Abstract
Increasing evidence suggests that epigenetic dysfunction may account for the alteration of gene transcription present in neuropsychiatric disorders such as schizophrenia (SZ), bipolar disorder (BP) and autism. Here, we studied the expression of the ten-eleven translocation (TET) gene family and activation-induced deaminase/apolipoprotein B mRNA-editing enzymes (AID/APOBEC) in the inferior parietal lobule (IPL) (BA39-40) and the cerebellum of psychotic (PSY) patients, depressed (DEP) patients and nonpsychiatric (CTR) subjects obtained from the Stanley Foundation Neuropathology Consortium Medical Research Institute. These two sets of enzymes have a critical role in the active DNA demethylation pathway. The results show that TET1, but not TET2 and TET3, mRNA and protein expression was increased (two- to threefold) in the IPL of the PSY patients compared with the CTR subjects. TET1 mRNA showed no change in the cerebellum. Consistent with the increase of TET1, the level of 5-hydroxymethylcytosine (5hmC) was elevated in the IPL of PSY patients but not in the other groups. Moreover, higher 5hmC levels were detected at the glutamic acid decarboxylase67 (GAD67) promoter only in the PSY group. This increase was inversely related to the decrease of GAD67 mRNA expression. Of 11 DNA deaminases measured, APOBEC3A mRNA was significantly decreased in the PSY and DEP patients, while APOBEC3C was decreased only in PSY patients. The other APOBEC mRNA studied failed to change. Increased TET1 and decreased APOBEC3A and APOBEC3C found in this study highlight the possible role of altered DNA demethylation mechanisms in the pathophysiology of psychosis.
Keywords: post-mortem brain, TET, AID/APOBECs, 5hmC, epigenetic
Introduction
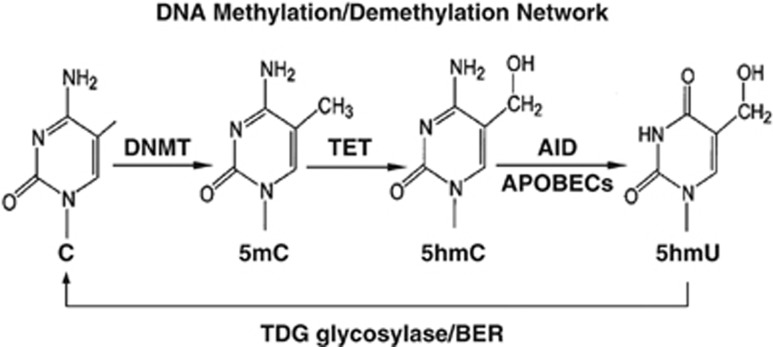
Epigenetic modifications of chromatin, including methylation of genomic DNA, and acetylation or methylation of histones, have a critical role in orchestrating the expression of specific genes in postmitotic neurons. Among these epigenetic modifications, DNA (cytosine) methylation at specific gene promoter CpG-rich regions is an example of a stable epigenetic modification that ensures and maintains neuronal phenotype identity.1 Increasing evidence suggests that epigenetic dysfunction of the genome, in particular alteration of histone and DNA methylation marks, may account for the misregulation of gene expression present in neuropsychiatric disorders, such as bipolar disorder (BP), schizophrenia (SZ) and autism.2, 3, 4, 5, 6, 7, 8, 9 Patients with psychosis, including BP and SZ patients, express an increase of DNA methyltransferase (DNMT)10, 11, 12, 13 and a wide variety of DNA methylation alterations.6 We recently reported that the reduction of brain-derived neurotrophic factor (BDNF) expression in the parietal cortex of SZ patients was associated with an increase of 5-methyl cytosine (5mC) at a BDNF promoter.4 Furthermore, reelin, a trophic factor expressed selectively in telencephalic GABAergic neurons, is downregulated in SZ patients and this dowregulation is associated with increased methylation at specific CpG promoter sites.2, 3, 14 However, inconsistent results with studies of reelin promoter methylation in relation to mental illness6, 15, 16, 17suggest that the DNA methylation status of specific promoter regions in postmitotic neurons is not stable, but in contrast is a rapidly reversible process maintained by the equilibrium between the DNA-methylating action of DNA methyltransferases and the demethylating action of an active DNA demethylation process18, 19 (Figure 1). This theory has been supported by several independent and particularly interesting findings showing that 5mC mark on promoter CpG-rich regions of specific genes can be oxidized to form 5-hydroxymethylcytosine (5hmC) by ten-eleven translocation (TET) proteins in mammalian brain.20, 21, 22, 23, 24 Recent evidence suggests that TET proteins, important contributors in DNA demethylation process, have essential roles in neurodevelopment and aging25 and are involved in human cancers.26
Figure 1.
Schematic representation of putative DNA methylation/demethylation pathways. AID/APOBEC, activation-induced deaminase/apolipoprotein B RNA-editing catalytic component; BER, base excision repair; C, cytosine; DNMT, DNA methyltransferase; 5hmC, 5-hydroxylmethylcytosine; 5hmU, 5-hydroxylmethyluracil; 5mC, 5-methylcytosine; TDG, thymine DNA glycosylase; TET, ten-eleven translocation protein.
Further, it has been proposed that 5hmC undergoes two successive processing steps: (a) a deamination step that is catalyzed by the AID (activation-induced deaminase)/APOBEC (apoliporotein B mRNA-editing enzyme complex) family of cytosine deaminases, turning 5hmC into 5-hydroxylmethyluracil (5hmU); and (b) the base excision repair pathway, in which 5hmU can be removed and replaced by unmethylated cytosine by a group of glycosylases such as MBD4 and TDG.4, 27, 28, 29 Rapidly inducible growth arrest and DNA damage-inducible proteins 45 (GADD45)30, 31, 32 are thought to be master coordinators of this process by recruiting deaminases and glycosylases to 5mC and 5hmC-rich promoter regions4 (Figure 1).
