Abstract
Although the development of sea urchin embryos has been studied extensively and clearly involves both cell adhesion and cell migration, rather little is known about the adhesion receptors and extracellular matrix molecules involved. The completion of the genome of Strongylocentrotus purpuratus allows a comprehensive survey of the complement of cell-cell and cell-matrix adhesion molecules in this organism. Furthermore, the phylogenetic position of echinoderms offers the opportunity to compare the complement of adhesion proteins between protostome and deuterostome invertebrates and between invertebrate and vertebrate deuterostomes. Many aspects of development and cell interactions differ among these different taxa and it is likely that analysis of the spectrum of adhesion receptors and extracellular matrix proteins can open up new insights into which molecules have evolved to suit particular developmental processes. In this paper, we report the results of an initial analysis along these lines. The echinoderm adhesome (complement of adhesion-related genes/proteins) is similar overall to that of other invertebrates although there are significant deuterostome-specific innovations and some interesting features previously thought to be chordate- or vertebrate-specific.
INTRODUCTION
Cell-cell and cell-matrix adhesion play essential roles in many aspects of development. The genes and proteins mediating cell-cell and cell-matrix adhesion are among the most rapidly evolving in deuterostome genomes. Vertebrates have many more cell adhesion genes than do protostome invertebrates – a few of the more obvious examples include the immunoglobulin superfamily (IgSF) genes involved in interactions among cells of the immune and nervous systems and the cadherin/protocadherin superfamily. There are also many more extracellular matrix molecules in vertebrates, including novel protein architectures not found in protostomes such as insects and nematodes. It is, therefore, of interest to explore when these genes evolved during the deuterostome lineage and echinoderms, as invertebrate deuterostomes, whose development has been extensively studied over many years, offer an excellent opportunity for such an exploration.
Cell adhesion proteins have characteristic domains and domain combinations (Copley et al, 1999; Hynes and Zhao, 2000; Hohenester and Engel, 2002; Whittaker and Hynes, 2002). Since most of these domains are shared among many genes, simple homology searches such as BLAST are not very informative, giving many “hits.” Therefore, one of our main strategies was to search for diagnostic domains and domain organizations as an initial basis for gene identification. This was then supplemented by extensive BLAST and BLAT comparisons using adhesion sequences of known adhesion proteins from other invertebrates or from vertebrates and, in a few cases, previously described echinoderm sequences. Since many adhesion genes are very large with many exons, the original gene predictions were frequently incomplete. More complete gene predictions were developed for many genes (although by no means all) by exploring proximity among gene predictions that might comprise fragments of a complete adhesion gene and by Genewise gene predictions around the original predicted gene fragments. We concentrated on obtaining as complete a picture as possible for relatively small (dozens) gene families (integrins, cadherins, collagens, laminins and several others) including phylogenetic analyses. Larger families (hundreds) of adhesion receptors and extracellular matrix proteins such as those containing IgSF, FN3, EGF or LRR domains, could not be analyzed in the same detail but we were able to extract reasonable estimates of the extent and complexity of those families together with more detailed analyses of some of the more interesting members.
Most adhesion genes common to protostomes and vertebrates are, as expected, also present in sea urchins and we discuss those genes here. We were particularly interested in exploring the presence of adhesion genes that have previously been detected only in chordates or only in vertebrates. Many of those could not be found, although there are some very intriguing exceptions to that generalization. Given the current state of the genome assembly, it is clear that some genes may well have been missed and any statements as to the absence of a particular class of genes/proteins may be subject to revision based on future data. Nonetheless, it seems clear from the analyses that we will discuss below, that sea urchins lack many adhesion genes that are present in chordates and we discuss the implications of these apparent absences as well as of the complement of adhesion genes (the echinoderm adhesome) that we were able to annotate.
METHODS
Gene Model Improvement Methods
When possible, we used the interim BAC-based assembly (BAC-WGS) to improve upon original GLEAN3 gene models. The original GLEAN3 models were mapped to the BAC-WGS using blat. Scaffolds containing GLEAN3 models of interest were used as query sequence in a BLAST search of a database containing all known proteins related to the one being annotated. New gene models based on BAC-WGS were created using Geneid (Human3iso parameters; Parra et al., 2000) and Genewise (using best BLAST hits as protein substrate; Birney et al., 2004). All these data were visualized by SMART (Letunic et al., 2006) and/or pfam (Finn et al., 2006) and plausible gene models were created using argo (http://www.broad.mit.edu/annotation/argo/). In general, an attempt was made to incorporate into the final gene model all exons predicted by the various gene-finding algorithms that were supported by relevant BLAST hits. Non-canonical splice junctions were allowed in order to repair open reading frames. The protein sequences of substantially modified gene models are provided in Supplemental Material 17.
Phylogenetic Analysis
Sequences were aligned using clustalX (Chenna et al., 2003) with the Gonnet250 protein weight matrix. Alignments were manually inspected and only regions of high quality alignment were considered for downstream analysis. Positions containing gaps were stripped from the alignments using GAPSTREEZE (http://hiv-web.lanl.gov/content/hiv-db/GAPSTREEZE/gap.html). For the α and β integrins, phylogenetic analysis was performed using the proml, protpars and fitch programs from the PHYLIP package (Felsenstein, 2005). Other phylogenetic analyses were done with the Neighbor Joining method included in clustalX, gaps were excluded and the Kimura correction for multiple substitution was employed. Cladograms were created using N-J plot (Perrière and Gouy, 1996) and unrooted trees were created using the PHYLIP program Drawtree. Reported bootstrap values correspond to the number out of 100 bootstrap trials that support the indicated branch.
Analysis of Embryonic Expression
Samanta et al., 2006 used a whole-genome tiling array to identify regions of the sea urchin genome likely to be transcribed during embryonic development. Probes that overlapped with gene models annotated by our group were identified and an average intensity value for each gene was obtained by dividing the sum of the signals from all probes overlapping a gene by the total number of overlapping probes. These data are presented in Suppl. Table 18. Background hybridization yields a signal of approximately 1.5 or less and any gene with an average value over 4 is likely to be expressed in embryos.
RESULTS
Integrins
β Integrin Subunits
The sea urchin genome has predictions for 4 β subunits; βC, βG, βL and βD (Suppl. Table 1). Three of these subunits were known from cDNA cloning (Marsden and Burke, 1997; Murray et al, 2000); βD is novel. All of these β subunits are expressed in embryos and are reported from EST projects on adult coelomocytes. Embryonic expression of βC and βD begins during cleavage, and the βC and βD proteins are expressed by blastodermal cells (Burke et al, 2004; and unpublished). The βG and βL subunits are expressed during gastrulation in a range of cell types including mesenchyme (Marsden and Burke, 1997). The majority of integrin β subunits have at least one cytoplasmic NPxY motif that supports intracellular binding to talin and the actin cytoskeleton. Sea urchin βC, βD, βG and βL each have two of these motifs suggesting that they all bind talin via their cytoplasmic domains.
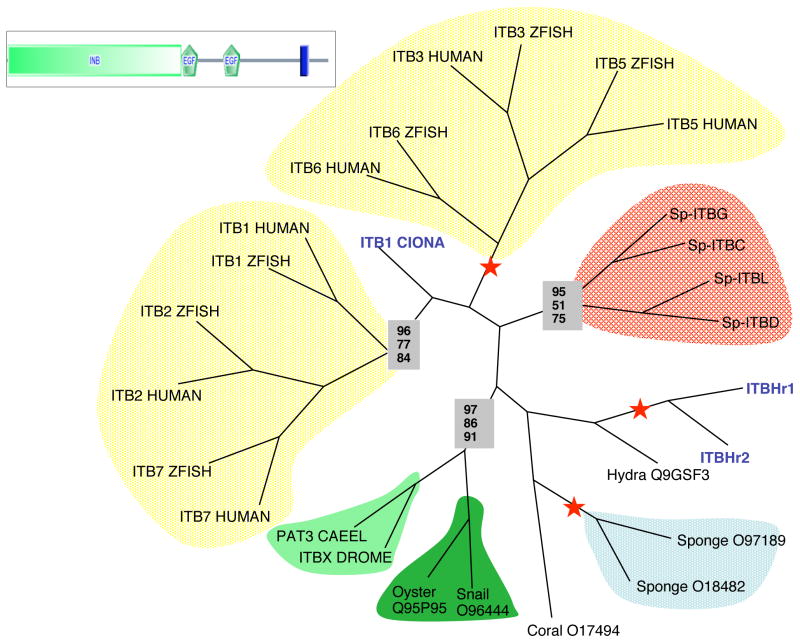
Genomic analyses have revealed integrin β subunits from vertebrates (8–9 genes), ascidians (5 genes), flies (2 genes), and nematodes (1 gene). In addition, there are cDNA clones for β subunits from a wide range of other metazoans. These subunits were combined with the four sea urchin molecules and phylogenetic analyses were performed. The results suggest a taxon-specific radiation of integrin β subunits in sea urchins (Figure 1). Vertebrate and protostome β subunits cluster in respective clades and β integrins from diploblastic organisms root at the base of the tree. (Ewan et al, 2005) reported an analysis of ascidian β subunits and suggested that Ci_b1 is the ascidian version of a deuterostome-specific β1-like subunit that has undergone independent radiations in urochordates and vertebrates. Our analysis of the sea urchin β subunits also demonstrates the occurrence of subphylum-specific expansions of β subunits but there is no evidence to support a grouping of Ci_b1 and the sea urchin βs with vertebrate β1. Vertebrates possess two clades (β1/2/7 and β3/5/6) plus two more divergent subunits (β4 and β8). This complement also fits the pattern of taxon-specific expansions of integrin β subunits but it is difficult to identify a true common orthologous primordial β subunit.
Figure 1. Phylogeny of Integrin β Subunits.
Phylogenetic analysis of NPxY motif-containing β integrin subunits (i.e., excluding β4 and β8 from vertebrates) was performed using the PHYLIP programs fitch (distance-based), protpars (maximum parsimony) and proml (maximum likelihood). The aligned portion of the β subunits is indicated by the domain diagram. Red stars indicate 100% bootstrap support for the branch in each method. Bootstrap support out of 100 for stable branches is indicated within boxes: fitch results are on top, protpars in the middle and proml on the bottom. Clades of vertebrate betas are highlighted in yellow, echinoderm in red, porifera in blue and protostomes in shades of green (lophotrochozoans in dark green, ecdysozoans in light green). Urochordate beta integrins are in blue text; not shown; all members of the urochordate-specific clade of β subunits containing ITBHr1/2 (Ewan et al, 2005; our unpublished analyses). For information about the SMART domains presented in this figure, see Suppl. Figure 16.
