Abstract
Most non-coding RNAs function properly only when folded into complex 3D structures, but the experimental determination of these structures remains challenging. Understanding of primary miRNA maturation is currently limited by a lack of solved structures for non-processed forms of the RNA. SHAPE chemistry efficiently determines RNA secondary structural information with single-nucleotide resolution, providing constraints suitable for input into the MC-Pipeline software for refinement of 3D structure models. Here we combine these approaches to analyze three structurally diverse primary miRNAs, revealing deviations from canonical dsRNA structure in the stem adjacent to the Drosha cut site for all three. The necessity of these deformable sites for efficient processing is demonstrated through Drosha processing assays. The structure models generated herein support the hypothesis that deformable sequences spaced roughly once per turn of A-form helix, created by non-canonical structure elements, combine with the necessary single-stranded RNA:double-stranded RNA junction to define the correct Drosha cleavage site.
Keywords: SHAPE, MC-Pipeline, chemical modification, RNA structure, microRNA, Drosha
Many non-coding RNAs undergo enzymatic processing to achieve their functional states, through mechanisms that are coupled to their three-dimensional structures. Therefore, generating atomic resolution structure models is necessary for complete characterization of non-coding RNA processing, though this is currently a daunting task to accomplish for most RNAs by crystallographic or NMR methods. For many smaller non-coding RNAs, such as microRNAs (miRNAs), adequate models are realized through computational prediction of secondary structure, underscoring the need for computational approaches that yield highly accurate and biochemically validated structure models. However, available structure-mapping techniques face limitations when rare and under-characterized motifs are encountered (1). Here we show that structure models of primary miRNA (pri-miRNA) transcripts, suitable for generating testable mechanistic hypotheses, are created by combining secondary structure mapping through selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) chemistry (2) with structure calculation in the MC-Pipeline software (3). We apply these methods to three structurally diverse pri-miRNAs, revealing deviations from canonical A-form structure in each; we then demonstrate that these structural deformations significantly influence in vitro processing efficiency by the RNase III enzyme Drosha.
Mature miRNAs – of which more than 1,000 have been annotated in humans (4) – regulate development and tissue differentiation through their role in the RNA silencing pathway (5). Canonical pri-miRNA transcripts adopt imperfect stem-loop structures embedded within single-stranded regions (5). In the canonical miRNA biogenesis pathway, pri-miRNAs are excised co-transcriptionally from longer RNAs by the Microprocessor complex – consisting minimally of Drosha and the dsRNA binding protein DGCR8 – in a process that is tightly regulated (5, 6). Cleavage results in precursor miRNA (pre-miRNA) that is approximately 70 nucleotides in). length and typically characterized by a two-nucleotide 3′-overhang at the cut site (7 To date, no atomic resolution structures of pri-miRNA models have been reported; regarding pre-miRNAs, only the structure of pre-mir-30a bound to Exportin-5 has been solved (8). Consensus mechanistic proposals emphasize a role for the ssRNA:dsRNA junction and pri-miRNA structural heterogeneity, from bulges and internal loops, in Microprocessor positioning and cut-site recognition (7, 9, 10). In a recent bioinformatic study, Warf et al. (11) predicted that most pri-miRNAs harbor a helical distortion at the Drosha cleavage site, with a majority of the distortions being symmetric internal loops of two nucleotides (i.e., single-nucleotide mismatches).
Generating a complete mechanistic model for miRNA processing requires the determination of structures of representative pri-miRNAs (12). SHAPE chemistry has emerged as a powerful method to define base-pairing status with single-nucleotide resolution (2). Hairpin RNAs similar to pri-miRNAs have been studied using SHAPE chemistry, yielding results that compare well to their previously determined secondary structures (13). However, these previous studies made no attempt to generate SHAPE-constrained three-dimensional structure models.
Here, we analyze the structures of three RNAs: pri-mir-16-1, pri-mir-30a, and pri-mir-107. Of the three, pri-mir-16-1 and pri-mir-30a were chosen because of their extensive prior use as models for in vitro processing studies (7–10, 14–16). We analyzed pri-mir-107, which contains a 1-by-3 asymmetric loop at the cleavage site (17), because inclusion of the scissile bonds in bulges and internal loops is predicted to produce inconsistent length pre-miRNA molecules (18). Secondary structure constraints were generated by SHAPE and the data were then incorporated in MC-Pipeline calculations (3), producing low resolution atomic structure models of the RNA stem-loops in a relatively high-throughput manner. Surprisingly, normalized SHAPE reactivity profiles indicate that many of the small helical imperfections in the RNA stems are not disruptive to the A-form helix – results that are corroborated by ribonuclease cleavage assays. In all three pri-miRNAs, the MC-Pipeline structure ensembles feature an extensive ability to deform the dsRNA stem between the ssRNA:dsRNA junction and the Drosha cut site. Drosha processing assays performed in vitro confirm that the presence of these deformable “hot spots” near the cut site enhances cleavage efficiency. Overall, we have developed an approach for generating structure models of small RNAs and applied it to pri-miRNAs to reveal an important structural aspect of Drosha processing.
Materials and Methods
RNA Preparation
All DNAs were purchased from Geneart. Template DNA for SHAPE reactions was inserted into a SHAPE cassette (19) with an inverted BsaI cut site at the 3′-end. All DNAs were cloned into pUC19 (New England Biolabs) and transformed into DH5α competent cells, which were grown in LB media at 37°C to an approximate OD600 of 3.75. Template DNA for ribonuclease structure mapping was prepared identically, except that the SHAPE cassette sequences were not present. Preparation of template DNA, transcription by T7 RNA polymerase, and purification of the transcribed RNA were all performed as previously described (20).
RNA Modification by 1M7
RNA (4 pmol) in 5 μL of sterile water was heated at 85°C for 1 minute and cooled to 4°C for 2 minutes prior to adding 5 μL of folding buffer (50 mM HEPES pH 7.6, 50 mM KCl), incubation at 37°C for 10 minutes, and treatment with 1 μL 1M7 (100 mM 1M7 in anhydrous DMSO) at 37°C for 70 seconds. Control and sequencing reactions were treated with 1 μL anhydrous DMSO. All reactions were cooled to 4°C and the RNA recovered by ethanol precipitation (1 μL 5 M NaCl, 0.5 μL 20 mg/mL glycogen, 12.5 μL sterile water, and 75 μL ethanol added to each reaction).
Primer Extension
All reactions were mixed with 0.5 μL Cy5 fluorescently labeled DNA primer (10 μM in sterile water, 5′-cy5-GAACCGGACCGAAGCCCGATTTGG-3′, purchased from Sigma, HPLC purified). The primers were annealed to the RNA by heating at 65°C for 5 minutes and cooling at 35°C for 5 minutes prior to addition of 3 μL reverse transcription buffer (167 mM Tris-HCl pH 8.3, 250 mM KCl, 1.67 mM each dNTP, 17 mM DTT, 10 mM MgCl2). Following heating to 52°C for 3 minutes, 1 μL of 5 mM ddNTP was added to the sequencing reactions and 1 μL of sterile water was added to all other reactions. Primer extension was initiated by the addition of 0.5 μL Superscript III (Invitrogen) and incubation at 52°C for 10 minutes. Immediately thereafter, 0.5 μL of 2 M NaOH was added and the solutions heated to 95°C for 7 minutes prior to cooling at 4°C for 5 minutes. cDNA was treated with 9 μL neutralizing gel loading solution (100 mM Tris-HCl pH 7.5, 20 mM EDTA in formamide), heated to 90°C for 90 seconds, and separated on a denaturing polyacrylamide gel (8% 29:1 acrylamide:bisacrylamide, 8.4 M urea, 1X TBE; 95 W, 1 hour). The gels were visualized on a Typhoon imager.
