In eukaryotic cells, PA2 is a central precursor for the synthesis of major glycerophospholipids, DAG and TAG, as well as a major signaling lipid. In mammalian cells, PA is implicated as an activator of cell growth and proliferation, vesicular trafficking, secretion, and endocytosis (1–7). In plants, PA is implicated in seed germination and response to stress induced by drought, salinity, and low temperature (3, 4).
The signaling roles of PA in the yeast Saccharomyces cerevisiae have not received as much attention as they have in higher eukaryotic cells. However, it is known that PA production via phospholipase D-mediated turnover of PC is necessary for suppression of growth and membrane trafficking defects in mutants defective in Sec14p, an essential PI/PC-binding protein (8–10). In addition, PA production via phospholipase D is implicated in Spo20p-mediated fusion of vesicles with the pre-spore membrane during sporogenesis (11, 12). The best studied regulatory function of PA in S. cerevisiae is its role as a signaling molecule in the transcriptional regulation of glycerophospholipid synthesis itself, the major topic of this review. We will focus on the pathways generating and utilizing PA in yeast and the evidence that PA plays a central role in the transcriptional regulation of glycerophospholipid synthesis.
The Role of PA in de Novo Lipid Synthesis
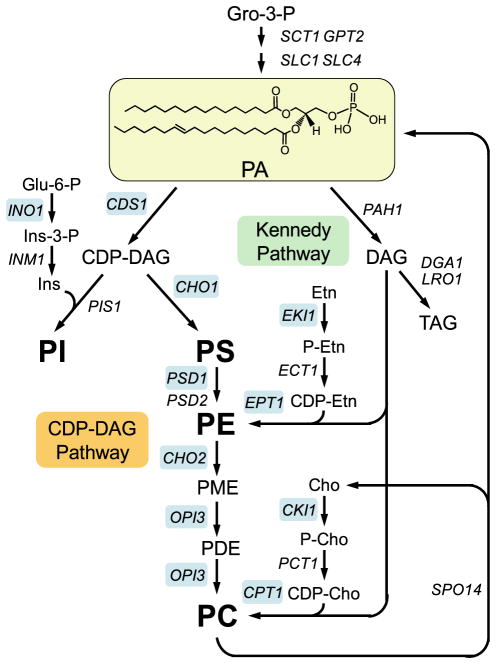
PA is the precursor of all membrane glycerophospholipids and the storage lipid TAG in S. cerevisiae (Fig. 1) (13–16). In the de novo pathway, PA is synthesized from glycerol 3-phosphate and then used for the synthesis of glycerophospholipids via the liponucleotide intermediate CDP-DAG (i.e. CDP-DAG pathway) (Fig. 1). PA is also used for the synthesis of PE and PC via DAG in the Kennedy pathway (CDP-ethanolamine and CDP-choline branches, respectively) (Fig. 1). The DAG derived from PA is also used for the synthesis of TAG (Fig. 1). Cells grown in the absence of choline synthesize PC primarily via the CDP-DAG pathway (17). In contrast, cells supplemented with choline synthesize PC primarily via the Kennedy pathway (17). Nonetheless, PC is synthesized from PA by both the CDP-DAG and Kennedy pathways regardless of whether choline is added to the growth medium (8, 18 –22). The choline required is derived from the phospholipase D-mediated turnover of the PC synthesized via the CDP-DAG pathway (Fig. 1) (8, 10). The PA produced is reincorporated into glycerophospholipids (8).
FIGURE 1. Structure of PA and its role in lipid synthesis.
The structure of PA is shown with fatty acyl groups of 16:0 (sn-1) and 18:1 (sn-2). The pathways shown for the synthesis of glycerophospholipids include the relevant steps discussed in this review. A more comprehensive pathway of lipid metabolism may be found in Ref. 34. The genes that encode the enzymes catalyzing individual steps in the pathway are indicated. The UASINO-containing genes that are subject to regulation by inositol supplementation and to nutrient depletion are highlighted in blue. Gro, glycerol; Ins, inositol; PME, phosphatidylmonomethylethanolamine; PDE, phosphatidyldimethylethanolamine; Etn, ethanolamine; P-Etn, phosphoethanolamine; Cho, choline; P-Cho, phosphocholine.
