Abstract
Background
Genome-wide association studies of European and East Asian populations have identified lung cancer susceptibility loci on chromosomes 5p15.33, 6p22.1-p21.31 and 15q25.1. We investigated whether these regions contain lung cancer susceptibly loci in African-Americans refined previous association signals by utilizing the reduced linkage disequilibrium observed in African-Americans.
Methods
1308 African-American cases and 1241 African-American controls from three centers were genotyped for 760 single nucleotide polymorphisms spanning three regions, and additional SNP imputation was performed. Associations between polymorphisms and lung cancer risk were estimated using logistic regression, stratified by tumor histology where appropriate.
Results
The strongest associations were observed on 15q25.1 in/near CHRNA5, including a missense substitution (rs16969968: OR = 1.57, 95% CI = 1.25–1.97, P = 1.1 × 10−4) and variants in the 5′-UTR. Associations on 6p22.1-p21.31 were histology-specific and included a missense variant in BAT2 associated with squamous-cell carcinoma (rs2736158: OR = 0.64, 95% CI = 0.48–0.85, P = 1.82 × 10−3). Associations on 5p15.33 were detected near TERT, the strongest of which was rs2735940 (OR = 0.82, 95% CI = 0.73–0.93, P = 1.1 × 10−3). This association was stronger among cases with adenocarcinoma (OR = 0.75, 95% CI = 0.65–0.86, P = 8.1 × 10−5).
Conclusions
Polymorphisms in 5p15.33, 6p22.1-p21.31 and 15q25.1 are associated with lung cancer in African-Americans. Variants on 5p15.33 are stronger risk factors for adenocarcinoma and variants on 6p21.33 associated only with squamous-cell carcinoma.
Impact
Results implicate the BAT2, TERT and CHRNA5 genes in the pathogenesis of specific lung cancer histologies.
Keywords: Lung cancer, adenocarcinoma, squamous-cell carcinoma, fine-mapping, African-American, genetic association
INTRODUCTION
Recent genome-wide association studies (GWAS) have identified lung cancer susceptibility loci on chromosomes 5p15.33 (1–5), 6p22.1-p21.31 (1–3, 6), and 15q25.1 (1–3, 6, 7). Thus far, lung cancer GWAS have been conducted only on European (1–4, 6, 7) or East Asian (5) populations, despite the fact that African-Americans have higher lung cancer incidence rates and lower lung cancer survival rates than other ethnic groups in the U.S. (8). Genetic effects are likely to differ in African-Americans because the previously associated single nucleotide polymorphisms (SNPs) have different allele frequencies and linkage patterns across populations (9, 10). As a result, how genetic variation in these three regions influences lung cancer susceptibility among African-Americans remains poorly understood.
The associated region on 5p15.33 contains two candidate lung cancer susceptibility genes, telomerase reverse transcriptase (TERT) and cleft lip and palate transmembrane 1-like (CLPTM1L). SNPs in this region have consistently been associated with both lung cancer and lung cancer histology. Specifically, rs2736100 in TERT has been shown to be significantly more common in cases with adenocarcinoma than with other lung cancer histologies (3). There is also evidence from GWAS that 5p15.33 is a susceptibility locus for multiple cancer sites, including: the pancreas (11), bladder (12), prostate (13) and brain (14).
The 6p22.1-p21.31 locus is part of the HLA region and is highly polymorphic. Association studies of lung cancer susceptibility have found inconsistent associations in this region, with several GWAS identifying significant associations (1–3, 6) and several follow-up replication studies failing to do so (10, 15, 16). Of note, multiple risk SNPs identified in European populations are non-polymorphic in East Asian populations (10). Because previously associated SNPs in the HLA region may represent European-specific susceptibility loci, studies conducted in other ethnic groups should assay additional SNPs in this region to capture genetic variation not seen in Europeans.
The 15q25.1 region associated with lung cancer contains six genes, three of which (CHRNA5, CHRNA3 and CHNB4) are nicotinic receptor subunit genes. SNPs in these genes have been associated with smoking behavior, with risk alleles conferring a propensity to smoke with greater frequency and intensity (17–19). This would suggest an indirect effect of genetic variation at 15q25.1 on lung cancer risk, where variants influence cancer risk through a causal pathway in which smoking is an obligate intermediate. However, the effects of SNPs in this region on lung cancer risk appear more complex than this. Multiple studies have found that SNPs at 15q25.1 also influence lung cancer risk among never smokers (6, 20). Functional analyses have further demonstrated that one of the six genes in this region, proteasome alpha 4 subunit isoform 1 (PSMA4), plays a role in cancer cell proliferation and apoptosis (21). Additional functional studies identified a lung cancer risk SNP on 15q25.1 that increases IREB2 expression (22). Identifying the causal variants at 15q25.1 has been challenging because SNPs in the region are in high linkage disequilibrium (LD) among Europeans (23). Fine mapping of the associated region in African-Americans, where LD is reduced, provides an opportunity to refine the location of lung cancer risk alleles by exploiting the shorter haplotype blocks (24, 25).
To better understand the role of genetic variation in these three regions, we conducted a multi-center case-control study of African-Americans to identify genetic variants associated with an increased risk of lung cancer. Samples were genotyped on an Illumina Golden Gate custom panel containing SNPs selected for fine-mapping of previous GWAS hits on chromosomes 5, 6 and 15. The goals of the study were to replicate SNP associations previously detected in European populations, identify novel or African American-specific lung cancer susceptibly SNPs in these regions, and refine previous association signals by exploiting the reduced LD observed in African-Americans.
METHODS
Study Population
A multi-center case-control study was designed to include African-American lung cancer cases and controls from three collaborating institutions: The University of California, San Francisco (UCSF) (447 cases, 453 controls), Wayne State University (WSU) (459 cases, 460 controls) and the MD Anderson Cancer Center (MDA) (479 cases, 376 controls). All participating institutions received IRB approval, and appropriate written informed consent was obtained from human subjects. All study participants reported being of African-American ethnicity.
