molecules. This heterogeneity can have dramatic implications for biochemistry and biology. For example, if these protein molecules are enzymes, each molecule may have a slightly different rate or specificity. Even small proteins, where some molecules will have identical primary sequences, will exhibit different conformations. Naturally-occurring isotopes give the same molecular heterogeneity, which can result in different rates of reactions of different molecules in the population. Only by detecting and analyzing these single molecules will we be able to fully characterize a population of molecules and thereby understand their behavior.
This Feature describes single molecule detection and analysis methods but focuses on optical methods. Moerner2 is credited with performing the first spectroscopic measurements of single molecules. Much of his seminal work and of others is in the realm of basic biophysics and will not be discussed in this Feature.
Historical—Rotman wins by a mile
The first single molecule measurement was performed in 1961 by Boris Rotman at Stanford Medical School3. Rotman sprayed a solution containing β-galalactosidase and a fluorogenic substrate over a silicone oil preparation to create droplets in oil. By waiting for several hours, Rotman was able to detect and measure the presence of individual enzyme molecules by observing which droplets became fluorescent.
A second demonstration of optical single molecule detection was performed by Hirschfeld 4. Hirschfeld labeled MW 20000 polyethyleneimine with 80–100 molecules of fluorescein isothiocyanate and then conjugated this highly fluorescent polymer to γ-globulin. Using total internal fluorescence detection, the labeled proteins were illuminated by a high intensity laser with sufficient power to photobleach the labeled molecules. This approach was taken in order to collect as many photons as possible as the detectors were not nearly as sensitive as today’s integrating detectors. By carrying out the detection at a known concentration, the expected number of fluorescent detection events was observed. This work was important in that it was the first single molecule measurement that did not employ an enzyme, it used immobilization to localize the molecules, and was the first to use total internal reflection for single molecule measurements.
Years later, Xie and Yeung5, followed soon thereafter by Dovichi and coworkers6, isolated single enzyme molecules (lactate dehydrogenase and alkaline phosphatase, respectively) in a capillary column and observed the formation of a fluorescent product. Dovichi used a capillary electrophoresis tube and was able to apply a pulse to move the product away from the enzyme so repeated kinetic measurements could be carried out on the same enzyme molecule. They observed that different enzyme molecules exhibited different activities. This observation has been corroborated many times.
Optical Methods for Single Molecule Analysis—General Considerations
Analog vs. Digital
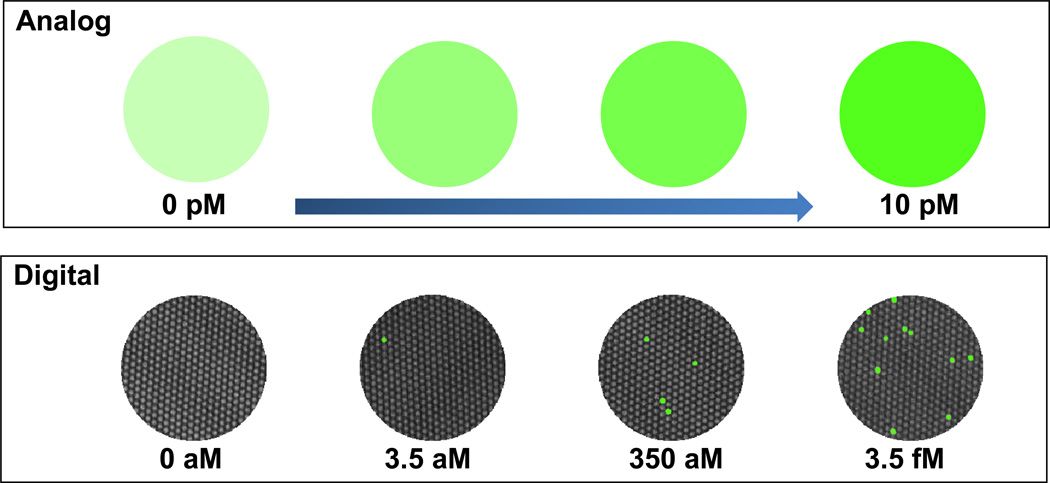
Optical methods for analysis are based on either absorbance or fluorescence. Fluorescence is more sensitive because it is intrinsically amplified as each fluorophore emits thousands to perhaps a million photons before it is photobleached. With either absorbance or fluorescence, the more of the absorbing or fluorescing species present, the higher the absorbance or fluorescence signal. One can think of this type of measurement as an analog signal—the higher the signal the higher the concentration of the species being measured (Figure 1, top). In contrast, single molecule measurements are digital in nature. Each molecule generates a signal that can be counted (Figure 1, bottom). It is much easier to measure the presence or absence of signal than to detect the absolute amount of signal—i.e. counting is easier than integrating. In fact, one could argue that the future of all analytical measurements will be molecule counting. It cannot get any better.
