Abstract
Background
Occupational exposure to nickel (Ni) is associated with an increased risk of lung and nasal cancers. Ni compounds exhibit weak mutagenic activity, alter the cell’s epigenetic homeostasis, and activate signaling pathways. However, changes in gene expression associated with Ni exposure have only been investigated in vitro. This study was conducted in a Chinese population to determine whether occupational exposure to Ni was associated with differential gene expression profiles in the peripheral blood mononuclear cells (PBMCs) of Ni-refinery workers when compared to referents.
Methods
Eight Ni-refinery workers and ten referents were selected. PBMC RNA was extracted and gene expression profiling was performed using Affymetrix exon arrays. Differentially expressed genes between both groups were identified in a global analysis.
Results
There were a total of 2756 differentially expressed genes (DEG) in the Ni-refinery workers relative to the control subjects (FDR adjusted p<0.05) with 770 up-regulated genes and 1986 down-regulated genes. DNA repair and epigenetic genes were significantly overrepresented (p< 0.0002) among the DEG. Of 31 DNA repair genes, 29 were repressed in the high exposure group and two were overexpressed. Of the 16 epigenetic genes 12 were repressed in the high exposure group and 4 were overexpressed.
Conclusions
The results of this study indicate that occupational exposure to Ni is associated with alterations in gene expression profiles in PBMCs of subjects.
Impact
Gene expression may be useful in identifying patterns of deregulation that precede clinical identification of Ni-induced cancers.
Keywords: nickel, nickel refinery workers, gene expression, metals, carcinogenesis
INTRODUCTION
Nickel (Ni) compounds are ubiquitous pollutants in occupational and environmental settings. In the environment, Ni is found as a contaminant in the air and drinking water, or as either a constituent or contaminant in a variety of foods including chocolate, peanuts, and grains (1). Environmental exposure to Ni also occurs by dermal contact with industrial and commercial products containing nickel alloys including jewelry, household and cooking utensils, orthodontic appliances, dental tools, orthopedic implants, and batteries (1). Occupational exposure to Ni occurs by inhalation in the mining, refining, alloy production, electroplating, and welding industries (2–4).
Human exposure to Ni-containing products can result in a variety of adverse health effects. Nickel allergy is one of the most common causes of contact allergic dermatitis (5). Ni is of great environmental concern, since epidemiological studies of Ni compounds from occupationally exposed populations have reported an increased incidence of lung and nasal cancers and elevated risks of acute respiratory syndromes, most clearly demonstrated in Ni refinery workers (2, 3). Ni compounds have been classified as class 1A human carcinogens by the International Agency for Research on Cancer (IARC) (6).
To date, the precise molecular mechanism(s) of nickel carcinogenesis has not been clearly defined. Using animal and in vitro cell models, nickel compounds have been shown to promote the generation of reactive oxygen species (ROS) (7); interact directly or indirectly with nucleic acids and cause genotoxic damage (8); activate signaling pathways such as PI3K/AKT (9), HIF-1α (10), NF-κB (11–13), AP-1 (12, 13), and NFAT (14). Additionally, chromatin structural alterations and changes in epigenetic marks have been implicated in nickel carcinogenesis (15–20). Previous studies have shown that in vitro exposure to Ni induces global DNA hypermethylation and can perturb global and gene specific levels of DNA methylation and post-translational histone modifications. Ni-induced changes in post-translational histone modifications can include loss of acetylation on histones H2A, H2B, H3, and H4, as well as increases in histone H3K9 dimethylation, H3K4 trimethylation, and H2A and H2B ubiquitylation (16, 17, 21, 22).
The changes in gene expression patterns that occur in human populations occupationally exposed to Ni compounds have not been previously studied. Gene expression may be useful in identifying patterns of deregulation that precede clinical identification of Ni-induced cancers. Several studies have suggested that gene expression data from peripheral blood mononuclear cells (PBMCs) can provide valuable information about exposure levels (23–27). The use of PBMCs has tremendous potential in a therapeutic setting since the use of blood as a surrogate for the primary tissue of interest greatly facilitates sample collection and analysis. Here, we have compared the differential gene expression profiles between subjects with occupational exposure to Ni (Ni-refinery workers) and referent subjects with low level environmental exposures to identify gene expression changes associated with exposure to high levels of Ni.
METHODS
Study Site and Subject Recruitment
This study was conducted among Ni refinery workers in Jinchang, China, and local residents in Gansu, China. Jinchang City, where the nickel refinery is located, is within the providence of Gansu. Study selection was based on the fact that the subjects from the high exposure group were workers of the Ni refinery and the referent subjects were local residents of the same providence, but with no known occupational exposure to nickel. The human subject protocol for this study was approved by the institutional review boards of both the New York University School of Medicine and the Lanzhou University School of Public Health. Written informed consent was obtained from all participating subjects.