UCSF cases and controls were enrolled as part of The Northern California Lung Cancer Study, which has been described in detail elsewhere (26). Cases and controls older than 18 years of age were identified during two collection periods, spanning September 1998–March 2003 and July 2005–March 2008. Cases were Northern California residents presenting with previously untreated, histologically confirmed lung cancer. Cases in the first accrual period were identified primarily through the Northern California Cancer Center (NCCC) rapid case ascertainment program and Alta Bates/Summit Hospital. Cases in the second accrual period were identified through both the NCCC and the Kaiser Permanente Medical Care Program (KPMCP). Control participants ascertained in the first accrual period were recruited through three sources: random-digit dialing, Health Care Financing Administration records, and community-based recruitment. Controls in the second accrual period were recruited through the KPMCP.
Wayne State University cases were identified through the population-based Metropolitan Detroit Cancer Surveillance System, an NCI-funded SEER registry, as part of the EXHALE study (27). Rapid case ascertainment was used to identify histologically-confirmed cases within several months of diagnosis. African-Americans diagnosed with a first primary lung cancer from November 1, 2005 through June 30, 2010 were recruited for the study. Controls were recruited through community-based methods and were frequency matched on race, sex and five-year age group.
MDA cases were recruited from The University of Texas M. D. Anderson Cancer Center and the Michael E. DeBakey VA Medical Center in Houston. All cases with newly diagnosed, histopathologically confirmed lung cancer were eligible. Case exclusion criteria were prior chemotherapy or radiotherapy or recent blood transfusion. African-American controls were recruited from Houston-area community centers and the Kelsey-Seybold Foundation, Houston’s largest multi-specialty physician group practice. Controls were matched to the cases on age (±5 years), sex, and African-American ethnicity.
SNP Selection
Three regions found in previous GWAS to be associated with an increased risk for lung cancer were selected for fine-mapping in African-American subjects. The custom SNP panel included 120 ancestry-informative markers (AIMs) for the calculation of % sub-Saharan African ancestry and 760 SNPs chosen to perform fine-mapping of the 5p15.33, 6p22.1-p21.31, and 15q25.1 regions. This includes 138 SNPs on 5p15.33, 356 SNPs on 6p22.1-p21.31 and 266 SNPs on 15q25.1. Markers used in analyses were selected based upon: known functional effect on activity of nicotinic acetylcholine receptors, previously associated in East Asian or European populations, allele frequency > 0.01 in African populations, position across the region, predicted effect on function, r-square value with respect to other markers <0.70 as determined by SNP browser version 4.0 (28), inclusion in one of three previous studies of African-American lung cancer susceptibility (24, 25, 29), or discovery by targeted sequencing (30).
Genotyping
UCSF samples were genotyped at the University of California, San Francisco Genome Center using an Illumina custom panel of 1536 SNPs. Unamplified genomic DNA samples extracted from whole blood (n = 750) were genotyped along with whole genome-amplified (WGA) blood or buccal DNA samples (n = 150), prepared as previously described (31). Genotypes for unamplified DNA and WGA DNA samples were clustered separately. A GenCall genotype quality threshold of 0.25 was used. Genotype reproducibility was verified with twelve duplicate samples with average concordance of 99.97% (range: 99.68–100%). Ceph Trios were genotyped to assess the accuracy of assigned genotype clusters. The average heritability for nine Parent-Parent-Child trios was 99.88% (99.19–100%). All cluster plots were visually inspected.
Wayne State University samples were genotyped at the Applied Genomics Technology Center (AGTC) at Wayne State University using the same Illumina Golden Gate Custom panel of 1536 SNPs. All WSU samples were unamplified genomic DNA, extracted from whole blood. Genotype reproducibility was verified with thirty duplicate samples, each with >99% concordance. Ten CEPH controls were genotyped and checked for concordance with published HapMap SNP genotypes at loci overlapping with those assayed by the Illumina custom panel.
MDA samples were genotyped at the MD Anderson Cancer Center using the same Illumina Golden Gate Custom panel as the other sites. Genotyping was performed on unamplified genomic DNA derived from peripheral whole blood. Genotype reproducibility was verified with seven duplicate samples, with concordance ranging from 99.93%–100%.
For all study sites, samples with genotyping call rate < 95% were excluded from analysis. SNPs with genotyping call rates <95% in more than one study site were excluded from analyses. To exclude poorly genotyped SNPs, any SNP with a Hardy-Weinberg Equilibrium (HWE) p-value <1.0 × 10−4 in controls, stratified by site, was removed from analysis. All SNP quality-control was performed using Plink v1.07 (32).
Calculation of % sub-Saharan African ancestry
The genetic structure of African-American subjects was evaluated using Structure v2.3.1 to estimate percent membership in three distinct founder populations: sub-Saharan African, European, and East Asian (33), where East Asian population ancestry was also used as a surrogate for Amerindian descent. Founder population allele frequencies were defined using SNP data from 102 unlinked (r2<0.20) ancestry informative markers (AIMs), genotyped in 502 unrelated HapMap individuals (167 Yoruban Africans, 165 Europeans, 84 Chinese and 86 Japanese) (34). These same AIMs were genotyped in our study participants for use with the Structure program.
Collection of covariates
Cases and controls recruited at all study sites completed interviews conducted by trained interviewers. Data on sex, age and smoking behaviors were collected as covariates for this study. Never smokers were defined as those who had smoked <100 cigarettes in their lifetimes; former smokers were those who had quit smoking >1 year before diagnosis (cases) or interview (controls); current smokers included those who had quit smoking within the past 12 months. Pack-years for former and current smokers were calculated as the years smoked times the average number of cigarettes per day divided by 20.
Cancer histology was determined using ICD-O codes abstracted from SEER (Surveillance Epidemiology and End Results) data from the California Cancer Registry (UCSF cases) or Detroit Cancer Registry (WSU cases). For MDA cases, histology was determined by extraction from medical records. The following ICD-O groupings were made: adenocarcinoma (ICD-O: 8140, 8230, 8250–8255, 8260, 8310, 8333, 8470, 8480, 8481, 8490, 8550), squamous cell carcinoma (8052, 8070–8073, 8083, 8084), and small cell carcinoma (8041–8045).