Figure 1.
Top: Analog measurements give increasing intensity as the concentration increases. Bottom: In contrast, digital measurements are independent of intensity and simply rely on a signal/no signal readout.
Size and Volume Considerations
The earliest method to detect single molecules was to interrogate extremely small volumes. Detection was performed in a serial fashion either by allowing a laser to scan a sample or by flowing a solution through a tightly focused laser spot. As a molecule transits the volume illuminated by the laser spot, it can be detected as a burst of fluorescence. A simple thought experiment is depicted in Table 1. For example, if a single molecule is confined to a cubic micron, corresponding to a femtoliter volume, the concentration is 1.5 nM. At that concentration, most fluorophores can be detected easily. So it’s not that difficult to detect a single molecule as long as one knows where to look. But there is a catch. At a concentration of 1 femtomolar, there are 600 molecules per microliter (Table 2) but there are 109 femtoliter volumes per microliter. So one may have to look at over a million femtoliter volumes before detecting a single molecule! Consequently, most flow-based single molecule detection systems work in the picomolar concentration range, which enables a statistically meaningful number of events to be measured while still keeping things dilute enough such that it is improbable for multiple molecules to simultaneously occupy the interrogation volume. These types of measurements take a long time and cannot access extremely low concentrations of analyte.
Table 1.
The relationship between volume and concentration for a small number of molecules
| Size | Volume | 60 molecules | 1 molecule |
|---|---|---|---|
| 0.1 µm3 | 1 aL | 100 µM | 1.5 µM |
| 1 µm3 | 1 fL | 100 nM | 1.5 nM |
| 10 µm3 | 1 pL | 100 pM | |
| 100 µm3 | 1 nL | 100 fM |
Table 2.
Number of molecules in a given volume at different concentrations
| 1uL | 100uL | |
|---|---|---|
| 1nM | 6 108 | 6 1010 |
| 1pM | 6 105 | 6 107 |
| 1fM | 600 | 60,000 |
Numbers vs. Concentration
A related, but one of the most confusing aspects of single molecule detection methods, is the difference between measuring concentration and detecting an absolute number of molecules. As can be seen from Table 2, 100 µL of a 1 fM solution contains 60000 molecules—plenty of molecules to detect. The problem is that many serial methods that interrogate small volumes cannot measure 100 µL—it simply would take too long to interrogate such a large volume when there is only 1 fL in the focal volume. Even if one were able to measure one thousand 100 fL volumes per second, it would take a million seconds to interrogate the entire 100µL volume. Consequently, most serial measurements are performed using higher (nanomolar to picomolar) concentrations. In this way a statistically significant number of molecules can be detected in a short time frame. In such cases, assay limits of detection are sometimes given in absolute numbers of molecules detected or an absolute amount of analyte—for example, if 10 nL of a 1 nM analyte solution are analyzed, one can claim a limit of detection of 10 attomoles. But the really important measurement and the biggest challenge is the concentration. Many single molecule measurements get around the low concentration problem by performing a pre-concentration step prior to the detection step. In this way, a dilute solution containing a few molecules can be converted into a smaller volume with a much higher concentration.