The vertebrate β4 subunit is distinctive structurally in having a large cytoplasmic domain that associates with intermediate filaments. The ascidian genome reveals that this is a shared feature of chordates (Ewan et al, 2005; our unpublished analyses). There is no similar subunit predicted from the sea urchin genome, suggesting that the β4 subunit is a derived, shared feature of chordates.
α Integrin Subunits
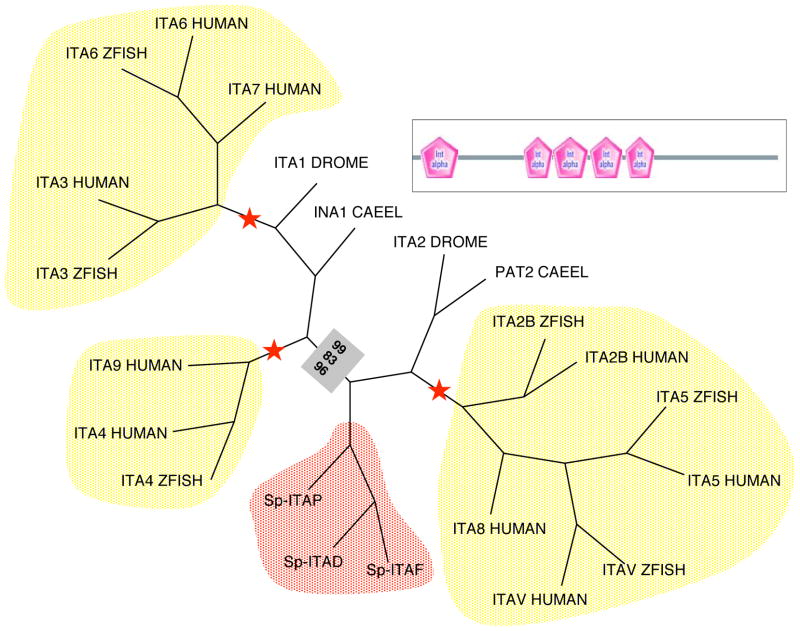
There are 8 gene predictions for α integrin subunits from the current assembly of the S. purpuratus genome – many of these predictions are incomplete because of the low conservation of α integrin subunits (Suppl. Table 2). One group comprises αD, αF, and αP; the αP subunit cDNA sequence was previously reported from two species of sea urchins (αSU2, Hertzler and McClay, 1999; Susan et al. 2000). In phylogenetic analyses, this group of subunits clusters with the clade including α5, αV, α8 subunits and with two protostome integrins, albeit with rather low bootstrap values (Figure 2). Thus, there appears to be an expansion of the RGD-binding sub-family of α integrins. Integrin subunits αD and αF are tandem genes that share a higher degree of similarity than other members of this clade. The vertebrate α integrins also have an expansion in the RGD-binding clade, but the ascidians do not (Ewan et al, 2005; our unpublished analyses). It is unclear what the potential RGD ligands for these receptors might be. Candidates include sea urchin homologs of nidogen, thrombospondin, perlecan, fibrillar collagens and laminins, many of which have RGD sequences. Curiously, the αP subunit has previously been shown to be expressed in basolateral domains of blastomeres and when expressed in mammalian cells it mediates adhesion to laminin, but not fibronectin, or collagen (Hertzler and McClay, 1999). There is a weak association of the αC subunit with vertebrate α3, 6, and 7, the laminin-binding subunits, but this is supported by a very low bootstrap value (data not shown). It is possible that this unconvincing association may improve with a more complete and accurate sequence. The phylogenetic relationships of the four remaining α subunits (αG, αK, αH, αJ) to known α integrins are unclear due to incomplete information.
Figure 2. Phylogeny of Integrin α Subunits.
Phylogenetic analysis of selected sea urchin and other non-VWA domain-containing α integrin subunits was performed using the PHYLIP programs fitch (distance-based), protpars (maximum parsimony) and proml (maximum likelihood). The aligned portion of the α subunits is indicated by the domain diagram. Only three out of the eight sea urchin alpha gene predictions are included in these analyses because it was not possible to obtain sufficiently accurate gene models representing the other five. Red stars indicate 100% bootstrap support for the branch in each method. Bootstrap support out of 100 for stable branches is indicated within boxes: fitch results are on top, protpars in the middle and proml on the bottom. Note the echinoderm-specific clade (red), distinct from, but proximal with, the vertebrate RGD-specific clade (5/8/IIb). Other clades are the laminin-specific(3/6/7) and vertebrate-specific (4/9) subunits. For information about the SMART domains presented in this figure, see Suppl. Figure 16.
The VWA/I domain-containing integrins are a distinctive group of collagen-binding and leukocyte integrins, including the complement and phagocytic receptors of vertebrates (Whittaker and Hynes, 2002). Analysis of the ascidian genome indicates that there is a deuterostome-derived clade of VWA/I domain containing α subunits (Ewan et al. 2005, Huhtala et al. 2005; our unpublished analyses). However, our analysis of the sea urchin genome revealed no sea urchin α subunits that contain a VWA/I domain, making this appear to be a chordate innovation. Members of the α4/α9 clade are absent from ascidians and appear to be absent also from sea urchins; this appears to be a vertebrate-specific expansion.
Phylogenetic analyses of α subunits have previously identified a small number of integrin subunits (a common β subunit and two α subunits, one apparently RGD-specific and one laminin-binding) that appear widely shared in the bilaterian metazoans (Hynes and Zhao, 2000; Hynes, 2002). These evolutionarily ancient subunits appear to have been supplemented by diversification of taxon-specific clades. The addition of the α subunit predictions from the ascidian and sea urchin genomes has made this analysis somewhat less clear. While additional taxon-specific clades occur in both, clear members of the presumed basal RGD- and laminin-binding families are less well defined in sea urchins. As mentioned, this could be a consequence of the low homology and incomplete nature of some of the gene predictions and further research using complete cDNA sequences will be needed to clarify the situation.
Cadherins
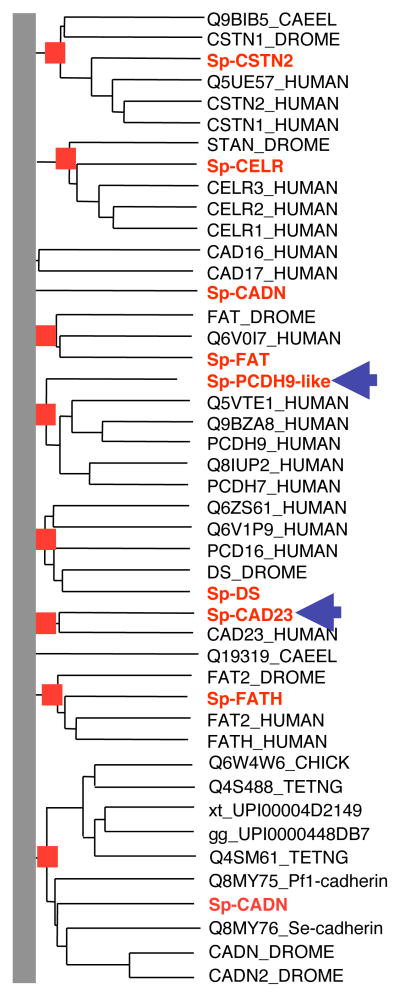
The complement of cadherins in the sea urchin genome is surprisingly small – fewer than a dozen well defined representatives; fewer even than flies or worms (Suppl. Table 3). This limited set of cadherins has some surprising features. There are only two genes encoding catenin-binding cytoplasmic domains characteristic of typical classical type I and atypical type II cadherins of vertebrates and neither of the sea urchin genes encodes such a typical vertebrate-type classical cadherin. One of these two sea urchin catenin-binding cadherins (Sp-CADN) is orthologous to Lytechinus variegates G-cadherin (Miller and McClay, 1997) and is most like protostome-type cadherins, containing LamG and EGF domains as well as cadherin domains (Figure 3). It turns out that non-mammalian vertebrates (fish, amphibians and birds) do have such cadherins, termed type III cadherins by Tanabe et al (2004). Therefore, Sp-CADN represents a class of cadherins found in both protostomes and deuterostomes but missing in mammals – an unusual phylogenetic distribution. The second sea urchin catenin-binding cadherin appears to be an echinoderm ortholog of Dachsous/protocadherin 16 (found in bilateria). The sea urchin genome also contains apparent orthologs of other large cadherin superfamily molecules common to protostomes and deuterostomes (bilateria); namely, fat, fatH, flamingo/CELSR and calsyntenin (Figures 3 and 4). In contrast, two sea urchin cadherins, sp_CAD23 and sp_PCDH9-like, both good homologs of vertebrate counterparts, were previously known only in chordates (Figure 4, blue arrows). Sp_PCDH9-like is homologous with the vertebrate protocadherins 1, 7, 9, 11 and 20. Four other gene predictions with cadherin domains, several with only one or two domains, are difficult to assign clear orthologies, although several give protocadherin 15 as their top BLAST hits (see below). Figure 3 shows some representative examples of the sea urchin cadherin gene set and Figure 4 shows the phylogenetic analysis.
Figure 3. Representative Sea Urchin Cadherins.
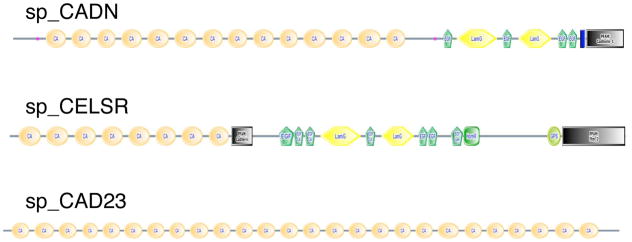
The figure shows domain structures for three of the sea urchin cadherins (see Suppl. Table 3 for the complete set). Sp-CELSR is an example of a structure shared with both major classes of bilateria (protostomes and deuterostomes). Sp-CADN is a cadherin with a structure common in protostomes, present in lower vertebrates (fish, amphibia and birds) but absent from mammals. Sp-CADH23 is an example of a deuterostome-specific cadherin with important functions in vertebrates (see text). For information about the SMART domains presented in this figure, see Suppl. Figure 16.
Figure 4. Phylogenetic Analysis of Sea Urchin Cadherins.
Sea urchin cadherins were combined with insect, nematode and a subset of human cadherins. The excluded human cadherins were in clades that clearly lacked an echinoderm homolog; these included the desmosomal cadherins, vertebrate classical/Type-I cadherins, atypical/Type-II cadherins and the clustered protocadherins (Nollet et al., 2000). Information at the root of the tree was discarded for the purpose of presentation (grey bar). Clades with 100% bootstrap support are indicated by red boxes. The 8 named sea urchin cadherins are in red. Sp-CAD23 and Sp-PCDH9-like (indicated by blue arrows) are homologous to cadherins previously found only in chordates. The remaining sea urchin cadherins are homologous with protostome/deuterostome cadherins.
