Abstract
Tourette syndrome (TS) is a highly heritable neuropsychiatric disorder characterised by motor and vocal tics. Despite decades of research, the aetiology of TS has remained elusive. Recent successes in gene discovery backed by rapidly advancing genomic technologies have given us new insights into the genetic basis of the disorder, but the growing collection of rare and disparate findings have added confusion and complexity to the attempts to translate these findings into neurobiological mechanisms resulting in symptom genesis. In this review, we explore a previously unrecognised genetic link between TS and a competing series of trans-synaptic complexes (neurexins (NRXNs), neuroligins (NLGNs), leucine-rich repeat transmembrane proteins (LRRTMs), leucine rich repeat neuronals (LRRNs) and cerebellin precursor 2 (CBLN2)) that links it with autism spectrum disorder through neurodevelopmental pathways. The emergent neuropathogenetic model integrates all five genes so far found to be uniquely disrupted in TS into a single pathogenetic chain of events described in context with clinical and research implications.
Keywords: autism spectrum disorder, genetics, neurexin, neurodevelopment, Tourette syndrome, trans-synaptic complexes
Introduction
Tourette syndrome (TS) is characterised by motor and vocal tics, with a pre-pubertal age of onset, a waxing and waning course, and improvement in symptoms in adulthood.1 Clinical and epidemiological studies point to an association with other childhood onset behavioural and developmental disorders such as obsessive-compulsive disorder (OCD), attention-deficit/hyperactivity disorder (ADHD) and to a lesser extent with autism spectrum disorder (ASD) (see Glossary).2 The remarkable complexity, high heritability and intriguing phenotype of TS linking genetics, brain, mind and behaviour has spawned an international research effort to discover the pathogenetic basis of this neurodevelopmental disorder. Although the genetic and pathophysiological basis of the disorder remains unresolved, there is converging evidence to suggest involvement of the cortical striatal–pallidothalamic–cortical circuitry that mediates the integration of movement, sensation, emotion and attention. It is suggested that the improvement with advancing age is the result of compensatory responses that come in line with maturation when the frontal cortices become more efficiently connected to the striatum and to the motor and sensorimotor cortices.1
The familial nature of TS was evident from the time of its original description by Gilles de la Tourette in 1885. Twin studies suggest a monozygotic to dizygotic concordance of 77 to 23%, whereas family studies have consistently shown a 10- to 100-fold increase in the rates of TS in first-degree relatives.3 Furthermore, it is suggested that chronic tics and OCD are manifestations of the same underlying genetic susceptibility as TS.4 ADHD is another significant co-morbidity and more recent studies have highlighted that ASD is over represented in TS, occurring in about 4 to 5% of the TS population.5, 6 Furthermore, Kadesjo and Gillberg7 found that while 5% of individuals with TS also had a diagnosis of Asperger's syndrome, 17% showed three or more autistic symptoms and 65% had deficits relating to the autism spectrum. There is emerging evidence to suggest an overlap between TS and ASD from phenomenological, epidemiological and pathogenetic perspectives.8 TS and ASD are both conditions that begin during childhood and mostly affect males. Clinically, symptoms such as obsessions, compulsive behaviours, involuntary movements (tics in TS and stereotypies in ASD), poor speech control and echolalia are common in both conditions. Genetic epidemiology studies also support the existence of common susceptibility genes in both disorders.8 In 1991, Sverd9 hypothesised that the TS gene in its homozygous form may lead to the co-occurrence of TS and ASD, while in its dominant heterozygous form may lead to poor socialisation or communication. Twenty years on, there has been little progress in understanding the pathogenetic basis of TS except that linkage and candidate gene analyses have now virtually ruled out any likelihood of common dominant mutations of large effect.
Attractive candidates
To contextualise the significance of the integrated synaptic model for TS presented in this review, it is important to cover with some detail the present state of uncertainty that exists regarding the genetic and molecular aetiology of TS. The depth of this uncertainty is reflected in the many candidate gene studies that populate the recent TS literature.3, 10 As dopamine antagonists represent the most effective medications for tic suppression, speculative candidate gene studies have largely focussed on those genes implicated within the dopaminergic pathway including the dopamine receptors DRD1-5, dopamine β-hydroxylase, dopamine-associated transporter SLC6A3, as well as the adrenergic receptors α1c-, α2c, α2a and β2-, various serotonin receptors and others.3 Sadly, the candidate gene approach has not yielded a single susceptibility gene of large effect for TS. However, together these studies do tend to suggest that if common biochemical pathways involving neurotransmitters such as serotonin, dopamine and glutamate are involved, they are likely positioned downstream of the primary molecular anomalies in TS.
Linkage promises
Two non-parametric linkage analyses sufficient in size to identify common variants of moderate to large effect have now been completed: a genome scan for Tourette disorder in affected-sibling-pair and multigenerational families;11 and a complete genome screen in sib pairs and multigeneration families by the Tourette Syndrome Association International Consortium for Genetics (TSAICG).12 One significant linkage region on 2p23 was identified11 but results were not reproducible between these two studies and no associated mutations have been identified.10, 11, 12 By comparison, parametric linkage analyses have been applied in the search for rare variants of large effect within single multigeneration pedigrees and isolated populations.11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 These latter studies have identified a number of impressive linkage regions within single pedigrees including peaks on 3q and 14q.13, 17 These two peaks overlap/replicate regions of interest identified in the 2007 TSAICG analysis of sib pairs,11 but no mutations have yet been reported from these loci either. The first and as yet the only linkage locus to yield a mutation of interest is 15q21.16 Subsequent sequence analysis within the critical linkage region on 15q identified a non-sense mutation in the L-histidine decarboxylase (HDC) gene that encodes the rate-limiting enzyme in histamine biosynthesis.16 This non-sense mutation co-segregates with the disorder in the original family composed of a father and his eight children with TS and OCD. Additional investigations suggest that dominant negative effects on histaminergic neurotransmission may be in play here with possible implications for the dysregulation of dopaminergic pathways.8, 23 Albeit, HDC mutations were absent from a large TS cohort screened by State et al.8 suggesting that the HDC association may be limited to this singular family.
Independent genomic rearrangements
Genomic rearrangements and copy number variations (CNVs) are the most common DNA lesions associated with TS and the most commonly rearranged locus in TS is chromosome 18q22. 2.6, 11, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47 Recent reviews and reports on the genetics of TS3, 45, 48 have identified at least 28 large independent genomic rearrangements and CNVs with unique breakpoints including deletions, insertions, duplications, inversions and inter-chromosomal translocations.6, 11, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47 Of the rearrangements that have been characterised, approximately one third have directly disrupted genes (Table 1), whereas the remainder have breakpoints within intergenic regions (Table 2).24, 25, 26, 28, 29, 31, 32, 37, 44, 45 Intergenic breakpoints are of great interest as they can lead to the dysregulation of a neighbouring gene(s) even when separated by very long distances.49 Candidate genes (Table 2) located near TS intergenic translocation breakpoints may be similarly affected by long-range dysregulation. For example, the SLITRK1 gene, which encodes a neuronal leucine-rich repeat transmembrane protein (LRRTM) involved in neurite outgrowth, is located immediately adjacent to a large gene desert on chromosome 13q. This gene desert was disrupted by a de novo inter-chromosomal translocation breakpoint in a child with TS and ADHD.24 Follow-on studies identified two rare functional sequence variants in SLITRK1 in unrelated TS patients. One of these mutations, a single nucleotide variation within a conserved region of the 3′UTR,24 appears to strengthen the binding of a micro RNA hsa-miR-189 with consequent downregulation of SLITRK1 expression. Unfortunately, a later study that identified the same variants in unaffected individuals50 indicates that SLITRK1 may be of limited effect in TS.10
Table 1. The five genes directly disrupted in Tourette syndrome by unique genomic lesions.
| Gene | Locus | Function | Co-morbidities | DNA lesions | Other |
|---|---|---|---|---|---|
| NRXN1a | 2p21 | Neurexin 1 synapse | ADHD | Two truncating deletions45 | T |
| NRXN4/CNTNAP2b | 7q35 | Neurexin superfamily | OCD, MR, SD | Intragenic insertion46 | c,d |
| IMMP2La (LRRN3) | 7q31 | (Neural development) | MR, SD | Two exonic deletions39, 40 | T, SA, R, Dup35 |
| CTNNA3a (LRRTM3) | 10q21 | (Neurexin ligand) | OCD, ADHD | Two intragenic deletions45 | SA |
| NLGN4Xb | Xp22.33 | Neurexin ligand | ASD | Truncating deletion36 | XM43 |
Abbreviations: ADHD, attention deficit/hyperactivity disorder; ASD, autism spectrum disorder; BD, block deletion associated with TS; Dup, duplication in TS; LS, linkage region of interest; MR, mental retardation; OCD, obsessive-compulsive disorder; R, most commonly duplicated locus in ASD; SA, strong polymorphic association with ASD; SCHZ, schizophrenia; SD, speech delay; T, TS translocation breakpoint association; TS, Tourette syndrome; XM, XXX chromosome mosaicism.43
Recurrent disruption of this gene in TS.
Gene disruption co-segregates with disorder in TS family.
2p21-23 block insertion that disrupted CNTNAP2 harbours a TSAICG critical linkage region.11
CNTNAP2 disrupted in family without TS.53
Bold type indicates protein implicated in Tourette syndrome.
Table 2. Candidate genes located near translocation breakpoints in Tourette syndrome.
| Candidate | Locus | Function | Proximity/distance | Co-morbidities |
|---|---|---|---|---|
| Support data | ||||
| CBLN2 | 18q22.2/18q21.1 | Neurexin ligand | Adjacent ∼2.0 Mb44 | OCD |
| BD33 | ||||
| CBLN2 | 18q22.2/7q31 | Neurexin ligand | Adjacent ∼400 kb25 | OCD |
| BD33 | ||||
| LRRTM1 | 2p12/18q22.2 | Neurexin ligand | Breakpoint region29 | OCD |
| Leucine-rich repeat | LS | |||
| LRTM1 | 3p21/8q24 | Neural development | Break point region26 | OCB |
| Leucine-rich-repeat | LS | |||
| SLITRK1 | 13q31/13q33 | Neural development | Adjacent ∼350 kb24 | ADHD |
| Leucine-rich repeat | ||||
| SLCO5A1 | 8q13/6p23 | Anion transport | Adjacent < 200 kb28 | OCB |
| T | ||||
| SLCO5A1 | 8q13/6q24 | Anion transport | Adjacent < 200 kb28 | ADHD, OCD |
| T | ||||
| SLC26A7 | 8q22.1/1q21.1 | Cl/HCO3 exchange | Adjacent ∼550 kb32, 37 | OCD, ADHD |
| LS, SCZ81 | ||||
| GRIK2 | 6q21/17p11 | Glutamate transporter | Break point region33 | Coprolalia |
| BD | ||||
| SLC1A1 | 9p23 recurrent del | Glutamate transporter | Within deleted region43, 47 | |
| BD43 | ||||
| CLIC6 | 21q22 | Neuronal Cl− channel | Adjacent to duplication45 | |
| Dopamine D2, 3 and 4R | ||||
| DGCR2 | 22q11.2 duplication | Mutated in SCZ81 | Adjacent inside boundary27 | Stereotypies |
| BD41 SCZ81 |
Bold type indicates protein implicated in Tourette Syndrome.