SNP imputation and association analyses
To refine association peaks from case-control analyses and identify additional risk variants, we performed SNP imputation in the 5p15.33, 6p22.1-p21.31 and 15q25.1 regions using the Impute2 v2.1.2 software and its standard Markov chain Monte Carlo algorithm using the default settings for targeted imputation (35). All 1000 Genomes Phase I interim release haplotypes were provided as the reference haplotype panel (36). Using a cosmopolitan set of reference haplotypes is currently recommended for imputation, and is especially critical for imputation of recently admixed populations, such as African-Americans (37).
SNPs with imputation quality (info) scores < 0.70 or posterior probabilities < 0.90 were excluded to remove poorly imputed SNPs. Any SNP with a minor allele frequency less than 1% in case subjects was excluded from association tests. All association statistics, both for imputed and for directly genotyped SNPs, were calculated using logistic regression in SNPTEST v2 assuming a log-additive model. A missing data likelihood score-test was applied to the imputed variants, as this produces standard errors which account for the additional uncertainty inherent in the analysis of imputed genotypes. The effect of individual SNPs on lung cancer risk was calculated while adjusting for sex, age, % African ancestry, % European ancestry, number of pack-years smoked, and study site. Adjustment for current smoking did not affect associations and this covariate was not included in the final models.
Because the 15q25.1 region contains nicotinic-receptor gene variants that have been associated with smoking behavior, and therefore SNPs in this region may influence lung cancer risk by modifying smoking behavior, these SNP associations were also calculated without adjustment for pack-years. All SNP associations were assessed in the full case-control sample, and also stratified by histology (adenocarcinoma vs. controls, squamous cell carcinoma vs. controls, small-cell lung cancer vs. controls). All associations are for an allelic additive model, adjusted for the indicated covariates, where odds ratios are for each additional copy of the minor allele.
Haplotype analyses
Haplotype analyses were performed using Haploview v4.2 to identify haplotype blocks and calculate R2 values (38). Associations between the identified haplotypes and lung cancer risk were calculated in Plink while controlling for appropriate covariates.
RESULTS
After excluding samples with call rates <95%, 2549 African-American participants remained for analysis (1308 cases, 1241 controls). Compared to controls, cases were more likely to be male, to be older, to smoke, and to have smoked a greater number of pack-years (Table 1). Cases and controls had similar African ancestry and similar Caucasian ancestry (Table 1, Figure S1). After excluding monomorphic SNPs and SNPs that did not meet call-rate or HWE thresholds, 660 genotyped SNPs remained for analysis. This included 111 SNPs on 5p15.33, 320 SNPs on 6p22.1-p21.31 and 242 SNPs on 15q25.1. An additional 103,928 SNPs were imputed in the three regions, of which 38.5% were excluded for having a MAF <1% in cases, 20.7% were excluded for imputation quality (info) scores < 0.70, and 4.6% were excluded for posterior probabilities < 0.90. This left 37,660 imputed SNPs for analysis.
Table 1.
Demographic, smoking and tumor histology characteristics of African-American lung cancer cases and controls appearing in the association analyses
| UCSF (N=879) | MDA (N=839) | WSU (N=831) | Total (N=2549) | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Case | Control | Case | Control | Case | Control | Case | Control | |
| Sample Size | 432 | 447 | 473 | 366 | 403 | 428 | 1308 | 1241 |
| Mean ± SE | ||||||||
| Age | 64.36 ± 0.53 | 64.17 ± 0.54 | 62.40 ± 0.47 | 55.41 ± 0.59 | 62.24 ± 0.51 | 62.04 ± 0.48 | 63.0 ± 0.29 | 60.86 ± 0.33 |
| % African Ancestry | 0.75 ± 0.007 | 0.76 ± 0.006 | 0.76 ± 0.007 | 0.76 ± 0.008 | 0.78 ± 0.006 | 0.77 ± 0.006 | 0.76 ± 0.004 | 0.76 ± 0.004 |
| % EuropeanAncestry | 0.19 ± 0.006 | 0.19 ± 0.007 | 0.18 ± 0.006 | 0.18 ± 0.008 | 0.17 ± 0.006 | 0.17 ± 0.006 | 0.18 ± 0.004 | 0.18 ± 0.004 |
| Pack-years smokeda | 33.39 ± 1.15 | 27.39 ± 1.74 | 39.92 ± 1.37 | 24.66 ± 1.32 | 39.17 ± 1.45 | 30.65 ± 3.52 | 37.45 ± 0.77 | 27.65 ± 1.41 |
| n (%) | ||||||||
| Male | 196 (45.37) | 204 (45.64) | 251 (53.07) | 154 (42.08) | 193 (47.89) | 195 (45.56) | 640 (48.93) | 553 (44.56) |
| Ever Smoker | 403 (93.29) | 289 (64.65) | 397 (83.93) | 270 (73.77) | 382 (94.79) | 294 (68.69) | 1182 (90.37) | 853 (68.73) |
| Current smoker | 181 (41.90) | 134 (29.98) | 246 (52.01) | 143 (39.07) | 241 (59.80) | 171 (39.95) | 668 (51.07) | 448 (36.10) |
| Adenocarcinoma | 176 (40.74) | NA | 215 (45.45) | NA | 167 (41.44) | NA | 558 (42.66) | NA |
| Squamous cell | 107 (24.77) | NA | 131 (27.70) | NA | 105 (26.05) | NA | 343 (26.22) | NA |
| Small Cell | 34 (7.87) | NA | 24 (5.07) | NA | 29 (7.20) | NA | 87 (6.65) | NA |
| Other/Not specified | 115 (26.62) | NA | 103 (21.78) | NA | 102 (25.31) | NA | 320 (24.46) | NA |
among ever-smokers
Abbreviations: UCSF, University of California San Francisco; MDA, M. D. Anderson Cancer Center; WSU, Wayne State University
Figures 1–2 show results of the association analyses for the 5p15.33, 6p22.1-p21.31 and 15q25.1 regions, respectively, with triangles identifying SNPs previously determined to be associated with lung cancer risk according to the NHGRI Catalog of Published Genome-Wide Association Studies. Table S1 provides association results for all the imputed SNPs included in Figures 1–2 that had P-values <0.01. Histology-specific association results are also provided.