Poisson sampling
The last major issue with making ultra-low concentration measurements using these single molecule techniques is molecular shot noise, which is a function of Poisson sampling. Poisson sampling has several implications. First, for extremely low concentration analytes, such as viruses that may be present at 100 copies per milliliter of blood, a microliter sample is more likely to not contain a virus than to contain one. It is therefore important that the sample volume is sufficiently large to ensure that there are sample molecules present or that a negative result is truly negative and not a consequence of sampling. Second, as pointed out by Chen and Dovichi for electrophoretic analysis of single molecules7, molecular shot noise due to sampling affects precision. These authors recognized that 10000 molecules must be detected in order to obtain a 1% relative precision (the square root of the number of molecules measured). The molecular shot noise due to these sampling processes affected characterization of the peak center and width in electropherograms, using single molecule detection of β-phycoerythrin. This sampling issue also makes single molecule detection a challenging proposition for measuring rare events. With small interrogation volumes, one can observe single molecules transiting across a laser beam. As illustrated above, even a microliter contains a billion femtoliter volumes, which takes time to interrogate. In such cases, it is therefore necessary to concentrate the analytes into small volumes before interrogating them with a laser.
A commercial platform that uses this preconcentration approach is the Erenna system from Singulex8. In this system, a sandwich immunoassay approach is used. Protein analytes are captured on magnetic particles using capture antibodies. A labeled detection antibody that recognizes a different epitope on the protein is then used to bind to the protein molecules captured on the magnetic particles. The labeled detection antibodies contain a fluorophore. After washing to remove non-specifically bound labeled Ab, the labeled Abs are released from the magnetic particle by urea to disrupt the protein Ab interactions. The released labeled Ab is then put into a capillary system and the number of fluorescent labels is counted as they transit a laser focal spot.
Amplification
Amplification is an essential part of many single molecule detection techniques. One method of amplification is to use a label—a fluorescent dye or quantum dot, that emits many photons per label instead of relying on some intrinsic optical property of the analyte molecule. An alternative label is an enzyme that can be used to generate many molecules of detectable product by catalyzing the conversion of substrate molecules into detectable product molecules. For example, β-galactosidase catalyzes the formation of hundreds of molecules per second of highly fluorescent resorufin molecules from non-fluorescent substrate molecules. This reaction has the double advantage in that it amplifies the label (one enzyme molecule produces thousands to millions of product molecules) and the product molecules are fluorescent, which are intrinsically amplified. Another way to amplify single molecules is by replicating the molecules of interest. This approach is the basis for many nucleic acid detection schemes in which a single molecule is amplified using, for example, the polymerase chain reaction (PCR).
Virtually all optical methods for single molecule detection and analysis are indirect in that they are not detecting the molecule of interest but a label that is associated with the molecule. Single molecule optical methods are primarily based on fluorescence because of its intrinsic amplification. Most analyte molecules do not fluoresce so direct measurements cannot be performed. Absorbance based single molecule methods are more challenging due to the low absorption cross sections of a single molecule—think Beer’s Law where the path length is one molecule thick. Refractive index (RI) changes could also be considered optical methods and also suffer from the high sensitivity required to measure an RI change of a single molecule. Both absorbance and refractive index-based single molecule methods require much more sensitive and complicated instrumentation relegating them to sophisticated laboratory measurements.
Methods and Applications are Diverse
One of the most active areas for single molecule analysis and detection is in the area of nucleic acid (NA) measurements. Nucleic acids can be amplified by the polymerase chain reaction (PCR). PCR enables single molecule detection in a variety of ways. Digital PCR is a method in which DNA is diluted until there are either one or zero molecules in a given volume9,10. This method was originally performed in microtiter plates but is now performed by partitioning small volumes using water in oil emulsions or microfluidics (Quantalife, Fluidigm, RainDance). All the necessary PCR reagents are included in the sample and the sample is then partitioned into droplets. Once partitioning is completed, PCR is carried out by thermal cycling. Samples containing the DNA sequence of interest are amplified and the amplification products are detected using a variety of fluorescence methods. In this manner, the number of single NA molecules with a specific DNA sequence is determined by counting the number of fluorescent droplets relative to the number of droplets with no signal.
For single molecule sequencing, there are two optical methods that have been employed. In the first method, developed by Quake and commercialized by Helicos, single stranded DNA template molecules are first captured on a surface. For sequencing, a sequencing-by-synthesis (SBS) method is employed11. The captured template DNA is exposed to a primer and then to a solution containing DNA polymerase and a fluorescent nucleotide. If incorporation of a nucleotide occurs, it means that base has been incorporated into the growing complementary strand of the template. The fluorescence is read using a TIRF-based Heliscope™ that records the positions where the DNA strand has incorporated a fluorescent nucleotide. This process continues for the other three nucleotides. The fluorescent labels are removed and the process is continued for the next base. The advantage of this system is it enables billions of molecules to be captured and sequenced. The disadvantage is that the system needs to be sensitive to single molecules and is therefore expensive.