Gene disruptions
Five genes have been directly disrupted in TS by independent genomic rearrangements and CNVs with unique breakpoints, namely IMMP2L, NRNX1, CTNNA3, NLGN4X and CNTNAP2.36, 39, 40, 45, 46 Viewed in isolation, each of these novel genomic rearrangements can be difficult to interpret in relation to a complex disorder like TS, even when a gene has been disrupted. There is always the concern that the rearrangement may be incidental or conversely that more than one gene may be affected.10, 45, 48 For example, in the 2011 case report describing the disruption of the IMMP2L gene there were additional genes deleted.39 To complicate this finding further, IMMP2L is a most unlikely candidate for TS that has no obvious functional association with the neuropathology of TS. IMMP2L is localised to the mitochondrial membrane where it regulates levels of reactive oxygen species associated with aging, kyphosis, wasting and ataxia.51 Extensive screening of TS patients also failed to identify any coding mutations in IMMP2L.39, 52 Together these findings for IMMP2L only heightened uncertainty regarding other rearrangements and CNVs in TS.8, 48 For example, there was already a degree of uncertainty regarding the disruption of the neurexin 1 (NRXN1), NLGN4X and CNTNAP2 genes in TS as each of these genes had been previously disrupted and/or mutated in ASD patients without the presentation of tics.53, 54
Identification of nestlings hatches new perspective of TS
Nevertheless, this fore mentioned case report implicating IMMP2L was significant in a larger context as it represented the second disruption of the IMMP2L gene described in TS. This now meant that all five of the genes known to be disrupted in TS by unique rearrangements had been disrupted in multiple individuals with the disorder. Three of the five genes (IMMP2L, NRNX1 and CTNNA3) had now undergone recurrent disruption in unrelated individuals with TS and the other two gene disruptions (NLGN4X and CNTNAP2) co-segregate with the disorder within families (Table 1),36, 39, 40, 45, 46 which greatly strengthens the likelihood of their pathogenicity. The co-morbidities associated with these five gene disruptions also overlap in a representative manner the developmental and behavioural spectrum of disorders reported independently in TS: two of these gene disruptions were associated with OCD; two with ADHD; two with speech delay; and the disruption of NLGN4X was associated with ASD (Table 1). However, this series of co-morbidities does not reflect the full spectrum of phenotypic associations reported for these same five genes from other studies. For example, the IMMP2L, NRNX1, CTNNA3, NLGN4X and CNTNAP2 genes have all shown independent association with ASD (without tics), often through multiple means of enquiry10, 55 thus strengthening the case further for a pathogenic relationship between all the five genes. This overlap also lends a scientific support to the long-standing clinical and epidemiological finding of a phenotypic overlap between TS and ASD. Albeit, the functional nature of this relationship remained obscure until we reviewed it through the lense of a seemingly unrelated ASD association study that implicated two additional genes LRRN3 and LRRTM3.56
Polymorphisms in two structurally related genes LRRN3 and LRRTM3 show strong association with ASD susceptibility.55, 56 LRRN3 and LRRTM3 are both neuronal leucine-rich repeat transmembrane protein genes (Box 1) and both are curiously positioned/nested inside other genes (see Glossary).57, 58, 59, 60, 61 Herein lies a most revealing twist in the TS story; LRRN3 and LRRTM3 are actually nested within two of the genes that have been disrupted in TS, namely IMMP2L and CTNNA3, respectively (NCBI Build 37.2, 2011). Thus, ASD associations have now been reported for both IMMP2L and CTNNA355, 62, 63 and for both of their nested genes, LRRN3 and LRRTM3, respectively,55, 56 that suggests that these separate studies have identified the same functional associations. LRRN3 and LRRTM3 share a very close structure–function relationship with each other as do their larger gene families (Box 1)64, 65, 66, 67, 68 that are intimately involved in brain development (discussed below). This close structure–function relationship strongly implies that the original ASD associations reported for IMMP2L and possibly CTNNA3 may represent cryptic association assignments. Nevertheless, as host genes, any change in their transcription has the very real potential of directly impacting the transcription of their nested genes. Two scenarios exist whereby the disruption of the IMMP2L and CTNNA3 genes could alter the transcription of their nested genes: first through the introduction or disruption of a regulatory element(s) that modulates the transcription of the nested gene(s) or; second and more consistent with the data is that the disruption of transcription of the host gene directly affects the transcriptional efficiency of the nested gene on the opposing DNA strand. The latter scenario infers the disruption of a pre-existing regulatory relationship between the host gene and nested gene(s). In this context, the nested relationship of the two LRR genes has significant implications for understanding TS: first, it strengthens the case for a genuine functional relationship between TS and ASD and all five genes disrupted in TS; second, given the close structure–function relationships that exist between LRRN3 and LRRTM3 it provides an invaluable starting point to begin to understand the biological function of the IMMP2L-LRRN3 and CTNNA3-LRRTM3 transcription complexes in neurodevelopment as it pertains to TS and its relationship to ASD (Box 1); third, it helps resolve confounding associations like that of IMMP2L; and finally, it provides the basis of a pathogenetic framework on which to connect other seemingly disparate genetic findings for TS.
Box 1. Leucine-rich repeat transmembrane protein genes.
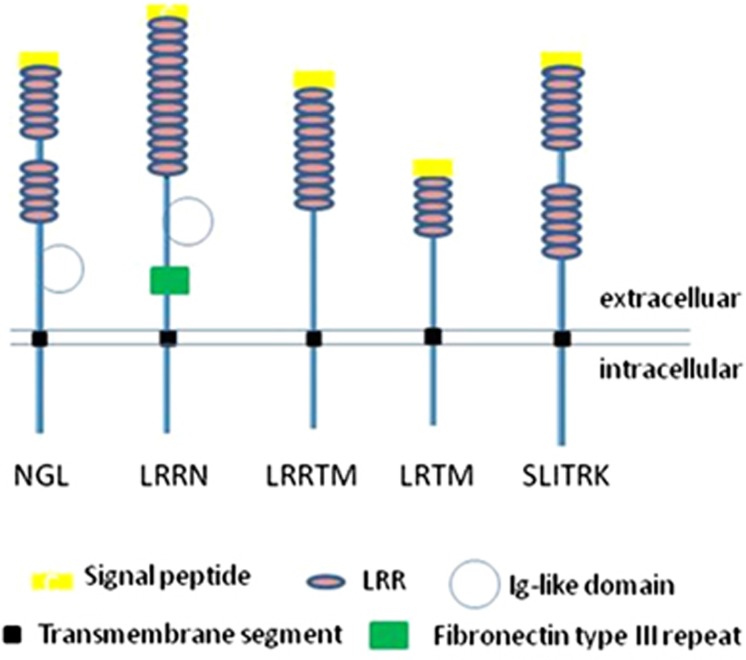
Leucine-rich repeats (LRRs) are common protein–protein interaction domains found in proteins with diverse structure and function. LRRs are typically 20–29 amino acids in length with a conserved consensus sequence LxxLxLxxN/CxL (where x can be any amino acid and L can be replaced by V, I or F).67, 68, 69, 70, 75, 76 There are several subgroups of LRR proteins differentiated by the consensus sequence and the inclusion of different combinations of supplementary domains (Figure 1). Among the ∼313 LRR coding genes in the human genome, the transmembrane subgroupings are brain enriched and/or highly expressed in the nervous system, with roles in neuronal development and/or synaptogenesis.64, 65, 66, 67, 77 The different families of transmembrane LRR proteins include AMIGO, NGL, LINGO, LRIG, FLRT, PAL, SALM, SLITRK, LRRN, LRRTM and LRTM (Figure 1).64, 65, 66, 67 The LRRN1, LRRN3, LRRTM1, LRRTM3 and LRTM1 genes implicated in the pathogenetic model for Tourette syndrome (Figure 2) all share the curious structural relationship of being nested in the antisense orientation within an intron of another gene (see ‘Nested genes' in Glossary). Evidence indicates convergent evolution of LRR gene nesting in different classes of genes.
The LRRTMs (leucine rich repeat transmembrane family) represent a highly conserved four-member gene family, which, with the exception of LRRTM4, are nested in the introns of different α-catenin genes.65 LRRTMs are enriched in the nervous system, each with a distinct and highly regulated pattern of expression. LRRTMs are synaptic cell adhesion organizing molecules initiating excitatory presynaptic differentiation and mediating post-synaptic specializations. LRRTM1 is located within intron 7 of CTNNA2 (α2-catenin) and is highly expressed within the brain and salivary gland.65 LRRTM1 appears to be associated with human handedness (relative hand skill), schizophrenia and language. LRRTM3, located within intron 7 of CTNNA3 (αT-catenin), expression is enriched within the cerebellum.
The LRRN (leucine rich repeat neuronal) gene family of four are all brain-enriched type I transmembrane protein genes. LRRN1 is nested within intron 8 of the extended form of the SUMF1 (sulphatase modifying factor 1) gene. LRRN1 regulates boundary formation within the brain. LRRN3 is nested within intron 3 of the IMMP2L (inner mitochondrial membrane peptidase-like) gene. Lrrn3 exhibits regulated expression in the developing ganglia and motor neurons of the neural system, and is upregulated during neuronal cortical injury.64
The LRTM (leucine-rich repeats and transmembrane domains) gene family is a highly conserved two-member family and both members are nested in introns of different CACNA2D (calcium channel, voltage-dependent, alpha 2/delta subunit) genes. LRTM1 is nested within intron 23 of CACNA2D3 and LRTM2 is nested within intron 23 of CACNA2D3.