Overall, this complement of cadherins is more invertebrate than vertebrate in character and major vertebrate-specific expansions (classical type I and atypical type II catenin-binding cadherins, CNR-type protocadherins, intermediate filament-binding cadherins – desmocollins and desmogleins) are all absent. The notable exceptions to this generalization are sp_PCDH9-like and Sp-cadherin 23 (and perhaps protocadherin 15), which will be discussed further below. It would appear that there has not been a major expansion of cadherin-mediated cell-cell adhesion in echinoderms (at least in S. purpuratus).
Ig Superfamily and FN3 domain adhesion receptors
A major class of cell adhesion receptors comprises those including immunoglobulin superfamily (IgSF) and/or fibronectin type III (FN3) domains. The sea urchin genome contains large numbers of such predicted genes (Suppl. Tables 4–6). The gene predictions include over 300 containing FN3 domains and around 400 containing IgSF domains. Given that many of these predictions are likely incomplete and/or inaccurate, it is not feasible to provide here a complete analysis of these gene sets. The tables contain organized lists of some of the more informative gene predictions – in addition to those listed there are almost 200 additional gene predictions that contain Ig domains alone (190) and over 100 with FN3 alone (137) – such genes are not straightforward to analyze and are not included here. Similarly, the myriad predicted genes (nearly 700) with EGF repeats are difficult to sort out and the tables include only those in which EGF repeats occur together with other adhesion domains in interpretable patterns (see Suppl. Tables 4–6).
There are around 80 predicted genes that contain various combinations of both IgSF and FN3 domains (Suppl. Table 4). Among these genes are good homologs of many previously described adhesion receptors (including N-CAM, DsCAM, DCC/neogenin and Robo, see Suppl. Table 4) based both on domain organization and phylogenetic trees. There are, in addition, sets of Ig/FN3 proteins that do not have close homologs in other taxa and appear to be taxon-specific expansions and there are further sets of proteins that have previously unreported combinations of Ig, FN3 and EGF domains (see Suppl. Tables 4 and 6). In addition to the genes listed in the tables, there are about two dozen receptor protein tyrosine kinases and ~18 receptor protein tyrosine phosphatases containing IgSF and/or FN3 domains (see articles by Bradham et al., LePage et al. and Byrum et al. in this issue).
LRR-containing adhesion receptors
There is a wide diversity of LRR receptors – both Tlrs (see Hibino et al., this issue) and others (Suppl. Tables 5, 6, 7). There are about 20 LRR/Ig receptors – more than in vertebrates - and other proteins with novel combinations of LRR repeats with kringle (SPU_010102) or SR domains (SPU_006659 and SPU_023934). Particularly notable is a large set of GPCRs with LRR repeats in their N-terminal extracellular domains (Suppl. Table 7). Preliminary analyses suggest around 40 of these receptors, more than in vertebrates. Several of these proteins appear related to the relaxin receptor (LDLa/LRR/7tm1) or other LRR/7tm1 receptors found in both protostomes and deuterostomes, but others have novel mixtures of additional domains not found elsewhere. One LRR-GPCR (SPU_012494) has a secretin-type 7TM segment (see below).
Adhesion-related GPCRs
This LRR/GPCR gene (SPU_012494), is a member of a group of about 90 GPCRs, all apparently sharing a secretin-type 7TM segment (7tm_2), usually with a membrane-proximal GPS cleavage site domain (Suppl. Table 8). This type of receptor is rare in protostomes, being confined to CELSR/flamingo, and a few lectin-containing forms. In the sea urchin genome there is a wide diversity of extracellular adhesion domains associated with this family, far more diverse and numerous than the vertebrate family of LNB-7TM receptors, which numbers around 36 in mammals (Fredriksson et al., 2003; Kwakkenbos et al., 2004). Examples of some of the novel structures found in sea urchins are shown in Figure 5. It is unclear whether or not these receptors are, in fact, involved in cell adhesion, although their domain structure certainly suggests that they are. The situation is similar for the vertebrate LNB-7TM receptor family – whatever the true function of these intriguing receptors in deuterostomes, it appears that sea urchins have a large taxon-specific expansion of this family. One of these genes (SPU_027371) encodes VLGR1, mutations in which are involved in human deafness syndromes (Weston et al, 2004; see further discussion below).
Figure 5. Representative Sea Urchin Adhesion GPCRs.
The examples depict novel domain architectures not previously reported in this class of GPCRs, which is specific to deuterostomes. Examples with and without GPS cleavage domains are shown – all have 7tm-2 transmembrane segments similar to secretin-type receptors. The full set of sea urchin members of this family is listed in Supplementary Table 8. For information about the SMART domains presented in this figure, see Suppl. Figure 16.
Basement membrane components and laminins
The entire basic “basement membrane ECM toolkit” common to protostomes and deuterostomes is present – two αIV collagen genes, laminin subunits, nidogen, perlecan, collagen XV/XVIII (Figure 5). The two αIV collagen genes are linked head-to-head as in most other taxa, suggesting coordinate regulation. There does not appear to be much expansion of these gene families in sea urchins. Thus, there are no additional αIV collagen genes as found in vertebrates and only a few additional laminin subunits (a single additional α and β subunit; see Suppl. Table 9), which contrasts with the large number of laminin α and β subunits in vertebrates. This general lack of elaboration of this gene set suggests less basement membrane diversity in sea urchins than in vertebrates.
Collagens
In addition to the basement membrane collagens (IVα1, IVα2, XV/XVIII), there are a number of fibrillar collagens. There are at least two, possibly as many as four, fibrillar collagen genes of the I/II/III type plus probably a pair of type V-like fibrillar collagen genes (see Suppl. Table 10). None of these represents a particular surprise, since collagens of all these types have been reported previously in protostomes and deuterostomes (including echinoderms). There is a representative member of a short collagen family with a C-terminal OLF domain, a structure reported in vertebrates and insects, and a homolog of the vertebrate-specific short collagen with an EMI domain (also known as collagen 26). However, in contrast, there appear to be no clear examples of the complex collagens prevalent in vertebrates (types VI, VII, XII, XIV, containing VWA and FN3 domains), although one sea urchin collagen gene (sp_col91) has some features of this family (see Suppl. Figure 11). There are several C1q homologs, although none has enough collagen repeats to be considered a homolog of collagen VIII or X and none contains an EMI domain as in emilin (an elastin linker protein of vertebrates). Thus, overall, the collagen complement of S. purpuratus is much simpler than that of vertebrates. Most of the collagen genes are common to many metazoans and only a few appear to be deuterostome-specific.
Neural adhesion
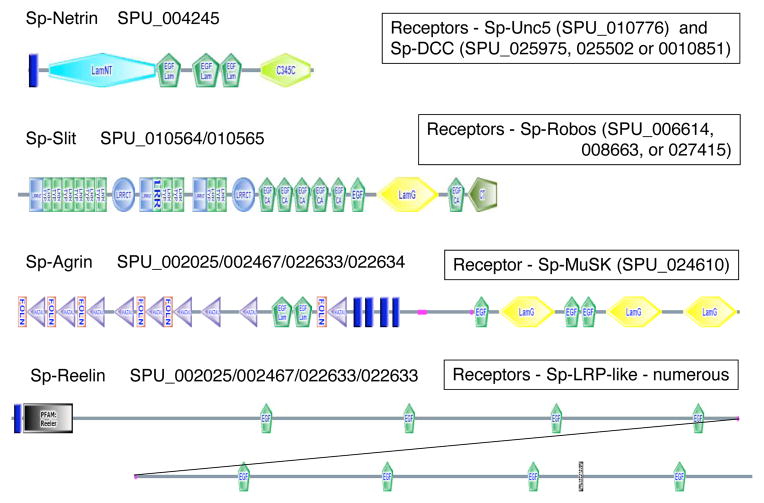
The sea urchin genome encodes good homologs of the axonal guidance matrix proteins, netrin and slit, and of the postsynaptic clustering matrix protein, agrin (Figure 7). That is not surprising because those are known in both protostomes and deuterostomes. In each of these cases, good candidates for the receptors for these axon guidance molecules are also found in the sea urchin genome (see Figure 7 for listings). Netrin is encoded in a single gene prediction, but the other proteins are each encoded by several initial gene predictions, because of the large size of their genes. In the cases of slit and agrin, linkage was established and additional analyses allowed development of the gene predictions shown in Figure 7. However, because of the state of genome assembly and the presence of haplotype pairs, these models will require validation by cDNA analyses. Nonetheless, it seems clear that close matches with the orthologs in other species are present in sea urchins and, in each case, it appears that there is a single haplotype pair – that is, there is no expansion of the gene families as seen for netrins and slits in vertebrates.
Figure 7. Neural Adhesion Systems.
The structures shown represent best estimates of the gene structures for four key ECM proteins involved in axonal guidance (netrin, slit), synapse formation (agrin) and neuroblast positioning (reelin). These predictions are based on the gene predictions listed, linkage in the current interim assembly and additional Genewise predictions on the scaffolds containing these predictions. Given the current state of the genome assembly and the presence of haplotype pairs, these predictions must be viewed as provisional and need confirmation by cDNA analyses. Nonetheless, it seems clear that good orthologs of each of these genes exist in the sea urchin genome. Also listed are the potential cellular receptors for each of these proteins. For information about the SMART domains presented in this figure, see Suppl. Figure 16.
The urchin homolog of agrin has FOLN/KAZAL, EGF and LAMG domains but, like other invertebrate agrin homologs, it is lacking NtA and SEA domains. MuSK is a transmembrane kinase receptor that functions as a receptor for Agrin in vertebrates and mediates acetylcholine receptor clustering in neuromuscular synapses. The sea urchin homolog contains Ig2, Fz, TM, and kinase domains and is predicted by SPU_024610. Agrin appears in an EST library from primary mesenchyme, suggesting that these adhesion molecules may have non-neural functions in sea urchins. Whereas agrin/MuSK mediate postsynaptic clustering, in vertebrates, β-neurexin binds neuroligin and is clustered to recruit pre-synaptic components (Washbourne et al. 2004). In sea urchins there is a single incomplete prediction (SPU_024416) for a neurexin containing six LamG domains. There are several predictions for neuroligins, the postsynaptic receptors for neurexins (SPU_015750, SPU_021785 and SPU_021786). These synaptic adhesion components are known from other invertebrates.