Mutants defective in PC synthesis via the CDP-DAG pathway require choline for growth; they synthesize PC via the CDP-choline branch of the Kennedy pathway (23–30). Mutants defective in the synthesis of PS (23, 24) and PE (25, 26) can synthesize PC if they are supplemented with ethanolamine. The ethanolamine is used to synthesize PE via the CDP-ethanolamine branch of the Kennedy pathway, which is then methylated to form PC in the CDP-DAG pathway (Fig. 1) (31, 32). Kennedy pathway mutants (e.g. cki1 eki1 and cpt1 ept1) defective in both the CDP-choline and CDP-ethanolamine branches must synthesize PC via the CDP-DAG pathway (18, 20, 33). However, unlike mutants defective in the CDP-DAG pathway (23–30), Kennedy pathway mutants do not exhibit any auxotrophic requirements (18, 33).
Inositol and Nutrient Depletion Regulate the Expression of UASINO-containing Glycerophospholipid Synthesis Genes
The synthesis of PC is coordinately regulated with the synthesis of PI (31, 32, 34, 35). This regulation is mediated by genetic and biochemical mechanisms (31, 32, 34 –37). In this review, we will focus on the transcriptional regulation of glycerophospholipid synthesis gene expression. Several glycerophospholipid synthesis genes are maximally expressed when inositol is absent from the growth medium and repressed when inositol is added to the growth medium. The repression by inositol requires PC synthesis and is enhanced by the inclusion of choline in the growth medium (15, 31, 32, 38, 39). The expression of glycerophospholipid synthesis genes is also repressed when cells enter the stationary phase of growth and when cells are depleted of nitrogen or zinc (31, 32, 35). The repression in the stationary phase and by nitrogen and zinc depletion occurs in the absence of inositol (31, 32, 35, 40).
The coordinate regulation of the glycerophospholipid synthesis genes by inositol supplementation, growth phase, and nutrient depletion is dependent on the transcriptional regulatory proteins Ino2p (41), Ino4p (42), and Opi1p (43), as well as a UASINO cis-acting element (44–47) present in the promoters of the glycerophospholipid synthesis genes (Fig. 2) (31, 32, 39, 48). The UASINO element contains a consensus binding site (CATGTGAAAT) for an Ino2p-Ino4p heterodimer complex that activates the expression of glycerophospholipid synthesis genes (31, 32, 39, 49–52). Repression of these genes is dependent on Opi1p (43, 53), which inhibits transcriptional activation of the Ino2p-Ino4p complex by binding to Ino2p (Fig. 2) (54). In fact, ino2 and ino4 mutants exhibit constitutively repressed levels of UASINO-containing genes, whereas opi1 mutants exhibit constitutively derepressed levels of UASINO-containing genes (15, 31, 38, 39). Because of the misregulation of INO1 expression, ino2 and ino4 mutants are auxotrophic for inositol, whereas opi1 mutants produce excessive amounts of inositol and excrete it into the growth medium (15, 31, 38, 39). Thus, inositol auxotrophy and inositol excretion are used as indicators of the misregulation of INO1 and other UASINO-containing genes (15, 31, 38, 39).
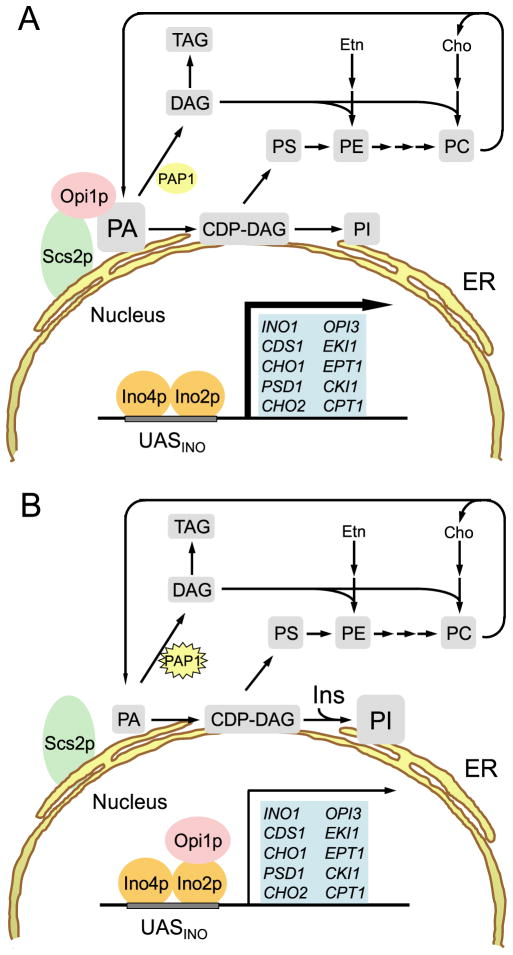
FIGURE 2. Model for the role of PA in the transcriptional regulation of UASINO-containing genes.