Processing of SHAPE Data
Each of the SHAPE reactions was carried out over 21 replicates and three background reactions. Gels were quantified using SAFA (21). After log-likelihood based processing, normalizing, and background subtraction, the average and standard deviation of the SHAPE reactivity was calculated using MATLAB scripts from the HiTRACE program (22, 23). The mean SHAPE reactivities with standard deviations larger than their respective means were discarded for further analysis, along with any negative values. Positive SHAPE reactivities were re-scaled in the range of 0 to 1.
Ribonuclease Structure Mapping
The RNAs were 5′-end labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs) and the RNA concentration determined by liquid scintillation. 5′-32P-end-labeled RNA (0.05 pmol total RNA, 4 μL in sterile water) was renatured at 60°C for 10 minutes followed by 25°C for 10 minutes prior to adding 1 μL folding buffer (50 mM HEPES pH 7.6, 50 mM KCl). The RNA was digested in the presence of 1 μL of single-strand-specific (0.11 ng/mL RNase A, 0.25 U/μL RNase T1) or double-strand-specific (1.7×10−3 U/μL RNase V1) nucleases (Ambion) under native conditions at 37°C for 30 minutes. The T1 ladder was generated by incubating the labeled RNA (0.05 pmol, 2 μL in sterile water) under denaturing conditions in 3.5 μL T1 digestion buffer (9.4 M urea, 28.3 mM sodium citrate pH 3.5, 1.4 mM EDTA) and 0.5 μL of 5 U/μL T1 at 50°C for 5 minutes. The hydrolysis ladder was generated by incubating the labeled RNA (0.03 pmol, 1.5 μL in sterile water) and 2 μL hydrolysis buffer (100 mM Na2CO3/NaHCO3 pH 9.0, 2 mM EDTA) at 90°C for 5 minutes. All reactions were quenched by addition of an equal volume of 100 mM Tris-HCl pH 7.5/20 mM EDTA/formamide loading buffer and boiled before fractionating on a denaturing polyacrylamide gel (both 8% and 12% 29:1 acrylamide:bisacrylamide, 8.4 M urea, 1X TBE; 100 W, 1 hour). The gels were visualized on a Typhoon imager.
MC-Fold and MC-Sym Simulation
MC-Fold and MC-Sym were used to simulate pri-miRNA structures, supplemented by the information provided by SHAPE. Initially, the 3′- and 5′-tails were constrained to be single-stranded in the MC-Fold simulations. MC-Fold output consisted of 1000 dot-bracket solutions generated to explore 15% of the sub-optimal structures and the solutions were ordered based on total energy calculation. The single-stranded probability of each nucleotide was calculated by the number of occurrences of dot over bracket in the solution set, which was used to color code the most stable secondary structure prediction from MC-Fold in the text. In summary, the MC-Fold output was used to create the secondary structure maps as these results are directly related to the base-pairing probabilities (24, 25).
SHAPE reactivity data was provided as input to the MC-Pipeline in order to further refine the single-strandedness predictions. Non-negative SHAPE reactivity was re-scaled to range from 0 (no reactivity) to 1 (maximum reactivity) and sub-divided into three categories: highly reactive nucleotides with normalized SHAPE reactivities > 0.22 (excepting pri-mi-107 mut2, where the high cutoff was 0.37) were assigned a “medium” constraint strength in MC-Fold, reflecting those residues in the top 5% of SHAPE reactivity; all other statistically significant reactivity was defined by a normalized SHAPE reactivity between 0.03 and 0.22 and assigned a “low” constraint strength in MC-Fold; any nucleotide with a reactivity < 0.03 was deemed not to have statistically significant reactivity and therefore was not assigned a reactivity based structure constraint in MC-Fold. We found in early iterations that assigning “high” constraint strengths to the strongest SHAPE hits resulted in over-weighting of the SHAPE data that seemed to overwhelm the influence of MC-Fold on the final output and thus chose to utilize the more conservative constraint strengths reported.
The most probable secondary structure model from MC-Fold analysis with low resolution SHAPE constraints was used to generate the input for the MC-Sym model generation. MC-Sym was configured to use the September 2011 snapshot of the fragment library to generate the 3D structures. The top ten structures explored by MC-Sym were configured to use probabilistic exploration of tertiary structure with the model diversity set to 1.0 Å. In all MC-Sym runs, a total of 1000 sub-optimal structures were generated in order to provide adequate sampling of three-dimensional space prior to terminating sampling. The top five moste probable models generated, based on free energy calculation, were selected for further analysis.
Drosha Processing Assays
RNA substrates for in vitro assays were transcribed as previously described (16). FLAG-Drosha (16) and FLAG-DGCR8 (AddGene) (referred to as Microprocessor) or FLAG-GFP were overexpressed in HEK-293T cells and FLAG-tagged proteins were isolated on M2-FLAG beads (Sigma). Briefly, 6 μg of plasmid was transfected into cells using Lipofectamine 2000 (Invitrogen) as per manufacturer’s instructions. Mock treatment was exposed to lipofectamine but no plasmids were transfected into the cells. Approximately 48 hours after transfection cells were washed with phosphate buffered saline, harvested, and lysed via sonication in lysis buffer (20 mM Tris-HCl pH 8.0, 100 mM KCl, and 0.2 mM EDTA). Lysate was combined with 10 fmol of labeled RNA, 10X reaction buffer (64 mM MgCl2), and RNasin (Promega). The reaction mix was incubated at 37°C for 5 min. Products were phenol/chloroform extracted and ethanol precipitated. RNA was analyzed on 12% denaturing PAGE gels.
Results
SHAPE-derived Primary miRNA Structures
More than half of the nucleotides in a typical RNA are found in Watson-Crick base-pairs, but a number of non-canonical structural motifs are commonly embedded within A-form secondary structures (26). One of the most prevalent motifs is the single-nucleotide mismatch, or 1-by-1 internal loop, which is often (nearly) isosteric with canonical Watson-Crick pairs (27). Also common are single-nucleotide bulges and small asymmetric loops, which often serve as recognition motifs for proteins (28). Deep sequencing in Caenorhabditis elegans shows an average of two 1-by-1 loops in a typical pre-miRNA, along with an average of two single-nucleotide bulges (11). The three human pri-miRNAs in our study contain a diverse set of both symmetric internal loops and small bulges. When the structures of pri-mir-16-1 and pri-mir-107 were predicted by mfold (29), both exhibited several alternative structures within the 10% suboptimality parameter range. These alternative structures displayed differences in the size and location of internal loops and/or bulges, which could lead to heterogeneous final miRNA length (18); and variation in the terminal loop size, which has implications for processing efficiency (15).
Because the structural imperfections in pri-miRNA transcripts have direct consequences for processing and function, it is necessary that they be accurately annotated. We subjected each of the three pri-miRNAs in our panel to modification by the SHAPE reagent 1-methyl-7-nitroisatoic anhydride (1M7) and analyzed the results by fractionating fluorescently labeled cDNA products on denaturing gels to identify RNA secondary structure with single-nucleotide resolution. Protection from modification by a SHAPE reagent is expected for a site that is canonically base-paired or for mismatch sites that are constrained at the level of secondary structure (19). The resulting normalized SHAPE reactivity for each pri-miRNA indicates that the majority of the symmetric internal loops do not provide the SHAPE reagent with access to the 2′-hydroxyl, implying that these defects do not significantly alter the A-form helix of the double-stranded stem (Fig. 1, left panels). As positive controls, we also looked at the reactivity of bulges and asymmetric loops in the same sequences. In contrast to the above, these motifs did react with the SHAPE reagent, albeit to different extents.