Reelin, a large extracellular matrix protein that affects organization of neurons in the mammalian brain, is mutated in the reeler mouse and mutations in the reeler gene in humans have been associated with Norman-Roberts-type lissencephaly syndrome. Reelin has a unique domain composition and organization (Reeler, EGF, BNR) that has not previously been found outside chordates but the sea urchin genome includes a very good homolog of reelin (Figure 7). ESTs indicate that the predicted gene is represented in early embryonic cDNA libraries and possibly expressed by primary mesenchyme. This suggests that reelin-like ECM molecules are not restricted to neural tissues in echinoids. A number of good candidate reelin receptors (LRP/LDLR-related, containing LDLa and LY domains) are also encoded in the urchin genome. The human homologs of these receptors are also involved in human disease (atherosclerosis).
Semaphorins and plexins play important roles in axon guidance in both protostomes and deuterostomes and also in vascular, cardiac and skeletal muscle development, as well as immune responses, tumor growth and metastasis in vertebrates (Behar et al. 1996; Hall et al. 1996; Miao et al. 1999; Christensen et al. 1998). There are at least two semaphorin genes in the sea urchin genome. Sp-SemaA (SPU_022057) has the domain structure of a Class 5 protein, Sp-SemaB (SPU_010703) is most like a Class 6 protein and two gene models (SPU_001892, SPU_010331) have semaphorin-like Sema domains but there is insufficient evidence to classify them in more detail (Suppl. Table 12; Semaphorin Nomenclature Committee, 1999). Thus, members of what have, until now, been considered vertebrate classes of semaphorins occur in invertebrate deuterostomes. The chordate receptors for semaphorins are homo- and heterodimeric complexes of neuropilins and plexins. The class 3 or soluble semaphorins bind homodimers of neuropilin whereas the transmembrane and GPI-anchored semaphorins bind heterodimers of a plexin and a neuropilin (Tamagnone et al. 1999). More than 10 vertebrate plexins are known and they are grouped into 4 subfamilies and there are 2 plexins in Drosophila (Fujisawa et al. 2004; Tamagnone et al. 1999). Semaphorins, plexins and Met receptors all contain a single Sema domain defined by a conserved set of cysteine residues, which form four disulfide bonds thought to stabilize a beta propeller-like structure. In S. purpuratus, there are about 30 distinct genes encoding proteins that contain Sema domains (Suppl. Table 12). Of these, four are the semaphorins and semaphorin-like genes mentioned above and the remaining 26 appear to be plexin-like based on phylogenetic analysis of the Sema domains alone (not shown). Among these 26 proteins, 17 fall in a closely related clade. These 17 are, on average, 92% identical to one another at the nucleotide level based on blat alignments (Kent, 2002) of the 17 mRNAs to the WGS-BAC assembly. The remaining 10 Sema-containing gene models are likely to represent at least 7 distinct genes, one of which may be an example of a novel Sema-containing domain architecture (GLEAN3_16041). This appears to be a substantial expansion in the number of genes containing this domain. A more detailed analysis of plexins in S. purpuratus requires better genome assemblies and cDNA sequences to determine relationships within this important group of axon guidance molecules. In contrast with this expansion of the plexin family, there are no clear homologs of neuropilins. Neuropilins also appear to be absent from the ascidian genomes and may be a vertebrate innovation.
In conclusion, the expected axonal guidance molecules - netrin, slit and their receptors, semaphorins and plexins - are present in sea urchins. The presence of reelin was unexpected. Synaptic adhesion systems - agrin/MuSK and neurexin/neuroligins - are present, again as expected. However, it is notable that, in vertebrates, cadherins and protocadherins both apparently play important roles in organization of the CNS and those families of genes have been greatly expanded in vertebrates. A similar expansion is not observed in sea urchins and, indeed, the linked clusters of protocadherin/CNR genes found in humans and mice are completely absent in the sea urchin genome. This would be consistent with their having evolved in higher deuterostomes in connection with the elaboration of the CNS.
Other ECM Proteins
Among the other matrix proteins that are present are thrombospondins (TSPs, both A and B subclasses), fibrillins, fibulin, osteonectin/SPARC, hemicentin and polydom (Table 1 ECM). Homologs of these matrix proteins are found in protostomes and deuterostomes, so their presence in sea urchins is not unexpected. In some cases, the sea urchin proteins have novel characteristics.
Table 1.
ECM proteins
| Present | Gene predictions | protostomesdeuterostomes | |||
|---|---|---|---|---|---|
| Thrombospondins | Sp-TSPA1 | SPU_001638 + SPU_023469 | A-type | N | Y |
| Sp-TSPB1 | SPU_012936 | B-type | Y | Y | |
| Sp-TSP_like-1 | SPU_022667 + SPU_017370 + SPU_013106 | ||||
| Sp-TSP_like-2 | SPU_009755 | ||||
| Fibrillins | Sp-Fibrillin A | SPU_001532 + SPU_001533 + SPU_020166 + SPU_021495 | Y | Y | |
| Sp-Fibrillin B | SPU_012550 | Y | Y | ||
| Osteonectin/SPARC | SPU_002473 + SPU_028275 | Y | Y | ||
| Hemicentin | Sp-Hemicentin | SPU_011693 | Y | Y | |
| Polydom | Sp-Polydom | SPU_022974 | Y | Y | |
| Fibulin | Sp-Fibulin | SPU_026629 + SPU_021630 | Y | Y | |
| Nidogen | Sp-Nidogen | SPU_016055 | missing N-term | Y | Y |
| Netrin | Sp-Netrin | SPU_004245 | Y | Y | |
| Slit | Sp-Slit | SPU_010564, SPU_010565 | Y | Y | |
| Agrin | Sp-Agrin | SPU_002025, SPU_002467, SPU_022633, SPU_022634 all on Scaffold 675 | Y | Y | |
| Reelin | Sp-Reelin | SPU_013071, SPU_008268, SPU_023409 Genewise prediction | N | Y | |
| Missing | Comments | protostomes | vertebrates |
|---|---|---|---|
| Fibronectin | No clear FN1 or FN2 domains | N | Y |
| von Willebrand factor | No VWA/VWD domain combination | N | Y |
| tenascins | No appropriate mix of EGF, FN3 and FBG domains | N | Y |
| cochlin | No clear LCCL domains | N | Y |
| vitrin | No clear LCCL domains | N | Y |
| matrilins | No VWA/EGF domain combinations | N | Y |
| Vitronectin | No SO/HX domain combinations | N | Y |
| VWA/FN3 collagens | No appropriate domain combinations | N | Y |
| LINK proteoglycans | No LINK domains coupled with other signature domains | N | Y |
In the case of the thrombospondins, sea urchins have two genes encoding thrombospondins with a TSPN domain at the N-terminus. One of these proteins also contains TSP1 repeats and is, therefore, a subgroup A TSP (Suppl. Table 13; Adams and Lawler 2004). Subgroup A thrombospondins were previously reported only from chordates but the presence of a TSP-1/2 homolog in sea urchins indicates that this is a deuterostome-specific trait. The other TSPN-containing TSP in urchins is a homolog of chordate-specific subgroubB TSPs; all TSPs from protostomes lack this N-terminal domain.
There are two genes encoding fibrillin homologs that cluster with vertebrate, honeybee and ascidian fibrillins (Suppl. Figure 14). The Latent-TGF-β binding protein class of fibrillin-related proteins is absent from both echinoderms and ascidians and may be a vertebrate-specific expansion There are 3 fibrillin genes in mammals (FBN-1, FBN-2, FBN-3) and a single fibrillin gene in C. elegans (fbn-1) and Drosophila (CG31999). The two fibrillin genes predicted from the sea urchin genome suggest a modest expansion of this family in deuterostomes. Mutations in fibrillin in humans cause Marfan syndrome (FBN-1) and the related disease, contractural arachnodactyly (FBN-2). EST data indicate that fibrillinA is expressed during cleavage and by primary mesenchyme cells, suggesting that it may be one of the fibrillar components of the blastocoel extracellular matrix that has ultrastructural features of fibrillin fibrils (Hardin, 1996).
In contrast, the following matrix proteins appear to be absent – fibronectin, tenascins, von Willebrand factor, vitronectin, matrilins, cochlin/vitrin, complex VWA/FN3 collagens (see Table 1). There may be a connection here with the absence of a vertebrate-type vasculature lined with endothelial cells, since some of these vertebrate-specific matrix proteins (fibronectin, von Willebrand factor, vitronectin) are involved in vascular adhesion functions. Related to that also may be the apparent absence of VWA/I domain integrins and selectins – involved in leukocyte/endothelial adhesion and traffic. Another vertebrate-specific function that also involves extensive cell-matrix adhesion is neural crest migration and that process might be another reason for the appearance of these proteins higher in the deuterostome lineage than echinoderms. Undoubtedly, some of these missing matrix proteins are involved in structural connective tissue, including cartilage, bones and teeth.
Proteoglycans
Proteoglycans (PGs) are also major components of the extracellular matrix involved in adhesion, motility, proliferation, differentiation and axon pathfinding (Kramer and Yost, 2003; Cattaruzza and Perris, 2005; Hacker et al., 2005; Holt and Dickson, 2005). PGs consist of core proteins decorated with one or more long repeating disaccharide chains called glycosaminoglycans (GAGs). GAGs are heavily sulfated molecules that interact with extracellular cytokines, growth factors and morphogens. Variations in GAG sulfation patterns can modulate their affinities for extracellular proteins (Mulloy, 2005). The status of GAG sulfation therefore plays an important role in regulating the function of PGs. Most of the enzymes necessary for synthesis and modification of the various GAGs are present in the sea urchin gene set (Suppl. Table 15). The chondroitin glucuronate C5-epimerase (EC 5.1.3.19) that transforms chondroitin to dermatan is not present, but NCAG1 – a possible dual epimerase and sulfotransferase involved in dermatan sulfate synthesis - was found (Maccarana et al, 2006). Additionally, neither of the beta-1,4-galactosyltransferases (EC 2.4.1.38) that initiate synthesis of keratan from N- or O-glycans (β4Gal-T1 and β4Gal-T4, respectively) were identified. Intriguingly though, the enzymes involved in subsequent keratan sulfation are present. There is documented evidence for the existence of GAGs in the urchin embryo; Nakatsuji and Lovtrup showed the existence of dermatan sulfate, heparan sulfate and keratan sulfate in the embryo (Nakatsuji and Lovtrup, 1978). Moreover, an antibody against chondroitin sulfate (CS-56) preferentially stains the lumen of the archenteron (Lane et al., 1993).