Although numerous studies of DNA methylation have been carried out in the post-mortem brain of psychotic (PSY) patients,2, 3, 4, 6 the different components of the DNA demethylation pathway, with the exceptions of GADD45 and MBD4 or TDG,4 remain largely unknown.
In this paper, we initiated an investigation on the expression of the TET gene family and AID/APOBEC deaminases in the inferior parietal lobule (IPL) and the cerebellum of a cohort of PSY patients, which includes BP patients with psychosis, and SZ patients, major depression (DEP) patients and nonpsychiatric (CTR) subjects obtained from the Stanley Foundation Neuropathology Consortium Medical Research Institute (Bethesda, MD, USA).
Materials and methods
Subjects
IPL (BA39-40) and cerebellar cortex were obtained from the Stanley Foundation Neuropathology Consortium. We selected IPL because this brain area is among the most seriously damaged cortical regions in SZ patients.33 Imaging studies have reported decreased volume and differences in hemispheric asymmetry in patients compared with controls.34 In addition, many of the IPL functions, such as sensory integration, body image, concept of self, visuospatial abilities and executive functions, have been found to be disturbed in SZ patients.33 Also, GADD45 proteins have been shown to be upregulated in this region of the cortex in PSY patients.4
The cerebellum was included in this study because this brain area contains high levels of TET.35 Cerebellar cortices were cut sagittally, lateral to the vermis and did not include the dentate nucleus.
The demographics associated with each group of subjects are listed in Supplementary Table 1. The cohort of ‘PSY patients' includes nine BP disorder patients with psychosis and 10 patients with SZ. The cohort of ‘depressed (DEP) patients' includes 12 subjects with major depression. Normal control (CTR) subjects include 11 non-psychiatric subjects who had no history of psychiatric disorders. Among CTR and the patient cohorts there are no significant differences in terms of the mean age (F(2, 41)=0.657, P=0.524) or the gender distribution (χ2=3.273, P=0.07). An one-way analysis of variance (ANOVA) did not reveal significant differences regarding brain pH (F(2,40)=1.572, P=0.220), post-mortem interval (F(2, 41)=1.604, P=0.213) or RNA integrity (F(3,38)=2.526, P=0.072) among the CTR subjects and the patient cohorts. Details regarding clinical characteristics and tissue handling for the post-mortem samples were described previously.36 To confirm the results obtained in the IPL of Stanley Foundation Neuropathology Consortium in a different cortical area (BA9) and a different cohort (Harvard Brain Tissue Resource Center, Belmont, MA, USA), we measured TET1 mRNA in the samples of 7 CTRs and 12 PSY patients (6 SZ and 6 BP). The samples were matched for age (CTR: 51.0±10.5; BP: 47.3±16.6; SZ: 54.3±7.5 years), gender (CTR: 4M/3F; BP: 3M/3F; SZ: 3M/3F), post-mortem interval (CTR: 20.2±3.6; BP: 21.5±3.8; SZ: 16.9±3.2 h) and pH (CTR: 6.6±0.2; BP: 6.3±0.3; SZ: 6.8±0.4).
Quantitative real-time polymerase chain reaction
Quantitative real-time polymerase chain reaction was carried out using the Applied Biosystems Real-Time PCR System with a SYBR green master mix (Fermentas, Glen Burnie, MD, USA). Total RNA, isolated using TRIZOL reagent (Life Technologies, Grand Island, NY, USA), was further purified using the Qiagen RNeasy kit (Qiagen, Valencia, CA, USA). The RNA integrity number was measured with an Agilent 2100 bioananlyzer (Agilent Technologies, Santa Clara, CA, USA). The primer sequences for the genes analyzed are summarized in Supplementary Table 2. Each sample was run in duplicate and repeated twice. For normalizing mRNA expression, several housekeeping genes (NSE, GAPDH and β-actin) were chosen as the internal control. For each housekeeping gene, we measured the gene stability ranking using the NormFinder algorithm.37 This procedure allows for the identification of the housekeeping gene best suited for normalization. Because each of the genes studied yielded similar results when normalized to either β-actin, GAPDH or NSE, and because β-actin has the highest housekeeping gene stability (NormFinder), we normalized our data to β-actin.
Detection and quantification of 5mC and 5hmC by immuno-dot-blot analysis
To measure 5mC and 5hmC expressed on genomic DNA, total DNA was isolated using a Puregene Core Kit A (Qiagen). The amount of DNA to be used in the dot-blot assay was determined by titration of the sample with various concentrations of standard DNA. In all, 25 ng of DNA, which is within linear range, were chosen for each sample. DNA extracted from the samples and 12.5, 25, 50 and 100 pg of 5mC or 5hmC standard DNA (EF-011-001 and EF-010-002; Diagenode, Denville, NJ, USA) were denatured by adding denaturing buffer (0.5 ℳ NaOH and 1.5 ℳ NaCl) and incubating at 65 °C for 45 min. After neutralization (0.5 ℳ Tris-HCl, 1.5 ℳ NaCl), the solution was loaded in duplicate into a well placed on a positively charged nylon membrane (Hybond; Amersham, Pittsburgh, PA, USA). The wells were then washed once with 2 × saturated sodium citrate. The membrane was removed and briefly washed with 2 × saline-sodium citrate buffer and dried. The crosslinking of the DNA with the membrane was carried out with a UV Stratalinker (Stratagene, Santa Clara, CA, USA) on the Auto Cross Link setting. To measure DNA recovery, the membrane was stained with SYBR gold dissolved in Tris-borate-EDTA buffer for 10 min. The membrane was scanned on a STORM 860 (Amersham). The fluorescence signal of DNA on each spot was measured and calculated based on the standard DNA of the same membrane. The membrane was washed with Tris-buffered saline buffer containing 5% milk for 1 h and incubated with a primary antibody of either 5mC or 5hmC (1:10 000; Active Motif, Carlsbad, CA, USA) for 2 h, and then incubated with the proper secondary antibody for 1 h. The immunoreactive dots were detected by the application of the ECL Plus chemiluminescence kit (Amersham Pharmacia, Pittsburgh, PA, USA) on a STORM 860 fluorescent imager (Molecular Dynamics, Sunnyvale, CA, USA) and quantified with IMAGEQUANT software. The linear regression standard curves of 5mC and 5hmC with r2=0.996 are shown in Supplementary Figure 1.