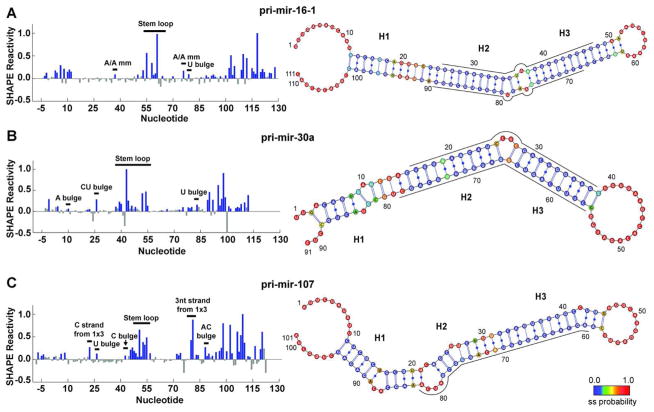
Figure 1.
SHAPE-constrained MC-Fold calculations yield secondary structures with embedded estimation of conformational dynamics. SHAPE reactivity traces (left) identify single-stranded nucleotides for (A) pri-mir-16-1, (B) pri-mir-30a, and (C) pri-mir-107. In the SHAPE reactivity traces, bar heights indicate the normalized mean reactivity constructed from 21 independent reactions. Blue filled bars indicate nucleotides with a positive mean reactivity and a magnitude greater than the uncertainty of the measurement. All grey bars indicate that the reactivity is either negative or has a mean magnitude below the uncertainty and are therefore considered insignificant. Addition of SHAPE-derived single-stranded constraints to MC-Fold calculations yields the combined probability of the nucleotide being single-stranded, which is mapped onto the most probable secondary structure (right). These probabilities are indicated in color as annotated in the color bar, ranging from most likely double-stranded (blue) to most likely single-stranded (red). Regions of high single-stranded probability divide the stems into three segments, labeled as H1, H2, and H3. Nucleotide numbering corresponds with the pri-miRNA numbering starting at 1 and the SHAPE cassette linkers (see Materials and Methods) being less than 1 and greater than the pri-miRNA length. Nucleotides corresponding to the mature miRNAs (as annotated in miRBase) are indicated by a line adjacent to the secondary structure diagram, which has been oriented such that the Drosha cut site is on the left in all three cases. Secondary. structure diagrams were generated in VARNA (54).
Symmetric internal loops, particularly 1-by-1 loops, are the most common base-pairing defect in pri-miRNAs. A total of six 1-by-1 internal loops are represented in the present data set (Table 1), but only the A/A mismatch in pri-mir-16-1 was modified above background by the SHAPE reagent. Consistent with this result, thermodynamic analysis indicates low helical stability for A/A mismatches as compared with most 1-by-1 loops (30). Chemical and enzymatic probing has also confirmed this result for the same mismatch in pre-mir-16-1 (14). Another symmetric internal loop of interest in our data set is the 2-by-2 AG•GA internal loop (i.e., GA tandem mismatch) located proximal to the Drosha cleavage site in pri-mir-16-1, for which Krol et al. propose a dramatically re-folded double-loop structure based on Ca2+ cleavage assays (14). This loop is not modified above background by the SHAPE reagent in the neighborhood of nucleotides 19–22, consistent with both an NMR structure reporting an AG•GA internal loop in a similar sequence context (31) and with thermodynamic data indicating high loop stability (32). It is noteworthy that there is a small but statistically significant amount of shape reactivity in the complementary strand, spanning nucleotides 92–95. Based on our SHAPE results and their consistency with findings in the literature, we conclude that most symmetric internal loops do not cause the average structure of pri-miRNAs to deviate significantly from A-form.
Table 1.
Complete annotation of secondary structure defects in the stem region of wild-type pri-mir-16-1, pri-mir-30a, and pri-mir-107
| Helical Imperfection | Found in | Sequence in RNA | SHAPE Detection | Single-stranded by Ribonucleases |
|---|---|---|---|---|
| U bulge | pri-mir-16-1 | 34-C--G-35 80-GUC-78 |
yes | no |
| U bulge | pri-mir-30a | 8-U--G-9 84-AUC-82 |
yes | no |
| U bulge | pri-mir-107 | 25-CUU-27 76-G--G-75 |
yes | no |
| A bulge | pri-mir-30a | 9-GAG-11 82-C--C-81 |
no | no |
| C bulge | pri-mir-107 | 41-CCU-43 61-G--A-60 |
no (small hit) | no |
| 2-nt bulge | pri-mir-30a | 25-CCUC-28 67-GG-66 |
yes | yes |
| 2-nt bulge | pri-mir-107 | 16-UG-17 89-ACAC-86 |
yes | no (small hit) |
| 1-by-1 symmetric internal loop* | pri-mir-16-1 | 23-CCU-25 91-GAA-89 |
no | no |
| 1-by-1 symmetric internal loop | pri-mir-16-1 | 36-UAA-38 77-AAU-75 |
yes | yes |
| 1-by-1 symmetric internal loop | pri-mir-16-1 | 45-GCG-47 68-CUC-66 |
no | no |
| 1-by-1 symmetric internal loop | pri-mir-16-1 | 47-GUU-49 66-CUA-64 |
no | no |
| 1-by-1 symmetric internal loop* | pri-mir-30a | 13-GAC-15 79-UCG-77 |
no | no |
| 1-by-1 symmetric internal loop | pri-mir-107 | 29-UUU-31 84-ACA-82 |
no(only C hit) | no (only C hit) |
| 2-by-2 symmetric internal loop | pri-mir-16-1 | 18-CAGU-21 96-GGAA-93 |
no | no for T1; yes for A |
| 1-by-3 asymmetric internal loop* | pri-mir-107 | 21-U--C--A-23 82-ACUAC-78 |
yes | yes |
Helical imperfections that surround the Drosha cleavage site are indicated by an asterisk.
While 1-by-1 loops were rarely modified by the SHAPE reagent, single-nucleotide bulges were often modified to a detectable extent. For example, each of the three U bulges were significantly modified (Table 1). There is also a weak SHAPE reactivity for the C bulge located near the terminal loop of pri-mir-107. The only single-nucleotide bulge not modified above background was the A bulge in pri-mir-30a (Fig. 1B, left), which is likely to be participating in purine-purine stacking on both sides. This result is consistent with previous studies monitoring protection from metal-induced catalytic cleavage at this site (33).
High normalized SHAPE reactivity indicates a lack of base-pairing and/or base stacking at the site of multiple-nucleotide bulges and asymmetric internal loops. Both two-nucleotide bulges present in the data set were strongly modified by the SHAPE reagent (Table 1), although one nucleotide in each pri-miRNA was modified much more heavily than the other (Fig. 1B and 1C; left). In the case of the 1-by-3 internal loop of pri-mir-107, both strands of the loop were heavily modified. Note that the single-nucleotide C-strand in this asymmetric loop was modified more heavily than any single-nucleotide bulge (Fig. 1C, left). The extensive SHAPE reactivity in this site is especially noteworthy, as similar patterns have been shown to correlate strongly with NMR observations of large amplitude conformational dynamics (34). As the predicted Drosha cleavage site is contained within this 1-by-3 loop, dynamic deformations in this region could inhibit the cleavage reaction. However, Drosha activity data shown below reveal that this is not the case.