The predicted sea urchin gene set is missing clear homologs of the lectican and small leucine-rich PG core protein families characteristic of vertebrates. The hyaluronan-binding LINK domain characteristic of many vertebrate PGs is not found in protostomes and is apparently very rare in the sea urchin genome (two gene predictions only) and is not found in domain combinations diagnostic of proteoglycans. Moreover, SPOCK/testican (a SPARC-related PG) seems to be absent from the echinoderm genome while homologs of SPARC and SMOC (another SPARC-related PG) are present. SPOCK/testican is also absent from the cnidarian, protostome and urochordate genomes sequenced to date, suggesting it is a vertebrate-specific gene. However, basement membrane PGs (perlecan, bamacan, agrin and SMOC) and membrane-associated PGs (betaglycan, dystroglycan, syndecan and glypicans) are present (Suppl. Table 15). The sea urchin has one syndecan and two glypican genes, one of the 1/2/4/6 class and one of the 3/5 class. These are expressed in distinct but overlapping radially symmetrical patterns in the S. purpuratus gastrula embryo as determined by in situ hybridization (K.F.B., unpublished). A similar pattern of syndecan expression was observed in Anthocidaris crassispina urchin blastula and gastrula embryos (Tomita et al., 2000).
Thus PGs with GAGs are present and expressed in sea urchins as in other bilateria - in basement membranes and as part of signal transduction systems - but large structural PGs are absent. This presumably correlates with the absence of hyaline cartilage and bone.
Echinoderm homologues of human deaf/blindness genes
One of the more surprising and striking observations to emerge from the analysis of potential adhesion genes/proteins encoded by the S. purpuratus genome is the discovery of a set of genes encoding homologs of human deaf/blindness genes. Human Usher syndromes are genetic diseases affecting hearing, balance and retinitis pigmentosa (retinal photoreceptor degeneration). Most of the genes associated with Usher syndrome have been identified and they encode a set of membrane and cytoskeletal proteins that have been shown to form an interacting network that controls the arrangement of mechanosensory stereocilia in hair cells of the mammalian inner ear (El-Amraoui and Patit, 2005; Adato et al, 2005a; van Wijk et al, 2006; Gillespie et al, 2006). Mutations in these genes in humans or mice lead to disorganization of the stereociliary bundle in the hair cells. These mutations also lead to progressive degeneration of photoreceptors of Usher syndrome patients and many or all of the same proteins play some roles in photoreceptor organization and/or maintenance. Remarkably, good homologs of virtually the entire set of membrane and cytoskeletal proteins of the Usher syndrome network are found in the sea urchin genome (Table 2). These include the very large membrane proteins, usherin (Adato et al, 2005b) and VLGR1 (Weston et al. 2004) and large cadherins (Cadh23 and possibly Pcad15), all of which participate in forming various links between stereocilia in mammalian hair cells (Figure 8). Also included are myosins 7 and 15, two PDZ proteins (harmonin and whirlin) and another adaptor protein (SANS), which participate in linking these membrane proteins to the cytoskeleton (Hoffman et al, 2006, this issue). In addition, two membrane transporters, NBC3 (a candidate Usher syndrome target known to interact with harmonin) and TrpA1 (the likely mechanosensory channel connected to the tip links containing cadherin 23), have been implicated in the same system. Both of these proteins also have homologs in the sea urchin genome (see Table 2).
TABLE 2.
Human Disease Genes with Homologs in Sea Urchins
| Protein | Characteristics | Human mutation | Mouse mutation | Urchin homologue | |
|---|---|---|---|---|---|
| Usherin | Large adhesion-type receptor, forms links between stereocilia | USH2A | SPU_020012 plus SPU_017733 Now assembled |
Figure 8 | |
| VLGR1 | Large adhesion-type receptor, forms links between stereocilia | USH2B | SPU_027371 | Figure 8 | |
| Cadherin 23 | Large adhesion receptor, forms tip links between stereocilia | USH1D | Waltzer | SPU_027133 SPU_013476 SPU_014606 SPU_015210 SPU_016980 SPU_004556 |
Figures 3, 4 & 8 Suppl. Table. 3 |
| Protocadherin 15 | Large adhesion receptor, forms links between stereocilia | USH1F | Ames Waltzer | Several possible homologs, full structure unclear | |
| NBC3 | Na/HCO3 cotransporter | Candidate for USH2B | SPU_014036 SPU_017882 SPU_025514 All linked |
||
| TPRA1 | Mechanosensory channel | SPU_010335 | |||
| Harmonin | PDZ adaptor protein | USH1C | Deaf circler | SPU_009191 SPU_028159 |
|
| Whirlin | PDZ adaptor protein | DFNB31 | Whirler | SPU_002363 SPU_021200 |
|
| SANS | ANK/SAM adaptor protein | USH1G | Jackson shaker | SPU_022872 | |
| Myosin 7A | Motor | USH1B | Shaker-1 | SPU_025990 SPU_013161 SPU_022040 |
|
| Myosin 15 | Motor | DFNB3 | SPU_022456 | ||
| Reelin | Brain extracellular matrix protein | Norman-Roberts-type lissencephaly syndrome | Reeler | SPU_013071 SPU_008268 SPU_023409 Now assembled |
Figure 7 |
| Fibrillins | Extracellular matrix proteins | Marfan syndrome and others | SPU_001533 SPU_001532 SPU_020166 SPU_021495 SPU_012550 |
Table 1 | |
| Nephrin | Adhesion receptor – forms slit diphragm in kidney glomerulus | Congenital nephrosis, Finnish type | SPU_024019 | ||
| Perlecan | Basement membrane proteoglycan, coreceptor for basic fibroblast growth factor | Dyssegmental dysplasia, Silverman-Handmaker type; Schwartz-Jampel syndrome | SPU_000937 SPU_012324 SPU_026338 SPU_028620 |
Figure 6 Suppl. Table 12 |
|
| Glypican-3 | GPI-linked proteoglycan, roles in morphogenesis and growth factor signaling | Simpson-Golabi-Behmel syndrome, type 1; Simpson dysmorphia syndrome | SPU_003874 SPU_010593 SPU_013086 |
Suppl. table 12 |
Figure 8. Echinoderm Homologs of Human Deaf/Blindness Genes.
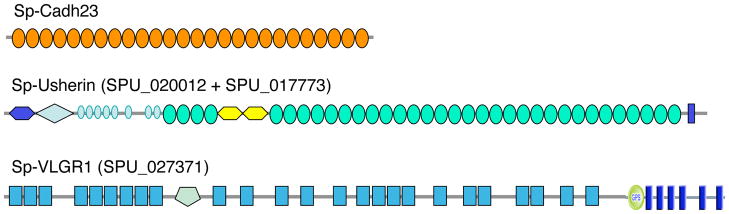
The figure depicts the structures of three large membrane proteins encoded in the sea urchin genome, which are orthologs of human genes involved in organization of stereocilia in hair cells of the ear. As described in the text and listed in Table 2, the proteins encoded by these genes are part of an interacting network of membrane and cytoskeletal proteins that organize the stereocilia in the vertebrate ear. Their presence in sea urchins suggests that echinoderms may use a similar interacting network to organize cellular protrusions, perhaps involved in mechanosensation or other sensory functions. The number and position of the repeated FN3 domains in Sp-usherin and Calx_beta in Sp-VLGR are indicated by parentheses and subscripts. For information about the SMART domains presented in this figure, see Suppl. Figure 16.
Thus, it appears that sea urchins share with mammals an interacting set of proteins, whose most prominent roles in mammals are in mechanosensation in hearing and balance and, at least in some cases, also in organization and maintenance of photoreceptors (modified cilia). Key members of this protein network (at least the large adhesion proteins) are deuterostome-specific. Sea urchins lack ears or eyes, so it is an interesting question whether they deploy these proteins in a concerted fashion as do mammals. The expression of nearly all of these genes can be detected in sea urchin embryos by microarray analysis (Samanta, et al. 2006). An attractive hypothesis is that the same proteins used in vertebrate hair cells and photoreceptor cells may organize stereocilia or sensory cilia of putative mechanosensory organs of echinoderms.
The behavior and anatomical features of echinoderms provide several possible candidate roles for these genes involved in hearing in vertebrates. Sea urchins are photosensitive, are able to sense and maintain body position and respond to a range of mechanosensory stimuli (reviewed by Hyman, 1955; Burke et al., 2006, this issue). The sensitivity to touch of echinoderms is thought to depend on the ciliated epithelium covering the test that includes interspersed cells having apparent sensory hairs. Echinoderm larvae can maintain a favored body orientation and respond rapidly to touch and other stimuli. Each ectodermal epithelial cell of sea urchin embryos and larvae has a motile cilium surrounded by a collar of tightly packed microvilli that may have a role in detecting the direction or extent of the stroke of the cilium (Crawford and Campbell, 1993). Many marine invertebrate larvae and adults have such ciliary collars that may have evolved into the stereocilia of more elaborate sensory systems such as those of vertebrate hair cells. Asteroids and echinoids have several types of pedicellaria, minute jawed appendages that bite and poison ectoparasites and aid in feeding and removal of debris. The cavities of some pedicellariae have hillocks of cells having long cilia thought to be sensory (Hyman, 1955). Adult echinoderms generally keep their oral surface apposed to the substrate and initiate a righting response when displaced from that position. This reaction may be influenced by gravity, but it is considered primarily a tactile response that is sometimes influenced by light. Sea urchins have sphaeridia, stalked bulbs on their tests, that may be organs for equilibrium (Hyman, 1955). Sea cucumbers are able to maintain their body orientation even when burrowing, so this cannot depend only on tactile stimuli. They have statocysts including statoliths in vesicular cavities that are thought to be involved in maintaining their equilibrium in relation to gravity (Hyman, 1955). Many invertebrates have statocysts/statoliths that have a role in geotaxis or body orientation and may have evolved into the macula of the vertebrate vestibular apparatus and eventually into the cochlea involved in hearing. It is, therefore, a plausible hypothesis that echinoderms have mechanosensory stereocilia that may be organized using the same proteins found in vertebrate hair cells or other cells having modified cilia and it would be interesting to test these hypotheses using the information from the genome annotation.
CONCLUSIONS
The echinoderm adhesome includes many hundreds of genes likely to be involved in cell-cell and cell-matrix adhesion. Sea urchins have the basic metazoan adhesion gene set but there are several apparently taxon-specific expansions of cell adhesion molecules. Overall, the echinoderm adhesome has more of an “invertebrate character” – that is, it is more similar to the adhesion gene sets of protostome invertebrates than to that of vertebrates. That said, there are some clear examples of genes/proteins previously “chordate-specific.”