Methylated DNA immunoprecipitation
Methylated DNA immunoprecipitation experiments were conducted using a previously published protocol.4 5mC monoclonal mouse (Diagenode) and 5hmC (Active Motif) antibodies were used in methylated DNA immunoprecipitation. We decided to use the procedure because bisulfite and most enzyme-dependent methods are incapable of distinguishing 5mC from the approximately 14% of methylcytosines in the brain that are 5hmC.8 Recent studies demonstrate that these 5mC and 5hmC may have very different functions and genomic locations.38, 39 The percentage of methylated vs unmethylated promoter was calculated using the following equation: % (meDNA−IP/total input)=2^[(Ct(10% input)−3.32)−Ct(meDNA−IP)] × 100% (adopted from MagMeDIP kit instruction manual; Diagenode).
Western blot analysis
The samples were homogenized directly in Laemmli buffer (100 μl/10 mg of tissue). The protein extract was separated by 4–12% Tris-glycine gel (Invitrogen, Grand Island, NY, USA) and transferred to nitrocellulose membranes (Invitrogen). The membrane was blocked for 1 h at room temperature with 5% non-fat milk in phosphate-buffered saline (10 mℳ phosphate-buffered saline, pH 7.4) and then reacted with 1:2000 anti-TET1 polyclonal antibody (GeneTex, Irvine, CA, USA). The immunoreactive bands were detected by using anti-rabbit IgG (Amersham) at 1:1000 for 1 h followed by the application of the ECL Plus chemiluminescence western blotting kit (Amersham). The immunoreactive bands were detected by the application of the ECL Plus chemiluminescence kit (Amersham) on a STORM 860 fluorescent imager (Molecular Dynamics) and analyzed with IMAGEQUANT software.
Statistical analysis
One-way ANOVA and analysis of covariance with Bonferroni post-hoc comparisons were performed using the mRNA content of the genes measured, 5mC or 5hmC levels, TET1 protein content, RNA integrity number, age, gender, post-mortem interval, substance use and alcohol abuse, to evaluate differences among the three groups (patients with psychosis (PSY), depression (DEP) and normal controls (CTR)), using PASW v.18 software (SPSS, Chicago, IL, USA). In all samples, P-values were two-tailed, and comparisons were considered to be statistically significant for P<0.05 level with the Bonferroni multiple comparison correction.
Results
mRNA levels of TET1, TET2 and TET3
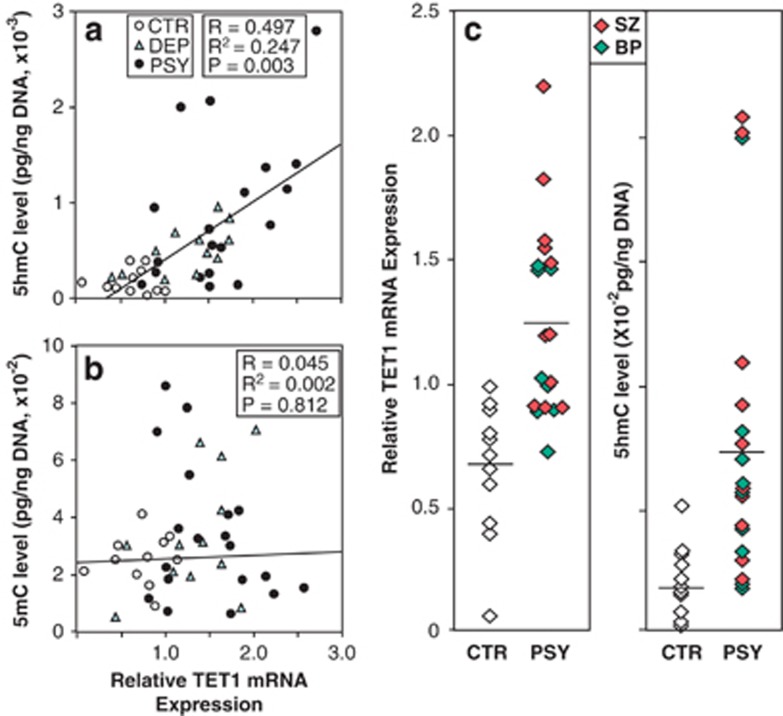
First, we examined the mRNA levels of TETs (TET1, TET2 and TET3) in the IPL of CTR subjects, and DEP and PSY patients. As shown in Table 1, ANOVA reveals that for TET1 mRNA, there is an overall significant difference among the three groups (F(2, 32)=7.43, P=0.002). We found an approximately twofold increase in the expression of TET1 mRNA in the PSY group compared with the CTR group (P=0.001, Dunnett's correction; P=0.002 Bonferroni correction). The increase of TET1 mRNA was not paralleled by a significant increase of TET2 and TET3 mRNA. Furthermore, the DEP group failed to show statistically significant increase in TET1, TET2 and TET3 mRNA compared with the CTR group. The increase of TET1 mRNA expression in the PSY group seemed to be region-specific because in the cerebellum of the same patients (Table 1) we failed to find an increase of TET1 mRNA compared with the CTR group (F(2, 37)=1.711, P=0.195). Table 1 also shows that the TET1 mRNA was expressed at a relatively higher level in the cerebellum than in the parietal cortex. The observation that TET1 is higher in the cerebellum than IPL is consistent with findings from other research groups.23, 40 The rational for combining SZ and BP disorder patients in a large PSY group was suggested by the observation that TET1 mRNA levels (SZ: 1.47±0.6; BP: 1.31±0.41) and protein levels (SZ: 0.34±0.08; BP: 0.30±0.07) were similar in SZ and BP disorder patients and higher than that in CTR subjects(mRNA: 0.64±0.09; protein: 0.07±0.01) (Figure 3c).