Alternative next generation sequencers using optical interrogation are based on SBS chemistry but generate “clusters” of DNA. Each cluster starts with a single DNA template molecule bound to a surface but uses a surface amplification method, similar to PCR in many respects, to make many copies of each molecule confined close to the original DNA template molecule. The only challenge is to keep the density of the template molecules low enough that the clusters do not grow into one another. SBS chemistry can then be conducted on these clusters and imaging is much easier because the signal emanates from many hundreds to thousands of identical molecules. While the method is not really single molecule detection, the origin of the signal can be traced to a single molecule. The advantage of this approach is the imaging sensitivity is significantly relaxed. The disadvantage is that there is a cluster generation step required and the fidelity of replication is not 100%, which can lead to errors in sequencing. On the other hand, there are many replicates of each template molecule in all SBS systems such that errors in reading one sequence are inconsequential because many reads of each sequence take place.
Another true single molecule detection sequencing method is based on work by Webb and coworkers and commercialized by Pacific Biosciences12. In this approach a zero mode waveguide is employed in which nanoscale wells are fabricated. The resulting optical signals are therefore limited to those signals that arise very close to the surface and confine the reaction to zeptoliter volumes. A single DNA polymerase is attached to each well and a DNA template molecule is then allowed to bind to the polymerase. Four differently labeled fluorescent nucleotides are then added and the polymerase begins to catalyze the incorporation of nucleotides complementary to the template strand. As the fluorescent nucleotides bind, the color of the complementary nucleotide resides in the ZMW long enough to be read during which the base is incorporated, the fluorescent label is cleaved and then diffuses away. Only the complementary nucleotide binds and provides a sufficient signal. Non-complementary nucleotides that diffuse into the interrogation volume do not reside in the volume long enough to provide a measurable signal and simply contribute to the background. The process continues with the next nucleotide. This system operates continuously and does not require sequential addition of nucleotides to read each base. The system provides long reads. The method is still an indirect one in that the sequence must be transduced into an optical signal.
A completely different approach to single molecule optical detection was taken by Fraser and coworkers13. They employed a high Q (quality) factor optical microcavity resonator—a whispering gallery mode structure consisting of a toroidal silica microring coupled to an optical fiber. In this system, the light “circulates” in the microring and samples the solution in the evanescent field just outside the ring structure. The resonant frequency of the resonator changes upon binding to the surface of the microring, which can be functionalized with binding reagents such as antibodies. Single molecule sensitivity is achieved because the bound molecule is heated and perturbs the resonant frequency via a thermo-optic effect. Single molecule binding events can be observed as stochastic changes in the resonant frequency when the microcavity toroid is immersed in a solution containing analyte molecules.