A, UASINO-containing genes (blue) are maximally expressed (thick arrow) when wild-type S. cerevisiae cells are grown in the absence of inositol and in the presence of sufficient amounts of essential nutrients. B, when inositol is added to the growth medium or when cells are depleted of an essential nutrient, transcription of UASINO-containing genes is attenuated (thin arrow) by the interaction of Opi1p with Ino2p. Dissociation of Opi1p from the nuclear/ER membrane is caused by a decrease in PA concentration, which is mediated by the stimulation of PI synthesis and by activation of Mg2+-dependent PA phosphatase (PAP1, denoted by the star). Etn, ethanolamine; Cho, choline; Ins, inositol.
Recent genome-wide studies using microarray analysis have revealed the full extent of genes responsive to addition of inositol and choline to the growth medium and dependent for this regulation on Ino2p, Ino4p, and Opi1p (55–57). These studies also revealed several other major classes of genes in addition to and independent of Ino2p, Ino4p, and Opi1p that respond to signals arising in the ER following inositol addition that are potentially related to lipid metabolism (57).
Genetic Clues Implicating PA as the Signaling Molecule That Regulates Transcription of UASINO-containing Glycerophospholipid Synthesis Genes
Mutations that block the utilization of PA for PC synthesis cause the derepression of INO1 and other UASINO-containing genes, whereas mutations that increase the utilization of PA cause the repression of UASINO-containing genes (31, 32). Mutants defective in the structural genes leading to PC from PA via the CDP-DAG pathway (Fig. 1) share in common the inositol excretion phenotype (31, 39). As indicated above, the inositol excretion phenotype results from the loss of Opi1p repressor function, which leads to the constitutive derepression and overexpression of INO1. Moreover, the remaining wild-type genes in a particular CDP-DAG pathway mutant cannot be repressed by inositol supplementation (31, 32). The inositol excretion phenotype of the CDP-DAG pathway mutants beginning with the formation of PS from CDP-DAG can be complemented if the mutants are supplemented with a water-soluble precursor that allows for the synthesis of glycerophospholipids via the Kennedy pathway (31, 32). The supplementation of the Kennedy pathway precursor also permits the inositol-mediated repression of the remaining wild-type genes in a particular CDP-DAG pathway mutant (31, 32). A complete block of the first step in the CDP-DAG pathway (e.g. cds1) is lethal (58). However, partial (59) or conditional (60) cds1 alleles derepress INO1 and excrete inositol (60). The inositol excretion phenotype of the cds1 mutants cannot be overcome by supplementation with a Kennedy pathway precursor, nor will inositol mediate the repression of other UASINO-containing genes in these strains (31, 32, 59). Thus, mutants defective at every step in the synthesis of PS, PE, and PC from CDP-DAG exhibit increases in PA levels and altered regulation of INO1 and have the inositol excretion phenotype. These observations led to the hypothesis that PA is the metabolic signal for INO1 derepression (31).
In contrast, mutants defective in PC synthesis via the CDP-choline branch of the Kennedy pathway do not excrete inositol and exhibit normal regulation of INO1 (61). The Kennedy pathway mutants do, however, exhibit a choline excretion phenotype (8). Interestingly, Kennedy pathway mutants that also possess a mutation in the SEC14 gene (e.g. cki1 sec14, pct1 sec14, and cpt1 sec14) exhibit extreme choline excretion in combination with an inositol excretion phenotype (8, 9). The excretion of choline and inositol by these double mutants is dependent on the phospholipase D-mediated turnover of PC (8, 9). The finding that excess PA produced by this reaction causes INO1 derepression even in the presence of inositol (8, 9) further supports the hypothesis that PA provides the signal for derepression of UASINO-containing genes (31).
The enzyme responsible for generating the DAG for glycerophospholipid synthesis via the Kennedy pathway is the PAH1-encoded Mg2+-dependent PA phosphatase (62). Mutations that affect the activity of this enzyme cause the misregulation of UASINO-containing genes (62–64). A pah1 mutant with reduced activity exhibits derepressed levels of INO1, OPI3, and INO2 (62, 63). However, the level of INO1 derepression is not sufficient to cause inositol excretion (62, 63). This may be the consequence of the significant amount of Mg2+-dependent PA phosphatase activity that remains in pah1 mutant cells (62). Pah1p is phosphorylated by cyclin-dependent Cdc28p kinase and dephosphorylated by the Nem1p-Spo7p phosphatase complex (63). Reduction in Pah1p phosphorylation elevates Mg2+-dependent PA phosphatase activity (64), indicating that phosphorylation inhibits activity. Cells that overexpress a phosphorylation-deficient enzyme are repressed for INO1 and OPI3 and require inositol for growth (64). The fact that the inositol auxotrophy of cells overexpressing Mg2+-dependent PA phosphatase activity can be suppressed by an opi1 mutation (64) indicates the role of Opi1p in this regulation.