The subjects for this study were randomly selected from a study population previously described (20). Eighteen healthy male subjects between 42 and 56 years of age were recruited; subjects diagnosed with chronic diseases, including cancer, were excluded. Eight subjects who had high occupational exposure to Ni having worked for at least 1 year at a Ni refinery in Jinchang, China in the flash smelting workshop where sulfidic Ni ores were processed and ten referent subjects from residents in Gansu, China who were maintenance or office workers, with no reported occupational exposure to Ni, were also selected. Ni refinery workers are exposed to ambient Ni concentrations as high as 1 mg/m3. Referent subjects are exposed to ambient air concentrations of 204.8 ± 268.6 ng/m3 of Ni. This study was conducted to determine whether Ni exposure was associated with differential gene expression profiles in the PBMCs of subjects.
Sample collection and handling
Blood collection, handling, and PBMC isolation for this study was performed as previously described (20). Fifty ml of urine was also collected from each subject. The isolated PBMC pellet was suspended in TRIzol (Invitrogen) and urine samples were stored at −80°C and hand-carried frozen on dry ice to New York University.
RNA isolation, amplification, and hybridization
Total RNA was extracted from each sample according to the TRIzol (Invitrogen) manufacturer’s protocol. cRNA probes were synthesized and labeled using GeneChip Whole Transcript Terminal Labeling Expression Kit (Affymetrix) and were subjected to hybridization with GeneChip Human Gene 1.0 ST Array (Affymetrix) that contains 28,869 well annotated genes. Hybridization and scanning of the arrays was performed using a standard procedure.
Data analysis to identify differentially expressed genes
Gene expression analyses were performed using R. Gene expression data were imported and normalized in batches using the Affymetrix package version 1.30.0 in R 2.13.2 GUI 1.42 Leopard build 32-bit and robust multichip average (RMA) (28, 29). Samples were then batch normalized and log-transformed using an empirical Bayes approach with the R package COMBAT run through SVA 3.0.2 in R 2.14.0 GUI 1.42 Leopard build 32-bit (30). Significance of gene expression changes between the exposed and referent populations was assessed using a gene-wise linear model approach with LIMMA 3.8.3, which utilizes an empirical Bayes approach to generate moderated t-statistics by taking into account the standard errors and estimated log-fold changes (31). P-values were subjected to FDR correction for multiple hypothesis testing and adjusted p-values with p<0.05 were considered significant (32). Genes that met significance thresholds via this approach were included in our study set for further analysis and are referred to as differentially expressed genes (DEG) or the “study set”. All microarray data is MIAME compliant and the raw data has been deposited in NCBIs Gene Expression Omnibus (GEO), and assigned Series accession number GSE40392.
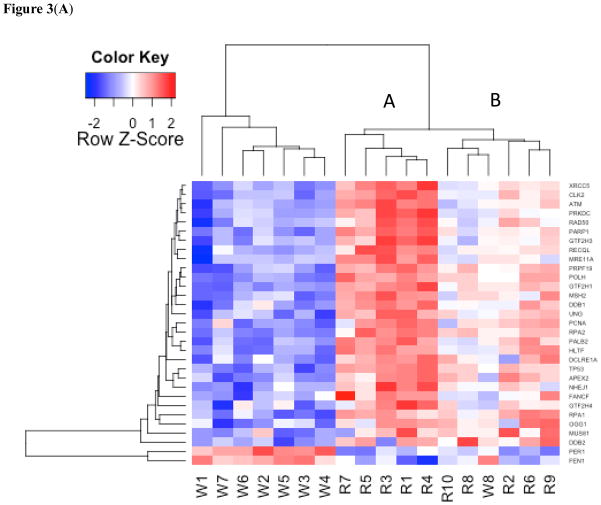
To investigate the biological relevance of the genes in our study set, we assessed the level of representation of previously established molecular signatures. Utilizing the molecular signatures database (Broad Institute Gene Set Enrichment Analysis) the DEG were evaluated for overlap with gene sets related to position, biological function, and disease states. This analysis led to the identification of a gene set involved in nasopharyngeal carcinoma (NPC) and further analysis was performed using this experimentally generated set (33). DNA repair genes were also investigated based on their previously established link with NPC and were tested for their overrepresentation in the DEG set using a hypergeometric distribution as previously described (34). Based on this overlap a list of 31 DNA repair genes were identified in the DEG and used in further analysis. Epigenetic genes were investigated based on nickel’s known role as an epigenetically active agent and were analyzed in the same manner as the DNA repair genes. Heat maps and PCA plots were generated using R. The heat map in Figure 1 was generated using the study set. The heat map in Figure 2 was generated based on the intersection of the study set and genes down-regulated in NPC (33). The heat map in Figure 3A was generated by taking the intersection of the study set and an extensive list of DNA repair genes to generate a gene set of significantly changed DNA repair genes (35–37). The heat map in Figure 3B was generated by taking the intersection of the study set and a compiled list of epigenetic related genes (38, 39). Correlation analysis for heat maps was performed using Pearson correlation analysis and hierarchical clustering along both axes was performed using the average method.
Figure 1. Gene expression profiles of PBMCs from Ni-refinery workers when compared with referents.