Ribonuclease Cleavage Structure Mapping
Chemical modification with SHAPE reagents rapidly showed, with single nucleotide resolution, that some loops and bulges are minimally disruptive to A-form helical structure in pri-miRNA. The functional significance of this finding prompted us to verify the SHAPE results through limited digestions with a panel of ribonucleases specific for single-stranded and double-stranded RNA. Figure 2 shows ribonuclease mapping under native conditions for pri-mir-16-1, pri-mir-30a, and pri-mir-107. The cleavage patterns for all RNAs are largely consistent with the SHAPE results (compare Table 1, columns 4 and 5). Substantial single-stranded specific cleavage is only indicated for bulges and asymmetric internal loops larger than a single-nucleotide and the A/A mismatch located in pri-mir-16-1, which were also sensitive to the SHAPE reagent. Interestingly, RNase A cleaves all of the surrounding nucleotides of the CU bulge located in pri-mir-30a (Fig. 2B), suggesting that this bulge is highly dynamic, although this could simply reflect RNase A’s tendency to cleave nucleotides adjacent to loops. The propensity for the CU bulge to also exist as an alternative UC bulge is consistent with prior metal-catalyzed cleavage assays (14) and with the crystal structure of pre-mir-30a bound to Exportin-5 (8).
Figure 2.
Ribonuclease structure mapping is consistent with the most probable secondary structure resulting from the SHAPE-constrained MC-Fold calculations for (A) pri-mir-16-1, (B) pri-mir-30a, and (C) pri-mir-107. For each RNA, a denaturing 12% polyacrylamide gel used in the analysis is shown, with lanes as follows: C is a control sample (no nuclease); OH− is a limited alkaline digest; and T1, A, and V1 are limited digests with ribonucleases specific for single-stranded G, single-stranded C and U, and 5′ to double-stranded or well stacked single-stranded regions, respectively. The reactions in lanes 2 and 3 were performed under RNA-denaturing conditions (denoted ‘Den.’) in order to provide a ladder correlating position in the gel with the nucleotide sequence; while the reactions in lanes 4–6 were performed under RNA-native conditions (denoted ‘Nat.’). Helical and loop regions of the RNA are indicated to the right of the gel. The highest probability secondary structure (see Fig. 1) with positions of cleavage by ribonucleases under native conditions indicated by symbols as described in the legend is displayed below each gel. Symbol size is proportional to cleavage intensity. In these secondary structure maps, proposed Drosha cleavage sites are identified with red arrows; regions near Drosha cleavage sites displaying high single-strand probability in our MC-Fold and SHAPE analysis are enclosed in grey boxes.
Secondary Structure Refinement by MC-Fold
The MC-Pipeline is a modeling program that utilizes a nucleotide cyclic motif library containing base-pair contextual information, derived from crystal structures deposited in the PDB, to ultimately predict 3D folds of RNA structures (3). SHAPE chemistry provides structural constraints suitable for input into the MC-Pipeline, which has the potential to improve structure quality when rare sequence motifs are encountered or when ambiguous base-pairing is possible. Therefore, we generated secondary structure predictions in MC-Fold by identifying the joint probability that a nucleotide is single-stranded based on the MC-Pipeline database and our experimental SHAPE constraints (as described in the Methods). An alternative approach in which a large number of decoy structures is generated in MC-Pipeline and then scored against low-resolution experimental constraints to generate a sub-ensemble consistent with the data has recently been reported (35), but was not used here. The most probable secondary structure resulting from our combined SHAPE and MC-Fold analysis is reported for each pri-miRNA in Figure 1 on the right and color coded to indicate the combined single-stranded probability (the ten most probable secondary structures for each pri-miRNA are displayed in Figures S1–S3 of the Supporting Information).
Importantly, some nucleotides provide only weak indications of ssRNA character by either SHAPE or MC-Fold alone; however, combining the two sources of data yields clear indications of base-pairing status (results from MC-Fold analysis performed without reference to the SHAPE data are summarized in Figures S4–S6 of the Supporting Information). When SHAPE and MC-Fold analysis are combined, the single-stranded probabilities of nucleotides near the Drosha cleavage site on the stem closer to the ssRNA:dsRNA junction of all three RNAs (labeled as the H1 stem in Figure 1, right) are considerably larger than those of other nucleotides in the stem. Helical disruptions near the Drosha cut site have previously been predicted by bioinformatics in C. elegans (11) and our data provide the first experimental validation of this structural feature in pri-miRNAs of human origin.
Another structural feature of pri- and pre-miRNA hairpins that has been heavily studied is the terminal loop of the hairpin. In some prior studies thermodynamic algorithms (e.g., mfold) have been observed to predict excess base-pairing near terminal loops; over 70% of the pre-miRNAs evaluated in a recent study were predicted by mfold to have short loops inconsistent with experimental results (14). Application of MC-Fold to the present set of pri-miRNA sequences also produces minimally sized terminal loops when SHAPE data is not added as an input constraint (Figures S4–S6 of the Supporting Information). Enforced shortening of the terminal loop in pri-mir-16-1 through mutagenesis decreased processing efficiency (15), and replacement with a thermostable tetraloop eliminated processing completely (15), thus establishing the necessity of properly defining loop size. Analysis of the SHAPE-constrained MC-Fold secondary structures in Figure 1 confirms the presence of large terminal loops for all three RNAs studied, suggesting that their loops should not impair Drosha processing.
3D Structure Modeling Using MC-Sym: Global Features
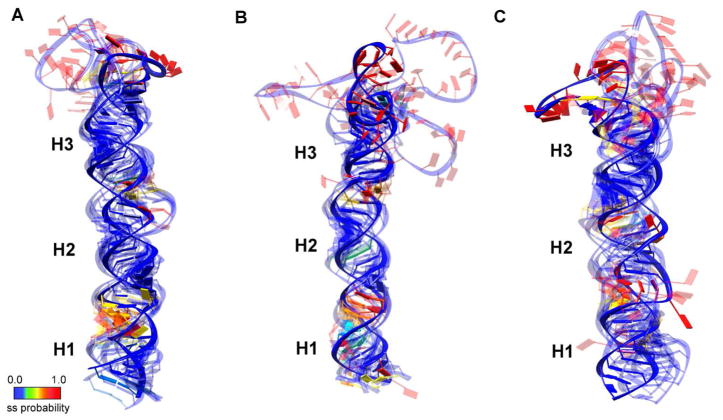
Developing mechanistic insight into biomolecular function often requires visualization of three-dimensional atomic structures. For example, coaxial stacking of helical segments to either side of disruptive motifs in the pri-miRNA stem, or the absence of it, influences the overall shape of the substrate recognized by the Microprocessor. The data discussed so far does not address these overall geometric features. Therefore, the highest scoring SHAPE-constrained MC-Fold secondary structures were provided as input for MC-Sym (3). The single-stranded flanking tails were excluded from the MC-Sym input because their inclusion resulted in stalling of the algorithm. It is noteworthy that the highly dynamic nature of such regions often results in their exclusion from high-resolution structure models (i.e., those derived from x-ray crystallography or NMR) as well. For similar reasons, caution is also required in attributing functional significance to the quantitative atomic positions in the terminal loops of the generated models, although the positioning of the single-stranded:double-stranded junction is robust. The five most probable structures of each pri-miRNA are superimposed in Figure 3, where they are aligned along the main stem of the RNA. The Drosha cleavage site is located between the regions labeled H1 and H2, directly above the portion of H1 with high single-stranded probability (red in Fig. 3), making the location of the cut site easily identifiable.
Figure 3.
Ensemble representation of the top five SHAPE-constrained models generated by MC-Sym for (A) pri-mir-16-1, (B) pri-mir-30a, and (C) pri-mir-107. All models are aligned along the main stem of the RNA with the sugar-phosphate backbone indicated by a blue ribbon. Planks representing the nucleotides are colored according to the probability of being single-stranded as reported in Figure 1 and indicated by the color bar, ranging from most likely double-stranded (blue) to most likely single-stranded (red). Regions of high single-stranded probability divide the stems into three segments, labeled as H1, H2, and H3. Inclusion of extended single-stranded tails renders MC-Sym calculations unstable; therefore, the expected tails are not represented in the models shown.