Expression of LRRN3 and closely related family member LRRN1 are both enriched within the brain.39, 56, 69 LRRN3 is localised within the genomic region most commonly duplicated in ASD62, 70 and it also appears duplicated in TS35 and LRRN1 is also duplicated in ASD,71 suggesting that dose increases for these two related molecules maybe pathogenic for ASD and TS. Consistent with the upregulation of LRRN3 in TS is the finding that the LRRN3 gene, nested within IMMP2L, was not disrupted in either of the TS cases where IMMP2L was disrupted.39, 40, 45 In addition, the relative high-level expression of LRRN3 in the adult brain is discordant with very low-level expression for IMMP2L when compared with other tissues.39 Together these findings suggest the discordant co-regulation of both genes. Such discordant regulation of nested genes being transcribed from opposing DNA strands has been hypothesised to occur via a mechanism of transcriptional interference.59 Discordant regulation of LRRN3 is also suggested by studies in the mouse where modification of the Immp2l transcription unit appears to result in the upregulation of Lrrn3 transcription.72 In this context, the discordant regulation of the IMMP2L-LRRN3 transcription complex ceases to be a confounding factor in TS aetiology and provides an all important element of structural and functional integrity to the TS story. Both LRRN3 and its closest relation LRRN1 are involved in brain development. LRRN1 is known to have a key role in regional boundary formation during brain development including an essential role in midbrain-hindbrain boundary (MHB) formation.73 Studies in the chick demonstrate that the MHB is established by the downregulation of Lrrn1 by Fgf8 on the posterior side of the future boundary73 by creating a differential cellular affinity between the two compartments. The molecular basis of this differential affinity appears likely to involve an as yet unspecified extracellular binding partner for LRRN1. During this process, Lrrn1 regulates the expression of another well-known developmental gene, Lunatic Fringe, which modulates Notch signalling to complete MHB formation. Experimental overexpression of Lrrn1 in cells positioned on the hindbrain side of the future MHB results in violation of the boundary and mixing of cells between midbrain and hindbrain compartments.73
The LRRTMs have also been implicated in brain development and disease: LRRTM1 expression levels have been implicated in schizophrenia, left right brain asymmetry and handedness.74 The LRRTMs and LRRNs also have important structural similarities: both are neuronal transmembrane proteins; both have short intracellular tails with putative PDZ-binding domains; and both have extracellular LRR domains. The LRR domain of the LRRTMs represents a ligand-binding site for the formation of trans-synaptic complexes with NRXNs1.64, 65, 66, 67, 75, 76 LRRTM1, LRRTM2 and LRRTM3 are all nested within other genes. However, in contrast to the discordant expression of LRRN3 and IMMP2L, the high-level expression of LRRTM's in the brain is concordant with the high-level expression of their host genes.77 Here it is interesting to note that the LRRTM1 and LRRTM2 genes have bidirectional promoters that they share with their host genes.77 The co-regulation of LRRTM3 with its host gene CTNNA3 could therefore be a factor in how the recurrent disruption of the CTNNA3 gene could affect and implicate LRRTM3 expression in TS.
The neurexin connection
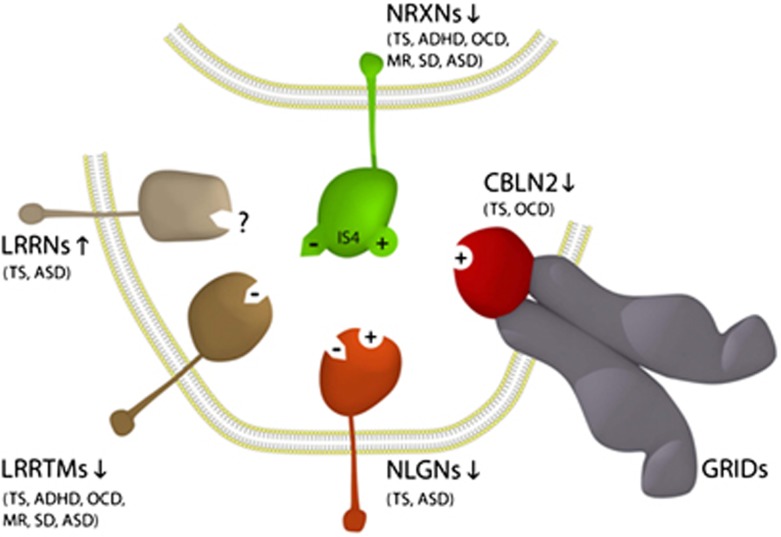
NRXNs 1–3 (NRXN1, NRXN2 and NRXN3) represent some of the largest genes in the human genome. The NRXN genes have dual promoters (α- and β-) and their transcripts are alternatively spliced into >1000 synaptic proteins. NRXNs are single-pass neuronal transmembrane proteins concentrated on the presynaptic side of the synapse. NRXNs appear to organise synapses by mediating cellular adhesion. The extracellular domain of presynaptic NRXNs binds to postsynaptic ligands (neuroligins (NLGNs), LRRTMs or cerebellin precursors (CBLNs)) to form trans-synaptic cell-adhesion complexes.8, 78 The three alpha-NRXNs 1, 2, 3 are essential for survival and have a pivotal role in neurodevelopment where their roles partially overlap.78 NRNX1 and NRNX4/CNTNAP2, two of the genes recurrently disrupted in TS, both encode members of the NRXN superfamily.53 NLGN4X, another of the genes recurrently disrupted in TS, is a member of the NLGN gene family that also encode single-pass neuronal transmembrane proteins. More importantly, NLGN4X functions as a postsynaptic cell-adhesion ligand for the NRXNs. In similar manner, LRRTM3, the gene nested within CTNNA3, is a member of the LRRTM gene family that also encode single-pass neuronal transmembrane proteins that function as postsynaptic cell-adhesion ligands for the NRXNs.8, 36, 39, 40, 45, 46 Similarly, the LRRN3 gene nested within IMMP2L encodes another neuronal transmembrane protein that has a close structure–function relationship with LRRTM3 (discussed above); however, extracellular binding partners have yet to be identified for the LRRNs and for NRXN4/CNTNAP2.73 As such, all five of the genes uniquely disrupted in TS encode neuronal transmembrane proteins of which at least four are members of the NRXN superfamily or form trans-synaptic connections with the NRXNs. We propose a neuropathogenetic model for TS (Figure 2) where an imbalance in the type and/or level of NRXN trans-synaptic connections triggers changes in the dynamics of synapse assembly, maintenance and function within the brain resulting in abnormalities in the neuronal circuitry.
Figure 1.
Schematic of domain architecture for a selection of neuronal leucine-rich repeat transmembrane protein families.
Figure 2.
Neuropathogenetic model for Tourette syndrome (TS) implicates the full complement of known neurexin (NRXN) trans-synaptic cell-adhesion ligand gene families through multiple means of enquiry: neuroligins (NLGNs); leucine-rich repeat transmembrane proteins (LRRTMs); and the cerebellin precursors (CBLNs). The presynaptic NRXNs form trans-synaptic complexes with postsynaptic ligands NLGNs, LRRTMs and CBLNs in the formation and/or maintenance of neuronal circuitry within the brain. Vertical arrows indicate putative pathogenic dose effects. Neurexin isoforms with (+) and without (−) the 30 amino-acid insert at splice site 4 (IS4) dictate the different/competitive binding of NRXNs between the ligands. Comorbidities listed are those associated with the TS translocations and copy number variations (CNVs) affecting the respective genes.
Using the neuropathogenetic model to interrogate other TS loci
The pathogenetic model for TS outlined in Figure 2 implicates an intersecting series of NRXN trans-synaptic complexes. We therefore searched the other TS translocation loci for genes that function within trans-synaptic signalling pathways. We revisited the locus most commonly rearranged in TS on 18q22.2.10, 25, 33, 44 In 2003, Mathew State and colleagues44 performed mutation analyses for the two genes located on either side of one of the TS translocation breakpoints on 18q22.2, namely CIS4 and GTSCR-1, but no mutations were identified in TS patients.44 With hindsight, this result was not altogether surprising given that CIS4 has since been shown to encode a regulator of T-cell activation and GTSCR-1 has recently been identified as a small pseudogene (<1 kb). More revealing is the absence of the GTSCR-1 pseudogene from mouse and rat indicating its relatively recent evolutionary retro-transposition within what is essentially a conserved gene desert (>2 Mb) that harbours numerous strongly conserved non-coding sequences (Vista Plot, NCBI Build 37.2). Unbeknown at the time of the original investigation, this 2-Mb gene desert on 18q22.2 is bordered at its distal end by the CBLN2 gene. CBLN2 was never screened for mutations in TS 44 but we can now clearly locate a second TS translocation breakpoint25, 44 within the same gene desert, albeit, much closer to CBLN2.44 The CBLN2 gene has also been deleted in TS.10, 25, 33, 44
CBLN2 is expressed widely in the brain and belongs to the CBLN subfamily (consisting of CBLN1-4) of the C1q/tumour necrosis factor superfamily, which serves diverse roles in intercellular signalling, neuronal cell adhesion, brain development and synaptogenesis. More specifically, CBLN2 encodes another ligand of the NRXNs. The full-length precursor of CBLN2 is secreted intact into the synaptic cleft in similar manner to its most closely related family member CBLN1.79 CBLN2 is an important synaptic organiser with similar synaptic connections to that of CBLN1, which forms a tripartite signalling complex with the NRXNs and the postsynaptic glutamate receptors delta 1 or delta 2 (GluD1/GRID1 or GluD2/GRID2).79, 80 Both CBLNs induce synaptogenesis in cerebellar, hippocampal and cortical neurons in vitro and the tripartite CBLN complex (NRXN-CBLN-GRID) actually competes with synaptogenesis mediated by NLGN179 (Figure 2). Thus, CBLN2 demonstrates an integrated competitive relationship with other members of the genetic model for TS (Figure 2) to provide the strongest of support for its role in the pathogenesis of TS. These findings expand the breadth of known NRXN trans-synaptic ligands/complexes now associated with TS and further strengthen the model (Figure 2) as a useful framework for understanding the broader pathogenesis of TS.
We searched other TS intergenic breakpoint loci24, 26, 27, 28, 29, 31, 32, 33, 35, 37, 41, 42, 43, 45 for genes that are structurally or functionally related to those within the model (Figure 2). Other LRR-coding genes LRRTM1, LRTM1 and SLITRK1 are located near breakpoints at 2p12, 3p21 and 13q31, respectively (Table 2).24, 26, 29, 37 Likewise, we searched critical TS linkage regions11, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 and found that the strongest linkage markers identified (D2S139 and D3S1289) from two separate parametric linkage analyses of single multi-generation TS pedigrees were positioned within the genes CTNNA2 and CACNA2D3, respectively (Table 3).17, 20 Surprisingly, both of these genes harbour nested neuronal LRR transmembrane coding genes, namely LRRTM1 and LRTM1, respectively. LRRTM1 and LRTM1 are the same two LRR candidates identified near TS translocation breakpoints (Table 2).24, 26, 29, 37 LRRTM1 and LRTM1 are structurally related to each other (Box 1) and to the two neuronal LRR transmembrane coding genes (LRRTM3 and LRRN3) nested within genes that are disrupted in TS (CTNNA3 and IMMP2L, respectively) (Table 1).64, 65, 66, 67 LRRTM1 is actually a member of the same gene family as LRRTM3. LRRTM1 has an important role in brain development (discussed earlier).74 Further independent support for LRRTM1 as a neurological disease gene comes from a separate non-parametric linkage analysis for schizophrenia where the strongest linkage marker was also located within the CTNNA2 gene.74 However, it was not the CTNNA2 gene that was demonstrated to be pathogenic for schizophrenia but rather the expression of its nested gene, LRRTM1.74 Could the differential regulation of LRRTM1 result in schizophrenia or TS?74, 81, 82 Viewed together, this stunning molecular correspondence between TS disrupted genes, intergenic breakpoints and parametric linkage regions, assorted ASD association studies and linkage analyses of schizophrenia provides impressive support for the pathogenic role for these nested LRR transmembrane coding genes in TS and for the broader application of our neuropathogenetic model (Figure 2). The association of LRRTM1 with TS further increases the number of NRXN postsynaptic cell-adhesion ligands now associated with TS (Figure 2).64, 67
Table 3. Candidate genes within linkage regions for Tourette syndrome.