PA Content Controls the Localization of Opi1p and Its Repressor Function
Loewen et al. (65–67) have shown that the Opi1p repressor, a protein lacking membrane-spanning domains (43), is found at the nuclear/ER membrane and within the nucleus. Opi1p associates with the nuclear/ER membrane through interaction with the integral membrane protein Scs2p (Fig. 2) (65). A connection between Scs2p and the regulation of glycerophospholipid synthesis is evident because scs2 mutants are auxotrophic for inositol due to the constitutive repression of INO1 (68–70). Thus, attenuation of the repressor activity of Opi1p depends on its association with the nuclear/ER membrane through interaction with Scs2p. Interestingly, the scs2 mutation also results in an increase in PC synthesis via the Kennedy pathway, and a block in the Kennedy pathway (e.g. cki1 scs2) restores normal INO1 expression (70). As discussed above, PC synthesis via the Kennedy pathway consumes PA via DAG, and a block in the Kennedy pathway should accumulate PA, thus potentially explaining the suppression of the inositol auxotrophy of scs2 mutants. Although this scenario has not been fully explored experimentally, it is consistent with the notion that PA plays a role in the regulation of INO1 and other UASINO-containing genes.
A major breakthrough in our understanding of glycerophospholipid synthesis regulation was the discovery that Opi1p is a PA-binding protein and that its association with the nuclear/ER membrane is stabilized by PA (66). Moreover, Loewen et al. (66) have shown that Opi1p translocates from the nuclear/ER membrane into the nucleus upon inositol supplementation, which results in a reduction in the level of PA coincident with its use as a precursor in the synthesis of PI via CDP-DAG (Fig. 2). The translocation of Opi1p in response to inositol supplementation coincides with the repression of INO1 expression (66). Thus, a key regulatory switch for controlling the expression of INO1 and other UASINO-containing genes is the localization of Opi1p, which is dependent on the presence of a certain amount of PA that is required for stabilizing the association of Opi1p at the nuclear/ER membrane (66). The establishment of this mechanism thus provided confirmation of the signaling role of PA predicted by the genetic experiments described above (31). Changes in PA concentration can be brought about by the action and regulation of several enzymes (e.g. CDP-DAG pathway enzymes, Kennedy pathway enzymes, Mg2+-dependent PA phosphatase, and phospholipase D) and thus are predicted to affect the location and repressor activity of Opi1p. As discussed below, this also explains why the expression of UASINO-containing genes is regulated by growth phase and nutrient depletion (e.g. nitrogen and zinc), conditions that lower PA levels, independent of inositol availability.
Model for the Transcriptional Regulation of UASINO-containing Genes
A model for the regulation of UASINO-containing genes by inositol and by nutrient depletion is presented in Fig. 2. According to this model, Opi1p is associated with the nuclear/ER membrane through interactions with PA and Scs2p when exponential phase cells are grown without inositol and in the presence of sufficient amounts of essential nutrients (e.g. zinc) (Fig. 2A). Upon inositol supplementation, exponential phase cells synthesize an elevated level of PI through increased substrate availability (71) and draw upon the pool of PA at the nuclear/ER membrane (Fig. 2B) (66). At the same time, inositol supplementation results in an increase in Mg2+-dependent PA phosphatase activity (72), which contributes to the decrease in the PA pool. TAG is synthesized at the expense of glycerophospholipids in stationary phase cells (73), and Mg2+-dependent PA phosphatase activity is elevated under this growth condition (74). The PAH1-encoded Mg2+-dependent PA phosphatase enzyme plays a major role in TAG synthesis, and its increased activity in stationary phase diminishes the PA pool (62). In zinc-depleted cells, PI synthesis increases (75), and this condition should draw upon the PA pool. The increase in PI synthesis in response to zinc depletion is not the result of elevated levels of inositol; INO1 is actually repressed under this growth condition (75). Instead, the increase in PI synthesis is due to the Zap1p-mediated induction of PIS1 gene expression (76). Furthermore, zinc depletion induces Mg2+-dependent PA phosphatase activity (77), and this would further reduce the PA pool. Thus, regardless of whether cells are supplemented with inositol or are depleted of an essential nutrient, a decrease in the PA level results in loss of Opi1p association with the nuclear/ER membrane, followed by Opi1p translocation into the nucleus. In the nucleus, Opi1p mediates repression of the UASINO-containing glycerophospholipid synthesis genes that are co-regulated through its interaction with Ino2p.