(A) Principal Components Analysis revealed distinct separation between Ni-refinery workers and referents. Red circles: Ni-refinery workers; blue circles: referents, subjects with environmental exposure. (B) Hierarchical clustering analysis of 2756 differentially expressed genes (DEG) (adjusted p-value <0.05) in which samples were sorted based on the similarity of gene expression revealed distinct separation between Ni-refinery workers (designated as W in figure) and referents (designated as R).
Figure 2. Heat map of genes associated with nasopharyngeal carcinoma.
See Table S2 for full list of genes.
Figure 3.
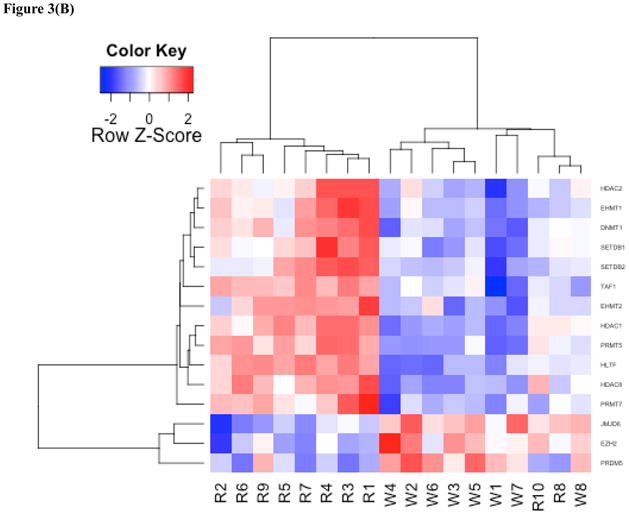
(A) Heat map of DNA repair genes. DNA repair genes include the following from top to bottom: XRCC5, CLK2, ATM, PRKDC, RAD50, PARP1, GTF2H3, RECQL, MRE11A, PRPF19, POLH, GTF2H1, MSH2, DDB1, UNG, PCNA, RPA2, PALB2, HLTF, DCLRE1A, TP53, APEX2, NHEJ1, FANCF, GTF2H4, RPA1, OGG1, MUS81, DDB2, PER1, FEN1. See Table S3 for the list of genes and associated p-values. (B) Heat map of epigenetic genes. Epigenetic genes include the following from top to bottom: HDAC2, EHMT1, DNMT1, SETDB1, SETDB2, MYST1, PMRT7, HDAC1, PRMT5, HDAC8, TAF1, EHMT2, MYST2, EZH2, JMJD6, and PRMD5. See Table S4 for the list of genes and associated p-values.
Real-time quantitative PCR
Total RNA was extracted from each sample using TRIzol Reagent (Invitrogen), and converted to single stranded cDNA using Superscript III (Invitrogen). Quantitative real-time PCR (q-RT-PCR) analysis was performed using SYBR green PCR system (Applied Biosystems) on ABI prism 7900HT system (Applied Biosystems). All q-RT-PCR reactions were performed in triplicate. Due to lack of sufficient biological material per sample, eight different RNA samples from the same study cohort that were not part of the original microarray analysis were utilized for the q-RT-PCR analysis. Quantitative real-time PCR was performed on CCL20, IL-6 and FOSB (up-regulated Ni-refinery workers) and CX3CR1 and MNDA (down-regulated in Ni-refinery workers) in four Ni-refinery workers and four referents. The data is presented as the absolute fold-change in gene expression of Ni-refinery worker samples relative to the referents. The expression of the target genes was first normalized to the expression of the house-keeping gene (GADPH) for the referent and Ni-refinery worker samples. Then the mean of the Ni-refinery worker samples was normalized to the mean of the referent samples.
Measurement of urinary nickel, cotinine, and creatinine
Urinary Ni was used to index the individual personal exposure to Ni and was analyzed for all study subjects by inductively coupled plasma mass spectroscopy (ICP-MS) (Elan DRCII, Perkin Elmer, USA) (40). Urinary cotinine, a major metabolite of nicotine and a valid biomarker of environmental tobacco smoke, was measured in each subject to confirm smoking status and control its potential confounding effects. Urinary cotinine was measured using a cotinine direct enzyme-linked immunosorbent (ELISA) kit (Immunalysis, Pomona, CA) (41). Urinary creatinine was measured in order to adjust the Ni and cotinine levels in urine samples. Urinary creatinines were determined using the creatinine incorporating dynamic stabilization technology assay kits (Fisher Scientific, Pittsburg, PA) according to the standard procedure.
Statistical analysis
Differences in age, self-reported smoking data (number of smokers, number of years smoking, and cigarettes per day smoked for smokers), urinary cotinine, and urinary Ni among groups were compared by two-sample Student’s t-test. Differences in number of smokers and nonsmokers between exposure groups were compared by chi-square test. All p-values were two sided, with p <0.05 considered statistically significant. Statistical analyses were performed in S-Plus statistical analysis software (TIBCO Software Inc., Palo Alto, CA, USA).