| Study | Pedigree | Linkage marker/locus | Candidate genes |
|---|---|---|---|
| Breedveld13 | Italian pedigree | D14S1000 | Linkage spans gene desert adjacent to NRXN3 |
| TSAICG11 | NPL pairs | 14q 31 | Suggested linkage region spans NRXN3 |
| TSAICG11 | NPL pairs | 3p 26 | Suggested linkage region spans SUMF1/LRRN1 |
| TSAICG11 | NPL pairs | 3q 26 | Suggested linkage region spans NLGN1 |
| Knight17 | Utah pedigree | D3S1289 | Marker located within CACNA2D3/LRTM1 |
| Simonic20 | Africana families | D2S139 | Marker located within CTNNA2/LRRTM1 |
| GATA28F12 | Marker near SLC26A7–Cl/HCO3 exchange | ||
| D11S1377 | GRIK 4–glutamate transport channel | ||
| Merette18 | Single Canadian pedigree | D11S1377 | GRIK 4–glutamate transport channel |
| Laurin22 | Single pedigree | D5S430 | SLC1A3–glutamate transport channel |
| Knight17 | Utah pedigree | D1S207 | Linkage spans ZnT7–zinc transporter |
| TSAICG11 | Non-parametric | D2S165 | Linkage spans ZnT3–synaptic zinc transporter |
| TSAICG11 | Multigeneration families | 5p | Linkage spans MSNP1AS–moesin antisense |
| Verkerk21 | Single Dutch pedigree | D3S1311 | Marker located within SAP97–synaptogenesis |
| Paschou19 | Two large pedigrees | D17S928/784 | Critical linkage region spans NPTX1 |
| Zhang14 | Sib pair families | D17S784 | Marker located near NPTX1 |
| Curtis15 | Single pedigree | D5S400 | Marker within SLIT3–LRR axonal guidance |
| D14S288 | Marker adjacent to LRFN5 | ||
| Ercan-Sencicek16 | Single pedigree | D15S126 | Non-sense mutation located within HDC |
Underline highlights linkage marker located with the candidate gene.
Also of interest are the three main suggestive linkage peaks reported by the TSAICG parametric study of affected sib pairs on 3p, 3q and 14q.11 These three peaks span additional neuronal transmembrane protein genes LRRN1, NLGN1 and NRXN3, respectively (Table 3). The gene desert 3′ of NRXN3 has also been linked to TS by parametric analysis within a large Italian kindred (Table 3).13 Impressively, all three of these genes are members of the same small specialised gene families now implicated in the TS model (Figure 2; Box 1).56, 67, 71 Furthermore, NLGN1 is a trans-synaptic ligand for NRXN3 and NRXN1;67, 83 NRXN3, NRXN1 and NRXN2 are mutated in ASD;54, 84 and LRRN1 like its close relation LRRN3 has been duplicated in ASD.71
Expanding the neuropathogenetic model for TS
Another gene of interest, ZnT3, is also worthy of mention here if only to demonstrate the utility of the new model to help interrogate broader aspects of TS pathogenicity (Figure 2). ZnT3 is one of the many genes located under the one and only significant linkage peak identified on chromosome 2p23 in the latest TSAICG non-parametric analysis of sib pairs and multi-generation families (Table 3).11 In light of our new pathogenic model for TS (Figure 2), ZnT3, a synaptic zinc transporter that controls Zn2+ concentrations within synaptic vesicles, now emerges as a most compelling candidate for TS on 2p23. The concentration of Zn2+ ions within the postsynaptic density (PSD), a specialised intracellular region of the excitatory synapse, is known to affect the recruitment of scaffolding proteins like SHANK2 and SHANK3, both of which have been mutated in ASD.85 The SHANK proteins mediate the attachment of the intracellular PDZ-binding domains of model trans-synaptic receptor/ligand complexes, like NRXN-NLGN and NRXN-LRRTM (Figure 2), to the local actin-based cytoskeleton within dendritic spines. In Purkinje cells, the postsynaptic clustering of SHANK2 with GluD2/GRID2 also appears to be dependent on the integrity of the tripartite NRXN-CBLN1-GRID2 trans-synaptic complex.79, 80 The LRRNs also have putative extracellular PDZ-binding domains.
A gene of related interest to ZnT3 is synapse-associated protein 97 (SAP97). Linkage analysis of a large Dutch pedigree with TS identified the highest linkage peak using a marker (D3S1311) located within the SAP97 gene (Table 3).46 A male individual with TS has also been identified with a duplication of the SAP97 gene locus (unpublished data), whereas micro-deletion of 3q inclusive of SAP97 is commonly characterised in schizophrenia.82 SAPs are thought of as scaffolds that organise the PSD of excitatory synapses with the ability to bind to membrane receptors, signalling molecules and the cytoskeleton. The SAP family consists of four homologues PSD93, PSD95/ SAP90, SAP102 and SAP97/DLG1. All SAPs contain three PDZ domains. SAP family proteins have been found to bind to AMPA, NMDA and kainate receptors at synapses86 and postsynaptic cell-adhesion molecules including NGLN1. There is also recent evidence that membrane-diffusing AMPARs can be rapidly trapped at PSD95 scaffolds assembled at nascent NRXN/NLGN adhesions, in competition with existing synapses.87 Overexpression of SAP97 enhances glutamatergic synaptic transmission.88
Synaptic model mechanisms
The synaptic model for TS outlined in Figure 2 had its genesis through the integration of all five of the genes recurrently disrupted in TS by novel breakpoints and is by all accounts an unbiased construction. In so doing, the model integrates two members of the NRXN superfamily (NRXN 1 and NRXN4) and two of the three known NRXN postsynaptic cell-adhesion ligand gene families, namely the NLGNs and LRRTMs. Further interrogation of other TS loci subsequently implicated another NRXN ligand gene CBLN2 located at the distal end of the gene desert most commonly rearranged in TS. As such, the synaptic model for TS (Figure 2) now includes the full complement of known NRXN postsynaptic cell-adhesion ligand gene families (NLGNs, CBLNs-GRIDs and LRRTMs) that regulate both the nature and strength of synaptic signalling, notwithstanding reports of NRXNs binding other molecules within the synapse including dystroglycan, GABAA-receptors and the secreted neurexophillins 1 and 3.89, 90, 91 In hindsight, this finding should not have been altogether unexpected, given the recurrent disruptions of NRXN1 in TS (Table 1).45 The resulting haploinsufficiency of NRXN1 would have the potential to impact cell-adhesion through the full range of competitive NRXN postsynaptic ligands including the NLGNs and CBLNs-GRIDs and more particularly the LRRTMs, as the latter have NRXN connections that appear to be restricted to NRXNs1 (Figure 2).67 This scenario is supported further by the comparable set of comorbidities associated with the disruptions of the NRXN1 gene and LRRTM3 gene in TS, including OCD, ADHD, speech delay, mental retardation and ASD (Table 1).
In the model (Figure 2) different affinities exist between the various ligands and the different NRXN isoforms which may provide important insight into the mechanisms at play in TS and ASD. For example, LRRTMs bind only NRXN1 isoforms that lack the 30 amino-acid insert at splice site 4 (NRXNs1IS4-).67 In contrast, CBLN1 and CBLN2 only bind NRXN isoforms that include this same insert at splice site 4 (NRXNsIS4+)79 compared with NLGNs, which appear to bind both of these isoform types but with different affinity (Figure 2).67 These common affinities in turn allow for competition for synaptogenesis, for example, competition exists between the NRXNs1IS4+-NLGN1 trans-synaptic complex and the tripartite NRXNs1IS4+-CBLN1-GRID2 complex79 and likely the NRXNs1IS4+-CBLN2-GRID1 complex (Figure 2). There is also clear potential for competition for synaptogenesis between the NRXNs1IS4--NLGN and NRXNs1IS4--LRRTM complexes (Figure 2).26, 65, 67, 75, 76 The outstanding question is to what degree do the patterns of expression of the NLGNs overlap with the other ligands. NLGNs are expressed throughout the brain but are differentially targeted to specific synapses:92 NLGN4X induces excitatory synapse formation and NLGN1 is specific for excitatory synapses; NLGN3 appears to be present in both inhibitory and excitatory synapses, whereas NLGN2 is restricted to inhibitory synapses.92 Together these findings suggest that loss of certain excitatory pathways may be pathogenic during development resulting in varying phenotypic presentations. Recent unexpected findings by Ko et al.92support this interpretation by their demonstration that NLGN1 and NLGN3 act redundantly with the LRRTMs in the maintenance of excitatory synapses.67, 79, 92 Excitatory synapses can apparently form in the absence of any and all of these ligands but are not maintained. However, any one of these ligands is sufficient to restore excitatory synapses to normal levels.92
From the findings in this review (Figure 2), it is clearly evident that many of the synaptic genes implicated in TS are also associated with ASD. Albeit, the novel association described here between CBLN2 and TS may provide valuable insight into the cellular and molecular features that distinguish TS from ASD. The CBLNs function as synaptic organisers through their binding of NRXNs and the GluD/GRID postsynaptic ligands, respectively.79, 93, 94 In contrast to the enrichment of CBLN1 and its receptor GRID2 within the cerebellum,95, 96, 97 CBLN2 expression patterns in the brain more closely overlap those of its receptor GRID1 including enrichment within the cortex.97, 98 This is of particular relevance given the common association of TS with ADHD and OCD and that the GRID1 knockout mouse presents with hyperactivity and aberrant emotional and social behaviours,98 which contrasts markedly with the ataxic presentation of the GRID2 knockout mouse.99 The behavioural phenotype of the CBLN2 knockout mouse100 is eagerly anticipated. At the molecular level GRID1's preferential role with CBLN2 in the induction of inhibitory presynaptic differentiation101 suggests that reduced inhibitory synaptogenesis may represent a distinguishing molecular feature of TS compared with ASD but this remains to be tested. Such a scenario, however, would be consistent with the fact that NLGN2, which is known to be restricted to inhibitory synapses, is the only NLGN that has not been linked with ASD.