RNase III enzymes are expected to cleave most efficiently in the interior of A-form helical regions. In all three structure bundles, the regions with low SHAPE reactivity (shown in blue) overlap well, supporting their non-dynamic nature. In contrast, the regions with high SHAPE reactivity (shown in the range from yellow to red) have a large spatial distribution owing to their dynamic nature. The models predominantly favor a coaxial stack of the stem regions with looped-out bulges. Again, the higher dynamics and lower base-pairing probability of nucleotides at the H1/H2 boundary of all three bundles identify the basal stem adjacent to the Drosha cut site. It is especially noteworthy that while this region is likely to be more deformed than the majority of the RNA, coaxial stacking of H1 and H2 minimizes disruption of the A-form helix – even in the case of pri-mir-107. This result is consistent with the expectation of proper cut-site recognition by the RNase III domains of the Drosha enzyme.
Primary miRNA Structural Heterogeneity
In addition to the increased single-stranded probability adjacent to the Drosha cut site, combined SHAPE and MC-Pipeline analysis indicates at least one other region of lower base-pairing probability at the H2/H3 boundary in the labeling scheme of Figures 1–3. Warf et al. propose that periodic structural distortions in the RNA stem, co-localized on a single side of the molecule, could enable the formation of a strong bend in the pri-miRNA structure spanning three turns of helix (11). A bend of this nature is necessary for the DGCR8 binding model proposed by Sohn et al. (36). Analysis of conformationally heterogeneous sites in the SHAPE-constrained MC-Sym ensembles provides insight into the mechanism of deformation necessary to support this hypothesis, and suggests a need for future molecular mechanics calculations in order to define the dynamics uniting the reported pri-miRNA conformational landscapes.
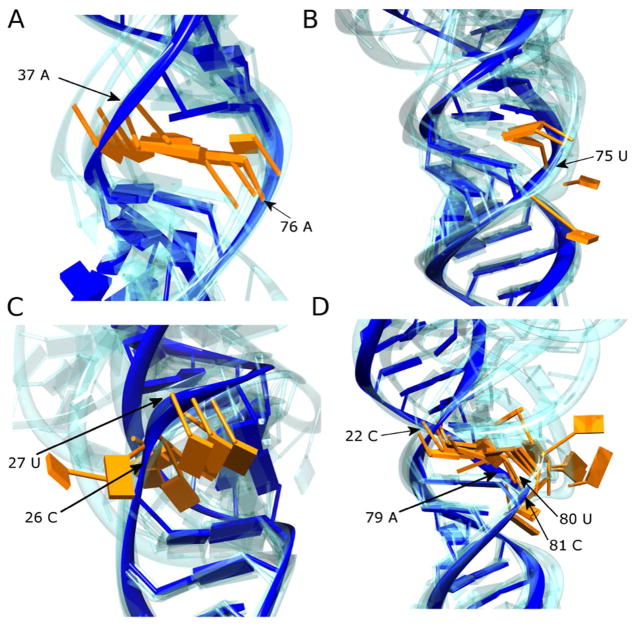
In pri-mir-16-1, an A/A mismatch found 11 nucleotides from the Drosha cut site (Fig. 4A), along with a nearby U bulge (Fig. 4B), is responsible for creating a second region of enhanced conformational variation (seen as the H2/H3 boundary in Figure 3a). Although both A nucleobases are stacked within the helix in each of the five most probable structures, base-pairing between them is poor. While the 5′UAA/3′AAU sequence containing the A/A mismatch found in pri-mir-16-1 was not present in the PDB at the time of writing, substantial evidence supports the dynamic nature of A/A mismatches in other sequence contexts. For example, when flanked by thermodynamically stable G/A sheared base-pairs, A/A was found to be highly dynamic by NMR (37). In addition, when flanked by Watson-Crick base-pairs, the A/A mismatch was found to be thermodynamically unstable (38). Moreover, the nearby U bulge in pri-mir-16-1 is flanked by a pyrimidine on its 5′-side, therefore yielding unfavorable 5′-pyrimidine-pyrimidine stacking (33) and further enhanced destabilization of this region.
Figure 4.
The secondary structures of pri-miRNA molecules harbor multiple dynamic bulges and internal loops. Expanded views of areas within the MC-Sym models that are highly dynamic are shown for (A) A/A mismatch in pri-mir-16-1, (B) U bulge in pri-mir-16-1, (C) CU bulge in pri-mir-30a, and (D) 1-by-3 asymmetric internal loop in pri-mir-107. The nucleotides involved in the imperfections are colored orange (their position in the nucleotide sequence is also annotated) and the most probable structure is shown otherwise in solid blue, with the models from the other four members of the ensemble reported in Figure 3 shown in transparent blue. All models are aligned to the nearest stable Watson-Crick base-pair neighboring the imperfection in the most probable model.
Asymmetric elements, such as the two-nucleotide bulge in pri-mir-30a and the 1-by-3 asymmetric loop in pri-mir-107, can feature either intercalative stacking or extrusion of the asymmetric base(s) from the duplex (28). The nucleobases of the two-nucleotide bulge in pri-mir-30a are extruded from the helix in all five structures, but deformation of the backbone imparts a bend to the overall structure (Fig. 4C). Consistent with the high SHAPE reactivity in pri-mir-107, the A and U nucleobases of the three-nucleotide strand in the asymmetric loop are extruded from the stack in all five structures (Fig. 4D), resulting in the introduction of a bend to the overall structure of the stem.
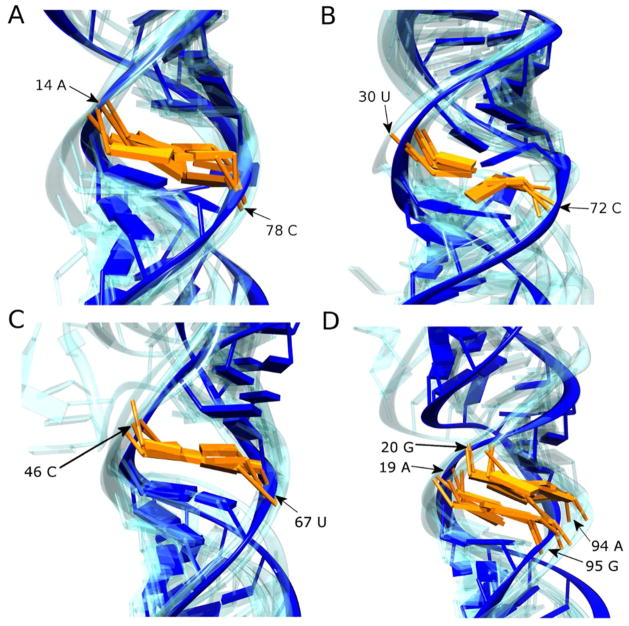
In contrast, many single-nucleotide mismatches do not significantly distort the RNA helix (39); thermodynamically, many do not even disrupt helix stability (40). For example, A/C mismatches are isosteric with G•U wobble pairs, although their stability is influenced by nearest-neighbor identity (41). In pri-mir-16-1, the A/C mismatch is neither reactive to the SHAPE reagent or RNase A cleavage (Table 1); nor is it predicted to be highly disordered by MC-Fold, which is consistent with the structure of an A/C mismatch from the large ribosomal subunit found in an identical sequence context (42). In pri-mir-30a, the A/C mismatch is similarly not reactive to the SHAPE reagent and RNase A, but the single-stranded probability from MC-Fold is high, suggesting deformability in the Drosha cut site (Fig. 1B, right). Analysis of the structure database shows that the strength of A/C base-pairing in similar sequence contexts is highly variable (27). The MC-Sym bundle for this mismatch in pri-mir-30a shows the most disorder of any non-canonical element with low SHAPE reactivity (Fig. 5A), suggesting weak base-pairing and a contribution to instability in this region.