Figure 1. Association of SNP on chromosome 5p15.33 and 6p22.1-p21.31 with lung cancer.
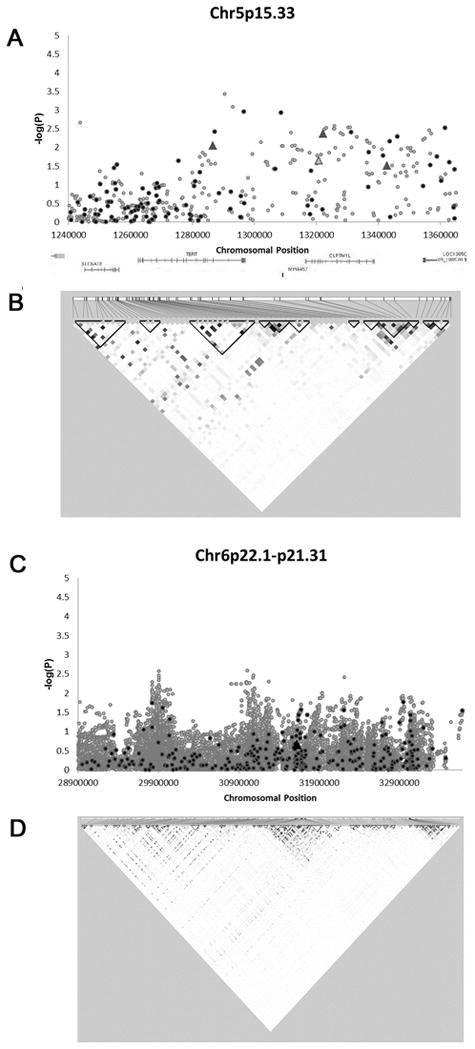
(A) Association of 111 directly genotyped SNPs (black) and 305 imputed SNPs (gray) with lung cancer, adjusted for sex, age, study site, % sub-Saharan African ancestry, % European ancestry, and number of pack-years smoked. Triangles denote SNPs which are associated with lung cancer in the NHGRI catalog. (B) Genes in the analyzed region and patterns of LD. Darker shading indicates higher r2 values and greater correlation between the SNPs. (C) Association of 320 directly genotyped SNPs (black) and 36,242 imputed SNPs (gray) with lung cancer, adjusted for sex, age, study site, % sub-Saharan African ancestry, % European ancestry, and number of pack-years smoked. Triangles denote SNPs which are associated with lung cancer in the NHGRI catalog. (D) Patterns of LD in the analyzed region. Darker shading indicates higher r2 values and greater correlation between the SNPs.
Figure 2. Association of SNPs on chromosome 15q25.1 with lung cancer.
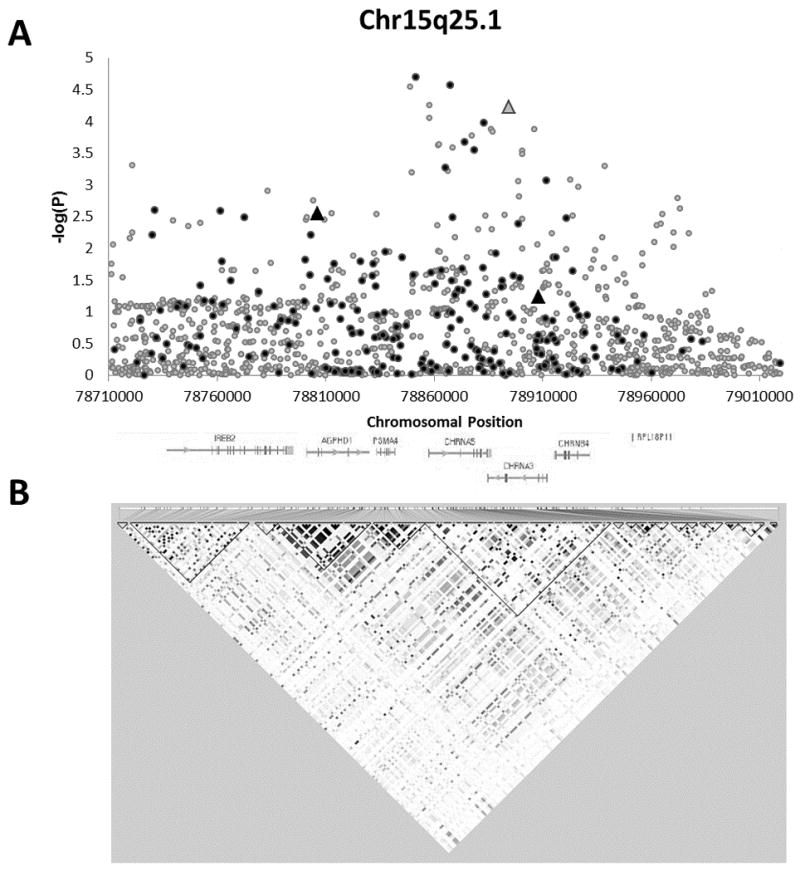
(A) Association of 242 directly genotyped SNPs (black) and 1,013 imputed SNPs (gray) with lung cancer, adjusted for sex, age, study site, % sub-Saharan African acnestry, % European ancestry, and number of pack-years smoked. Triangles denote SNPs which are associated with lung cancer the NHGRI catalog. (B) Patterns of LD in the analyzed region. Darker shading indicates higher r2 values and greater correlation between the SNPs.