Perspectives
The control of enzymes (e.g. CDP-DAG synthase, Mg2+-dependent PA phosphatase, and phospholipase D) that directly contribute to the PA pool at the nuclear/ER membrane should have a major impact on glycerophospholipid synthesis regulation by Opi1p. As indicated above, the phosphorylation state of Mg2+-dependent PA phosphatase plays a role in the Opi1p-mediated regulation of INO1 and OPI3 (64). However, it is unclear how various growth conditions (e.g. inositol supplementation and zinc depletion) affect the phosphorylation-dephosphorylation of the enzyme. Opi1p is phosphorylated by protein kinase A (78), protein kinase C (79), and casein kinase II (80). Phosphorylation by protein kinase A and casein kinase II stimulates Opi1p repressor function (78, 80), whereas phosphorylation by protein kinase C attenuates Opi1p repressor function (79). Whether these phosphorylation events influence the interaction of Opi1p with PA, Scs2p, or Ino2p is unclear.
Inositol auxotrophy has been described in conjunction with mutations in several major signaling pathways that include the unfolded protein response (81–83), the protein kinase C/mitogen-activated protein kinase (MAPK) pathway (84), and the glucose response pathway (85). These observations have led to the speculation that these signaling pathways might play a direct role in transmitting the signal controlling INO1 expression. The demonstration that Opi1p translocation into the nucleus following inositol supplementation in response to PA availability is the major signal for repression of UASINO-containing genes indicates that these signaling pathways must play a more peripheral role. Such roles could include modulation of Opi1p activity by phosphorylation, as suggested for protein kinase C (79), or by influencing histone phosphorylation and subsequent acetylation at the INO1 locus, as suggested for the glucose response pathway (86, 87). However, effects on histone modification at INO1 cannot fully explain the inositol auxotrophy of snf1 and snf4 mutants defective in the glucose response pathway because these phenotypes can be suppressed by direct inhibition of the enzymatic activity of acetyl-CoA carboxylase (85), the enzyme controlling the rate-limiting step in fatty acid synthesis. Acetyl-CoA carboxylase is a direct target of Snf1p (related to mammalian AMP-activated protein kinase) in both yeast and mammals (88). Thus, additional signals coming from fatty acid synthesis may also influence INO1 expression. It is quite possible that these signals are also coordinated through the de novo synthesis of PA because acyl-CoAs serve as immediate precursors. Future work on the coordination of lipid metabolism with other cellular processes, such as membrane trafficking and responses to stress and nutrient availability, via the major signaling pathways should provide new insights into the myriad roles of PA in the cell. These insights should provide models of metabolic regulation and lipid signaling testable in both animals and plants.
Acknowledgments
We thank Yu-Fang Chang, Lorena Egüez, M. Laura Gaspar, Gil-Soo Han, Stephen A. Jesch, and Manual J. Villa-Garcia for helpful comments on the manuscript.
Footnotes
The abbreviations used are: PA, phosphatidic acid; DAG, diacylglycerol; TAG, triacylglycerol; PC, phosphatidylcholine; PI, phosphatidylinositol; PE, phosphatidylethanolamine; PS, phosphatidylserine; UASINO, upstream activating sequence inositol-responsive element; ER, endoplasmic reticulum.
This minireview will be reprinted in the 2007 Minireview Compendium, which will be available in January, 2008. This work was supported in part by United States Public Health Service Grants GM-28140 (to G. M. C.) and GM-19629 (to S. A. H.) from the National Institutes of Health.