Figure 5.
The secondary structures of pri-miRNA molecules harbor multiple non-Watson-Crick mismatches that are predicted to be well-ordered by SHAPE reactivity. Expanded views of areas within the MC-Sym models representing these mismatches are shown for (A) A/C mismatch in pri-mir-30a, (B) U/C mismatch in pri-mir-107, (C) C/U mismatch in pri-mir-16-1, and (D) AG•GA internal loop in pri-mir-16-1. The nucleotides involved in the imperfections are colored orange (their position in the nucleotide sequence is also annotated) and the most probable structure is shown otherwise in solid blue, with the models from the other four members of the ensemble reported in Figure 3 shown in transparent blue. All models are aligned to the nearest stable Watson-Crick base-pair neighboring the imperfection in the most probable model.
The U/C (or C/U) mismatch is also found in two of the three pri-miRNA sequences studied. Crystal structures of RNA stems harboring U/C mismatches show that, with the assistance of bridging waters, a hydrogen bonding network capable of promoting a good fit of the nucleobases within the RNA helix is achievable (43, 44). In both pri-mir-107 (Fig. 5B) and pri-mir-16-1 (Fig. 5C), MC-Sym ensembles display well-ordered stacking of these mismatches that support the possibility of water-mediated hydrogen bonding between the nucleobases. It is noteworthy that the C of the pri-mir-107 U/C mismatch was somewhat sensitive to both the SHAPE reagent and RNase A (Table 1), and a slight distortion in the backbone along the 3′-side of the mismatch is observed in the MC-Sym bundles.
Unique to the set of pri-miRNAs, a 2-by-2 AG•GA internal loop is found in pri-mir-16-1 in the basal H1 stem, adjacent to the Drosha cut site (Fig. 5D). GA tandem mismatches are generally thermodynamically stable (45), although both the thermodynamics of loop closure and the structures they adopt are highly sequence dependent (45–49). Although the PDB contains no examples of the exact 5′CAGU/3′GGAA sequence found in pri-mir-16-1, the Watson-Crick face base-pairing observed in the MC-Sym bundles (Fig. 5D) is consistent with the general literature trend (47, 49). Despite the relatively high order seen in the MC-Sym bundles for the AG•GA internal loop and nearby A/C mismatch, these non-canonical elements collectively contribute to a high probability of single-strandedness in the H1 stem that identifies the Drosha cut site.
Drosha Processing of Primary miRNAs
To verify the hypothesis that imperfections in the A-form helix near the Drosha cut site are necessary for efficient cleavage by the Microprocessor, in vitro Drosha processing assays were conducted with wild-type pri-mir-16-1 and pri-mir-107. For both, mutants were designed to modify the flexible region near the Drosha cut site. Of note, Drosha activity is dependent on the presence of Mg2+ in the reaction buffer, whereas our SHAPE reactions were performed in the absence of divalent metals in order to minimize the risk of metal-catalyzed cleavage causing false positives. As specific metal binding is most likely to affect tertiary structure, which does not exist in our single-hairpin transcripts, we feel there is minimal risk that the structures are altered significantly by Mg2+ in the cleavage assays. Control SHAPE reactions with pri-mir-107 in the presence of Mg2+ showed cleavage profiles identical to those reported in Figure 1 (data not shown). On the whole, our results support a correlation between cut site flexibility and processing efficiency.
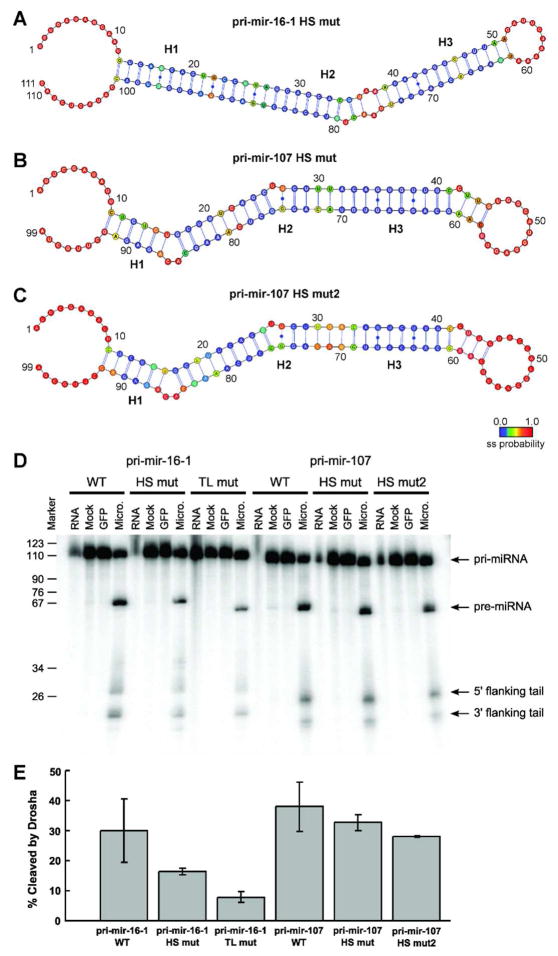
For pri-mir-16-1, the flexible hot spot adjacent to the Drosha cut site was eliminated by mutating both the AG•GA internal loop and the A/C mismatch to Watson-Crick base-pairs (labeled pri-mir-16-1-HS mut in Fig. 6). The hot spot region was successfully rigidified, as confirmed by combined SHAPE and MC-Fold analysis (Fig. 6A). While wild-type pri-mir-16-1 was processed efficiently in the in vitro assays, processing of the cut site mutant was reduced two-fold (Fig. 6E). As a negative control for processing, we created another mutant of pri-mir-16-1 in which the large terminal loop was replaced by a UUCG tetraloop (labeled pri-mir-16-1-TL mut in Fig. 6). Consistent with previous studies where similar tetraloops were shown to diminish processing (9, 15), pri-mir-16-1-TL processing was reduced approximately four-fold (Fig. 6E). Thus, deformability of the stem near the cut site is not the sole factor important for cleavage by the Microprocessor, but its presence significantly improves processing efficiency.
Figure 6.
Drosha processing of pri-miRNAs to pre-miRNAs in vitro confirms the necessity of hot spot flexibility for efficient cleavage. (A) The mutant pri-mir-16-1-HS has significantly reduced hot spot flexibility compared with wild-type, as established by combined SHAPE and MC-Fold analysis. (B) The mutant pri-mir-107-HS has the asymmetric 1-by-3 internal loop near the Drosha cleavage site replaced by a flexible C•C non-canonical base-pair, (C) while the mutant pri-mir-107-HS2 has only Watson-Crick base-pairs at the cleavage site, as established in both cases by combined SHAPE and MC-Fold analysis. (D) Denaturing gels for the processing of (from left to right) pri-mir-16-1 wild-type (WT), hot spot mutant (HS mut), and tetraloop mutant (TL mut) constructs; in addition to pri-mir-107 wild-type (WT), hot spot mutant (HS mut), and second hot spot mutant (HS mut2) constructs. In all six assays, lanes represent RNA collected prior to addition of Microprocessor (RNA), exposed to FLAG-beads with addition of cell lysate that did not express FLAG-tagged proteins for 5 minutes (Mock), exposed to GFP for 5 minutes (GFP), and 5 minutes after exposure to purified Microprocessor (Micro.). (E) The percentage of pri-miRNAs cleaved by the Microprocessor in vitro after 5 minutes averaged over three independent experiments. Cleavage is calculated as the sum of the intensities of the pre-miRNA product and the cleaved flanking tails divided by the sum of the intensities of the product, tails, and the remaining pri-miRNA substrate.