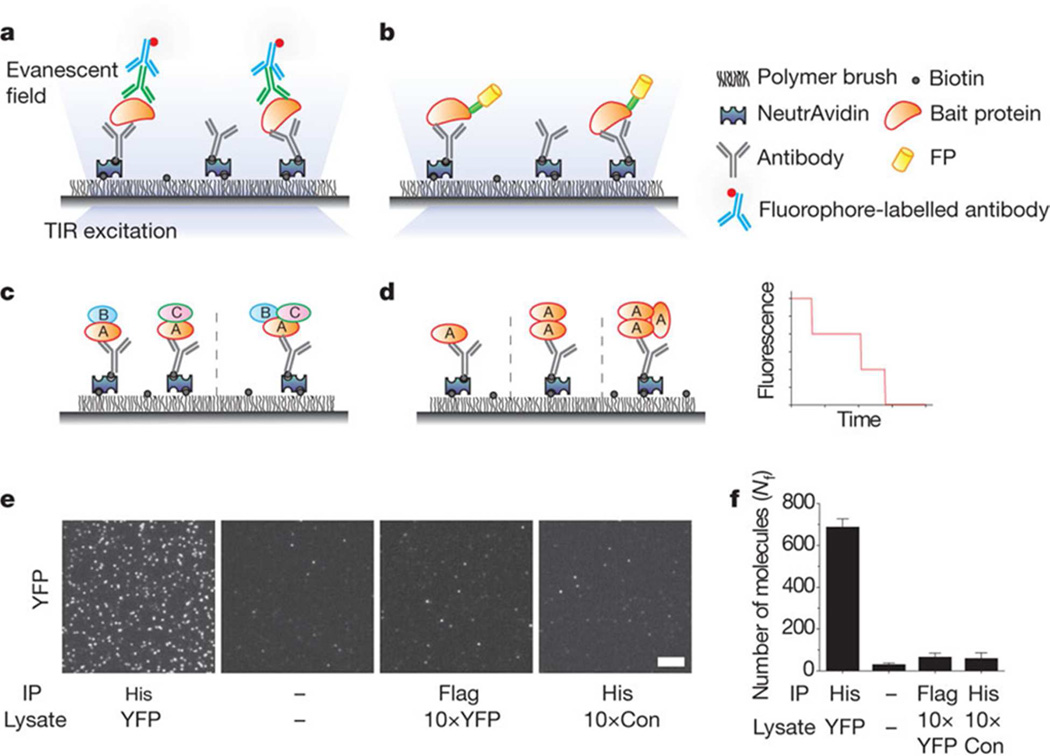
Another widely used method for single molecule detection is based on capturing molecules on surfaces and then counting the number of captured molecules after a given time. Total internal reflection fluorescence (TIRF) single molecule methods have been developed by Ha and coworkers including both FRET and pull down methods14,15. In these methods (Figure 2), a capture agent, such as a capture antibody attached to the surface, binds the analyte and then a fluorescent labeled detection antibody is added that recognizes a different epitope. The resulting labeled complex can be visualized using TIRF. This method uses sensitive detection to visualize the captured species. A different tack is taken by Schultz and coworkers16. They use colloidal silver plasmon resonant particles (PRPs) that scatter light in a highly efficient manner and can be designed to scatter at many different wavelengths. These highly visible labels can be used to visualize binding using sandwich format assays. In the case of TIRF, the sensitive detection of the instrument enables single molecule detection whereas with highly visible labels, the detection instrument does not need to be sensitive.
Figure 2.
a, b, Immunoprecipitated protein complexes are visualized using TIRF microscopy using fluorophore-labelled antibody (a) or fluorescent protein (FP) tags (b). TIR, total internal reflection. c, Multi-colour colocalization can distinguish between subcomplexes (for example, AB + AC versus ABC). d, Photobleaching analysis can provide stoichiometric information. A simulated photobleaching trajectory for a trimeric protein. e, TIRF images for YFP pulled down from cells expressing His6–YFP (YFP) and control cells (Con) using His-tag or a control (Flag-tag) antibody. Minus sign indicates no antibody or sample. IP, immunoprecipitate. Scale bar, 5 µm. f, Average number of fluorescent molecules per imaging area, Nf. Error bars denote standard deviation (s.d.) (n > 20). Reprinted by permission from Macmillan Publishers Ltd: NATURE 2011, 473, p484.
In my laboratory, we have developed a single molecule detection method based on using enzymatic amplification17. Using methods similar to the single molecule pioneers, we first capture the analyte onto a surface—in our case we use fiber optic microwell arrays containing 50000 wells. The microwells are approximately 4 µm in diameter and 3 µm deep giving a volume of approximately 40 fL. We first capture the analyte but we do so such that there is either zero or one captured analyte molecule in each microwell. A detection reagent that is labeled with an enzyme is then used to form the sandwich. We use β-galactosidase as the label. After washing to remove any non-specifically bound reagents, we seal the microwells with a fluorogenic substrate onto a PDMS membrane. Any well that contains an enzyme, as a consequence of the formation of a sandwich complex, will convert the substrate into a fluorescent product. The enzyme converts hundreds of molecules of substrate into product per second so within a few seconds there is nanomolar or higher concentrations of fluorescent product. The key to the success of this assay format is that the product cannot diffuse away because the microwells are sealed. After a few minutes, the wells containing enzyme-labeled analyte are highly fluorescent while the wells with no analyte are black. Simply counting the ratio of fluorescent wells to the unlabeled wells provides the concentration of analyte in the original sample solution. The key for all three of these capture methods (TIRF, PRP labels, enzyme labels) is that, for quantification, the incubation times need to be kept constant because longer times will give more bound molecules.