Four previously identified lung cancer risk SNPs on 5p15.33 were associated with lung cancer in this African-American sample (p<0.05, effect size in same direction as previous reports), but were not the most statistically significant associations observed in the region. The most significantly associated genotyped SNP in the region was rs2735940 (OR = 0.82, 95% CI = 0.73–0.93, P = 1.1 × 10−3) and the most significantly associated imputed SNP in the region was rs62332591 (OR = 0.78, 95% CI = 0.68–0.89, P = 3.7 × 10−4). Overall, the most significant associations were localized to the 5′ end of the TERT gene, just downstream of CLPTM1L (Figure 1A, Tables 2–3).
Table 2.
Top 10 directly genotyped SNP associations from the case-control analysis of lung cancer in African-Americans
| SNPa | Position | Gene | Alleleb | MAFc | OR (95% CI)d | P-valuee |
|---|---|---|---|---|---|---|
|
| ||||||
| rs2036527 | Chr15:78851615 | 6.3kb upstream of CHRNA5 | C/T | 0.2102 | 1.34(1.17–1.54) | 2.0 × 10−5 |
| rs17486278 | Chr15:78867482 | intron 1 of CHRNA5 | A/C | 0.2802 | 1.31 (1.15–1.48) | 2.7 × 10−5 |
| rs16969968 | Chr15:78882925 | exon 5 of CHRNA5 (missense Asp → Asn) | G/A | 0.06008 | 1.57(1.25–1.97) | 1.1 × 10−4 |
| rs7180002 | Chr15:78873993 | intron 2 of CHRNA5 | A/T | 0.102 | 1.40(1.17–1.68) | 2.1 × 10−4 |
| rs951266 | Chr15:78878541 | intron 2 of CHRNA5 | C/T | 0.1031 | 1.39 (1.16–1.66) | 2.8 × 10−4 |
| rs17486195 | Chr15:78865197 | intron 1 of CHRNA5 | A/G | 0.114 | 1.36 (1.14–1.61) | 5.4 × 10−4 |
| rs4243084 | Chr15:78911672 | intron 1 of CHRNA3 | C/G | 0.1753 | 1.28 (1.11–1.48) | 8.5 × 10−4 |
| rs2735940 | Chr5:1296486 | 1.3kb upstream of TERT | T/C | 0.479 | 0.82 (0.73–0.93) | 1.1× 10−3 |
| rs4635969 | Chr5:1308552 | 13.3kb upstream of TERT | C/T | 0.3328 | 0.81 (0.72–0.92) | 1.2 × 10−3 |
| rs17405217 | Chr15:78731149 | intron 1 of IREB2 | C/T | 0.05842 | 1.43(1.14–1.81) | 2.5× 10−3 |
ordered by P-value;
minor allele listed second;
MAF in controls only;
OR for each additional copy of the minor allele, estimated in a logistic regression model adjusted for: age, sex, study site, % sub-Saharan African Ancestry, %European ancestry, and number of pack-years smoked;
P-value in an allelic additive logistic regression model, adjusted for: age, sex, study site, % sub-Saharan African Ancestry, %European ancestry, and number of pack-years smoked
Table 3.
Top three directly genotyped SNP associations in each lung cancer histologic subgroupa
| Adenocarcinoma (N=538) | Squamous cell (N=343) | Small cell (N=87) | All histologies (N=1308) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Histologic subtype | SNP | Position | Gene | Alleleb | ORc | P-valued | ORc | P-valued | ORc | P-valued | ORc | P-valued |
| Adenocarcinoma | rs2735940 | Chr5:1296486 | 1.3kb upstream of TERT | T/C | 0.75 | 8.1 × 10−5 | 0.81 | 0.024 | 0.87 | 0.3991 | 0.82 | 1.1× 10−3 |
| rs7725218 | Chr5:1282414 | intron 2 of TERT | G/A | 1.33 | 2.0× 10−4 | 1.08 | 0.43 | 0.91 | 0.5635 | 1.13 | 0.047 | |
| rs2736100 | Chr5:1286516 | intron 1 of TERT | T/G | 1.31 | 2.7 × 10−4 | 1.13 | 0.17 | 1.12 | 0.4955 | 1.17 | 8.9× 10−3 | |
|
| ||||||||||||
| Squamous cell | rs401681 | Chr5:1322087 | intron 13 of CLPTM1L | T/C | 1.12 | 0.12 | 1.35 | 1.5× 10−3 | 1.16 | 0.37 | 1.18 | 4.3 × 10−3 |
| rs2844463 | Chr6:31615167 | intron 7 of BAT3 | C/T | 0.92 | 0.26 | 0.74 | 1.8× 10−3 | 1.0 | 0.99 | 0.91 | 0.13 | |
| rs2736158 | Chr6:31600304 | exon 16 of BAT2 (missense Ala → Gly) | C/G | 1.06 | 0.61 | 0.64 | 1.8× 10−3 | 1.16 | 0.53 | 1.04 | 0.66 | |
top three associations for small-cell carcinoma are not included due to limited sample size. All three SNPs were located on 15q25.1
minor allele listed second
OR for each additional copy of the minor allele, estimated in a logistic regression model adjusted for: age, sex, study site, % sub-Saharan African Ancestry and number of pack-years smoked
P-value in an allelic additive logistic regression model, adjusted for: age, sex, study site, % sub-Saharan African Ancestry and number of pack-years smoked
In analysis of all tumor histologies, the most significantly associated genotyped SNP in the 6p22.1-p21.31 region was rs184054 (P = 0.0172). No genotyped or imputed SNPs had a P-value below 1.0 × 10−3. A previously identified lung cancer risk SNP, rs3117582, was not associated with case-control status in these African-Americans (P = 0.2181) (Figure 1C).