Table 1. Summary of the TET1, TET2 and TET3 mRNA expression in the IPL (parietal cortex) and the cerebellum of CTR subjects and DEP and PSY patients.
| Variable (mRNA) | Patient cohort | Overall ANOVA | Multiple comparison | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 CTR | 2 DEP | 3 PSY | F | d.f. | P-value |
DEP vs |
PSY vs |
|||
| |
|
|
|
|
|
|
1 |
3 |
1 |
2 |
| Parietal cortex | ||||||||||
| TET1 | 0.64±0.09 | 1.02±0.18 | 1.29±0.10 | 7.656 | 2 | 0.002 | — | — | 0.00.1 | — |
| TET2 | 1.38±0.15 | 2.0±0.24 | 1.93±0.23 | 1.786 | 2 | NS | — | — | — | — |
| TET3 | 0.91±0.16 | 1.23±0.14 | 1.28±0.11 | 2.187 | 2 | NS | — | — | — | — |
| Cerebellum | ||||||||||
| TET1 | 3.45±0.49 | 2.95±0.44 | 3.28±0.27 | 1.843 | 2 | NS | — | — | — | — |
Abbreviations: CTR, nonpsychiatric; DEP, depressed; d.f., degree of freedom; IPL, inferior parietal lobule; NS, nonsignificant; PSY, psychotic; TET, ten-eleven translocation protein.
TET1 mRNA expression was significantly increased in the PSY group compared with the CTR group. Each value represents the mean±s.e.m. of relative quantity (2−ΔCt × 10−2) of TET mRNA vs β-actin mRNA
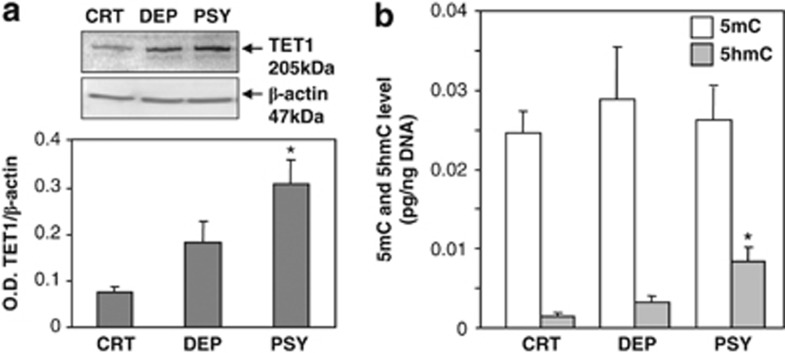
To determine if elevated TET1 mRNA expression is associated with TET1 protein expression, we examined the TET1 protein level in the same samples. As shown in Figure 2a, the expression of TET1 protein was increased in the PSY and the DEP cohorts, but only the PSY group showed statistical significance compared with the CTR group (F(2, 39)=5.89, P=0.006).
Figure 2.
(a) Ten-eleven translocation (TET)1 protein expression in the nonpsychiatric (CTR), depressed (DEP) and psychotic (PSY) group. In the inset is the representative gel image of western blot of TET1 and β-actin. The TET1 protein expression was increased in the PSY group compared with the CTR group: *P<0.01 (one-way analysis of variance (ANOVA)). Each value represents mean±s.e.m. of the optical density (OD) ratio of TET1 protein vs β-actin. (b) Levels of 5-methylcytosine (5mC) and 5-hydroxylmethylcytosine (5hmC) expression in total DNA isolated from the CTR, DEP and PSY groups. Each value represents the OD ratios of 5mC and 5hmC immunostaining (calculated from their corresponding standard curves) and the DNA loaded on the membrane (calculated from standard curve). The 5hmC level was increased in the PSY group compared with the CTR group (*P<0.01), while 5mC levels failed to change. The data represent the mean±s.e.m.
5mC and 5hmC levels in total DNA extracts
5hmC is the oxidized product of 5mC catalyzed by the TET family of proteins. To examine if the levels of 5hmC in the DNA are affected by an increase in TET1 levels in the tissue, we measured 5mC and 5hmC levels by immunoblotting using specific 5mC and 5hmC antibodies (see Materials and methods for details). Figure 2b shows that 5mC levels in total DNA are substantially higher than the levels of 5hmC in each of the three groups; however, 5hmC levels are significantly higher in PSY patients when compared with CTR subjects, but not in the DEP group (F(2, 36)=5.47, P=0.009; post hoc test: P=0.01). 5mC showed no significant differences among the three groups (F(2, 36)=0.3, P=nonsignificant) (Figure 2b).