The biggest problem with capturing analyte molecules on a surface is the long time required to capture molecules at low concentration. Robert Corn of the University of California-Irvine calls this slow binding the “tyranny of the Langmuir binding isotherm”. For 1 fM analyte concentrations binding to a capture antibody immobilized on a surface, it takes nearly a year to come to equilibrium. One obviously can’t wait a year to make a measurement but for single molecule detection methods, an hour of binding is often sufficient to capture a sufficient number of molecules to detect. Even the initial part of the binding curve provides enough molecules for these methods.
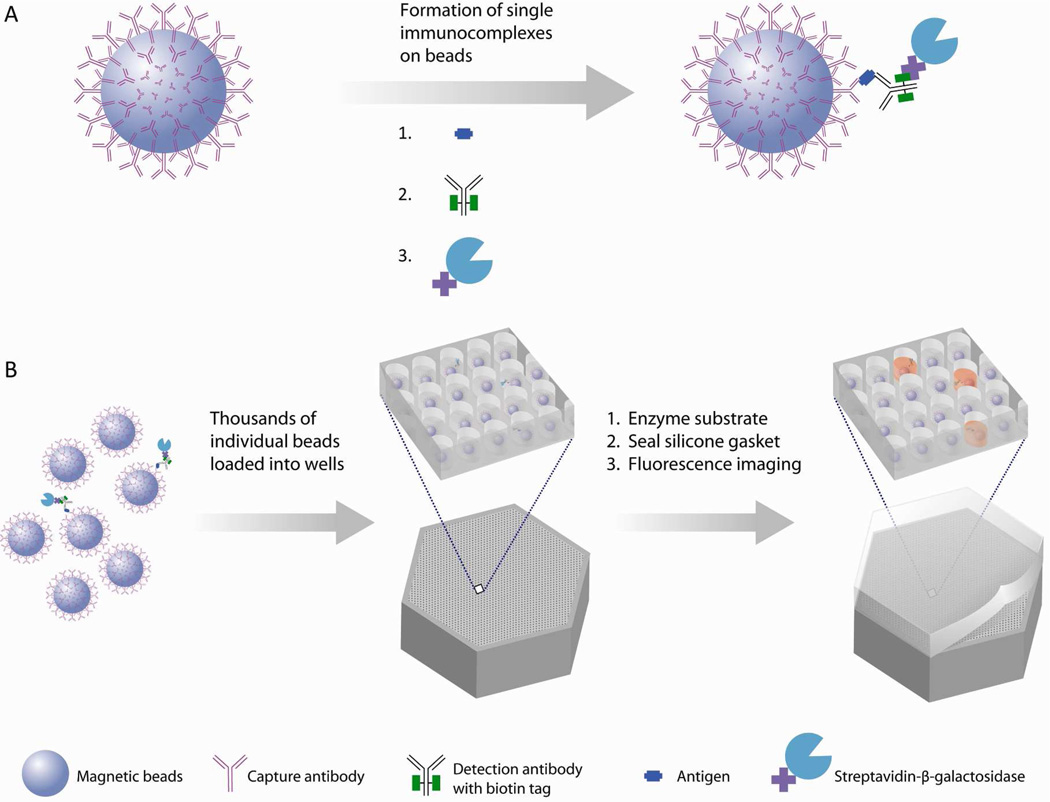
To solve the inefficiencies with binding, Duffy and coworkers use a sample preparation step that concentrates the molecules (Figure 3)18. Microspheres with capture agents attached to the surface can be used to concentrate a dilute solution of molecules. Capture reagents (DNA probes or antibodies) are attached to the surface of microspheres. 3 µm beads typically contain approximately 250,000 attachment sites so one can think of each bead as having a “lawn” of capture molecules. The beads are added to the sample solution such that there are more beads than molecules. As can be seen in the table above, 100 µL of a 1fM solution contains 60000 molecules. Typically 500,000 beads will be added to a 100 µL sample. There are two advantages for adding so many beads. First, at a roughly 10:1 bead to molecule ratio, which is in the Poisson regime, each bead captures either one or zero molecules. Second, with so many beads in solution, the bead-to-bead distance is short such that every molecule encounters a bead in less than a minute. Diffusion of the target analyte molecules, even large proteins, occurs on a timescale such that with only a 30 minute incubation, all the molecules should have multiple collisions with multiple beads. In this manner, the slow binding to a fixed capture surface is avoided and the efficiency of binding increases dramatically. The remainder of the steps is the same—beads are washed to remove non-specifically bound proteins, incubated with biotinylated detection antibody, and then with β-gal labeled streptavidin. In this manner, each bead that has captured a single protein molecule is labeled with an enzyme. Beads that do not capture a molecule remain label free. The beads are then loaded into the same microwells as described above, sealed with substrate, and then the substrate is converted to a fluorescent product by captured enzyme label. The ratio of the number of wells containing a bead with an enzyme label to the total number of wells containing a bead corresponds to the analyte concentration in the sample.