Basic Adhesion Toolkit
The main classes of adhesion receptors (integrins, cadherins, Ig superfamily, FN3, LRR, C-type lectins) are all represented in the S.purpuratus genome. The integrin family is larger than that in insects or nematodes but lacks several chordate-specific expansions (see below). The cadherin gene set is surprisingly small and more like that of protostome invertebrates. The basement membrane “toolkit” (collagens IV, XV/XVIII, laminins, nidogen, perlecan) is present as in all metazoa whose genomes have been analyzed but it is little expanded. The basic set of neural guidance matrix proteins and their receptors is present, as are homologs of many known adhesion receptors used in development of the nervous system (N-CAM, DsCAM, neogenin etc), much as in protostomes.
Taxon-specific Expansions
Several families of adhesion proteins show expansions. Both integrin subunit families include what appear to be echinoderm-specific expansions, as is characteristic of other taxa. There are subsets of Ig/FN3 adhesion receptors that appear to be echinoderm-specific expansions. Various families of LRR-domain-containing receptors appear expanded, including but not limited to the Tlrs involved in innate immune defenses (see Hibino et al, 2006). It could be that the LRR domains have been used in other families of adhesion receptors for purposes of defense against pathogens. The scavenger receptor (SR) domain family is also expanded (Hibino et al, 2006, this issue). One notable expansion is of GPCRs with extended extracellular domains, many of them containing adhesion domains. This class of proteins is significantly larger in S.purpuratus than in vertebrates and it will be interesting to investigate the functions of these enigmatic receptors.
Interesting Absences
S.purpuratus lacks specific subfamilies of integrins, particularly those with VWA domains, including those that are involved in adhesion to collagen. This could be related to the absence of complex collagens (see below). There is a paucity of cadherins and many vertebrate cadherin subfamilies are missing – interestingly there are neither integrins nor cadherins of the intermediate-filament-linked subclasses although S.purpuratus does have intermediate filaments (Hoffman et al, 2006, this issue).
The absence of the more complex vertebrate-type collagens and proteoglycans is perhaps to be expected, given absence of cartilage and bones. Echinoderms are known to have collagenous tissues that undergo rapid, neurally mediated changes in mechanical properties (Motokawa, 1984; Wilkie, 2002). The stiffening of sea urchin spines in response to mechanical stimulation is a well studied example of this (Wilkie, 1996; Takemae and Motokawa, 2005). The low diversity of collagen types and similarity to the collagens of other metazoans are consistent with hypotheses that the changes in mechanical properties of collagenous connective tissues are mediated through accessory molecules in the matrix rather than the collagens themselves (Motokawa, 1984; Wilkie, 2002; Trotter et al. 1996). Many vertebrate accessory ECM proteins are not found in S. purpuratus but others are present.
Other absences may reflect the absence of endothelial-lined vasculature. Many vertebrate ECM proteins are missing, including matrix proteins involved in assembly, maintenance or functions of a high-pressure, high-shear, lined vascular system. Examples include fibronectin and von Willebrand factor, both involved in adhesion of vascular cells during hemostasis; fibronectin is also involved in angiogenesis. Thrombospondins are also involved in regulating angiogenesis in vertebrates but are present in sea urchins consistent with their having additional functions. Echinoderm coelomocytes are macrophage-like cells that appear to be mediators of the innate immune responses of sea urchins. The biology of these cells is poorly understood, but they are probably the equivalent of vertebrate thrombocytes and leukocytes. It would be interesting to know which adhesion receptors are expressed on those cells – there are none of the characteristic vertebrate leukocyte integrins (β2 or α4 subclasses) and no selectins – presumably a low-shear circulation would impose fewer adhesion demands. However, echinoderm coelomocytes likely express some or many of the candidate innate immune adhesion receptors.
Another potential reason for the absence of many adhesive ECM proteins may be the lack of neural crest migration. Lack of expansion of the laminin and type IV collagen families suggests a lack of basement membrane diversity. As mentioned, some of the basic neural adhesion systems are present but it is notable that there is very little expansion of the cadherin superfamily, believed to play major roles in establishing vertebrate nervous systems. There are few protocadherins and none of the clustered protocadherin multigene loci found in vertebrates.
Surprising Presences
In contrast with these absent adhesion proteins, the presence of reelin mentioned earlier is something of a surprise since this gene is involved in controlling organization of neurons within mammalian brains. Its presence and embryonic expression in sea urchins suggest that it plays some different role here but it will be of interest to explore its expression in the sea urchin nervous system..
Other surprises include several genes known in vertebrates for their involvement in genetic diseases leading to loss of hearing – not generally a skill attributed to echinoderms. These include usherin, a large ECM protein and VLGR1, a very large GPCR as well as cadherin 23 and several cytoskeletal proteins involved in a network of protein interactions in stereocilia and photoreceptors. As discussed earlier, it will be interesting to explore whether this set of proteins is expressed in sea urchin sensory organelles (stereocilia, sensory cilia). These sea urchin homologs of mammalian deaf/blindness genes and reelin represent only some of the adhesion genes involved in human disease that have homologs in echinoderms (see Table 2). Given the experimental tractability of the sea urchin system, it is reasonable to anticipate that studies of these “disease gene homologs” in this system will yield new insights.
In conclusion, the sea urchin adhesome includes some surprises but overall reflects the phylogenetic position of echinoderms. The complexity of the adhesome is somewhat larger than that of protostome invertebrates and contains some genes characteristic of deuterostomes. There are intriguing taxon-specific expansions of some gene families, that merit further investigation. However, many of the elaborations of cell adhesion receptors and extracellular matrix seen in vertebrates are absent, many of them possibly reflecting the absence of processes requiring cell adhesion and migration such a neural crest development and leukocyte traffic in a high-shear vasculature and the absence of a complex central nervous system. Some but not all of those vertebrate expansions are seen in urochordates. It will soon be possible to compare the adhesomes of all the major deuterostome sub-phyla, which should yield additional hypotheses as to the evolutionary consequences of the appearance of new members and families of adhesion-related genes. The sea urchin will provide one of the experimental systems in which to test those hypotheses.
Supplemental tables 1–3, 9,10,12 and 13 all contain the column “Sequence”. If the identifier in this column begins with “SPU_”, the annotated sequence is identical to the original GLEAN3 gene model. If it is listed as “Hand Edited”, the protein sequence is reported in the Suppl. Material 17 FASTA file. In the case of previously known genes, the Uniprot identifiers are listed.
Supplementary Material
The sea urchin genome contains four β integrin genes. Three of the four were previously characterized by cDNA sequencing. The common name, gene prediction identifier, representative sequence identifier and domain architecture of the predicted proteins are displayed in the table.
The sea urchin genome contains 8 α integrin genes. Sp_ITAP is the only previously known protein. This α integrin plus the Sp_ITAD and Sp_ITAF genes are nearly full length and phylogenetic analysis indicated that they cluster with the RGD-binding subfamily of α subunits (Figure 2). Incomplete gene models represent the remaining five genes and, as a result of their incomplete structure, their phylogenetic position is unclear. The common name, gene prediction identifier, representative sequence identifier and domain architecture of the predicted proteins are displayed in the table.
The sea urchin genome contains approximately 14 distinct genes encoding cadherin family members. Eight of these genes encode proteins that are clearly homologous with previously known cadherins (above yellow line; Figure 4). The phylogenetic relationships of the remaining six urchin cadherins to known cadherins are unclear (below yellow line). In some cases this is due to incomplete gene models, in others it may be because the protein is a urchin-specific cadherin species. The common name, gene prediction identifier, representative sequence identifier and domain architecture of the predicted protein are displayed in the table.
Gene predictions including both Ig and FN3 domains. The table is split into four sections. In each section, the order and numbers of domains is listed in N- to C-terminal order with presence of predicted transmembrane segment indicated. Genes that clearly encoded Ig/FN3 receptor kinases or phosphatases were omitted because they are covered in other articles in this issue. No attempt has yet been made to extend these gene predictions.
A. Genes encoding homologs of known adhesion proteins, based on domain structure and preliminary phylogenetic tree analyses. Homologs are named where clear, other genes listed clustered with vertebrate homologs in phylogenetic trees but data were insufficient to assign a specific homolog.
B. Genes which clustered in sea-urchin-specific clades in phylogenetic trees – could represent taxon-specific expansions.
C. Novel mixtures of EGF, Ig and FN3 domains not previously reported.
D. Receptors with mixtures of LRR, Ig/Igc2 and FN3 domians.
Gene predictions including Ig but, in most cases, no FN3 domains. The table is split into two sections. In each section, the order and numbers of domains is listed in N- to C-terminal order with presence of predicted transmembrane segment indicated.
A. Gene predictions including Ig domains (but no FN3 or LRR domains) often with EGF domains as indicated, along with miscellaneous other domains characteristic of adhesion molecules. Some of these proteins (e.g., the proteases) are unlikely to be membrane proteins but are included here for completeness.
B. Ig/LRR proteins. All are likely to be receptors, although some lack TM domains in the current assembly/predictions
Gene predictions including FN3 but, in most cases, no Ig domains. The table is split into three sections. In each section, the order and numbers of domains is listed in N- to C-terminal order with presence of predicted transmembrane segment indicated.
A. C-type-lectin/FN3 receptors.
B. EGF/FN3 receptors.
C. Novel domain organizations – except for SPU_027145 which encodes an Eph receptor.
Gene predictions including LRR and, in most cases, seven transmembrane segments. The order and numbers of domains are listed in N- to C-terminal order with presence of predicted seven transmembrane segments indicated. Also indicated is whether or not the domain composition/organization has been reported in protostomes and/or deuterostomes or whether it is novel.
Gene predictions including adhesion domains and, in most cases, seven transmembrane segments. The order and numbers of domains is listed in N- to C-terminal order with presence of predicted seven transmembrane segment indicated. Also indicated is whether or not the domain composition/organization has been reported in protostomes and/or deuterostomes or whether it is novel. Two examples of sea urchin homologues of (a) stn/fmi in Drosophila – CELSR in vertebrates and (b) VLGR-1 in vertebrates, are indicated and further discussed in the text. The later entries are included because of their GPS/7TM2 domain structure even in the absence of predited adhesion domains – these are less firmly implicated in cell adhesion but because this gene set is poorly understood in any species, they are included here for reference, pending further study.
The sea urchin genome encodes six complete laminins (three α chains, two β chains and one γ chain). One laminin α chain (Sp_LAMAdrome) is located on two different scaffolds and cannot be assembled with the data currently available. In addition, there are two gene fragments that are likely to be laminins based on BLAST analysis but their type is unclear. The common name, gene prediction identifier, representative sequence identifier and domain architecture of the predicted proteins are displayed in the table.
The sea urchin genome contains at least seven fibrillar collagen genes, two type IV collagens in a head-to-head arrangement in the genome and a single homolog of collagen 15/18, a homolog of an uncharacterized collagen-olfactomedin domain protein and col26a1(EMID2) homolog. There is also a non-fibrillar, non-VWA-containing TSPN domain-containing collagen (Sp_col91). With the exception of a honeybee collagen, this type of molecule is only found in chordates. The domain architecture suggests that Sp_col91 may be a homolog of human col9a1 but the phylogenetic analysis is inconclusive (Suppl. Figure 11). There are also six gene models that contain at least one collagen repeat but further characterization is difficult due to incomplete information.