Having confirmed that flexibility near the Drosha cut site is favorable for efficient processing, we next determined whether asymmetric loops, which are flexible, mitigate these positive effects. Despite the presence of the 1-by-3 internal loop at the cut site, wild-type pri-mir-107 was processed just as efficiently as pri-mir-16-1 in our assay. In order to determine the effect of an asymmetric loop versus a symmetric internal loop on the overall cleavage efficiency, we created a mutant (labeled pri-mir-107-HS in Fig. 6) with a two-base deletion on the 3-nucleotide side of the loop, leaving a symmetric 1-by-1 loop (i.e., a C/C mismatch). The SHAPE reactivity profile and MC-Pipeline results for pri-mir-107-HS confirm the formation of a C/C mismatch and show that the mutant pri-mir-107 is still deformable near the Drosha cut site (Fig. 6B). Cleavage efficiency was slightly decreased for the mutant pri-mir-107 in comparison to the wild-type (Fig. 6E). Based on the migration of the pre-mir-107 band for the mutant, which migrates as a species two nucleotides shorter than wild-type (compare WT and HS mut in Fig. 6D), creating a symmetric 1-by-1 loop eliminated two nucleotides from the predicted pre-mir-107 product, but otherwise did not change the position of Drosha cleavage. In addition to this mutant, another mutant of pri-mir-107 was created to test the impact of a complete lack of deformability at the cleavage site as was done with pri-mir-16-1. The second mutant for pri-mir-107 had the asymmetric internal loop replaced with a Watson-Crick base-pair (labeled pri-mir-107-HS2 in Fig. 6), whose structure was again confirmed by the combined SHAPE/MC-Pipeline analysis (Fig. 6C). As with pri-mir-16-1, a decrease in Drosha cleavage efficiency was seen when all flexibility at the cleavage site was abolished (Fig. 6E). Therefore, we conclude that the presence of an asymmetric loop near the Drosha cut site is less disruptive to processing than the total absence of flexibility in the stem.
Discussion
Previous conclusions made about pri-miRNA structure-function relationships have been based on the RNA secondary structure either predicted by mfold (29), derived from deep-sequencing analysis (11), or in some cases based on biochemical data (9, 15, 18). There are currently no solved atomic resolution pri-miRNA structures available, which impedes further progress. In this study, we have shown that providing SHAPE-derived base-pairing constraints to the MC-Pipeline produces pri-miRNA atomic structure models that are consistent with existing biochemical data and support prevailing mechanistic hypotheses. This study employed pri-miRNA sequences of human origin and is broadly consistent with C. elegans deep-sequencing analysis – both validating the generalization of previous conclusions and encouraging future in vivo genetics studies utilizing this attractive model organism. Moreover, the helical distortions quantified in our analysis plausibly resolve inconsistencies in accepted thermodynamic secondary structure models, while accounting for the structural heterogeneity predicted from bioinformatic analysis.
In the prevailing model for Dicer cut-site recognition, the 3′-overhang left by Drosha cleavage is bound by the Dicer PAZ domain and simple steric measurement of the expected A-form helix length from that reference point provides a “molecular ruler,” identifying the cleavage site for Dicer (50, 51). Demonstrating an equivalent molecular ruler for Drosha has been elusive. As can be seen in the SHAPE-constrained MC-Fold secondary structures in Figure 1 (right panels), there is a high degree of conformational variation in the H1 stem adjacent to the Drosha cut site. These deformable “hot spots” arise in the data due to a combination of SHAPE reactivity and the probability of being single-stranded as determined directly by MC-Fold. The deformable hot spot region located adjacent to the Drosha cut site was shown to enhance processing via in vitro Drosha cleavage assays (Fig. 6). This result suggests that the degree of base-pairing surrounding the Drosha cut site affects how the Microprocessor locates and/or cleaves the pri-miRNA.
The ssRNA:dsRNA junction at the base of the stem, one turn of A-form helix removed from the Drosha cut site, has previously been identified as a necessary structural element for cut-site recognition by DGCR8 (9). Warf et al. propose the existence of an unstable face on the pri-miRNA stem featuring strong structural distortions at the cleavage site that may serve as a marker, negating the need for a molecular ruler in Drosha capable of measuring from a fixed reference (11). We propose that our MC-Sym predicted models support previous biochemical models in which DGCR8 binds proximal to the ssRNA:dsRNA junction (5) – identifying the basal stem in analogy to the role of the PAZ domain in Dicer – and senses the periodic deformable sites that occur approximately once per turn of A-form helix, as evident in Figure 3. The periodic deformable sites therefore could permit bending of the pri-miRNA, which is needed to engage both DGCR8-dsRBD binding faces in the Sohn et al. (36) model. Collectively, the Microprocessor is able to specifically identify this set of features in only those nuclear dsRNAs that require cutting by Drosha. Our Drosha processing assays support the general hypothesis that cut site deformability contributes to processing efficiency; future testing of this model through DGCR8 and Drosha binding assays will clarify the molecular mechanism of this effect.
A potential consequence of inaccurate cut site identification by the Microprocessor is variability in pre-miRNA length and, ultimately, mature miRNA sequence – resulting in markedly dramatic effects if the seed sequence is altered. Warf et al. suggest that Drosha is capable of producing both typical (e.g., 2-nt 3′-overhang) and atypical overhangs based on the observation of variable overhang lengths in C. elegans pre-miRNAs (11). Improper overhang lengths may be the result of improper Drosha positioning caused by large imperfections at the cleavage site, as could be the case for pri-mir-107 – a hypothesis supported by the numerous mature miRNA sequences reported from deep sequencing reads for miR-107 in miRBase. On the other hand, an intriguing recent study by Park et al. shows that Dicer may be able to use 5′-end binding as an alternative to 3′-end recognition in anomalous cases like this (52). While the models in our MC-Sym structure bundles show a remarkable extent of coaxial stacking of the H1 and H2 stems surrounding the Drosha cut site in this molecule, it remains possible that looping out of the 3-nucleotide side of the bulge will produce an atypical overhang length. Moreover, the asymmetric loop in pri-mir-107 may exert a stronger influence on the kinetics of processing by Drosha than on the ultimate sequence composition of the mature miRNA.
Resolving the remaining questions surrounding the miRNA maturation process will require experimental determination of atomic resolution structure models for pri-miRNAs and pre-miRNAs both in isolation and when bound to their processing complexes. Combining SHAPE chemistry with analysis in MC-Pipeline enables the generation of high quality structure models that are both predictive and capable of unifying previous biochemical hypotheses. Minor imperfections within the A-form helix central in the pri-miRNA stem result in minimal disruption of the A-form structure, but impart significant deformability that may be sufficient to guide the Microprocessor to the appropriate cleavage site. Combination of our methodology with recent advances in high-throughput SHAPE analysis – so-called SHAPE-seq methods (53) – offers the possibility to generate hundreds of comparable structure bundles in the future from which the present findings can be generalized. Application to other problems in RNA metabolism, such as the evaluation of structures adopted by riboswitches and other regulatory elements, should be straight-forward.
Supplementary Material
Acknowledgments
Funding Sources:
This work was supported by the US National Institutes of Health grant R01GM098451 and startup funds from the Pennsylvania State University to SAS. MLH was supported in part by the US National Institutes of Health grant R01NS069759 and MAH was supported in part by F31NS076237.
This work was supported by the US National Institutes of Health grant R01GM098451 and start-up funds from the Pennsylvania State University to SAS. MLH was supported in part by the US National Institutes of Health grant R01NS069759 and MAH was supported in part by F31NS076237. We thank Phil Bevilacqua, Durga Chadalavada, and Kit Kwok for helpful discussion while developing the experimental protocols.