References
- 1.Waggoner DW, Xu J, Singh I, Jasinska R, Zhang QX, Brindley DN. Biochim Biophys Acta. 1999;1439:299–316. doi: 10.1016/s1388-1981(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 2.Sciorra VA, Morris AJ. Biochim Biophys Acta. 2002;1582:45–51. doi: 10.1016/s1388-1981(02)00136-1. [DOI] [PubMed] [Google Scholar]
- 3.Testerink C, Munnik T. Trends Plant Sci. 2005;10:368–375. doi: 10.1016/j.tplants.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Devaiah SP, Zhang W, Welti R. Prog Lipid Res. 2006;45:250–278. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Brindley DN. J Cell Biochem. 2004;92:900–912. doi: 10.1002/jcb.20126. [DOI] [PubMed] [Google Scholar]
- 6.Howe AG, McMaster CR. Can J Physiol Pharmacol. 2006;84:29–38. doi: 10.1139/Y05-138. [DOI] [PubMed] [Google Scholar]
- 7.Foster DA. Cancer Res. 2007;67:1–4. doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- 8.Patton-Vogt JL, Griac P, Sreenivas A, Bruno V, Dowd S, Swede MJ, Henry SA. J Biol Chem. 1997;272:20873–20883. doi: 10.1074/jbc.272.33.20873. [DOI] [PubMed] [Google Scholar]
- 9.Sreenivas A, Patton-Vogt JL, Bruno V, Griac P, Henry SA. J Biol Chem. 1998;273:16635–16638. doi: 10.1074/jbc.273.27.16635. [DOI] [PubMed] [Google Scholar]
- 10.Xie ZG, Fang M, Rivas MP, Faulkner AJ, Sternweis PC, Engebrecht J, Bankaitis VA. Proc Natl Acad Sci U S A. 1998;95:12346–12351. doi: 10.1073/pnas.95.21.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudge SA, Sciorra VA, Iwamoto M, Zhou C, Strahl T, Morris AJ, Thorner J, Engebrecht J. Mol Biol Cell. 2004;15:207–218. doi: 10.1091/mbc.E03-04-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakanishi H, Morishita M, Schwartz CL, Coluccio A, Engebrecht J, Neiman AM. J Cell Sci. 2006;119:1406–1415. doi: 10.1242/jcs.02841. [DOI] [PubMed] [Google Scholar]
- 13.Rattray JB, Schibeci A, Kidby DK. Bacteriol Rev. 1975;39:197–231. doi: 10.1128/br.39.3.197-231.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henry SA. In: The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Strathern JN, Jones EW, Broach JR, editors. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1982. pp. 101–158. [Google Scholar]
- 15.Paltauf F, Kohlwein SD, Henry SA. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Jones EW, Pringle JR, Broach JR, editors. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1992. pp. 415–500. [Google Scholar]
- 16.Czabany T, Athenstaedt K, Daum G. Biochim Biophys Acta. 2007;1771:299–309. doi: 10.1016/j.bbalip.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 17.McMaster CR, Bell RM. J Biol Chem. 1994;269:14776–14783. [PubMed] [Google Scholar]
- 18.Morash SC, McMaster CR, Hjelmstad RH, Bell RM. J Biol Chem. 1994;269:28769–28776. [PubMed] [Google Scholar]
- 19.McGee TP, Skinner HB, Whitters EA, Henry SA, Bankaitis VA. J Cell Biol. 1994;124:273–287. doi: 10.1083/jcb.124.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMaster CR, Bell RM. J Biol Chem. 1994;269:28010–28016. [PubMed] [Google Scholar]
- 21.Ostrander DB, O’Brien DJ, Gorman JA, Carman GM. J Biol Chem. 1998;273:18992–19001. doi: 10.1074/jbc.273.30.18992. [DOI] [PubMed] [Google Scholar]
- 22.Kim K-H, Voelker DR, Flocco MT, Carman GM. J Biol Chem. 1998;273:6844–6852. doi: 10.1074/jbc.273.12.6844. [DOI] [PubMed] [Google Scholar]
- 23.Atkinson K, Fogel S, Henry SA. J Biol Chem. 1980;255:6653–6661. [PubMed] [Google Scholar]
- 24.Atkinson KD, Jensen B, Kolat AI, Storm EM, Henry SA, Fogel S. J Bacteriol. 1980;141:558–564. doi: 10.1128/jb.141.2.558-564.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trotter PJ, Voelker DR. J Biol Chem. 1995;270:6062–6070. doi: 10.1074/jbc.270.11.6062. [DOI] [PubMed] [Google Scholar]
- 26.Trotter PJ, Pedretti J, Yates R, Voelker DR. J Biol Chem. 1995;270:6071–6080. doi: 10.1074/jbc.270.11.6071. [DOI] [PubMed] [Google Scholar]
- 27.Kodaki T, Yamashita S. J Biol Chem. 1987;262:15428–15435. [PubMed] [Google Scholar]