The TSPN domains from a selection of known collagens and one sea urchin collagen were subjected to phylogenetic analysis using clustalX. The urchin TSPN domain clusters with vertebrate non-fibrillar TSPN collagens (col9a1, col19a1), a honeybee protein (col9a1_Bee) and a group of vertebrate VWA domain-containingTSPN collagens (col12a1, col14a1, coll20a1, col21a2 and col22a1). The urchin protein is in red, branches with >95% bootstrap support are indicated by red stars. The portion of the proteins used in the alignment is indicated by the inset domain diagram.
The sea urchin genome contains about 30 genes that encode proteins with Sema domains. These include two semaphorins, two semaphorin-like fragments and 27 plexin-like proteins and one homolog of the tyrosine kinase receptor Met/Ron.
The sea urchin genome contains four thrombospondin genes. Sp-TSPA1 contains TSP1 domains and, therefore, is a subgroup A TSP. This represents the first non-chordate example of this TSP subtype. TSPB1 is a chordate-type subgroup B TSP due to N-terminal TSPN domain and is the first non-chordate example of this type of TSP. Sp-TSP-like-1 resembles vertebrate COMP and the protostome TSP homologs in that it lacks the TSPN and TSP1 domains. Sp-TSP-like-2 is likely to be an incomplete gene model and phylogenetic analysis (not shown) suggests that it is related to Sp-TSP-like-1. The common name, gene prediction identifier, representative sequence identifier and domain architecture of the predicted proteins are displayed in the table.
The two sea urchin fibillins (see Table 1) were combined with fibrillins from Ciona intestinalis, human and honeybee and phylogenetic analysis was performed using clustalX. There are two sea urchin fibrillin genes and both are of the type found in Ciona and honey bee. Homologs of the vertebrate latent TGF β binding proteins were not detected. The sea urchin proteins are in red, branches with > 95% bootstrap support are indicated by red stars.
The sea urchin genome encodes five membrane-associated proteoglycans, four basement membrane proteoglycans and a variety of enzymes involved in GAG synthesis and modification. This collection of proteoglycans more closely resembles the set found in protostomes than that of chordates.
All the domains presented in the manuscript figures as well as hyperlinks to the domain annotation pages are shown in tabular format.
Sequences for substantially edited gene models are available in this file. The information in the FASTA definition line is equivalent to the Common Name field in the Supplemental Tables.
The embryonic expression levels of the 850 adhesome genes identified in this study as reported by the genomic tiling experiment of Samanta et al., 2006. Genes with an average expression level greater than 4 are likely to be expressed during embryonic development.
Figure 6. Basement Membrane ECM Toolkit.
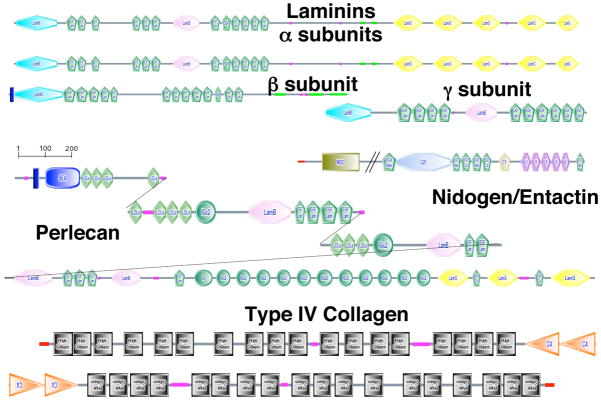
To date, all metazoan genomes encode a basic set (“toolkit”) of extracellular matrix proteins that form the core of basement membranes. This set includes a pair of type IV collagen genes (collagen repeats are indicated by black boxes), typically arranged head-to-head and probably sharing a promoter, collagen XV/XVIII, a set of laminin genes (2α, 1β, 1γ), nidogen and perlecan. The sea urchin genome encodes a similar set as shown. Laminin and collagen genes are listed in Supplementary Tables 9 and 10. The nidogen gene comprises gene prediction, SPU_016055 plus a missing N-terminal NIDO domain, and the perlecan gene comprises gene predictions, SPU_000937; SPU_012324; SPU_026338; SPU_028620 (see Suppl. Table 2). In the present state of the genome assembly, it is not possible to derive complete gene predictions for these two genes but it seems clear that good orthologs do exist in the sea urchin genome. For information about the SMART domains presented in this figure, see Suppl. Figure 16.
Acknowledgments
Grants from Howard Hughes Medical Institute (ROH), National Cancer Institute (CAW), Science and Engineering Canada and Canadian Institutes of Health Research (RDB), NSERC to BPB, NSF Fellowship to JW
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JC, Lawler J. The thrombospondins. Int J Biochem Cell Biol. 2004;36:961–8. doi: 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adato A, Lefevre G, Delprat B, Michel V, Michalski N, Chardenoux S, Weil D, El-Amraoui A, Petit C. Usherin, the defective protein in Usher syndrome type IIA, is likely to be a component of interstereocilia ankle links in the inner ear sensory cells. Hum Mol Genet. 2005a;14:3921–32. doi: 10.1093/hmg/ddi416. [DOI] [PubMed] [Google Scholar]
- Adato A, Michel V, Kikkawa Y, Reiners J, Alagramam KN, Weil D, Yonekawa H, Wolfrum U, El-Amraoui A, Petit C. Interactions in the network of Usher syndrome type 1 proteins. Hum Mol Genet. 2005b;14:347–56. doi: 10.1093/hmg/ddi031. [DOI] [PubMed] [Google Scholar]
- Behar O, Golden JA, Mashimo H, Schoen FJ, Fishman MC. Semaphorin III is needed for normal patterning and growth of nerves, bones and heart. Nature. 1996;383:525–8. doi: 10.1038/383525a0. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–95. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradham C, Foltz KR, Beane WS, Arnone MI, Rizzo F, Coffman JA, Mushegian A, Goel M, Morales J, Geneviere A, Lapraz F, Robertson AJ, Kelkar H, Loza-Coll M, Townley IK, Raisch M, Roux MM, Lapage T, Gache C, McClay DR, Manning G. The Sea Urchin Kinome: A First Look. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.074. This issue. [DOI] [PubMed] [Google Scholar]
- Burke RD, Angerer LM, Elphick MR, Humphrey GW, Yaguchi SS, Kiyama T, Liang S, Mu X, Agca C, Klein WH, Brandhorst BP, Rowe M, Wilson K, Churcher AM, Taylor JS, Chen N, Murray G, Wang DY, Mellott D, Hallbook F, Olinski R, Thorndyke MC. A Genomic View of the Sea Urchin Nervous System. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.007. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke RD, Murray G, Rise M, Wang D. Integrins on eggs: the betaC subunit is essential for formation of the cortical actin cytoskeleton in sea urchin eggs. Dev Biol. 2004;265:53–60. doi: 10.1016/j.ydbio.2003.09.025. [DOI] [PubMed] [Google Scholar]
- Byrum CA, Walton KD, Robertson AJ, Carbonneau S, Thomason RT, Coffman JA, McClay DR. Protein Tyrosine and Serine-Threonine Phosphatases in the Sea Urchin, Strongylocentrotus purpuratus: Identification and Potential Functions. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.050. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaruzza S, Perris R. Proteoglycan control of cell movement during wound healing and cancer spreading. Matrix Biol. 2005;24:400–17. doi: 10.1016/j.matbio.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CR, Klingelhofer J, Tarabykina S, Hulgaard EF, Kramerov D, Lukanidin E. Transcription of a novel mouse semaphorin gene, M-semaH, correlates with the metastatic ability of mouse tumor cell lines. Cancer Res. 1998;58:1238–44. [PubMed] [Google Scholar]
- Committee SN. Unified nomenclature for the semaphorins/collapsins. Cell. 1999;97:551–2. doi: 10.1016/s0092-8674(00)80766-7. [DOI] [PubMed] [Google Scholar]
- Copley RR, Schultz J, Ponting CP, Bork P. Protein families in multicellular organisms. Curr Opin Struct Biol. 1999;9:408–15. doi: 10.1016/S0959-440X(99)80055-4. [DOI] [PubMed] [Google Scholar]
- Crawford BJ, Campbell SS. The microvilli and hyaline layer of embryonic asteroid epithelial collar cells: a sensory structure to determine the position of locomotory cilia? Anat Rec. 1993;236:697–709. doi: 10.1002/ar.1092360414. [DOI] [PubMed] [Google Scholar]
- El-Amraoui A, Petit C. Usher I syndrome: unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J Cell Sci. 2005;118:4593–603. doi: 10.1242/jcs.02636. [DOI] [PubMed] [Google Scholar]
- Ewan R, Huxley-Jones J, Mould AP, Humphries MJ, Robertson DL, Boot-Handford RP. The integrins of the urochordate Ciona intestinalis provide novel insights into the molecular evolution of the vertebrate integrin family. BMC Evol Biol. 2005;5:31. doi: 10.1186/1471-2148-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) 2005. [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–51. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Gloriam DE, Hoglund PJ, Lagerstrom MC, Schioth HB. There exist at least 30 human G-protein-coupled receptors with long Ser/Thr-rich N-termini. Biochem Biophys Res Commun. 2003;301:725–34. doi: 10.1016/s0006-291x(03)00026-3. [DOI] [PubMed] [Google Scholar]
- Fujisawa H. Discovery of semaphorin receptors, neuropilin and plexin, and their functions in neural development. J Neurobiol. 2004;59:24–33. doi: 10.1002/neu.10337. [DOI] [PubMed] [Google Scholar]
- Gillespie PG, Dumont RA, Kachar B. Have we found the tip link, transduction channel, and gating spring of the hair cell? Curr Opin Neurobiol. 2005;15:389–96. doi: 10.1016/j.conb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–41. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- Hall KT, Boumsell L, Schultze JL, Boussiotis VA, Dorfman DM, Cardoso AA, Bensussan A, Nadler LM, Freeman GJ. Human CD100, a novel leukocyte semaphorin that promotes B-cell aggregation and differentiation. Proc Natl Acad Sci U S A. 1996;93:11780–5. doi: 10.1073/pnas.93.21.11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin J. The cellular basis of sea urchin gastrulation. Curr Top Dev Biol. 1996;33:159–262. doi: 10.1016/s0070-2153(08)60339-7. [DOI] [PubMed] [Google Scholar]
- Hertzler PL, McClay DR. alphaSU2, an epithelial integrin that binds laminin in the sea urchin embryo. Dev Biol. 1999;207:1–13. doi: 10.1006/dbio.1998.9165. [DOI] [PubMed] [Google Scholar]
- Hibino T, Loza-Coll M, Messiera C, Majeske AJ, Cohen A, Terwilliger DP, Buckley KM, Brockton V, Nair SV, Berney K, Fugmann SD, Anderson MK, Pancer Z, Cameron RA, Smith LC, Rast JP. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.065. This issue. [DOI] [PubMed] [Google Scholar]