Those regions of the brain where the expression of CBLN2 and GRID1 are enriched are likely to be of particular importance to the pathogenesis of TS. In contrast to the NLGNs, the other molecules within the synaptic model (Figure 2) display both mixed and restricted patterns of expression in different regions of the brain. For example, CBLN2 is widely expressed in the brain but it has a distinctive pattern of expression in cortical laminae II, III, V and VI.80, 97 GRID1 is also widely expressed in the brain and cortex, while CBLN1, CBLN3 and GRID2 are more selectively enriched within the cerebellum.79, 97 All LRRTMs are expressed within the dentate gyrus while their expression is distinct and complimentary within the different laminae of the cortex.56, 65, 67 These studies of the adult brain, however, are not informative of expression patterns within the developing brain. As indicated earlier, studies during embryogenesis in the chick indicate a dynamic pattern of expression for Lrrn1 during embryogenesis that is of fundamental importance in establishing regional boundaries within the brain. Likewise, CBLN2 has a particularly dynamic pattern of expression during development in the chick.97 In this context, the synaptic model for TS (Figure 2) presented herein overlaps with a range of neuropsychiatric disorders with and without TS and its various co-morbidities including ASD, ADHD, OCD and to a lesser extent schizophrenia. In this broader context, the synaptic model for TS (Figure 2) provides direction for both the genetic stratification of patients and the elucidation of new avenues for improved treatment.
TS model perspective on brain anatomy, neuronal circuitry and synaptic signalling
This study, the first to implicate LRRNs, LRRTMs and CBLN2 in TS (Figure 2), provides an invaluable window for improved understanding of the molecular basis of higher brain functions affected by changes to brain anatomy, neural circuitry and synaptic signalling in TS.
LRRN regulation of neuronal migration and brain pathology
The extracellular binding affinity of LRRN1 appears certain to restrict cellular migration between brain compartments as the priori basis for boundary formation in the hindbrain.73 The close familial relationship between LRRN1 and LRRN3 and the similarity between the extracellular LRR ligand-binding domains of the LRRNs and LRRTMs suggests that comparable mechanisms of affinity-regulated cellular migration during embryogenesis may be of broader pathological significance for TS and ASD. Abnormal increases in brain volume are commonly observed early in the postnatal life of individuals with ASD, including the frontal lobes and cerebellar vermis.102 As described earlier, the FGF8 signalling that is so central to boundary formation and regionalisation of the hindbrain and midbrain is mediated through the FGF8-dependent downregulation of LRRN1.73 FGF8 appears to have a similar role in the regionalisation and growth of the frontal cortex.102 Loss of hypomorphic mutations in Fgf8 in the mouse result in small and unpatterned telencephalons, particularly of the dorsomedial frontal cortex—the region that shows the largest increase in size in ASD.102 For ASD, loss of parvalbumin (PV) expressing interneurons has been reported as the hallmark of ASD-like dysfunctions. The physiological formation of synaptic connections between PV-positive interneurons and principal pyramidal neurons has been implicated in functional maturation of the postnatal cerebral cortex, and deficits in this process have been proposed as a pathogenic mechanism of ASD.103 In the case of TS, postmortem basal ganglia tissue from individuals with TS and normal controls has revealed markedly higher total neuron number in the globus pallidus pars interna (GPi) of TS with a lower neuron number and density in the globus pallidus pars externa and in the caudate.104 These investigators also observed an increased number and proportion of the GPi neurons positive for the calcium-binding protein PV in tissue from TS subjects, whereas lower densities of PV-positive interneurons were observed in both the caudate and putamen of TS subjects,48, 105 suggesting abnormal neuronal migration during development. In fact, small caudate volume in childhood, perhaps due to the reduction in the interneurons as described above, is one of the prognostic indicators of TS severity in adulthood.106 These anatomical changes are consistent with a developmental defect in the tangential migration of some GABAergic neurons. Different anatomical and functional deficits of the GABAergic system have also been discovered in ASD mouse models and CNTNAP2 knock-out mice, which present with hyperactivity and repetitive behaviours, display anomalies in neuronal migration and reduced number of interneurons, as well as abnormal neuronal network activity.107
LRRTMs compete for neural circuitry
The imbalance in striatal- and GPi-inhibitory neuron distribution described above suggests that the functional dynamics of cortico–striato–thalamic circuitry are fundamentally altered in severe, persistent TS. LRRTM1's role in left right brain asymmetry and handedness also belies a role for the LRRTMs in regulating important aspects of brain circuitry. The LRRTMs compete with the NLGNs for trans-synaptic NRXN binding as do the CBLNs (Figure 2). LRRTMs have similar ligand-binding sites to those of the LRRNs, indicating further potential for competition in brain circuitry formation Figure 2). As described earlier, the downregulation of NLGN1 can alter the balance between excitatory and inhibitory neurotransmission leading to changes in the synapse dynamics affecting synaptic production, organisation and patterning. In TS, it has been proposed that the involvement of the dopaminergic striatal pathways results in tics, whereas that of the serotonergic striatal–limbic minicircuits results in OCD, and the involvement of frontal cortical circuits results in ADHD and socially inappropriate behaviours. When the entire cortical striatal–pallidothalamic–cortical circuitry is involved this results in a number of co-morbidities and psychopathology in addition to tics. Thus, the clinical phenotype and the severity of symptoms, as well as the associated psychopathology, observed in TS may be influenced by the nature and extent of involvement of the above circuitry.1 Similarly, a dimensional model has been suggested for ASD, with autism on one end of the spectrum and language, social-cognitive and other developmental difficulties including mental retardation on the other end108, 109 mediated by the extent of circuitry involvement.
In the competitive and dynamic molecular environment of trans-synaptic cell-adhesion, it is not surprising to find that dose effects associated with disruptions, duplications and dysregulation of the genes/ligands described here can render profound pathogenic yet variable consequences for brain, mind and behaviour (Figure 2).56, 67, 79, 83 These events may be sufficient but not essential for the pathogenesis of TS. Phenotypic variability may be related to the redundancy described above. In addition, variable expression of the different genes is likely to impact on the penetrance of the different co-morbidities. For example, the sex-specific imprinting of NRXN4/CNTNAP2, CTNNA3 and LRRTM1 is known to have dramatic and variable effects on levels of gene expression and the parent-of-origin phenotypic inheritance patterns.74, 110 Such variations may also be caused by other modifying genes, perinatal events and other environmental factors. Thus, a particular phenotypic co-morbidity may present based on the type and level of involvement of the different neurotransmitter pathways that in turn may be based on the extent (which may be dose dependent) or the timing of events, as different circuits develop at different time points in neurodevelopment. For example, an early environmental insult could alter the epigenetic programming with consequent changes in neural function.111 Furthermore, as evident from animal models, phenotypic characterisation can show large modifying effects of genetic background and complex and unpredictable epistatic interactions. Zhang and Meaney,112 using rodent studies, suggested that environmental signals can activate intracellular pathways leading to epigenetic changes that can result in neural function changes during early development.
Phenotypic variability can also be affected by non-genetic factors, or ‘second hits' such as prematurity, perinatal trauma, injury, hypoxia, oxidative stress, infections, inflammations and autoimmunity, neural and psychosocial stressors or other modulators including gender.108 It has been shown that there are sex-specific differences in the topographic segregation and functionality of GABA-A systems in the substantia nigra and that the presence of circulating testosterone is essential for the development of the substantia nigra region in the neonatal period and to a lesser extent in the final maturation in the peripubertal period.113 In this regard, the role for testosterone in the extreme male brain hypothesis has been suggested in ASD.114 Similarly, OCD has been proposed as an alternative phenotypic expression of the TS genes with a gender-dependent difference in the expression leading to male members of the family exhibiting more tic behaviours and the female members exhibiting OCD.4, 115 An imbalance in the excitatory/inhibitory ratio in local and extended neuronal circuits could therefore have a role.
CBLN inhibitory synaptic signalling model
The present study is the first to identify a disease association for any of the CBLNs. Loss of CBLN2 is associated with reduced mediation of inhibitory synaptogenesis101 that appears in opposition with reduced number of excitatory synapses associated with the downregulation of the LRRTMs and NLGN4X (Figure 2),67, 75 albeit the downregulation/disruption of NRXN1 infers loss of both excitatory and inhibitory synaptic connections. Neural circuits utilise a number of homeostatic mechanisms to regulate the strength of excitation, inhibition and intrinsic excitability thereby maintaining synaptic homeostasis. In most networks, small changes in the balance between excitation and inhibition can have a significant impact on the neuronal firing and there is compelling evidence to suggest that the balance between excitation and inhibition is tightly regulated.116, 117 When the balance is upset, two distinct mechanisms have been proposed for restoring synaptic homeostasis: one mediated by the strength of excitatory and inhibitory synaptic inputs and the second by the balance of inward and outward voltage-dependent conductances. Thus, the neurons can compensate by using synaptic mechanisms to modify the balance between excitatory and inhibitory inputs or they can use intrinsic mechanisms to modify the balance of inward and outward voltage-dependent current.118 When there is an imbalance in the excitatory/inhibitory ratio in the neuronal circuits, this could in turn affect neuronal development.
NRXNs, NLGNs, LRRTMs and CBLN-GRIDs are neuronal adhesion molecules located at the pre- or postsynaptic region and promote synapse formation and/or maintenance bi-directionally in the glutamatergic and GABA-ergic nerve system that may result in subtle differences in neuronal connectivity and synapse patterning:75, 76, 78, 119 synapses being specialised intercellular junctions that connect the presynaptic machinery for neurotransmitter release to the postsynaptic machinery for receptor signalling. It has been shown that synapses are formed even when αNRXN l is deleted from the mouse genome; however, this compromises synaptic function.78 Release of neurotransmitters like glutamate requires the presynaptic co-assembly of Ca2+ channels with the secretory apparatus and this Ca2+ channel function is impaired in αNRXN knockout mice with consequent reductions in neurotransmitter release.78 NRXN-NLGN and NRXN-LRRTM connections, which are sensitive to extracellular Ca2+ concentrations, appear to trigger postsynaptic differentiation and control the balance of inhibitory GABA-ergic and excitatory glutamatergic inputs.79 By comparison, LRRTM1 null mice have altered distribution of the excitatory presynaptic vesicular glutamate transporter VGLUT1.75, 76 Glutamate, the main excitatory neurotransmitter in the vertebrate brain, has a major role in cortico–striatal–thalamo–cortical circuits and several lines of evidence support the role of glutamate in TS including: the TS association of glutamate receptors that are localised in the cellular membranes of both neurons and glia; the recognised extensive interaction between glutamate and dopamine systems; results of familial genetic studies (Table 3); and data from neurochemical analyses of postmortem brain samples. Loss of excitatory synaptic connections resulting in a hypo-glutamatergic state is consistent with a loss in the synaptic weight important for reinforcing circuit strength via repeated stimulation as required by language. However, due to the competition and redundancy for trans-synaptic cell-adhesion outlined in the TS model (Figure 2) and the involvement of both excitatory and inhibitory pathways in TS,101 there remains insufficient data to determine whether TS is definitively associated with a hyper- or hypo-glutamatergic state.