Pearson's correlation analysis shows that the 5hmC levels were positively correlated with the level of TET1 mRNA in each of the three groups (Figure 3a) (P=0.003). On the other hand, the levels of 5mC failed to show any correlation with the amount of TET1 mRNA (P=0.812) (Figure 3b). As most of the PSY patients had TET1 mRNA relative values over 0.8 and high levels of 5hmC (Figure 3c), these variables when considered together using binary regression analysis reinforce the ability of the two measurements to identify the PSY patients. The binary logistic regression takes into account both the TET1 mRNA and the 5hmC levels in classifying correctly all the CTR subjects and the PSY patients (χ2=25.5, d.f.=2, P<0.00001), except for one PSY patient misclassified as a control. Given the small number of samples, this finding must await replication, and it would take several large N samples to calculate statistically a more exact classification function.
Figure 3.
Pearson's correlation analysis between relative ten-eleven translocation (TET)1mRNA expression and 5-hydroxylmethylcytosine (5hmC) (a) or 5-methylcytosine (5mC) (b) levels of all groups. (c) Plots of TET1 mRNA and 5hmC levels from nonpsychiatric (CTR) subjects and psychotic (PSY) patients. The line represents median value. BP, bipolar disorder; DEP, depressed; SZ, schizophrenia.
Higher 5hmC level at the GAD67 promoter in psychosis
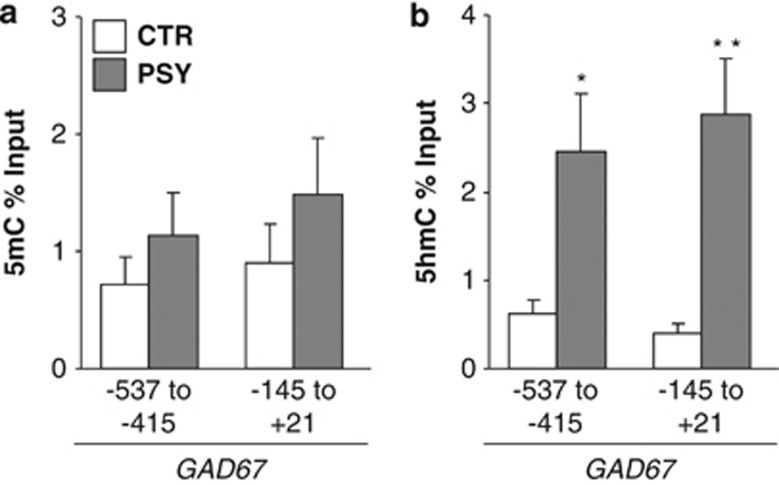
A map of the genome-wide expression of 5hmC suggests that 5hmC is primarily expressed at CG dinucleotides located in the body (particularly exons) of the genes; however, 5hmC has also been found enriched in CpG islands located in the proximity of the promoter transcriptional star sites (TSS) where it may have an important role in transcription.25 Here, we studied whether specific promoter regions of glutamic acid decarboxylase67 (GAD67), a gene whose transcription is downregulated in PSY subjects,41, 42, 43, 44 express an enrichment of 5mC and 5hmC at its CpG-rich TSS. We used methylated DNA immunoprecipitation to measure 5hmC and 5mC at the GAD67 promoter using previously published procedures.4 As shown in Figure 4, we found significantly higher 5hmC levels at the GAD67 gene at −537 to −415 and −145 to +21 in PSY vs CTR. In the same patients,4 we reported an increase in 5hmC at the BDNF IXabcd gene at −60 to +50 in PSY vs CTR, but not within BDNF exon IXd at +1185 to +1305. 5mC is also increased at the BDNF IXabcd promoter at −60 to +50 in PSY vs CTR.4 However, we found no differences in 5mC at either the GAD67 promoter site or within the BDNF IXd exon. As expected, there was a significant negative correlation between GAD67 mRNA expression and GAD67 5hmC at −145 to +21 relative to the putative TSS (Spearman=−0.351; P=0.04) and a trend for a negative correlation at −537 to −415 (Spearman=−0.309; P=0.07).
Figure 4.
Increased 5-hydroxymethylcytosine (5hmC) at the glutamic acid decarboxylase 67 (GAD67) promoter in psychotic (PSY) patients. Binding of 5-methylcytosine (5mC) (a) and 5hmC (b) to GAD67 promoter was measured by immunoprecipitation using specific 5hmC and 5mC antibodies with previously published procedures.4 We find significantly increased 5hmC, but not 5mC, in PSY patients ( ) compared with nonpsychiatric (CTR) subjects (□) at both regions of the GAD67 promoter examined. *P<0.05 vs CTR; **P<0.01 vs CTR.
) compared with nonpsychiatric (CTR) subjects (□) at both regions of the GAD67 promoter examined. *P<0.05 vs CTR; **P<0.01 vs CTR.
mRNA levels of AID/APOBEC deaminases
We next measured the gene expression of AID/APOBEC family genes because they are important cytidine deaminases that have an important role in the DNA demethylation pathway.24, 45 The AID/APOBEC family is comprised of the AID, APOBEC1, APOBEC2, APOBEC3 and APOBEC4 subgroups.46 Quantitative real-time polymerase chain reaction failed to detect any reliable signal for AID, APOBEC1 and APOBEC2 in these samples, indicating a very low gene dosage in our IPL samples. In contrast, APOBEC3 and APOBEC4 mRNA are easily detectable. As shown in Table 2, ANOVA analysis revealed a significant decrease in the expression of APOBEC3A mRNA (F(2, 25)=15.38, P=0.0001) in both the PSY and DEP groups (post hoc test: P=0.0001 and 0.001, respectively). In addition, a dramatic mRNA downregulation was also observed in APOBEC3C in the PSY group compared with the CTR group (F(2, 23)=5.718, P=0.01; post hoc test: P=0.01), but not in the DEP group. Although APOBEC3B, APOBEC3F and APOBEC3H levels in DEP and PSY groups, and APOBEC3D levels in DEP group tended to be lower than in CTR group, the decrease did not reach statistical significance. No changes in APOBEC4 mRNA expression were found in any of the three groups (Table 2).