Abbreviations
- miRNA
microRNA
- pri-miRNA
primary microRNA
- SHAPE
selective 2′-hydroxyl acylation analyzed by primer extension
- RNase
ribonuclease
- DGCR8
DiGeorge critical region 8
- pre-miRNA
precursor microRNA
- ss
single-stranded
- ds
double-stranded
- 1M7
1-methyl-7-nitroisatoic anhydride
Footnotes
Author Contributions:
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Figures S1–S6. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Mathews DH, Moss WN, Turner DH. Folding and finding RNA secondary structure. Cold Spring Harb Perspect Biol. 2010;2:a003665. doi: 10.1101/cshperspect.a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weeks KM, Mauger DM. Exploring RNA Structural Codes with SHAPE Chemistry. Acc Chem Res. 2011;44:1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 6.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 8.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 9.Han JJ, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho YJ, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 10.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warf MB, Johnson WE, Bass BL. Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer. RNA. 2011;17:563–577. doi: 10.1261/rna.2432311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sashital DG, Doudna JA. Structural insights into RNA interference. Curr Opin Struct Biol. 2010;20:90–97. doi: 10.1016/j.sbi.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bindewald E, Wendeler M, Legiewicz M, Bona MK, Wang Y, Pritt MJ, Le Grice SF, Shapiro BA. Correlating SHAPE signatures with three-dimensional RNA structures. RNA. 2011;17:1688–1696. doi: 10.1261/rna.2640111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem. 2004;279:42230–42239. doi: 10.1074/jbc.M404931200. [DOI] [PubMed] [Google Scholar]
- 15.Zeng Y, Zhang XX. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010;38:7689–7697. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–4640. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang GL, Huang QW, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, Krzyzosiak WJ. Structural basis of microRNA length variety. Nucleic Acids Res. 2011;39:257–268. doi: 10.1093/nar/gkq727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 20.Wostenberg C, Quarles KA, Showalter SA. Dynamic Origins of Differential RNA Binding Function in Two dsRBDs from the miRNA “Microprocessor” Complex. Biochemistry. 2010;49:10728–10736. doi: 10.1021/bi1015716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kladwang W, Vanlang CC, Cordero P, Das R. Understanding the Errors of SHAPE-Directed RNA Structure Modeling. Biochemistry. 2011;50:8049–8056. doi: 10.1021/bi200524n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon S, Kim J, Hum J, Kim H, Park S, Kladwang W, Das R. HiTRACE: high-throughput robust analysis for capillary electrophoresis. Bioinformatics. 2011;27:1798–1805. doi: 10.1093/bioinformatics/btr277. [DOI] [PubMed] [Google Scholar]
- 24.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathews DH. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagaswamy U, Larios-Sanz M, Hury J, Collins S, Zhang Z, Zhao Q, Fox GE. NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res. 2002;30:395–397. doi: 10.1093/nar/30.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis AR, Kirkpatrick CC, Znosko BM. Structural characterization of naturally occurring RNA single mismatches. Nucleic Acids Res. 2011;39:1081–1094. doi: 10.1093/nar/gkq793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermann T, Patel DJ. RNA bulges as architectural and recognition motifs. Structure. 2000;8:R47–54. doi: 10.1016/s0969-2126(00)00110-6. [DOI] [PubMed] [Google Scholar]
- 29.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kierzek R, Burkard ME, Turner DH. Thermodynamics of single mismatches in RNA duplexes. Biochemistry. 1999;38:14214–14223. doi: 10.1021/bi991186l. [DOI] [PubMed] [Google Scholar]
- 31.Wu M, SantaLucia J, Jr, Turner DH. Solution structure of (rGGCAGGCC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry. 1997;36:4449–4460. doi: 10.1021/bi9625915. [DOI] [PubMed] [Google Scholar]
- 32.Wu M, McDowell JA, Turner DH. A periodic table of symmetric tandem mismatches in RNA. Biochemistry. 1995;34:3204–3211. doi: 10.1021/bi00010a009. [DOI] [PubMed] [Google Scholar]
- 33.Ciesiolka J, Michalowski D, Wrzesinski J, Krajewski J, Krzyzosiak WJ. Patterns of cleavages induced by lead ions in defined RNA secondary structure motifs. J Mol Biol. 1998;275:211–220. doi: 10.1006/jmbi.1997.1462. [DOI] [PubMed] [Google Scholar]
- 34.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisien M, Major F. Determining RNA three-dimensional structures using low-resolution data. Journal of Structural Biology. 2012;179:252–260. doi: 10.1016/j.jsb.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y. Crystal structure of human DGCR8 core. Nature Struct Mol Biol. 2007;14:847–853. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- 37.Chen G, Kennedy SD, Qiao J, Krugh TR, Turner DH. An alternating sheared AA pair and elements of stability for a single sheared purine-purine pair flanked by sheared GA pairs in RNA. Biochemistry. 2006;45:6889–6903. doi: 10.1021/bi0524464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis AR, Znosko BM. Thermodynamic characterization of single mismatches found in naturally occurring RNA. Biochemistry. 2007;46:13425–13436. doi: 10.1021/bi701311c. [DOI] [PubMed] [Google Scholar]
- 39.Nagaswamy U, Voss N, Zhang Z, Fox GE. Database of non-canonical base pairs found in known RNA structures. Nucleic Acids Res. 2000;28:375–376. doi: 10.1093/nar/28.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroeder SJ, Burkard ME, Turner DH. The energetics of small internal loops in RNA. Biopolymers. 1999;52:157–167. doi: 10.1002/1097-0282(1999)52:4<157::AID-BIP1001>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 41.Tran T, Disney MD. Molecular recognition of 6′-N-5-hexynoate kanamycin A and RNA 1×1 internal loops containing CA mismatches. Biochemistry. 2011;50:962–969. doi: 10.1021/bi101724h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 43.Cruse WB, Saludjian P, Biala E, Strazewski P, Prange T, Kennard O. Structure of a mispaired RNA double helix at 1.6-A resolution and implications for the prediction of RNA secondary structure. Proc Natl Acad Sci USA. 1994;91:4160–4164. doi: 10.1073/pnas.91.10.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holbrook SR, Cheong C, Tinoco I, Jr, Kim SH. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 1991;353:579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- 45.SantaLucia J, Jr, Kierzek R, Turner DH. Effects of GA mismatches on the structure and thermodynamics of RNA internal loops. Biochemistry. 1990;29:8813–8819. doi: 10.1021/bi00489a044. [DOI] [PubMed] [Google Scholar]
- 46.Christiansen ME, Znosko BM. Thermodynamic characterization of the complete set of sequence symmetric tandem mismatches in RNA and an improved model for predicting the free energy contribution of sequence asymmetric tandem mismatches. Biochemistry. 2008;47:4329–4336. doi: 10.1021/bi7020876. [DOI] [PubMed] [Google Scholar]
- 47.Walter AE, Wu M, Turner DH. The stability and structure of tandem GA mismatches in RNA depend on closing base pairs. Biochemistry. 1994;33:11349–11354. doi: 10.1021/bi00203a033. [DOI] [PubMed] [Google Scholar]
- 48.Tolbert BS, Kennedy SD, Schroeder SJ, Krugh TR, Turner DH. NMR structures of (rGCUGAGGCU)2 and (rGCGGAUGCU)2: probing the structural features that shape the thermodynamic stability of GA pairs. Biochemistry. 2007;46:1511–1522. doi: 10.1021/bi061350m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammond NB, Tolbert BS, Kierzek R, Turner DH, Kennedy SD. RNA internal loops with tandem AG pairs: the structure of the 5′GAGU/3′UGAG loop can be dramatically different from others, including 5′AAGU/3′UGAA. Biochemistry. 2010;49:5817–5827. doi: 10.1021/bi100332r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 51.Lau PW, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ. The molecular architecture of human Dicer. Nat Struct Mol Biol. 2012;19:436–440. doi: 10.1038/nsmb.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.