Research perspective
The identification of the ligand(s) for the LRRNs is eagerly anticipated as is the identification of any comparable restrictions to cellular migration regulated by the other cell-adhesion molecules of the neuropathogenetic model for TS (Figure 2). In this respect, the CBLN2 and GRID1 KO mouse models may prove invaluable for identifying those brain-affected regions that overlap within the NRXN1, NRXN3 and NRXN4/CNTNAP2 KO mouse models. The same mouse models may also be helpful in determining those brain regions and neuronal circuits most sensitive to the haploinsufficiency of heterozygotes as represented in the TS model. In this respect, patient screening should be expanded to include mutations that regulate levels of expression or loss of function of the relevant NRXNs, NLGNs, LRRTMs, LRRNs and CBLN2 including those mutations that affect the expression levels of harbouring genes including IMMP2L. Stratification of more patients through mutation screening should precede the pre-emption of any pharmacological strategies for the treatment of tics and related comorbidities.
Glossary
Motor and vocal tics: A simple motor tic is a sudden, brief, involuntary, repetitive, nonpurposeful movement of a single muscle group, such as an eye blink, face twitch, shoulder shrug, arm or leg jerk. Complex tics include forced touching, pulling clothes, a whole body jump or an abnormal walk. Vocal tics are involuntary sounds produced by moving air through the nose, mouth or throat, or vocalizations. These are also called phonic tics and examples include throat clearing, grunting and coughing. Tourette syndrome affects ∼1% of the school-aged population and ∼10% of these require lifelong therapy.
Echolalia: The automatic repetition of vocalizations made by another person.
Stereotypies: These are repetitive and ritualistic movements or posture, such as body rocking, swaying movements, or crossing and uncrossing of legs.
Obsessive–compulsive disorder (OCD): A disorder characterized by intrusive, persistent thoughts (obsessions) and/or repetitive, intentional behaviours (compulsions) that result in significant distress or dysfunction. It affects 1–3% of the general population.
Attention-deficit hyperactivity disorder (ADHD): A disorder characterized by inattention and/or hyperactivity and impulsivity affecting around 5% of school-aged children and causing impairment in social and academic performance; the symptoms may persist into adult life.
Autism spectrum disorder (ASD): A developmental disorder characterized by abnormalities in social interactions and communication, as well as restricted interests and repetitive behaviours.
Synapse: Synapse formation is the key step in the development of neural networks. Synapses are specialized intercellular junctions in which cell adhesion molecules connect the presynaptic machinery of neurons for neurotransmitter release to the postsynaptic machinery for receptor signalling.
Striatum: A subcortical structure of the brain, which is part of the basal ganglia system and is divided into the caudate nucleus and putamen by a white matter tract called the internal capsule.
Cortices: The outer layer of the cerebral cortex composed of gray matter.
Nested genes: A nested gene is any gene located wholly within another gene. Nested genes are usually located within an intron of the host gene.57, 58, 59, 60, 61 Nested genes are relatively common within the genome and are most often coded on the complementary strand and transcribed in an antisense direction relative to the host gene. Nested genes often display high levels of tissue-specific expression and overlapping genes more generally are four times more likely to be co-expressed than expected by random probability; however, little is known regarding the mechanism of co-regulation57 or whether co-regulation and transcriptional interference operate simultaneously, thereby constraining gene expression within the normal range. Two hypotheses have been proposed for interactive expression of nested gene pairs. The functional co-regulation hypothesis predicts a positive correlation between levels of expression of nested genes in different tissues (for example, BMCC1 and PCA3)120 and the transcriptional collision/interference hypothesis predicts a negative correlation as proposed in this study between LRRN3 and IMMP2L (Table 1). Transcriptional interference between the gene pairs has been investigated in bacteria and might take place by direct competition for the transcription apparatus and/or by formation of double-stranded RNAs.
The authors declare no conflict of interest.
Supplementary Material
References
- Eapen V, Crncec R. Tourette syndrome in children and adolescents: special considerations. J Psychosom Res. 2009;67:525–532. doi: 10.1016/j.jpsychores.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Robertson MM, Eapen V, Cavanna AE. The international prevalence, epidemiology, and clinical phenomenology of Tourette syndrome: a cross-cultural perspective. J Psychosom Res. 2009;67:475–483. doi: 10.1016/j.jpsychores.2009.07.010. [DOI] [PubMed] [Google Scholar]
- O'Rourke JA, Scharf JM, Yu D, Pauls DL. The genetics of Tourette syndrome: a review. J Psychosom Res. 2009;67:533–545. doi: 10.1016/j.jpsychores.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen V, Pauls DL, Robertson MM. Evidence for autosomal dominant transmission in Tourette's syndrome. United Kingdom cohort study. Br J Psychiatry. 1993;162:593–596. doi: 10.1192/bjp.162.5.593. [DOI] [PubMed] [Google Scholar]
- Burd L, Li Q, Kerbeshian J, Klug MG, Freeman RD. Tourette syndrome and comorbid pervasive developmental disorders. J Child Neurol. 2009;24:170–175. doi: 10.1177/0883073808322666. [DOI] [PubMed] [Google Scholar]
- Freeman RD, Fast DK, Burd L, Kerbeshian J, Robertson MM, Sandor P. An international perspective on Tourette syndrome: selected findings from 3,500 individuals in 22 countries. Dev Med Child Neurol. 2000;42:436–447. doi: 10.1017/s0012162200000839. [DOI] [PubMed] [Google Scholar]
- Kadesjo B, Gillberg C. Tourette's disorder: epidemiology and comorbidity in primary school children. J Am Acad Child Adolesc Psychiatry. 2000;39:548–555. doi: 10.1097/00004583-200005000-00007. [DOI] [PubMed] [Google Scholar]
- State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68:254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sverd J. Tourette syndrome and autistic disorder: a significant relationship. Am J Med Genet. 1991;39:173–179. doi: 10.1002/ajmg.1320390212. [DOI] [PubMed] [Google Scholar]
- State MW. The genetics of Tourette disorder. Curr Opin Genet Dev. 2011;21:302–309. doi: 10.1016/j.gde.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAICG Genome scan for Tourette disorder in affected-sibling-pair and multigenerational families. Am J Hum Genet. 2007;80:265–272. doi: 10.1086/511052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAICG A complete genome screen in sib pairs affected by Gilles de la Tourette syndrome. The Tourette Syndrome Association International Consortium for Genetics. Am J Hum Genet. 1999;65:1428–1436. doi: 10.1086/302613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breedveld GJ, Fabbrini G, Oostra BA, Berardelli A, Bonifati V. Tourette disorder spectrum maps to chromosome 14q31.1 in an Italian kindred. Neurogenetics. 2010;11:417–423. doi: 10.1007/s10048-010-0244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Leckman JF, Pauls DL, Tsai CP, Kidd KK, Campos MR. Genomewide scan of hoarding in sib pairs in which both sibs have Gilles de la Tourette syndrome. Am J Hum Genet. 2002;70:896–904. doi: 10.1086/339520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Brett P, Dearlove AM, McQuillin A, Kalsi G, Robertson MM, et al. Genome scan of Tourette syndrome in a single large pedigree shows some support for linkage to regions of chromosomes 5, 10 and 13. Psychiatr Genet. 2004;14:83–87. doi: 10.1097/01.ypg.0000107927.32051.f5. [DOI] [PubMed] [Google Scholar]
- Ercan-Sencicek AG, Stillman AA, Ghosh AK, Bilguvar K, O'Roak BJ, Mason CE, et al. L-histidine decarboxylase and Tourette's syndrome. N Engl J Med. 2010;362:1901–1908. doi: 10.1056/NEJMoa0907006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Coon H, Johnson M, Leppert MF, Camp NJ, McMahon WM. Linkage analysis of Tourette syndrome in a large Utah pedigree. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:656–662. doi: 10.1002/ajmg.b.31035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merette C, Brassard A, Potvin A, Bouvier H, Rousseau F, Emond C, et al. Significant linkage for Tourette syndrome in a large French Canadian family. Am J Hum Genet. 2000;67:1008–1013. doi: 10.1086/303093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschou P, Feng Y, Pakstis AJ, Speed WC, DeMille MM, Kidd JR, et al. Indications of linkage and association of Gilles de la Tourette syndrome in two independent family samples: 17q25 is a putative susceptibility region. Am J Hum Genet. 2004;75:545–560. doi: 10.1086/424389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonic I, Nyholt DR, Gericke GS, Gordon D, Matsumoto N, Ledbetter DH, et al. Further evidence for linkage of Gilles de la Tourette syndrome (GTS) susceptibility loci on chromosomes 2p11, 8q22 and 11q23-24 in South African Afrikaners. Am J Med Genet. 2001;105:163–167. doi: 10.1002/ajmg.1192. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Cath DC, van der Linde HC, Both J, Heutink P, Breedveld G, et al. Genetic and clinical analysis of a large Dutch Gilles de la Tourette family. Mol Psychiatry. 2006;11:954–964. doi: 10.1038/sj.mp.4001877. [DOI] [PubMed] [Google Scholar]
- Laurin N, Wigg KG, Feng Y, Sandor P, Barr CL. Chromosome 5 and Gilles de la Tourette syndrome: Linkage in a large pedigree and association study of six candidates in the region. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:95–103. doi: 10.1002/ajmg.b.30779. [DOI] [PubMed] [Google Scholar]
- Haas HL, Sergeeva OA, Selbach O. Histamine in the nervous system. Physiol Rev. 2008;88:1183–1241. doi: 10.1152/physrev.00043.2007. [DOI] [PubMed] [Google Scholar]
- Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Boghosian-Sell L, Comings DE, Overhauser J. Tourette syndrome in a pedigree with a 7;18 translocation: identification of a YAC spanning the translocation breakpoint at 18q22.3. Am J Hum Genet. 1996;59:999–1005. [PMC free article] [PubMed] [Google Scholar]
- Brett PM, Curtis D, Robertson MM, Dahlitz M, Gurling HM. Linkage analysis and exclusion of regions of chromosomes 3 and 8 in Gilles de la Tourette syndrome following the identification of a balanced reciprocal translocation 46 XY, t(3:8)(p21.3 q24.1) in a case of Tourette syndrome. Psychiatr Genet. 1996;6:99–105. doi: 10.1097/00041444-199623000-00001. [DOI] [PubMed] [Google Scholar]