Table 2. APOBEC mRNA expressions.
| Variable (mRNA) | Relative APOBEC mRNA expressiona | Overall ANOVA | Multiple comparison | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 CTR | 2 DEP | 3 PSY | F | d.f. | P-value |
DEP vs |
PSY vs |
|||
| |
|
|
|
|
|
|
1 |
3 |
1 |
2 |
| APOBEC3A | 1.0±0.23 | 0.15±0.06 | 0.31±0.12 | 15.38 | 2 | 0.0001 | 0.001 | — | 0.0001 | — |
| APOBEC3B | 1.0±0.05 | 0.74±0.07 | 0.42±0.02 | 1.26 | 2 | NS | — | — | — | — |
| APOBEC3C | 1.0±0.02 | 0.58±0.04 | 0.50±0.03 | 5.718 | 2 | 0.01 | — | — | 0.01 | — |
| APOBEC3D | 1.0±0.01 | 0.69±0.01 | 1.02±0.01 | 0.782 | 2 | NS | — | — | — | — |
| APOBEC3F | 1.0±0.12 | 0.80±0.08 | 0.88±0.07 | 0.173 | 2 | NS | — | — | — | — |
| APOBEC3G | 1.0±0.13 | 1.08±0.11 | 0.97±0.04 | 0.081 | 2 | NS | — | — | — | — |
| APOBEC3H | 1.0±0.05 | 0.86±0.04 | 0.68±0.03 | 0.561 | 2 | NS | — | — | — | — |
| APOBEC4 | 1.0±0.10 | 1.02±0.17 | 1.09±0.11 | 0.198 | 2 | NS | — | — | — | — |
Abbreviations: AID, activation-induced deaminase; APOBEC, apolipoprotein B mRNA-editing enzyme; CTR, nonpsychiatric; DEP, depressed; d.f., degree of freedom; IPL, inferior parietal lobule; NS, nonsignificant; PSY, psychotic.
Note that AID, APOBEC1 and APOBEC2 were not detected in the IPL of our cohort, but were considerably abundant in NT2 cell line (human neuronal progenitor cell), indicating that their expressions are extremely low in terminally differentiated neurons or glial cells. The relative basic expression levels (2-ΔCt × 10−3 vs β-actin) of APOBECs measured in the IPL of the CTR group are: 0.71±0.23 (APOBEC3A); 0.11±0.05 (APOBEC3B); 2.5±0.5 (APOBEC3C); 0.11±0.04 (APOBEC3D); 0.42±0.97 (APOBEC3F); 0.55±0.16 (APOBEC3G); 0.15±0.07 (APOBEC3H) and 1.6±0.36 (APOBEC4), indicating that the APOBEC3 subgroup genes are expressed heterogeneously in the IPL.
The expression of APOBEC3A was decreased both in the PSY and the DEP groups compared with the CTR group, while the expression of APOBEC3C mRNA was decreased only in the PSY group. Each value represents the mean±s.e.m. of relative quantity (2−ΔΔCt) of APOBECs vs β-actin.
Potential confounding variables
To establish that the observed effects on the levels of TET1 mRNA, TET1 protein, APOBEC3A, APOBEC3C and 5hmC could not be attributed to potential confounding factors (age, brain pH, RNA integrity number, post-mortem interval), we analyzed the data using ANOVA and analysis of covariance. We found no significant differences among the three groups (the PSY or DEP patients and CTR subjects) with ANOVA. When the effect of these variables on the group differences of 5hmC, TET1 mRNA or TET1 protein was examined by entering them as covariates (analysis of covariance), we found the observed differences among the three groups were still significant. In addition, similar analyses of history of substance or alcohol abuse, antipsychotic, antidepressant or mood stabilizer treatment entered as covariant failed to influence group differences observed for 5hmC, TET1 mRNA and TET1 protein.
Discussion
The major finding presented in this paper is that TET1 (mRNA and protein) is markedly increased (two- to threefold) in the parietal cortex of PSY patients and this increase associates with an increase of 5hmC level at genomic DNA and with specific increases of 5hmC levels at GAD67 (Figure 4) and BDNF4 promoters in proximity of their TSS. Importantly, the increase of TET1 mRNA observed in the parietal cortex from the Stanley Foundation Neuropathology Consortium cohort was repeated in a small number of prefrontal cortex sections obtained from the HBTRC. We now are replicating these findings in a larger HBTRC cohort comparing 32 CTR and 42 PSY patients.
The increase may be specific to the cortex because the cerebellum of the same patients failed to show significant TET1 changes, even though the levels of TET1 in the cerebellum were 3–4 times higher than in the cortex. The reason why the cortex, but not the cerebellum, shows an increase in TET expression was not addressed in this study and remains only a matter of speculation. Other brain areas such as the hippocampus also require study.