Figure 3.
Proteins are captured on antibody coated beads. One or zero molecules of analyte are captured on each bead. A labeled detection antibody creates a sandwich complex . When the beads are loaded into microwells along with substrate and sealed, enzyme-labeled beads covert the substrate into a fluorescent product that can be easily detected. Reprinted by permission from Macmillan Publishers Ltd: NATURE BIOTECHNOLOGY, 2010, 28, p596.
One of the biggest challenges with all these protein detection methods is non-specific binding (NSB)—non-target molecules that bind and are counted. Non-target molecules include high concentration proteins in the sample, proteins that resemble the target molecule, or enzyme molecules or other labels that ultimately are detected. NSB sets a background “floor”. The number of background molecules requires that one must detect three times the standard deviation of the background before one obtains a meaningful analytical result. Consequently, many single molecule methods, while intrinsically able to detect a single molecule, cannot provide particularly sensitive detection limits because NSB is high. Techniques for reducing NSB include many surface modification approaches that attempt to passivate the surface so that it does not attract proteins. A related issue is the non-specific binding of the analyte molecule to surfaces during the assay protocol. Glass and plastic adsorb proteins and many other molecules to some extent. When analyte concentrations are in the femtomolar and lower concentration range, loss of analyte to these surfaces can deplete the sample and produce erroneous results. Consequently, care must be taken to passivate these surfaces and to minimize transferring samples from one vessel to another.
Another issue with single molecule detection is when a low absolute number of molecules is being detected, one encounters a serious problem with Poisson noise. In this situation, the uncertainty of the measurement is √n where n is equal to the number of molecules counted. For example, if four molecules are counted, then the uncertainty of the measurement is 4±2, which makes for a CV that is analytically unacceptable. Consequently, the intrinsic uncertainty of the measurement at low concentrations may relegate the measurements to semi-qualitative or qualitative. Such measurements are not without value—for example, if a cancer marker or virus protein are present that shouldn’t be present, a high sensitivity single molecule based method may be able to detect it and the absolute concentration wouldn’t matter—just the presence of the marker would signify that there was a problem. In such cases, the earlier the detection, the better.
Summary and Future Directions
As stated previously, single molecule detection is as sensitive as it gets. Counting molecules is the most accurate method for measuring the concentration of molecules in solution. As discussed in this Feature, optical methods for single molecule detection typically amplify the target molecule, as in the case of DNA, or use labels for signal amplification. Of course optical methods for measuring single molecules comprise only one approach. Electrical detection methods are also incredibly powerful. Measuring the conductance signals through single nanopores as pioneered by Branson and Bayley is beginning to be used for DNA sequencing. Today we are beginning to see a next generation of methods in which single molecules can be detected directly when they bind to a surface. These ultra-sensitive technologies will undoubtedly continue to advance. There is every reason to believe that such techniques will eventually be able to interrogate samples in a way that enables both the number and identity of multiple target molecules to be determined.