- 28.Kodaki T, Yamashita S. Eur J Biochem. 1989;185:243–251. doi: 10.1111/j.1432-1033.1989.tb15109.x. [DOI] [PubMed] [Google Scholar]
- 29.Summers EF, Letts VA, McGraw P, Henry SA. Genetics. 1988;120:909–922. doi: 10.1093/genetics/120.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGraw P, Henry SA. Genetics. 1989;122:317–330. doi: 10.1093/genetics/122.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry SA, Patton-Vogt JL. Prog Nucleic Acid Res. 1998;61:133–179. doi: 10.1016/s0079-6603(08)60826-0. [DOI] [PubMed] [Google Scholar]
- 32.Carman GM, Henry SA. Prog Lipid Res. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 33.Kim K, Kim K-H, Storey MK, Voelker DR, Carman GM. J Biol Chem. 1999;274:14857–14866. doi: 10.1074/jbc.274.21.14857. [DOI] [PubMed] [Google Scholar]
- 34.Gaspar ML, Aregullin MA, Jesch SA, Nunez LR, Villa-Garcia M, Henry SA. Biochim Biophys Acta. 2007;1771:241–254. doi: 10.1016/j.bbalip.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 35.Carman GM, Han G-S. Biochim Biophys Acta. 2007;1771:322–330. doi: 10.1016/j.bbalip.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carman GM, Zeimetz GM. J Biol Chem. 1996;271:13293–13296. doi: 10.1074/jbc.271.23.13293. [DOI] [PubMed] [Google Scholar]
- 37.Carman GM, Kersting MC. Biochem Cell Biol. 2004;82:62–70. doi: 10.1139/o03-064. [DOI] [PubMed] [Google Scholar]
- 38.Carman GM, Henry SA. Annu Rev Biochem. 1989;58:635–669. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- 39.Greenberg ML, Lopes JM. Microbiol Rev. 1996;60:1–20. doi: 10.1128/mr.60.1.1-20.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griac P, Henry SA. Nucleic Acids Res. 1999;27:2043–2050. doi: 10.1093/nar/27.9.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikoloff DM, McGraw P, Henry SA. Nucleic Acids Res. 1992;20:3253. doi: 10.1093/nar/20.12.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoshizaki DK, Hill JE, Henry SA. J Biol Chem. 1990;265:4736–4745. [PubMed] [Google Scholar]
- 43.White MJ, Hirsch JP, Henry SA. J Biol Chem. 1991;266:863–872. [PubMed] [Google Scholar]
- 44.Kodaki T, Nikawa J, Hosaka K, Yamashita S. J Bacteriol. 1991;173:7992–7995. doi: 10.1128/jb.173.24.7992-7995.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopes JM, Hirsch JP, Chorgo PA, Schulze KL, Henry SA. Nucleic Acids Res. 1991;19:1687–1693. doi: 10.1093/nar/19.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuller HJ, Hahn A, Troster F, Schutz A, Schweizer E. EMBO J. 1992;11:107–114. doi: 10.1002/j.1460-2075.1992.tb05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuller HJ, Richter K, Hoffmann B, Ebbert R, Schweizer E. FEBS Lett. 1995;370:149–152. doi: 10.1016/0014-5793(95)00818-t. [DOI] [PubMed] [Google Scholar]
- 48.Ambroziak J, Henry SA. J Biol Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- 49.Hirsch JP, Henry SA. Mol Cell Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bachhawat N, Ouyang Q, Henry SA. J Biol Chem. 1995;270:25087–25095. doi: 10.1074/jbc.270.42.25087. [DOI] [PubMed] [Google Scholar]
- 51.Loewy BS, Henry SA. Mol Cell Biol. 1984;4:2479–2485. doi: 10.1128/mcb.4.11.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwank S, Ebbert R, Rautenstrauss K, Schweizer E, Schuller HJ. Nucleic Acids Res. 1995;23:230–237. doi: 10.1093/nar/23.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greenberg M, Reiner B, Henry SA. Genetics. 1982;100:19–33. doi: 10.1093/genetics/100.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner C, Dietz M, Wittmann J, Albrecht A, Schuller HJ. Mol Microbiol. 2001;41:155–166. doi: 10.1046/j.1365-2958.2001.02495.x. [DOI] [PubMed] [Google Scholar]
- 55.Santiago TC, Mamoun CB. J Biol Chem. 2003;278:38723–38730. doi: 10.1074/jbc.M303008200. [DOI] [PubMed] [Google Scholar]
- 56.Jesch SA, Zhao X, Wells MT, Henry SA. J Biol Chem. 2005;280:9106–9118. doi: 10.1074/jbc.M411770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jesch SA, Liu P, Zhao X, Wells MT, Henry SA. J Biol Chem. 2006;281:24070–24083. doi: 10.1074/jbc.M604541200. [DOI] [PubMed] [Google Scholar]