The increase of TET1 in the cortex of PSY patients is not associated with an increase of TET2 and TET3 mRNA and cannot be attributed apparently to confounding demographic variables, nor to the type and dose of antipsychotic used. Furthermore, the increase of TET1 and of 5hmC may not generalize to all major psychiatric disorders because it was absent in DEP patients. In this study, we analyzed the data by combining SZ and BP disorder patients in the PSY patient group, because the increase in TET1 mRNA was similar in the two groups (Figure 3) and because the pathogenesis and the symptomathology of these two diseases are related. This categorization is supported by recent findings showing that lower expression of GAD6741, 42, 43, 47 and BDNF,48, 49 and overexpression of DNMT11and GADD45β4 are common biochemical alterations in both of these psychiatric disorders.
Recent DNA demethylation studies suggest that TET1, by hydroxylating 5mC residues at CpG-rich gene bodies or at TSS of specific promoters, could have an important role in triggering neuronal activity-induced enzymatic removal of the repressive epigenetic DNA mark, 5mC, in mammalian brain.22
The role of TET1 may be to facilitate the removal of 5mC from CpG-rich gene regions via formation of the intermediate 5hmC to favor the deamination catalyzed by AID/APOBEC family of cytidine deaminases and the removal of 5hmU by the base excision repair pathway. In an animal model, evidence is accumulating that TET1 and APOBEC are involved in neuronal activity-induced, region-specific, active DNA demethylation and subsequent activation of gene expression in adult mouse brain in vivo.4, 22, 25, 31
Although the TET1-dependent active DNA demethylation and gene expression upregulation may occur in the brain under physiological conditions,22 in the IPL of our PSY patient cohort we found an increase of TET1 that positively correlates with an increase of 5hmC at genomic DNA (Figure 3a) and at gene promoters such as GAD67 (Figure 4) and BDNF, which have been consistently associated with downregulation in the expression of these target genes.41, 42, 43, 47, 48, 49 A possible explanation for the unexpected finding is that an increase of TET1 associated with gene expression downregulation is associated with a downregulation of some of the main APOBEC-deaminating enzymes. Our studies on APOBEC show that the most abundant isoforms (3A and 3C) of these cytidine-deaminating enzymes may be reduced significantly in psychosis preventing the active demethylation of the gene promoters, resulting in an accumulation of 5hmC, which by recruiting MBD3–NURD complex appears to be a DNA mark that suppresses transcription.50, 51
Up to now there have been 13 human cytidine deaminases identified.46 In our parietal cortex cohorts, AID, APOBEC1 and APOBEC2 were virtually undetectable, and APOBEC3B, APOBEC3D, APOBEC3F, APOBEC3G and APOBEC3H were expressed at a very low level. Only APOBEC3A, APOBEC3C and APOBEC4 were detectable at a significant level. The expression of APOBEC3A and APOBEC3C were significantly decreased in the cortex of PSY patients (Table 2).
In a previous study, we found no changes in the expression of the base excision repair pathway enzymes in PSY patients (that is, TDG, XPG, MBD4).4 Hence, the decrease of APOBEC cytidine deaminase enzymes in the cortex of PSY patients may be the limiting factor that reduces the rate of conversion of 5hmC into 5hmU preventing demethylation of 5hmC genes and inducing the gene transcriptional repression that may underlie the pathophysiology of major psychosis.
An alternative explanation for the role of TET1 in transcriptional repression is that the repressive function of TET1 is independent of its catalytic activity. In fact, it has been found that TET1, which contains CXXC DNA binding sites,52, 53 recruits the polycomb repressive complex 2 and SIN3A co-repressor proteins at target genes, suggesting that this repressive protein and SIN3A may have an important function in TET1-mediated gene repression.54 Our analyses of history of substance or alcohol abuse, antipsychotic, antidepressant or mood stabilizer treatment, entered as covariants in analysis of covariance, failed to reveal an influence on group differences in 5hmC, TET1 mRNA and TET1 protein, but our conclusion of no effect of these variables requires further validation. Although our statistical analysis with medications as a covariant failed to show an influence of drug treatment on TET and 5hmC expression, medication factors on gene expression in post-mortem studies cannot be excluded. In fact, the medication history in the demographic records lacks detail and precision. In addition, there is evidence that different antipsychotics have different effects on epigenetic mechanisms.41
In summary, the increase of TET1 and the decreases in APOBAC3A and APOBAC3C found in this study together with previous studies showing an increase in DNMT and GADD45β in the cortex of PSY patients support the hypothesis that an epigenetic misregulation of gene expression may be operative in the pathogenesis of PSY disorders. Clearly, to test the hypothesis there is a need for a more detailed investigation of the binding of TET1, APOBEC3A and APOBAC3C, and the presence of 5hmC across the genome. In addition to consolidating the findings from human samples and to study drugs that may normalize the altered epigenetic mechanisms, we need to develop a rodent model that will allow us to investigate changes in TET1, APOBEC, DNMT and GADD45β similar to the changes detected in the brain of PSY patients.
Table 3. Increased 5hmC at GAD67 and BDNF promoters in psychotic patients.
| Gene (position relative to TSS) | 5mC | 5hmC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
CTR |
PSY |
t-Value |
d.f. |
P-Value |
CTR |
PSY |
t-Value |
d.f. |
P-value |
| GAD67 (−537 to −415) | 0.71±0.24 | 1.13±0.37 | 0.799 | 39 | NS | 0.62±0.16 | 2.45±0.90 | 2.004 | 39 | 0.05 |
| GAD67 (−145 to +21) | 0.90±0.33 | 1.48±0.49 | 0.819 | 39 | NS | 0.39±0.12 | 2.87±0.89 | 2.801 | 39 | 0.009 |
| BDNF IXabcd (−60 to +50) | 1.99±0.68 | 9.42±2.26 | 3.14 | 39 | 0.005 | 2.32±0.81 | 9.86±2.50 | 2.873 | 39 | 0.008 |
| BDNF IXabcd (+1185 to +1305) | 8.83±2.65 | 10.78±2.49 | 0.499 | 39 | NS | 11.10±2.58 | 12.85±2.59 | 0.438 | 39 | NS |
Abbreviations: APOBEC, apolipoprotein B mRNA-editing enzyme; BDNF, brain-derived neurotrophic factor; CTR, nonpsychiatric; DEP, depressed; d.f., degree of freedom; GAD67, glutamic acid decarboxylase67; 5hmC, 5-hydroxymethylcytosine; 5-methyl cytosine; IPL, inferior parietal lobule; NS, nonsignificant; PSY, psychotic; TSS, transcriptional star site.