Analysis of the 15q25.1 region identified a number of strongly associated SNPs (Figure 2), including two previously reported to influence lung cancer risk. These associations were replicated in our African-American case-control sample at both the genotyped SNP rs8034191 (P = 0.0028, effect in same direction as previous reports) and the imputed SNP rs1051730 (P = 5.8 × 10−5, effect in same direction as previous reports). The most significantly associated SNP in 15q25.1 was rs2036527 (OR = 1.34, 95% CI = 1.17–1.54, P = 2.0 × 10−5). This SNP was also the most significantly associated SNP in the analysis of all three regions. Indeed, of the ten most strongly associated genotyped SNPs in the full analysis of the three regions, eight are located on 15q25.1 (Table 2). These eight SNPs are primarily located in or upstream of CHRNA5. One of these, rs16969968, is a functional SNP located in the fifth exon of CHRNA5 (OR = 1.57, 95% CI = 1.25–1.97, P = 1.1 × 10−4). The A>G substitution changes an aspartic acid residue to an asparagine residue and is found at a frequency of 8.8% in cases. None of the 1,013 imputed SNPs on 15q25.1 were more significantly associated with lung cancer than the two most significant genotyped SNPs, although both rs55853698 and rs55781567 in the 5′ untranslated region of CHRNA5 were strongly associated with lung cancer (P = 8.9 × 10−5 and 5.6 × 10−5 respectively). Both SNPs are located within predicted transcription-factor binding sites generated from ENCODE data.
Modeling all 15q25.1 SNP associations conditional on rs16969968 genotype (i.e. including the number of rs16969968 risk alleles as an ordinal covariate in the sex, age, study site, ancestry, and smoking-adjusted model) attenuated associations (Figure S2). However, the two most significant SNPs from Table 2 (rs2036527 and rs17486278) remained associated with lung cancer in the conditional analysis (P = 0.0056 and 0.0047, respectively). Overall, the most strongly associated SNPs in the conditional analysis were rs149156593 in an intron of CHRNB4 (OR = 2.31, 95% CI = 1.45–3.69, P = 4.5 × 10−4) and rs111819086 in an intron of AGPHD (OR = 1.92, 95% CI = 1.31–2.82, P = 7.9 × 10−4). Analysis without adjustment for pack-years modestly increased the strength of the 15q25.1 associations (Figure S3).
In haplotype analyses of the three chromosomal regions, no significant haplotype (p<0.05) was more statistically significant than the most significant SNP within that haplotype. As a result, haplotype analyses did not outperform single SNP analyses.
Performing association tests after stratifying by tumor histology produced striking results for the 5p15.33 and 6p22.1-p21.31 regions (Figure 3), whereas associations were very consistent across histologic subtypes in the 15q25.1 region (data not shown). Several SNPs in the 5p15.33 region are strongly associated with adenocarcinoma of the lung, but have weaker associations with other lung cancer types. As seen in Table 3 and Figure 3A, SNPs near the promoter and first exons of TERT were more strongly associated with adenocarcinoma than with other histologies, resulting in a dampening of the association signal in unstratified analyses. The strongest of these associations was rs2735940, with an odds ratio of 0.75 comparing adenocarcinoma patients to controls (95% CI = 0.64–0.86, P = 8.1 × 10−5) and an odds ratio of 0.82 (95% CI = 0.73–0.93, P =1.1 × 10−3) comparing all lung cancer histologies to controls.
Figure 3. Association of SNPs on chromosome 5p15.33 and 6p21.33 with lung cancer, by histologic subtype.
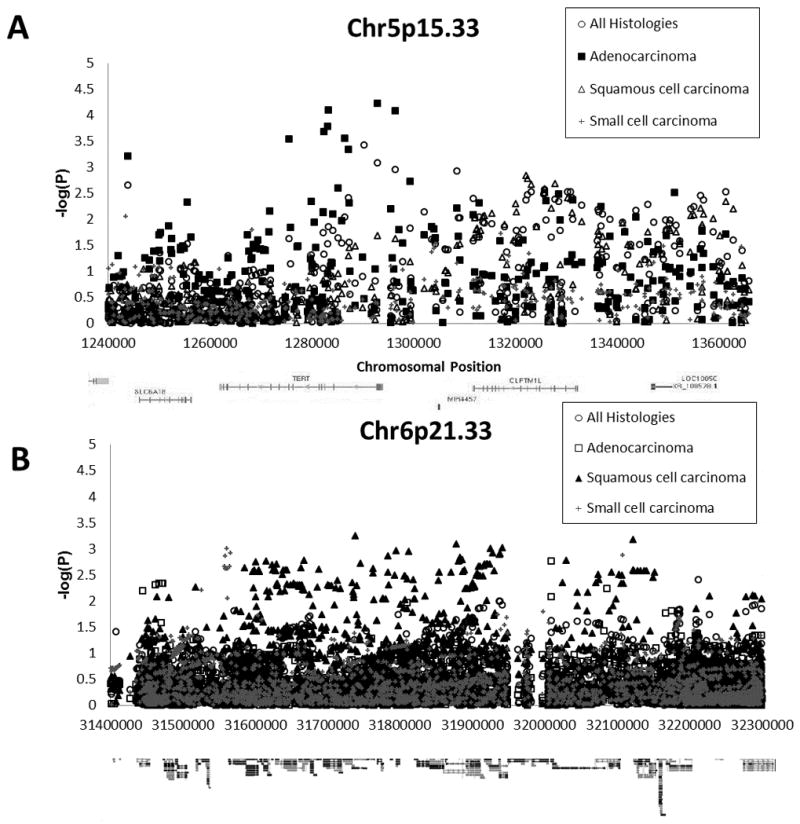
(A) Association of SNPs at 5p15.33 with lung cancer, adjusted for sex, age, study site, % sub-Saharan African ancestry, % European ancestry, and number of pack-years smoked. Circles denote associations for all lung cancer histologies compared to controls. Squares denote associations for adenocarcinoma cases compared to controls. Triangles denote associations for squamous-cell carcinoma cases compared to controls. Crosses denote associations for squamous cell carcinoma cases compared to controls. (B) Association of SNPs at 6p21.33 with lung cancer, adjusted for sex, age, study site, % sub-Saharan African ancestry, % European ancestry, and number of pack-years smoked. Circles denote associations for all lung cancer histologies compared to controls. Squares denote associations for adenocarcinoma cases compared to controls. Triangles denote associations for squamous-cell carcinoma cases compared to controls. Crosses denote associations for squamous cell carcinoma cases compared to controls.