- 58.Shen H, Heacock PN, Clancey CJ, Dowhan W. J Biol Chem. 1996;271:789–795. doi: 10.1074/jbc.271.2.789. [DOI] [PubMed] [Google Scholar]
- 59.Klig LS, Homann MJ, Kohlwein SD, Kelley MJ, Henry SA, Carman GM. J Bacteriol. 1988;170:1878–1886. doi: 10.1128/jb.170.4.1878-1886.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen H, Dowhan W. J Biol Chem. 1996;271:29043–29048. doi: 10.1074/jbc.271.46.29043. [DOI] [PubMed] [Google Scholar]
- 61.Griac P, Swede MJ, Henry SA. J Biol Chem. 1996;271:25692–25698. doi: 10.1074/jbc.271.41.25692. [DOI] [PubMed] [Google Scholar]
- 62.Han G-S, Wu W-I, Carman GM. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Hara L, Han G-S, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S. J Biol Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loewen CJR, Roy A, Levine TP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loewen CJR, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 67.Loewen CJR, Levine TP. J Biol Chem. 2005;280:14097–14104. doi: 10.1074/jbc.M500147200. [DOI] [PubMed] [Google Scholar]
- 68.Nikawa J, Murakami A, Esumi E, Hosaka K. J Biochem (Tokyo) 1995;118:39–45. doi: 10.1093/oxfordjournals.jbchem.a124889. [DOI] [PubMed] [Google Scholar]
- 69.Kagiwada S, Hosaka K, Murata M, Nikawa J, Takatsuki A. J Bacteriol. 1998;180:1700–1708. doi: 10.1128/jb.180.7.1700-1708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kagiwada S, Zen R. J Biochem (Tokyo) 2003;133:515–522. doi: 10.1093/jb/mvg068. [DOI] [PubMed] [Google Scholar]
- 71.Kelley MJ, Bailis AM, Henry SA, Carman GM. J Biol Chem. 1988;263:18078–18085. [PubMed] [Google Scholar]
- 72.Morlock KR, Lin Y-P, Carman GM. J Bacteriol. 1988;170:3561–3566. doi: 10.1128/jb.170.8.3561-3566.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taylor FR, Parks LW. Biochim Biophys Acta. 1979;575:204–214. doi: 10.1016/0005-2760(79)90022-5. [DOI] [PubMed] [Google Scholar]
- 74.Hosaka K, Yamashita S. Biochim Biophys Acta. 1984;796:110–117. [PubMed] [Google Scholar]
- 75.Iwanyshyn WM, Han G-S, Carman GM. J Biol Chem. 2004;279:21976–21983. doi: 10.1074/jbc.M402047200. [DOI] [PubMed] [Google Scholar]
- 76.Han S-H, Han G-S, Iwanyshyn WM, Carman GM. J Biol Chem. 2005;280:29017–29024. doi: 10.1074/jbc.M505881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iwanyshyn WM. PhD dissertation. Rutgers University; New Brunswick, NJ: 2005. Regulation of Phospholipid Synthesis in Saccharomyces cerevisiae by Zinc. [DOI] [PubMed] [Google Scholar]
- 78.Sreenivas A, Carman GM. J Biol Chem. 2003;278:20673–20680. doi: 10.1074/jbc.M300132200. [DOI] [PubMed] [Google Scholar]
- 79.Sreenivas A, Villa-Garcia MJ, Henry SA, Carman GM. J Biol Chem. 2001;276:29915–29923. doi: 10.1074/jbc.M105147200. [DOI] [PubMed] [Google Scholar]
- 80.Chang Y-F, Carman GM. J Biol Chem. 2006;281:4754–4761. doi: 10.1074/jbc.M513064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nikawa J, Yamashita S. Mol Microbiol. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 82.Cox JS, Shamu CE, Walter P. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 83.Chang HJ, Jesch SA, Gaspar ML, Henry SA. Genetics. 2004;168:1899–1913. doi: 10.1534/genetics.104.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nunez LR. PhD dissertation. Cornell University; Ithaca, NY: 2006. Phospholipid Synthesis in Yeast: The Role of the PKC1-MPK1 Signal Transduction Pathway. [Google Scholar]
- 85.Shirra MK, Patton-Vogt J, Ulrich A, Liuta-Tehlivets O, Kohlwein SD, Henry SA, Arndt KM. Mol Cell Biol. 2001;21:5710–5722. doi: 10.1128/MCB.21.17.5710-5722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 87.Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL. EMBO J. 2005;24:997–1008. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Woods A, Munday MR, Scott J, Yang X, Carlson M, Carling D. J Biol Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]