These methods should find use for both fundamental studies and important applications. There are already a large number of fundamental studies in which single molecules are observed in action. Exemplary research in this area includes early work of Xie and coworkers in which single turnovers of cholesterol oxidase were observed using the intrinsic fluorescence of flavin nucleotides19 or the work of Bustamante and coworkers in which single DNA molecules were tethered to beads, optically trapped, and observed to detect the activity of single molecules of RNA polymerase20. There are many research groups now focused on making single cell measurements. The small absolute numbers of various molecules in a single cell make it difficult to accurately measure their concentrations. Single molecule measurements should make it possible to detect these molecules much more efficiently and provide accurate readings of cell-to-cell variation. Measuring the “dark space” in clinical samples is another opportunity where single molecule measurements should shine. For example, there are less than 150 proteins of clinical utility with FDA approval in use today21, yet the human proteome contains over 2500 secreted proteins22. Most of the “missing” proteins are simply below the detection limit of the best ELISAs. Consequently, more sensitive measurements will likely result in earlier cancer detection, earlier detection of infectious disease, and identification of a host of new biomarkers with utility for both diagnostic screening and environmental monitoring. The future has arrived!
Acknowledgements
Support from NIH (1R01HG006021, NHGRI), DARPA, and the Breast Cancer Research Program administered through the Army Research Office is gratefully acknowledged.
Biography
David R. Walt is Robinson Professor of Chemistry, Professor of Biomedical Engineering, Professor of Genetics, and Professor of Oral Medicine at Tufts University and is a Howard Hughes Medical Institute Professor. His research is focused on analytical applications of single molecule detection, single molecule enzymology, and single cell analysis.
References Cited
- 1.Bregeon D, Colot V, Radman M, Taddei F. Genes and Development. 2001;15:2295–2306. doi: 10.1101/gad.207701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moerner WE. The Journal of Physical Chemistry B. 2002;106:910–927. [Google Scholar]
- 3.Rotman B. Proceedings of the National Academy of Sciences. 1961;47:1981–1991. doi: 10.1073/pnas.47.12.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirschfeld T. Appl. Opt. 1976;15:2965–2966. doi: 10.1364/AO.15.002965. [DOI] [PubMed] [Google Scholar]
- 5.Xie XS, Dunn RC. Science. 1994;265:361–364. doi: 10.1126/science.265.5170.361. [DOI] [PubMed] [Google Scholar]
- 6.Craig DB, Arriaga EA, Wong JCY, Lu H, Dovichi NJ. Journal of the American Chemical Society. 1996;118:5245–5253. [Google Scholar]
- 7.Chen D, Dovichi NJ. Analytical Chemistry. 1996;68:690–696. [Google Scholar]
- 8.Todd J, Freese B, Lu A, Held D, Morey J, Livingston R, Goix P. Clinical Chemistry. 2007;53:1990–1995. doi: 10.1373/clinchem.2007.091181. [DOI] [PubMed] [Google Scholar]
- 9.Vogelstein B, Kinzler KW. Proceedings of the National Academy of Sciences. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 11.Braslavsky I, Hebert B, Kartalov E, Quake SR. Proceedings of the National Academy of Sciences. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 13.Armani AM, Kulkarni RP, Fraser SE, Flagan RC, Vahala KJ. Science. 2007;317:783–787. doi: 10.1126/science.1145002. [DOI] [PubMed] [Google Scholar]
- 14.Roy R, Hohng S, Ha T. Nat Meth. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz S, Smith DR, Mock JJ, Schultz DA. Proceedings of the National Academy of Sciences. 2000;97:996–1001. doi: 10.1073/pnas.97.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Hayman RB, Walt DR. J. Am. Chem. Soc. 2008;130:12622–12623. doi: 10.1021/ja8053018. [DOI] [PubMed] [Google Scholar]
- 18.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. Nat Biotech. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu HP, Xun L, Xie XS. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 20.John R, Davenport, Wuite GJL, Landick R, Bustamante C. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 21.Anderson NL, Anderson NG. Molecular & Cellular Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Klee EW, Carlson DF, Fahrenkrug SC, Ekker SC, Ellis LBM. Nucleic Acids Research. 2004;32:1414–1421. doi: 10.1093/nar/gkh286. [DOI] [PMC free article] [PubMed] [Google Scholar]