For the 6p22.1-p21.31 region, the association pattern seen for participants with squamous-cell tumors differed from those with other tumor histologies, including a noteworthy association peak spanning from ~31.60–32.10 Mb and overlapping with previous GWAS hits in BAT3 (Table 3 and Figure 3B). The most strongly associated genotyped SNPs in the region were rs2844463 in an intron of BAT3 (OR = 0.74, 95% CI = 0.62–0.90, P = 1.79 × 10−3) and rs2736158 in exon 16 of BAT2 (OR = 0.64, 95% CI = 0.48–0.85, P = 1.82 × 10−3). This missense C>G substitution changes an alanine residue to a glycine residue, and is found at a frequency of 12% in controls where it acts as a protective allele decreasing the risk of squamous-cell carcinoma. Imputation of an additional 3447 SNPs in the region provides additional evidence that a squamous-cell risk variant is located on 6p21.33. As a whole, these results provide evidence that previous GWAS hits in and around BAT2/BAT3 are valid associations, but that at least in African-Americans the association is specific to squamous-cell lung cancer.
DISCUSSION
Our findings provide evidence that inherited variation in the three regions most frequently associated with lung cancer in European and East Asian populations are also associated with lung cancer in African-Americans. Despite this consistency, there remains heterogeneity across ethnicities in which particular SNPs have the strongest associations, particularly on 15q25.1 where the association peak in African-Americans is more strongly localized to CHRNA5 than it is among other ethnicities. We further demonstrate that SNPs which confer risk for specific lung cancer histologies are not necessarily risk factors for all types of lung cancer, as 5p15.33 associations were driven by risk for adenocarcinoma and 6p21.33 associations appear specific to risk for squamous-cell carcinoma. Imputation of an additional 305 SNPs on 5p15.33 and 3447 SNPs on 6p21.33 further support the histology-specific nature of these associations. Finally, a list of the mostly strongly associated SNPs in the analysis includes functional variants on 15q25.1 and 6p21.33 which may directly influence lung cancer risk in African-Americans. Histology-specific lung cancer associations are being reported with increasing frequency, as researchers begin to refine case-definitions in their association studies (39, 40).
We observed a moderate association between lung cancer and variants at 5p15.33 in TERT and CLPTM1L, expanding on previous reports of associations in this region in African-Americans (29). These associations appear largely to be confined to risk for adenocarcinoma, as several SNP associations in and near TERT were greatly strengthened when analysis was performed only on cases with adenocarcinoma, despite the reduction in sample size. The association in adenocarcinoma cases was well-localized to the region immediately upstream and in the first two introns of TERT, suggesting that TERT is likely the gene in this region functionally involved in lung cancer pathogenesis.
In analysis of the full case-control dataset, variants in the HLA region on 6p22.1-p21.31 showed little association with lung cancer. However, when stratified by histology, a previously associated region in BAT3 was replicated in the squamous-cell carcinoma subgroup and an associated missense variant in BAT2 was identified. Associations in the 6p21.33 region have not been consistently replicated, in part due to the European risk SNPs having low allele frequencies in East Asian populations. We offer another possible reason for the inconsistent associations of SNPs in this region–namely, that the associations appear to be histology-specific and may be difficult to detect when squamous-cell carcinomas are analyzed with other tumor histologies. Just as the TERT associations are strongest in cases of adenocarcinoma, in African-Americans the 6p21.33 associations appear strongest in cases of squamous-cell carcinoma. Indeed, of the three GWAS identifying an association peak in/near BAT3, all contained a substantial proportion of cases with squamous-cell tumors (range: 19.5%–38%) (1–3). The histology-specific association of 6p21 SNPs with squamous-cell tumors was also recently detected in a meta-analysis of Caucasian lung cancer patients (39).
The strongest associations in this African-American lung cancer study were located on 15q25.1, in or just upstream of CHRNA5. Several associated SNPs in CHRNA5 may be functionally relevant, including strongly associated variants in the 5′ UTR and in exon 5. We genotyped many more SNPs in the 15q25.1 region than previous studies of African-Americans (41), and imputed additional variants using data from the 1000 Genomes project. Whereas studies in European populations have had difficulty mapping the 15q25.1 signal to a particular gene because of large LD blocks in the region, the reduced LD in African-Americans has allowed functional and tagging variants in CHRNA5 to emerge as the likeliest candidates in our analysis.
Controlling for the number of pack-years smoked modestly attenuated CHRNA5 associations, but they remained the strongest association signals in the analyses. Because SNPs in this region are associated with smoking behaviors, and pack-years is an imperfect measurement of smoking behavior, the effects observed here may be mediated by smoking. Conditioning association tests on missense SNP rs16969968 revealed additional association signals in CHRNB4 and AGPHD. Whether these associations are due to the effects of additional genes on 15q25.1, or simply result from linkage between SNPs in CHRNB4 and AGPHD with other CHNRA5 risk variants remains to be determined. Whether differences in the association patterns on 15q25.1 seen in African-Americans, compared to other ethnicities, are due to differences in LD patterns or differences in smoking behaviors remains to be determined.
The current analysis represents the largest genetic study of lung cancer conducted in African-Americans to date and has identified functional variants which may contribute to the pathogenesis of this devastating disease. While multi-center studies pose certain challenges, we believe that the close similarity of the % sub-Saharan African ancestry estimates and the similar sample size across study sites is a particular strength of the study. Our study provides evidence that regions which are associated with lung cancer in Europeans and East Asians are also associated in African-Americans, but that there is incomplete overlap of the associated SNPs. We further conclude that tumor histology is an important consideration in genetic association analyses of lung cancer, especially in African-Americans where there is both a different distribution of tumor histology (42) and of SNP allele frequencies compared to European populations.
Supplementary Material
Acknowledgments
FUNDING:
This work was supported by National Institutes of Health grants: R01CA52689, R01ES06717, R01CA121197, R01CA121197S2, R01CA14176, R01CA060691, N01PC35145, HHSN261201000028C, R25CA112355 (KMW) and K02DA021237 (LJB).