RESULTS
In the present study, a total of eighteen subjects, eight Ni-refinery workers and ten referents, were selected from a previously described study to determine whether occupational exposure to Ni induces alterations in gene expression patterns in PBMCs (20). As shown in Table 1, no significant difference was found between Ni-refinery workers and referents, with respect to age, self-reported data on smoking habits, or urinary cotinine levels. As expected, urinary Ni was elevated in Ni-refinery workers (6.86 ± 1.60) when compared to referents (3.41 ± 3.30) (p = 0.0119). Although a large percentage of subjects reported that they were smokers in both groups, it is widely accepted that the Ni concentrations in blood plasma and urine are similar among smokers and non-smokers (42).
Table 1.
Characteristics of the Ni-refinery workers and referents selected to assess gene expression profiles in peripheral blood mononuclear cells (PBMCs).
| Parameter | Referent Subjectsa | Ni-refinery workersb | p-value |
|---|---|---|---|
| mean ± SD | mean ± SD | ||
| Age | 43.90 ± 8.84 | 45.50 ± 5.45 | 0.6441 |
| Smoking (self-reported) | |||
| Smokers [N/(%)] | [7/70%] | [6/75%] | 0.8262 |
| Smoking years | 10.70 ± 10.91 | 12.75 ± 10.69 | 0.6943 |
| Cigarettes/day | 11.70 ± 15.35 | 9.25 ± 9.44 | 0.6833 |
| Urinary cotinine (μg/g creatinine) | 8115.82 ± 10345.48 | 9034.08 ± 13552.52 | 0.8767 |
| Urinary Ni (μg/g creatinine) | 3.41 ± 3.30 | 6.86 ± 1.60 | 0.0119 |
| Median | 2.37 | 6.32 | |
| Min | 1.19 | 5.23 | |
| Max | 12.58 | 9.44 | |
| 10th percentile | 1.19 | 5.23 | |
| 25th percentile | 2.30 | 5.37 | |
| 75th percentile | 3.25 | 8.38 | |
| 90th percentile | 12.58 | 8.38 | |
In order to characterize the differences in gene expression profiles in PBMCs of subjects with occupational exposure compared to referents, we analyzed the gene expression profiles of PBMCs from both Ni-refinery workers and referents using Affymetrix Human Gene 1.0 ST Array containing 28,869 well-annotated genes. There were a total of 2756 differentially expressed genes (DEG) in the Ni-refinery workers relative to the control subjects (FDR adjusted p<0.05) with 770 up-regulated genes and 1986 down-regulated genes. A total of 2158 genes in samples from Ni-refinery workers displayed a greater than 1.25 fold-change as compared with referents. When the cut-off threshold was increased to 1.50 and 2.0 fold-change, the numbers decreased to 578 and 58, respectively. Principal Components Analysis (PCA) of the microarray data revealed a clear separation between Ni-refinery workers and referents (Figure 1A). Similar results were observed with a hierarchical clustering analysis of DEG (adjusted p< 0.05) in which samples were sorted based on the similarity of gene expression (Figure 1B). To validate the microarray results, q-RT-PCR was performed in four Ni-refinery workers and four referents on CCL20, IL-6, FOSB, (up-regulated in Ni-refinery workers), CX3CR1 and MNDA (down-regulated in Ni-refinery workers). As shown in Table 2, the up- or down-regulated patterns for these four genes obtained from q-RT-PCR were similar to those in the microarray study. In comparing the fold-change values between the microarray data and the q-RT-PCR data (Table 2), we see the changes are comparable in that both sets of data reflect fold changes occurring in the same direction with similar magnitudes. It is important to note that this comparison is occurring across different types of data, where the fold-change values for the microarray data are qualitative and the fold-change values for the q-RT-PCR are quantitative, and therefore we do not expect such values to be exactly the same. Thus, the match in direction and the similarity in magnitude constitute consistency in the findings.
Table 2.
Validation of microarray results by quantitative real-time PCR. Data is represented as absolute fold change values for the exposed group (Ni-refinery workers) relative to the referents for both the microarray and q-RT-PCR.
| Gene name | Gene symbol | Microarray | qRT-PCR |
|---|---|---|---|
| Chemokine (C-C motif) ligand 20 | CCL20 | 4.20 | 8.79 |
| Interleukin-6 (interferon, beta 2) | IL6 | 4.02 | 4.8 |
| FBJ murine osteosarcoma viral oncogene homolog B | FOSB | 2.14 | 1.75 |
| Chemokine (C-X3-C motif) receptor 1 | CX3CR1 | 0.26 | 0.51 |
| Myeloid cell nuclear differentiation antigen | MNDA | 0.35 | 0.40 |
To assess the biological relevance of the DEG, (adjusted p<0.05) those genes exhibiting more than a 1.15 fold-change difference in gene expression (Table S1), including a total of 2734 genes, with 756 up-regulated and 1978 down-regulated genes, were to identify the functional categories that were significantly over-represented (p<0.05) in Ni-refinery workers. Gene families overrepresented with the list of genes differentially expressed between Ni-refinery workers and referents are listed in Table 3. The largest group of gene families, in terms of number of genes, was transcription factors, followed by protein kinases, cell differentiation markers, oncogenes, translocated cancer genes, tumor suppressors, and homeodomain proteins, respectively.