Acknowledgments
We thank Drs A Guidotti, DR Grayson and P Tueting for their critical reading and comments on the manuscript. We gratefully acknowledge the support of ROAA fund from University of Illinois at Chicago.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B. 2005;134B:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- Gavin DEP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. 2012;2:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S. Epigenetics of schizophrenia. Curr Top Behav Neurosci. 2010;4:611–628. doi: 10.1007/7854_2010_38. [DOI] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Akbarian S. Epigenetic and post-transcriptional dysregulation of gene expression in schizophrenia and related disease. Neurobiol Dis. 2012;46:255–262. doi: 10.1016/j.nbd.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Shulha HP, Cheung I, Whittle C, Wang J, Virgil D, Lin CL, et al. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch Gen Psychiatry. 2012;69:314–324. doi: 10.1001/archgenpsychiatry.2011.151. [DOI] [PubMed] [Google Scholar]
- Veldic M, Caruncho HJ, Liu WS, Davis J, Satta R, Grayson DR, et al. DNA methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci USA. 2004;101:348–353. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E, Davis JM, Costa E. In psychosis, cortical interneurons overexpress DNA-methyltransferase 1. Proc Natl Acad Sci USA. 2005;102:2152–2157. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Kadriu B, Maloku E, Agis-Balboa RC, Guidotti A, Davis JM, et al. Epigenetic mechanisms expressed in basal ganglia GABAergic neurons differentiate schizophrenia from bipolar disorder. Schizophr Res. 2007;91:51–61. doi: 10.1016/j.schres.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry. 2007;4:385–397. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Stary JM, Earle JA, Araghi-Niknam M, Eagan E. GABAergic dysfunction in schizophrenia and mood disorders as reflected by decreased levels of glutamic acid decarboxylase 65 and 67 kDa and Reelin proteins in cerebellum. Schizophr Res. 2005;72:109–122. doi: 10.1016/j.schres.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Tochigi M, Iwamoto K, Bundo M, Komori A, Sasaki T, Kato N, et al. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol Psychiatry. 2008;63:530–533. doi: 10.1016/j.biopsych.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Dempster EL, Mill J, Craig IW, Collier DA. The quantification of COMT mRNA in post mortem cerebellum tissue: diagnosis, genotype, methylation and expression. BMC Med Genet. 2006;7:10. doi: 10.1186/1471-2350-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintas C, Persico AM, Neocortical RELN. promoter methylation increases significantly after puberty. NeuroReport. 2010;2:114–118. doi: 10.1097/WNR.0b013e328334b343. [DOI] [PubMed] [Google Scholar]
- Szyf M. Epigenetics. DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- Dong E, Chen Y, Gavin DP, Grayson DR, Guidotti A. Valproate induces DNA demethylation in nuclear extracts from adult mouse brain. Epigenetics. 2010;8:730–735. doi: 10.4161/epi.5.8.13053. [DOI] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F, Dong E, Gavin DP, Nicoletti F, Guidotti A. Activation of group-II metabotropic glutamate receptors promotes DNA demethylation in the mouse brain. Mol Pharmacol. 2011;80:174–182. doi: 10.1124/mol.110.070896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey EF. Schizophrenia and the inferior parietal lobule. Schizophr Res. 2007;97:215–225. doi: 10.1016/j.schres.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Goldstein JM, Goodman JM, Seidman LJ, Kennedy DN, Makris N, Lee H, et al. Cortical abnormalities in schizophrenia identified by structural magnetic resonance imaging. Arch Gen Psychiatry. 1999;56:537–547. doi: 10.1001/archpsyc.56.6.537. [DOI] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The stanley foundation brain collection and neuropathology consortium. Schizophr Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci USA. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Sodhi M, Kleinman JE. Epigenetic mechanisms in schizophrenia. Biochim Biophys Acta. 2009;1790:869–877. doi: 10.1016/j.bbagen.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229.1–229.10. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T, Oki Y, Yanagawa Y, Fukuda A. A heterozygous deletion in the glutamate decarboxylase 67 gene enhances maternal and fetal stress vulnerability. Neurosci Res. 2011;69:276–282. doi: 10.1016/j.neures.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Buckley PF, Pillai A, Howell KR. Brain-derived neurotrophic factor: findings in schizophrenia. Curr Opin Psychiatry. 2011;24:122–127. doi: 10.1097/YCO.0b013e3283436eb7. [DOI] [PubMed] [Google Scholar]
- Weickert CS, Hyde TM, Lipska BK, Herman MM, Weinberger DR, Kleinman JE. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8:592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- Robertson J, Robertson AB, Klungland A. The presence of 5-hydroxymethylcytosine at the gene promoter and not in the gene body negatively regulates gene expression. Biochem Biophys Res Commun. 2011;411:40–43. doi: 10.1016/j.bbrc.2011.06.077. [DOI] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.