ABBREVIATIONS
- SNP
single nucleotide polymorphism
- GWAS
genomewide association study
- UCSF
University of California San Francisco
- MDA
M.D. Anderson
- WSU
Wayne State University
- LD
linkage disequilibrium
Footnotes
CONFLICT OF INTEREST STATEMENT:
Laura J. Bierut served as a consultant for Pfizer Inc. in 2008 and is an inventor on the patent “Markers for Addiction” (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. Other authors do not have any conflicts of interest, financial or otherwise.
References
- 1.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, et al. Common 5p15.33 and 6p21. 33 variants influence lung cancer risk. Nat Genet. 2008;40(12):1407–9. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85(5):679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69(16):6633–41. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, et al. Lung cancer susceptibility locus at 5p15. 33. Nat Genet. 2008;40(12):1404–6. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsiung CA, Lan Q, Hong YC, Chen CJ, Hosgood HD, Chang IS, et al. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 2010;6(8) doi: 10.1371/journal.pgen.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 7.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25. 1. Nat Genet. 2008;40(5):616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61(4):212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11(5):356–66. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truong T, Hung RJ, Amos CI, Wu X, Bickeboller H, Rosenberger A, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst. 2010;102(13):959–71. doi: 10.1093/jnci/djq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15. 33. Nat Genet. 2010;42(3):224–8. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42(11):978–84. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43(8):785–91. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, Hu L, Shen H, Dong J, Shu Y, Xu L, et al. Candidate variants at 6p21.33 and 6p22. 1 and risk of non-small cell lung cancer in a Chinese population. Int J Mol Epidemiol Genet. 2010;1(1):11–8. [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12. 2 in Han Chinese. Nat Genet. 2011;43(8):792–6. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 17.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–7. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiraishi K, Kohno T, Kunitoh H, Watanabe S, Goto K, Nishiwaki Y, et al. Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis. 2009;30(1):65–70. doi: 10.1093/carcin/bgn257. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Liu P, Wen W, James MA, Wang Y, Bailey-Wilson JE, et al. Haplotype and cell proliferation analyses of candidate lung cancer susceptibility genes on chromosome 15q24–25. 1. Cancer Res. 2009;69(19):7844–50. doi: 10.1158/0008-5472.CAN-09-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fehringer G, Liu G, Pintilie M, Sykes J, Cheng D, Liu N, et al. Association of the 15q25 and 5p15 lung cancer susceptibility regions with gene expression in lung tumor tissue. Cancer Epidemiol Biomarkers Prev. 2012;21(7):1097–104. doi: 10.1158/1055-9965.EPI-11-1123-T. [DOI] [PubMed] [Google Scholar]
- 23.Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69(17):6848–56. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen HM, Xiao Y, Rice T, Bracci PM, Wrensch MR, Sison JD, et al. Fine mapping of chromosome 15q25. 1 lung cancer susceptibility in African-Americans. Hum Mol Genet. 2012;19(18):3652–61. doi: 10.1093/hmg/ddq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amos CI, Gorlov IP, Dong Q, Wu X, Zhang H, Lu EY, et al. Nicotinic acetylcholine receptor region on chromosome 15q25 and lung cancer risk among African Americans: a case-control study. J Natl Cancer Inst. 2010;102(15):1199–205. doi: 10.1093/jnci/djq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabral DN, Napoles-Springer AM, Miike R, McMillan A, Sison JD, Wrensch MR, et al. Population- and community-based recruitment of African Americans and Latinos: the San Francisco Bay Area Lung Cancer Study. Am J Epidemiol. 2003;158(3):272–9. doi: 10.1093/aje/kwg138. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz AG, Wenzlaff AS, Bock CH, Ruterbusch JJ, Chen W, Cote ML, et al. Admixture mapping of lung cancer in 1812 African-Americans. Carcinogenesis. 2011;32(3):312–7. doi: 10.1093/carcin/bgq252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De La Vega FM, Isaac HI, Scafe CR. A tool for selecting SNPs for association studies based on observed linkage disequilibrium patterns. Pac Symp Biocomput. 2006:487–98. [PubMed] [Google Scholar]
- 29.Van Dyke AL, Cote ML, Wenzlaff AS, Abrams J, Land S, Iyer P, et al. Chromosome 5p Region SNPs Are Associated with Risk of NSCLC among Women. J Cancer Epidemiol. 2009;2009:242151. doi: 10.1155/2009/242151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei C, Han Y, Spitz MR, Wu X, Chancoco H, Akiva P, et al. A case-control study of a sex-specific association between a 15q25 variant and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2011;20(12):2603–9. doi: 10.1158/1055-9965.EPI-11-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen HM, Wiemels JL, Wrensch M, Wiencke JK. DNA quantification of whole genome amplified samples for genotyping on a multiplexed bead array platform. Cancer Epidemiol Biomarkers Prev. 2007;16(8):1686–90. doi: 10.1158/1055-9965.EPI-06-1024. [DOI] [PubMed] [Google Scholar]
- 32.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The International HapMap Project. Nature. 2003;426(6968):789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 34.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howie B, Marchini J, Stephens M. Genotype Imputation with Thousands of Genomes. G3: Genes, Genomes, Genetics. 2012;1(6):457–70. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 39.Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeboller H, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi J, Chatterjee N, Rotunno M, Wang Y, Pesatori AC, Consonni D, et al. Inherited variation at chromosome 12p13. 33, including RAD52, influences the risk of squamous cell lung carcinoma. Cancer Discov. 2012;2(2):131–9. doi: 10.1158/2159-8290.CD-11-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz AG, Cote ML, Wenzlaff AS, Land S, Amos CI. Racial differences in the association between SNPs on 15q25. 1, smoking behavior, and risk of non-small cell lung cancer. J Thorac Oncol. 2009;4(10):1195–201. doi: 10.1097/JTO.0b013e3181b244ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gadgeel SM, Severson RK, Kau Y, Graff J, Weiss LK, Kalemkerian GP. Impact of race in lung cancer: analysis of temporal trends from a surveillance, epidemiology, and end results database. Chest. 2001;120(1):55–63. doi: 10.1378/chest.120.1.55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