Table 3.
Gene families represented with list of genes differentially expressed between Ni-refinery workers and referents (p<0.05).
| Gene Family | Number of genes |
|---|---|
| Oncogenes | 46 |
| Tumor suppressors | 21 |
| Transcription factors | 233 |
| Cytokines/Growth Factors | 37 |
| Protein Kinases | 98 |
| Translocated cancer genes | 46 |
| Cell differentiation markers | 82 |
| Homeodomain protein | 12 |
Analysis of molecular signatures revealed that the DEG had significant levels of overlap with gene sets categorically classified as cancer modules (i.e. nasopharyngeal carcinoma), cancer neighborhoods (i.e. BRCA1 network), and genes related to chemical and genetic perturbations. Our Ni exposure related gene set demonstrated highly significant (p< 0.00E+00) overlap with a gene set comprised of genes down-regulated in nasopharyngeal carcinoma tumors (33), where 253 genes from the study set were present in the nasopharyngeal carcinoma set (Table S2). Of these 253 genes, 51 were up-regulated and 202 were down-regulated. Cluster analysis of the genes related to nasopharyngeal carcinoma revealed cluster formation that is consistent with the full study set (Figure 2).
DNA repair genes were significantly overrepresented among the DEG two-fold (p< 0.0002). Of 31 DNA repair genes, 29 were repressed in the Ni-refinery workers and two were overexpressed (Table S3). Cluster analysis with DNA repair genes yielded two primary clusters that largely correspond to the occupational and referent groups, but unlike the cluster formation in the study set, there were two main sub-branch clusters within the referent population, where the left sub-branch (A) was characterized largely by overexpression and the right sub-branch (B) was characterized by expression levels near the mean (Figure 3A). Epigenetic genes were also significantly overrepresented in the DEG by 2 fold (p< 0.02). Of these 16 epigenetic genes, 12 were repressed in the Ni-refinery workers and 4 were overexpressed (Table S4). Similar results were observed with a hierarchical clustering analysis of epigenetic genes in which samples were sorted based on the similarity of gene expression (Figure 3B).
DISCUSSION
Ni is a well-established human carcinogen associated with an increased risk of lung and nasal cancers. However, the molecular mechanisms and changes in gene expression that mediate Ni’s toxicity and carcinogenicity are not well understood. In the present study, we found that differential patterns of gene expression in PBMC exist among Ni-refinery workers compared to referent subjects, who are only environmentally exposed to Ni. There were a total of 2756 differentially expressed genes (DEG) in the Ni-refinery workers relative to the control subjects (FDR adjusted p<0.05) with 770 up-regulated genes and 1986 down-regulated genes. Although various mechanisms for the carcinogenesis of nickel compounds have been postulated, the changes in gene expression associated with occupational exposure to Ni have not been previously investigated. Here we have found that the function of the majority of the nickel-induced and repressed genes by in vivo exposure to nickel were related to cytokine–cytokine interaction and chemokine signaling (IL1A, CCL20, CCR2, CX3CR1, IL6, IL1RN, TNFSF10, IL8RB, and IL8RA). Of particular interest is IL-1, a cytokine known to be elevated in many tumor types and which has been implicated as a factor in tumor progression by its ability to increase the expression of metastatic, angiogenic, and growth factor genes (43). Interestingly, IL1A was the most up-regulated gene in the PBMCs of the Ni-refinery workers. These results suggest that high exposure to nickel compounds was associated with increased expression of cancer related genes. In this case, the high expression of IL1A and other genes in Ni-refinery workers suggests that these genes should be further studied in order to identify their role in nickel-induced adverse health effects.
Moreover, the genes aberrantly expressed in this study share a significant similarity with those aberrantly expressed in nasopharyngeal carcinoma tumors as described in Dodd et. al. (2006). Nasal and sino-nasal cancers have been historically established as an endpoint of occupational nickel exposure (2, 3), and the similarity in gene deregulation between the refinery workers in our study and NPC tumors, suggests that gene expression profiling can be used to link human in vivo nickel exposures to cancerous endpoints by identifying gene expression patterns that precede clinical presentations of cancer. Though the two cancer endpoints in discussion here are not synonymous, their anatomical proximity and the significance of our p-value merits further consideration. While genes from NPC were significantly overrepresented in our study, there was not a complete overlap and the direction of deregulation was only partially consistent with the Dodd study set. These results likely reflect the similar endpoints, cancers of the nasal and nasopharynx regions, but also point towards the different etiologies likely involved in these related but distinct cancers. The Dodd study emphasizes that NPC is often associated with Epstein-Barr virus (EBV) and that factor is not one that we would expect to be represented in our study. Both etiologies demonstrate a strong involvement of the DNA repair genes, but involve different groups of genes with only UNG, MSH2, PRKDC, and PCNA in common. The Dodd et. al. study found DNA repair genes to be up-regulated, while we observed that DNA repair genes were largely repressed in the exposed population. The repression of DNA repair genes observed in our study may be explained by nickel’s role as an epigenetically active agent that can actively silence genes. The repression of DNA repair genes was associated with high levels of exposure, and may be due to nickel induced silencing, a process which is supported by the significant over-representation of epigenetic genes in the study set, their aberrant expression patterns relative to the referents, and a previous study which demonstrated that occupational exposure to nickel induces consistent changes in histone modifications (16, 17, 20–22, 44). Together, the changes observed in epigenetic regulation and suppression of DNA repair can be detrimental to the genetic stability of cells, lead to increased spontaneous mutations, and promote conditions in which carcinogenesis is favored.