- Clarke RA, Fang ZM, Diwan AD, Gilbert DL.Tourette syndrome and Klippel-Feil anomaly in a child with chromosome 22q11 duplication Case Reports Med 20092009361518, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford FC, Ait-Ghezala G, Morris M, Sutcliffe MJ, Hauser RA, Silver AA, et al. Translocation breakpoint in two unrelated Tourette syndrome cases, within a region previously linked to the disorder. Hum Genet. 2003;113:154–161. doi: 10.1007/s00439-003-0942-4. [DOI] [PubMed] [Google Scholar]
- Cuker A, State MW, King RA, Davis N, Ward DC. Candidate locus for Gilles de la Tourette syndrome/obsessive compulsive disorder/chronic tic disorder at 18q22. Am J Med Genet A. 2004;130A:37–39. doi: 10.1002/ajmg.a.30066. [DOI] [PubMed] [Google Scholar]
- Curtis D, Robertson MM, Gurling HM. Autosomal dominant gene transmission in a large kindred with Gilles de la Tourette syndrome. Br J Psychiatry. 1992;160:845–849. doi: 10.1192/bjp.160.6.845. [DOI] [PubMed] [Google Scholar]
- Dehning S, Riedel M, Muller N. Father-to-son transmission of 6;17 translocation in Tourette's syndrome. Am J Psychiatry. 2008;165:1051–1052. doi: 10.1176/appi.ajp.2008.07111828. [DOI] [PubMed] [Google Scholar]
- Devor EJ, Magee HJ. Multiple childhood behavioral disorders (Tourette syndrome, multiple tics, ADD and OCD) presenting in a family with a balanced chromosome translocation (t1;8)(q21.1;q22.1) Psychiatr Genet. 1999;9:149–151. doi: 10.1097/00041444-199909000-00007. [DOI] [PubMed] [Google Scholar]
- Donnai D. Gene location in Tourette syndrome. Lancet. 1987;1:627. doi: 10.1016/s0140-6736(87)90263-7. [DOI] [PubMed] [Google Scholar]
- Hanna PA, Janjua FN, Contant CF, Jankovic J. Bilineal transmission in Tourette syndrome. Neurology. 1999;53:813–818. doi: 10.1212/wnl.53.4.813. [DOI] [PubMed] [Google Scholar]
- Kroisel PM, Petek E, Emberger W, Windpassinger C, Wladika W, Wagner K. Candidate region for Gilles de la Tourette syndrome at 7q31. Am J Med Genet. 2001;101:259–261. doi: 10.1002/1096-8628(20010701)101:3<259::aid-ajmg1374>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Lawson-Yuen A, Saldivar JS, Sommer S, Picker J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur J Hum Genet. 2008;16:614–618. doi: 10.1038/sj.ejhg.5202006. [DOI] [PubMed] [Google Scholar]
- Matsumoto N, David DE, Johnson EW, Konecki D, Burmester JK, Ledbetter DH, et al. Breakpoint sequences of an 1;8 translocation in a family with Gilles de la Tourette syndrome. Eur J Hum Genet. 2000;8:875–883. doi: 10.1038/sj.ejhg.5200549. [DOI] [PubMed] [Google Scholar]
- Miranda DM, Wigg K, Kabia EM, Feng Y, Sandor P, Barr CL. Association of SLITRK1 to Gilles de la Tourette Syndrome. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:483–486. doi: 10.1002/ajmg.b.30840. [DOI] [PubMed] [Google Scholar]
- Patel C, Cooper-Charles L, McMullan DJ, Walker JM, Davison V, Morton J. Translocation breakpoint at 7q31 associated with tics: further evidence for IMMP2L as a candidate gene for Tourette syndrome. Eur J Hum Genet. 2011;19:634–639. doi: 10.1038/ejhg.2010.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petek E, Windpassinger C, Vincent JB, Cheung J, Boright AP, Scherer SW, et al. Disruption of a novel gene (IMMP2L) by a breakpoint in 7q31 associated with Tourette syndrome. Am J Hum Genet. 2001;68:848–858. doi: 10.1086/319523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson MM, Shelley BP, Dalwai S, Brewer C, Critchley HD. A patient with both Gilles de la Tourette's syndrome and chromosome 22q11 deletion syndrome: clue to the genetics of Gilles de la Tourette's syndrome. J Psychosom Res. 2006;61:365–368. doi: 10.1016/j.jpsychores.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Shelley BP, Robertson MM, Turk J. An individual with Gilles de la Tourette syndrome and Smith-Magenis microdeletion syndrome: is chromosome 17p11.2 a candidate region for Tourette syndrome putative susceptibility genes. J Intellect Disabil Res. 2007;51 (Part 8:620–624. doi: 10.1111/j.1365-2788.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- Singh DN, Howe GL, Jordan HW, Hara S. Tourette's syndrome in a black woman with associated triple X and 9p mosaicism. J Natl Med Assoc. 1982;74:675–682. [PMC free article] [PubMed] [Google Scholar]
- State MW, Greally JM, Cuker A, Bowers PN, Henegariu O, Morgan TM, et al. Epigenetic abnormalities associated with a chromosome 18(q21-q22) inversion and a Gilles de la Tourette syndrome phenotype. Proc Natl Acad Sci USA. 2003;100:4684–4689. doi: 10.1073/pnas.0730775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram SK, Huq AM, Wilson BJ, Chugani HT. Tourette syndrome is associated with recurrent exonic copy number variants. Neurology. 2010;74:1583–1590. doi: 10.1212/WNL.0b013e3181e0f147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Mathews CA, Joosse M, Eussen BH, Heutink P, Oostra BA. CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82:1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]
- Taylor LD, Krizman DB, Jankovic J, Hayani A, Steuber PC, Greenberg F, et al. 9p monosomy in a patient with Gilles de la Tourette's syndrome. Neurology. 1991;41:1513–1515. doi: 10.1212/wnl.41.9.1513. [DOI] [PubMed] [Google Scholar]
- Bloch M, State M, Pittenger C. Recent advances in Tourette syndrome. Curr Opin Neurol. 2011;24:119–125. doi: 10.1097/WCO.0b013e328344648c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, Fang ZM, Hilton EN, McGaughran J, Zhao Z, de Bock CE, et al. Mutations in GDF6 are associated with vertebral segmentation defects in Klippel-Feil syndrome. Hum Mutat. 2008;29:1017–1027. doi: 10.1002/humu.20741. [DOI] [PubMed] [Google Scholar]
- Keen-Kim D, Mathews CA, Reus VI, Lowe TL, Herrera LD, Budman CL, et al. Overrepresentation of rare variants in a specific ethnic group may confuse interpretation of association analyses. Hum Mol Genet. 2006;15:3324–3328. doi: 10.1093/hmg/ddl408. [DOI] [PubMed] [Google Scholar]
- George SK, Jiao Y, Bishop CE, Lu B. Mitochondrial peptidase IMMP2L mutation causes early onset of age-associated disorders and impairs adult stem cell self-renewal. Aging Cell. 2011;10:584–594. doi: 10.1111/j.1474-9726.2011.00686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petek E, Schwarzbraun T, Noor A, Patel M, Nakabayashi K, Choufani S, et al. Molecular and genomic studies of IMMP2L and mutation screening in autism and Tourette syndrome. Mol Genet Genomics. 2007;277:71–81. doi: 10.1007/s00438-006-0173-1. [DOI] [PubMed] [Google Scholar]
- Belloso JM, Bache I, Guitart M, Caballin MR, Halgren C, Kirchhoff M, et al. Disruption of the CNTNAP2 gene in a t(7;15) translocation family without symptoms of Gilles de la Tourette syndrome. Eur J Hum Genet. 2007;15:711–713. doi: 10.1038/sj.ejhg.5201824. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, et al. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet. 2011;130:563–573. doi: 10.1007/s00439-011-0975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa I, Clark TG, Holt R, Pagnamenta AT, Mulder EJ, Minderaa RB, et al. Polymorphisms in leucine-rich repeat genes are associated with autism spectrum disorder susceptibility in populations of European ancestry. Mol Autism. 2010;1:7. doi: 10.1186/2040-2392-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solda G, Suyama M, Pelucchi P, Boi S, Guffanti A, Rizzi E, et al. Non-random retention of protein-coding overlapping genes in Metazoa. BMC Genomics. 2008;9:174. doi: 10.1186/1471-2164-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Ma D, Xu M. Nested genes in the human genome. Genomics. 2005;86:414–422. doi: 10.1016/j.ygeno.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Assis R, Kondrashov AS, Koonin EV, Kondrashov FA. Nested genes and increasing organizational complexity of metazoan genomes. Trends Genet. 2008;24:475–478. doi: 10.1016/j.tig.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib AA, Gulcher JR, Hognason T, Zheng L, Stefansson K. The OMgp gene, a second growth suppressor within the NF1 gene. Oncogene. 1998;16:1525–1531. doi: 10.1038/sj.onc.1201683. [DOI] [PubMed] [Google Scholar]
- Crampton N, Bonass WA, Kirkham J, Rivetti C, Thomson NH. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 2006;34:5416–5425. doi: 10.1093/nar/gkl668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestrini E, Pagnamenta AT, Lamb JA, Bacchelli E, Sykes NH, Sousa I, et al. High-density SNP association study and copy number variation analysis of the AUTS1 and AUTS5 loci implicate the IMMP2L-DOCK4 gene region in autism susceptibility. Mol Psychiatry. 2010;15:954–968. doi: 10.1038/mp.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls DL, Leckman JF. The inheritance of Gilles de la Tourette's syndrome and associated behaviors. Evidence for autosomal dominant transmission. N Engl J Med. 1986;315:993–997. doi: 10.1056/NEJM198610163151604. [DOI] [PubMed] [Google Scholar]
- Chen Y, Aulia S, Li L, Tang BL. AMIGO and friends: an emerging family of brain-enriched, neuronal growth modulating, type I transmembrane proteins with leucine-rich repeats (LRR) and cell adhesion molecule motifs. Brain Res Rev. 2006;51:265–274. doi: 10.1016/j.brainresrev.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Lauren J, Airaksinen MS, Saarma M, Timmusk T. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81:411–421. doi: 10.1016/s0888-7543(03)00030-2. [DOI] [PubMed] [Google Scholar]
- Proenca CC, Gao KP, Shmelkov SV, Rafii S, Lee FS. Slitrks as emerging candidate genes involved in neuropsychiatric disorders. Trends Neurosci. 2011;34:143–153. doi: 10.1016/j.tins.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GJ, Washbourne P. Neurexins, neuroligins and LRRTMs: synaptic adhesion getting fishy. J Neurochem. 2011;117:765–778. doi: 10.1111/j.1471-4159.2010.07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui TJ, Craig AM. Synaptic organizing complexes. Curr Opin Neurobiol. 2011;21:132–143. doi: 10.1016/j.conb.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CS, Wu-Chou YH, van Doeselaar M, Simons EJ, Chang HC, Breedveld GJ, et al. The LRRK2 Arg1628Pro variant is a risk factor for Parkinson's disease in the Chinese population. Neurogenetics. 2008;9:271–276. doi: 10.1007/s10048-008-0140-6. [DOI] [PubMed] [Google Scholar]
- Pagnamenta AT, Bacchelli E, de Jonge MV, Mirza G, Scerri TS, Minopoli F, et al. Characterization of a family with rare deletions in CNTNAP5 and DOCK4 suggests novel risk loci for autism and dyslexia. Biol Psychiatry. 2010;68:320–328. doi: 10.1016/j.biopsych.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LK, Meyer KJ, Rudd DS, Librant AL, Epping EA, Sheffield VC, et al. Novel copy number variants in children with autism and additional developmental anomalies. J Neurodev Disord. 2009;1:292–301. doi: 10.1007/s11689-009-9013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Poirier C, Gaspar T, Gratzke C, Harrison W, Busija D, et al. A mutation in the inner mitochondrial membrane peptidase 2-like gene (Immp2 l) affects mitochondrial function and impairs fertility in mice. Biol Reprod. 2008;78:601–610. doi: 10.1095/biolreprod.107.065987. [DOI] [PubMed] [Google Scholar]