The importance of identifying genes through an in vivo format can be highlighted with a comparison of our in vivo data and pilot data from an in vitro study. We assessed the short-term gene changes associated with PBMC’s, exposed to nickel in vitro and found that gene changes at the 1.5 fold-change level were strikingly different from those observed in the in vivo study. PBMC’s were exposed to nickel chloride (NiCl2) at 1.0, 0.5, and 0.25 mM for 24 hours and single samples were then prepared for gene expression analysis and hybridized to Affymetrix HGU133a.2 arrays. For all three treatments there were 98 genes that changed 1.5 fold (Table S3). Of these genes only 9 were found among the genes that changed 1.5 fold in the in vivo study. These genes include: CD14, FUCA1, DAPK1, HMOX1, FGL2, IGF6, FCGR1B, TLR8, and CLEC7A. While the data from the in vitro study remains preliminary and was not subjected to statistical analysis due to a lack of replicates, it does suggest that in vitro exposures do not accurately replicate the gene expression changes found in vivo. Further, the gene changes observed in vitro do not share the similarity with NPC that was evident in vivo, again indicating the importance of conducting in vivo human studies to identify biomarkers that are linked with disease endpoints.
Identification of genes associated with high occupational exposure to nickel will help identify agents that combat the toxic effects of exposure to nickel or can be used as biomarkers for Ni exposure and effects. It is of interest that occupational exposure to Ni causes 29 of 31 DNA repair genes to be suppressed in expression, while in the referent group these same genes were up-regulated in the left sub-branch (Figure 3B, Branch A) and were expressed near the mean in the right sub-branch (Figure 3B, Branch B). Interestingly, the repression of six of the DNA repair genes (MUS81, PALB2, XRCC5, PRKDC, CLK2, HLTF, TP53) found in this study were also found suppressed in the Beas-2B cell line exposed to Ni in vitro (45). This same study also found the expression of six epigenetic genes (EHMT1, EHMT2, PRMT7, DETDB1, HLTF, JMD6) repressed as in our study (45). The formation of three primary clusters and three types of DNA repair response suggests the presence of a nickel induced dose-response mechanism. In subjects with occupational exposure to nickel, DNA repair genes were largely repressed, and in the referent population there were two types of low-level responses, in the first response type the DNA repair genes were highly up-regulated (Figure 3A, Branch A), while in the second there was no distinct response with values near the mean (Figure 3B, Branch B). This clear difference between the worker group and branch A of the referent population suggests there may be a dose response mechanism whereby high exposure leads to the repression of DNA repair genes and low exposure can lead to their overexpression. While the urinary nickel levels for these two groups (worker group and branch A of referents) are not significantly different, the means for the two groups do reflect this trend. Further study is needed to determine whether the two types of low-level response for branches and A and B of the referent population are reflective of a dose-response mechanism or of another factor such as a genetic polymorphism as there was no statistically significant difference in the urinary nickel levels for these two groups either. However, it should be noted that urinary Ni may not be a true measure of worker exposure to Ni, since inhaled particulate Ni compounds (i.e. nickel oxides etc.) may not be cleared from the lungs into the urine for months to years (46, 47). Although Ni-exposure resulted in the differential expression of a large number of genes, these differentially expressed genes may not only be Ni-response genes. Future experiments should focus on whether the gene expression changes found in this study are specific to Ni exposure and/or toxicity and determine whether changes are occurring as a result of exposures to other agents found in nickel refineries.
In summary, we have identified many novel changes in gene expression that were different from the immediate response genes identified from previous studies using acute in vitro exposure of tissue culture cells to Ni. These genes may be involved in Ni induced toxicity, carcinogenesis and can be utilized as potential biomarkers of exposure to Ni compounds. Furthermore, these changes also demonstrate strong similarity with changes observed in nasopharyngeal carcinoma, suggesting that biomarkers of disease may be developed from gene expression studies of PBMCs. Thus, further studies of the mechanism(s) of Ni induced alterations in gene expression patterns may be a useful a tool for identifying patterns of deregulation that precede clinical identification of Ni-induced cancers.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by grant numbers ES000260, ES010344, ES014454, ES005512, 5 T32 ES007324 from the National Institutes of Environmental Health Sciences, and support from grant number CA16087 from National Cancer Institute and RR029893 from National Center for Research Resources.
Footnotes
Competing financial interest declaration: The authors have no actual or potential competing financial interests.
References
- 1.Cempel M, Nikel G. Nickel: A review of its sources and environmental toxicology. Pol J Environ Stud. 2006;15:375–82. [Google Scholar]
- 2.Grimsrud TK, Berge SR, Haldorsen T, Andersen A. Exposure to different forms of nickel and risk of lung cancer. Am J Epidemiol. 2002;156:1123–32. doi: 10.1093/aje/kwf165. [DOI] [PubMed] [Google Scholar]
- 3.Doll R, Morgan LG, Speizer FE. Cancers of the lung and nasal sinuses in nickel workers. Br J Cancer. 1970;24:623–32. doi: 10.1038/bjc.1970.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimsrud TK, Berge SR, Resmann F, Norseth T, Andersen A. Assessment of historical exposures in a nickel refinery in Norway. Scand J Work Environ Health. 2000;26:338–45. doi: 10.5271/sjweh.551. [DOI] [PubMed] [Google Scholar]
- 5.Giordano-Labadie F, Rance F, Pellegrin F, Bazex J, Dutau G, Schwarze HP. Frequency of contact allergy in children with atopic dermatitis: results of a prospective study 137 cases. Contact Dermatitis. 1999;40:192–5. doi: 10.1111/j.1600-0536.1999.tb06032.x. [DOI] [PubMed] [Google Scholar]
- 6.IARC IAfRoC. Chromium, Nickel and Welding. Lyon: 1990. IARC Monograph on the Evaluation of Carcinogenic Risks to Humans; pp. 1–691. [PMC free article] [PubMed] [Google Scholar]
- 7.Landolph JR. Role of free radicals in metal-induced carcinogenesis. Met Ions Biol Syst. 1999;36:445–83. [PubMed] [Google Scholar]
- 8.Sunderman FW., Jr Search for molecular mechanisms in the genotoxicity of nickel. Scand J Work Environ Health. 1993;19 (Suppl 1):75–80. [PubMed] [Google Scholar]
- 9.Li J, Davidson G, Huang Y, Jiang BH, Shi X, Costa M, et al. Nickel compounds act through phosphatidylinositol-3-kinase/Akt-dependent, p70(S6k)-independent pathway to induce hypoxia inducible factor transactivation and Cap43 expression in mouse epidermal Cl41 cells. Cancer Res. 2004;64:94–101. doi: 10.1158/0008-5472.can-03-0737. [DOI] [PubMed] [Google Scholar]
- 10.Salnikow K, Blagosklonny MV, Ryan H, Johnson R, Costa M. Carcinogenic nickel induces genes involved with hypoxic stress. Cancer Res. 2000;60:38–41. [PubMed] [Google Scholar]
- 11.Chen F, Ding M, Castranova V, Shi X. Carcinogenic metals and NF-kappaB activation. Mol Cell Biochem. 2001;222:159–71. [PubMed] [Google Scholar]
- 12.Cruz MT, Goncalo M, Figueiredo A, Carvalho AP, Duarte CB, Lopes MC. Contact sensitizer nickel sulfate activates the transcription factors NF-kB and AP-1 and increases the expression of nitric oxide synthase in a skin dendritic cell line. Exp Dermatol. 2004;13:18–26. doi: 10.1111/j.0906-6705.2004.00105.x. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, Davidson G, Li J, Yan Y, Chen F, Costa M, et al. Activation of nuclear factor-kappaB and not activator protein-1 in cellular response to nickel compounds. Environ Health Perspect. 2002;110 (Suppl 5):835–9. doi: 10.1289/ehp.02110s5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C, Li J, Costa M, Zhang Z, Leonard SS, Castranova V, et al. Hydrogen peroxide mediates activation of nuclear factor of activated T cells (NFAT) by nickel subsulfide. Cancer Res. 2001;61:8051–7. [PubMed] [Google Scholar]
- 15.Chen H, Ke Q, Kluz T, Yan Y, Costa M. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol Cell Biol. 2006;26:3728–37. doi: 10.1128/MCB.26.10.3728-3737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang J, Zhang Y, Chen J, Chen H, Lin C, Wang Q, et al. Nickel-induced histone hypoacetylation: the role of reactive oxygen species. Toxicol Sci. 2003;74:279–86. doi: 10.1093/toxsci/kfg137. [DOI] [PubMed] [Google Scholar]
- 17.Ke Q, Ellen TP, Costa M. Nickel compounds induce histone ubiquitination by inhibiting histone deubiquitinating enzyme activity. Toxicol Appl Pharmacol. 2008;228:190–9. doi: 10.1016/j.taap.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Li Q, Arita A, Sun H, Costa M. Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicol Appl Pharmacol. 2009;236:78–84. doi: 10.1016/j.taap.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ke Q, Davidson T, Chen H, Kluz T, Costa M. Alterations of histone modifications and transgene silencing by nickel chloride. Carcinogenesis. 2006;27:1481–8. doi: 10.1093/carcin/bgl004. [DOI] [PubMed] [Google Scholar]