- Tossell K, Andreae LC, Cudmore C, Lang E, Muthukrishnan U, Lumsden A, et al. Lrrn1 is required for formation of the midbrain-hindbrain boundary and organiser through regulation of affinity differences between midbrain and hindbrain cells in chick. Dev Biol. 2011;352:341–352. doi: 10.1016/j.ydbio.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francks C, Maegawa S, Lauren J, Abrahams BS, Velayos-Baeza A, Medland SE, et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia Mol Psychiatry 2007121129–1139.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Fuccillo MV, Malenka RC, Sudhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhoff MW, Lauren J, Cassidy RM, Dobie FA, Takahashi H, Nygaard HB, et al. An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron. 2009;61:734–749. doi: 10.1016/j.neuron.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kask M, Pruunsild P, Timmusk T. Bidirectional transcription from human LRRTM2/CTNNA1 and LRRTM1/CTNNA2 gene loci leads to expression of N-terminally truncated CTNNA1 and CTNNA2 isoforms. Biochem Biophys Res Commun. 2011;411:56–61. doi: 10.1016/j.bbrc.2011.06.085. [DOI] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, et al. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Yuzaki M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci. 2011;33:1447–1461. doi: 10.1111/j.1460-9568.2011.07638.x. [DOI] [PubMed] [Google Scholar]
- Joo JY, Lee SJ, Uemura T, Yoshida T, Yasumura M, Watanabe M, et al. Differential interactions of cerebellin precursor protein (Cbln) subtypes and neurexin variants for synapse formation of cortical neurons. Biochem Biophys Res Commun. 2011;406:627–632. doi: 10.1016/j.bbrc.2011.02.108. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezato A, Kimura-Sato J, Yamamoto N, Iijima Y, Kunugi H, Nishikawa T. Further evidence for a male-selective genetic association of synapse-associated protein 97 (SAP97) gene with schizophrenia. Behav Brain Funct. 2012;8:2. doi: 10.1186/1744-9081-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottos A, Rissone A, Bussolino F, Arese M. Neurexins and neuroligins: synapses look out of the nervous system. Cell Mol Life Sci. 2011;68:2655–2666. doi: 10.1007/s00018-011-0664-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaags AK, Lionel AC, Sato D, Goodenberger M, Stein QP, Curran S, et al. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet. 2012;90:133–141. doi: 10.1016/j.ajhg.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabrucker AM, Knight MJ, Proepper C, Bockmann J, Joubert M, Rowan M, et al. Concerted action of zinc and ProSAP/Shank in synaptogenesis and synapse maturation. EMBO J. 2011;30:569–581. doi: 10.1038/emboj.2010.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Sia GM, Garner CC, Huganir RL. Synapse-associated protein-97 isoform-specific regulation of surface AMPA receptors and synaptic function in cultured neurons. J Neurosci. 2003;23:4567–4576. doi: 10.1523/JNEUROSCI.23-11-04567.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondin M, Labrousse V, Hosy E, Heine M, Tessier B, Levet F, et al. Neurexin-neuroligin adhesions capture surface-diffusing AMPA receptors through PSD-95 scaffolds. J Neurosci. 2011;31:13500–13515. doi: 10.1523/JNEUROSCI.6439-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MA, Elias GM, Elias LA, Swat W, Nicoll RA. The role of SAP97 in synaptic glutamate receptor dynamics. Proc Natl Acad Sci USA. 2010;107:3805–3810. doi: 10.1073/pnas.0914422107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Hammer RE, Sudhof TC. Neurexophilin binding to alpha-neurexins. A single LNS domain functions as an independently folding ligand-binding unit. J Biol Chem. 1998;273:34716–34723. doi: 10.1074/jbc.273.52.34716. [DOI] [PubMed] [Google Scholar]
- Sugita S, Saito F, Tang J, Satz J, Campbell K, Sudhof TC. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 2001;154:435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Atasoy D, Arac D, Yang X, Fucillo MV, Robison AJ, et al. Neurexins physically and functionally interact with GABA(A) receptors. Neuron. 2010;66:403–416. doi: 10.1016/j.neuron.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Soler-Llavina GJ, Fuccillo MV, Malenka RC, Sudhof TC. Neuroligins/LRRTMs prevent activity- and Ca2+/calmodulin-dependent synapse elimination in cultured neurons. J Cell Biol. 2011;194:323–334. doi: 10.1083/jcb.201101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Uemura T, Yoshida T, Mishina M. GluRdelta2 assembles four neurexins into trans-synaptic triad to trigger synapse formation. J Neurosci. 2012;32:4688–4701. doi: 10.1523/JNEUROSCI.5584-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzaki M. The delta2 glutamate receptor: a key molecule controlling synaptic plasticity and structure in Purkinje cells. Cerebellum. 2004;3:89–93. doi: 10.1080/14734220410028921. [DOI] [PubMed] [Google Scholar]
- Kano M, Hashimoto K, Kurihara H, Watanabe M, Inoue Y, Aiba A, et al. Persistent multiple climbing fiber innervation of cerebellar Purkinje cells in mice lacking mGluR1. Neuron. 1997;18:71–79. doi: 10.1016/s0896-6273(01)80047-7. [DOI] [PubMed] [Google Scholar]
- Miura E, Iijima T, Yuzaki M, Watanabe M. Distinct expression of Cbln family mRNAs in developing and adult mouse brains. Eur J Neurosci. 2006;24:750–760. doi: 10.1111/j.1460-9568.2006.04950.x. [DOI] [PubMed] [Google Scholar]
- Yadav R, Gupta SC, Hillman BG, Bhatt JM, Stairs DJ, Dravid SM. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS One. 2012;7:e32969. doi: 10.1371/journal.pone.0032969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Pang Z, Bao D, Miyazaki T, Li L, Miura E, et al. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat Neurosci. 2005;8:1534–1541. doi: 10.1038/nn1576. [DOI] [PubMed] [Google Scholar]
- Rong Y, Wei P, Parris J, Guo H, Pattarini R, Correia K, et al. Comparison of Cbln1 and Cbln2 functions using transgenic and knockout mice. J Neurochem. 2012;120:528–540. doi: 10.1111/j.1471-4159.2011.07604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura M, Yoshida T, Lee SJ, Uemura T, Joo JY, Mishina M. Glutamate receptor delta1 induces preferentially inhibitory presynaptic differentiation of cortical neurons by interacting with neurexins through cerebellin precursor protein subtypes. J Neurochem. 2012;121:705–716. doi: 10.1111/j.1471-4159.2011.07631.x. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL. Three hypotheses for developmental defects that may underlie some forms of autism spectrum disorder. Curr Opin Neurol. 2010;23:118–123. doi: 10.1097/WCO.0b013e328336eb13. [DOI] [PubMed] [Google Scholar]
- Di Cristo G. Development of cortical GABAergic circuits and its implications for neurodevelopmental disorders. Clin Genet. 2007;72:1–8. doi: 10.1111/j.1399-0004.2007.00822.x. [DOI] [PubMed] [Google Scholar]
- Kalanithi PS, Zheng W, Kataoka Y, DiFiglia M, Grantz H, Saper CB, et al. Altered parvalbumin-positive neuron distribution in basal ganglia of individuals with Tourette syndrome. Proc Natl Acad Sci USA. 2005;102:13307–13312. doi: 10.1073/pnas.0502624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka Y, Kalanithi PS, Grantz H, Schwartz ML, Saper C, Leckman JF, et al. Decreased number of parvalbumin and cholinergic interneurons in the striatum of individuals with Tourette syndrome. J Comp Neurol. 2010;518:277–291. doi: 10.1002/cne.22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch MH, Leckman JF, Zhu H, Peterson BS. Caudate volumes in childhood predict symptom severity in adults with Tourette syndrome. Neurology. 2005;65:1253–1258. doi: 10.1212/01.wnl.0000180957.98702.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen V. Genetic basis of autism: is there a way forward. Curr Opin Psychiatry. 2011;24:226–236. doi: 10.1097/YCO.0b013e328345927e. [DOI] [PubMed] [Google Scholar]
- Perche O, Laumonnier F, Baala L, Ardourel MY, Menuet A, Robin V, et al. [Autism, genetics and synaptic function alterations] Pathol Biol (Paris) 2010;58:381–386. doi: 10.1016/j.patbio.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Oudejans CB, Mulders J, Lachmeijer AM, van Dijk M, Konst AA, Westerman BA, et al. The parent-of-origin effect of 10q22 in pre-eclamptic females coincides with two regions clustered for genes with down-regulated expression in androgenetic placentas. Mol Hum Reprod. 2004;10:589–598. doi: 10.1093/molehr/gah080. [DOI] [PubMed] [Google Scholar]
- Herbert MR. Contributions of the environment and environmentally vulnerable physiology to autism spectrum disorders. Curr Opin Neurol. 2010;23:103–110. doi: 10.1097/WCO.0b013e328336a01f. [DOI] [PubMed] [Google Scholar]
- Zhang TY, Meaney MJ.Epigenetics and the environmental regulation of the genome and its function Annu Rev Psychol 201061439–466.C1-3. [DOI] [PubMed] [Google Scholar]
- Veliskova J, Moshe SL. Sexual dimorphism and developmental regulation of substantia nigra function. Ann Neurol. 2001;50:596–601. doi: 10.1002/ana.1248. [DOI] [PubMed] [Google Scholar]
- Baron-Cohen S.The extreme-male-brain theory of autism.In: Tager-Flusberg H (ed).Neurodevelopmental Disorders, 1999MIT Press: Cambridge, MA [Google Scholar]
- Eapen V, Robertson MM, Alsobrook JP, II, Pauls DL. Obsessive compulsive symptoms in Gilles de la Tourette syndrome and obsessive compulsive disorder: differences by diagnosis and family history. Am J Med Genet. 1997;74:432–438. doi: 10.1002/(sici)1096-8628(19970725)74:4<432::aid-ajmg15>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Atallah BV, Scanziani M. Instantaneous modulation of gamma oscillation frequency by balancing excitation with inhibition. Neuron. 2009;62:566–577. doi: 10.1016/j.neuron.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille F, Marin-Burgin A, Adesnik H, Atallah BV, Scanziani M. Input normalization by global feedforward inhibition expands cortical dynamic range. Nat Neurosci. 2009;12:1577–1585. doi: 10.1038/nn.2441. [DOI] [PubMed] [Google Scholar]
- Turrigiano G. Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harb Perspect Biol. 2012;4:a005736. doi: 10.1101/cshperspect.a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Clarke RA, Zhao Z, Guo AY, Roper K, Teng L, Fang ZM, et al. New genomic structure for prostate cancer specific gene PCA3 within BMCC1: implications for prostate cancer detection and progression. PLoS One. 2009;4:e4995. doi: 10.1371/journal.pone.0004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.