- 20.Arita A, Niu J, Qu Q, Zhao N, Ruan Y, Nadas A, et al. Global levels of histone modifications in peripheral blood mononuclear cells of subjects with exposure to nickel. Environ Health Perspect. 2012;120:198–203. doi: 10.1289/ehp.1104140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Ke Q, Kluz T, Yan Y, Costa M. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol Cell Biol. 2006;26:3728–37. doi: 10.1128/MCB.26.10.3728-3737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Li Q, Arita A, Sun H, Costa M. Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicol Appl Pharmacol. 2009;236:78–84. doi: 10.1016/j.taap.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bushel PR, Heinloth AN, Li J, Huang L, Chou JW, Boorman GA, et al. Blood gene expression signatures predict exposure levels. Proc Natl Acad Sci U S A. 2007;104:18211–6. doi: 10.1073/pnas.0706987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lobenhofer EK, Auman JT, Blackshear PE, Boorman GA, Bushel PR, Cunningham ML, et al. Gene expression response in target organ and whole blood varies as a function of target organ injury phenotype. Genome Biol. 2008;9:R100. doi: 10.1186/gb-2008-9-6-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umbright C, Sellamuthu R, Li S, Kashon M, Luster M, Joseph P. Blood gene expression markers to detect and distinguish target organ toxicity. Mol Cell Biochem. 2010;335:223–34. doi: 10.1007/s11010-009-0272-5. [DOI] [PubMed] [Google Scholar]
- 26.Sellamuthu R, Umbright C, Roberts JR, Chapman R, Young SH, Richardson D, et al. Blood gene expression profiling detects silica exposure and toxicity. Toxicol Sci. 2011;122:253–64. doi: 10.1093/toxsci/kfr125. [DOI] [PubMed] [Google Scholar]
- 27.Sellamuthu R, Umbright C, Roberts JR, Chapman R, Young SH, Richardson D, et al. Transcriptomics analysis of lungs and peripheral blood of crystalline silica-exposed rats. Inhal Toxicol. 2012;24:570–9. doi: 10.3109/08958378.2012.697926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 29.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 30.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 31.GKS, Gentleman RVC, Dudoit S, Irizarry R, Huber W. Bioinformatics and Computational Biology Solutions using R and Bioconductor. NewYork: 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 32.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 33.Dodd LE, Sengupta S, Chen IH, den Boon JA, Cheng YJ, Westra W, et al. Genes involved in DNA repair and nitrosamine metabolism and those located on chromosome 14q32 are dysregulated in nasopharyngeal carcinoma. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive. Oncology. 2006;15:2216–25. doi: 10.1158/1055-9965.EPI-06-0455. [DOI] [PubMed] [Google Scholar]
- 34.Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 35.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–9. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 36.Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577:275–83. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miremadi A, Oestergaard MZ, Pharoah PD, Caldas C. Cancer genetics of epigenetic genes. Human molecular genetics. 2007;16(Spec No 1):R28–49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]
- 39.Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer. 2006;94:179–83. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliveira JP, de Siqueira ME, da Silva CS. Urinary nickel as bioindicator of workers’ Ni exposure in a galvanizing plant in Brazil. Int Arch Occup Environ Health. 2000;73:65–8. doi: 10.1007/pl00007940. [DOI] [PubMed] [Google Scholar]
- 41.Hu Y, Li G, Xue X, Zhou Z, Li X, Fu J, et al. PAH-DNA adducts in a Chinese population: relationship to PAH exposure, smoking and polymorphisms of metabolic and DNA repair genes. Biomarkers. 2008;13:27–40. doi: 10.1080/13547500701671895. [DOI] [PubMed] [Google Scholar]
- 42.Torjussen W, Zachariasen H, Andersen I. Cigarette smoking and nickel exposure. J Environ Monit. 2003;5:198–201. doi: 10.1039/b209065c. [DOI] [PubMed] [Google Scholar]
- 43.Lewis AM, Varghese S, Xu H, Alexander HR. Interleukin-1 and cancer progression: the emerging role of interleukin-1 receptor antagonist as a novel therapeutic agent in cancer treatment. Journal of translational medicine. 2006;4:48. doi: 10.1186/1479-5876-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee YW, Klein CB, Kargacin B, Salnikow K, Kitahara J, Dowjat K, et al. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogens. Mol Cell Biol. 1995;15:2547–57. doi: 10.1128/mcb.15.5.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tchou-Wong KM, Kiok K, Tang Z, Kluz T, Arita A, Smith PR, et al. Effects of nickel treatment on H3K4 trimethylation and gene expression. PloS one. 2011;6:e17728. doi: 10.1371/journal.pone.0017728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernacki EJ, Parsons GE, Roy BR, Mikac-Devic M, Kennedy CD, Sunderman FW. Urine nickel concentrations in nickel-exposed workers. Annals of clinical and laboratory science. 1978;8:184–9. [PubMed] [Google Scholar]
- 47.Tola S, Kilpio J, Virtamo M. Urinary and plasma concentrations of nickel as indicators of exposure to nickel in an electroplating shop. Journal of occupational medicine: official publication of the Industrial Medical Association. 1979;21:184–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.