- Hohenester E, Engel J. Domain structure and organisation in extracellular matrix proteins. Matrix Biol. 2002;21:115–28. doi: 10.1016/s0945-053x(01)00191-3. [DOI] [PubMed] [Google Scholar]
- Holt CE, Dickson BJ. Sugar codes for axons? Neuron. 2005;46:169–72. doi: 10.1016/j.neuron.2005.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhtala M, Heino J, Casciari D, de Luise A, Johnson MS. Integrin evolution: insights from ascidian and teleost fish genomes. Matrix Biol. 2005;24:83–95. doi: 10.1016/j.matbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Hyman LH. Echinodermata. iv. McGraw-Hill; New York: 1955. The Invertebrates. [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–87. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Hynes RO, Zhao Q. The evolution of cell adhesion. J Cell Biol. 2000;150:F89–96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer KL, Yost HJ. Heparan sulfate core proteins in cell-cell signaling. Annu Rev Genet. 2003;37:461–84. doi: 10.1146/annurev.genet.37.061103.090226. [DOI] [PubMed] [Google Scholar]
- Kwakkenbos MJ, Kop EN, Stacey M, Matmati M, Gordon S, Lin HH, Hamann J. The EGF-TM7 family: a postgenomic view. Immunogenetics. 2004;55:655–66. doi: 10.1007/s00251-003-0625-2. [DOI] [PubMed] [Google Scholar]
- Lane MC, Koehl MA, Wilt F, Keller R. A role for regulated secretion of apical extracellular matrix during epithelial invagination in the sea urchin. Development. 1993;117:1049–60. doi: 10.1242/dev.117.3.1049. [DOI] [PubMed] [Google Scholar]
- Lepage T, Lapraz F, Rottinger E, Duboc V, Range R, Duloquin L, Walton K, Wu S, Bradham C, Wittaker C, Loza M, Hibino T, Wilson K, Poustka A, Mc Clay DR, Angerer LM, Gache C. Genes for Receptors Tyrosine Kinases and TGF-b signaling pathways encoded in the sea urchin genome. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.048. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–60. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarana M, Olander B, Malmstrom J, Tiedemann K, Aebersold R, Lindahl U, Li JP, Malmstrom A. Biosynthesis of Dermatan Sulfate: CHONDROITIN-GLUCURONATE C5-EPIMERASE IS IDENTICAL TO SART2. J Biol Chem. 2006;281:11560–11568. doi: 10.1074/jbc.M513373200. [DOI] [PubMed] [Google Scholar]
- Marsden M, Burke RD. Cloning and characterization of novel beta integrin subunits from a sea urchin. Dev Biol. 1997;181:234–45. doi: 10.1006/dbio.1996.8451. [DOI] [PubMed] [Google Scholar]
- Miao HQ, Soker S, Feiner L, Alonso JL, Raper JA, Klagsbrun M. Neuropilin-1 mediates collapsin-1/semaphorin III inhibition of endothelial cell motility: functional competition of collapsin-1 and vascular endothelial growth factor-165. J Cell Biol. 1999;146:233–42. doi: 10.1083/jcb.146.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, McClay DR. Characterization of the role of cadherin in regulating cell adhesion during sea urchin development. Dev Biol. 1997;192:323–39. doi: 10.1006/dbio.1997.8740. [DOI] [PubMed] [Google Scholar]
- Motokawa T. Connective-Tissue Catch in Echinoderms. Biological Reviews of the Cambridge Philosophical Society. 1984;59:255–270. [Google Scholar]
- Mulloy B. The specificity of interactions between proteins and sulfated polysaccharides. An Acad Bras Cienc. 2005;77:651–64. doi: 10.1590/s0001-37652005000400007. [DOI] [PubMed] [Google Scholar]
- Murray G, Reed C, Marsden M, Rise M, Wang D, Burke RD. The alphaBbetaC integrin is expressed on the surface of the sea urchin egg and removed at fertilization. Dev Biol. 2000;227:633–47. doi: 10.1006/dbio.2000.9910. [DOI] [PubMed] [Google Scholar]
- Nakatsuji N, Lovtrup S. Preliminary observations on the synthesis of glycosaminoglycans in the sea urchin embryo. Arch Anat Microsc Morphol Exp. 1978;67:185–9. [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–72. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- Parra G, Blanco E, Guigo R. GeneID in Drosophila. Genome Res. 2000;10:511–5. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–9. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Samanta M, Tongprasit W, Istrail S, Cameron A, Tu Q, Davidson EH, Stolc V. A high resolution transcriptome map of the sea urchin embryo. Dev Biol. 2006 doi: 10.1126/science.1131898. This issue. [DOI] [PubMed] [Google Scholar]
- Susan JM, Just ML, Lennarz WJ. Cloning and characterization of alphaP integrin in embryos of the sea urchin Strongylocentrotus purpuratus. Biochem Biophys Res Commun. 2000;272:929–35. doi: 10.1006/bbrc.2000.2878. [DOI] [PubMed] [Google Scholar]
- Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, Tessier-Lavigne M, Comoglio PM. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99:71–80. doi: 10.1016/s0092-8674(00)80063-x. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Takeichi M, Nakagawa S. Identification of a nonchordate-type classic cadherin in vertebrates: chicken Hz-cadherin is expressed in horizontal cells of the neural retina and contains a nonchordate-specific domain complex. Dev Dyn. 2004;229:899–906. doi: 10.1002/dvdy.10493. [DOI] [PubMed] [Google Scholar]
- Tomita K, Yamasu K, Suyemitsu T. Cloning and characterization of cDNA for syndecan core protein in sea urchin embryos. Dev Growth Differ. 2000;42:449–58. doi: 10.1046/j.1440-169x.2000.00529.x. [DOI] [PubMed] [Google Scholar]
- Trotter JA, Lyons-Levy G, Luna D, Koob TJ, Keene DR, Atkinson MA. Stiparin: a glycoprotein from sea cucumber dermis that aggregates collagen fibrils. Matrix Biol. 1996;15:99–110. doi: 10.1016/s0945-053x(96)90151-1. [DOI] [PubMed] [Google Scholar]
- van Wijk E, van der Zwaag B, Peters T, Zimmermann U, Te Brinke H, Kersten FF, Marker T, Aller E, Hoefsloot LH, Cremers CW, Cremers FP, Wolfrum U, Knipper M, Roepman R, Kremer H. The DFNB31 gene product whirlin connects to the Usher protein network in the cochlea and retina by direct association with USH2A and VLGR1. Hum Mol Genet. 2006;15:751–65. doi: 10.1093/hmg/ddi490. [DOI] [PubMed] [Google Scholar]
- Washbourne P, Dityatev A, Scheiffele P, Biederer T, Weiner JA, Christopherson KS, El-Husseini A. Cell adhesion molecules in synapse formation. J Neurosci. 2004;24:9244–9. doi: 10.1523/JNEUROSCI.3339-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Luijendijk MW, Humphrey KD, Moller C, Kimberling WJ. Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am J Hum Genet. 2004;74:357–66. doi: 10.1086/381685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. 2002;13:3369–87. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie IC. Mutable collagenous structure or not? A comment on the re-interpretation by del Castillo et al of the catch mechanism in the sea urchin spine ligament. Biological Bulletin. 1996;190:237–242. doi: 10.2307/1542544. [DOI] [PubMed] [Google Scholar]
- Wilkie IC. Is muscle involved in the mechanical adaptability of echinoderm mutable collagenous tissue? J Exp Biol. 2002;205:159–65. doi: 10.1242/jeb.205.2.159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sea urchin genome contains four β integrin genes. Three of the four were previously characterized by cDNA sequencing. The common name, gene prediction identifier, representative sequence identifier and domain architecture of the predicted proteins are displayed in the table.
The sea urchin genome contains 8 α integrin genes. Sp_ITAP is the only previously known protein. This α integrin plus the Sp_ITAD and Sp_ITAF genes are nearly full length and phylogenetic analysis indicated that they cluster with the RGD-binding subfamily of α subunits (Figure 2). Incomplete gene models represent the remaining five genes and, as a result of their incomplete structure, their phylogenetic position is unclear. The common name, gene prediction identifier, representative sequence identifier and domain architecture of the predicted proteins are displayed in the table.
The sea urchin genome contains approximately 14 distinct genes encoding cadherin family members. Eight of these genes encode proteins that are clearly homologous with previously known cadherins (above yellow line; Figure 4). The phylogenetic relationships of the remaining six urchin cadherins to known cadherins are unclear (below yellow line). In some cases this is due to incomplete gene models, in others it may be because the protein is a urchin-specific cadherin species. The common name, gene prediction identifier, representative sequence identifier and domain architecture of the predicted protein are displayed in the table.
Gene predictions including both Ig and FN3 domains. The table is split into four sections. In each section, the order and numbers of domains is listed in N- to C-terminal order with presence of predicted transmembrane segment indicated. Genes that clearly encoded Ig/FN3 receptor kinases or phosphatases were omitted because they are covered in other articles in this issue. No attempt has yet been made to extend these gene predictions.
A. Genes encoding homologs of known adhesion proteins, based on domain structure and preliminary phylogenetic tree analyses. Homologs are named where clear, other genes listed clustered with vertebrate homologs in phylogenetic trees but data were insufficient to assign a specific homolog.
B. Genes which clustered in sea-urchin-specific clades in phylogenetic trees – could represent taxon-specific expansions.
C. Novel mixtures of EGF, Ig and FN3 domains not previously reported.
D. Receptors with mixtures of LRR, Ig/Igc2 and FN3 domians.
Gene predictions including Ig but, in most cases, no FN3 domains. The table is split into two sections. In each section, the order and numbers of domains is listed in N- to C-terminal order with presence of predicted transmembrane segment indicated.
A. Gene predictions including Ig domains (but no FN3 or LRR domains) often with EGF domains as indicated, along with miscellaneous other domains characteristic of adhesion molecules. Some of these proteins (e.g., the proteases) are unlikely to be membrane proteins but are included here for completeness.
B. Ig/LRR proteins. All are likely to be receptors, although some lack TM domains in the current assembly/predictions
Gene predictions including FN3 but, in most cases, no Ig domains. The table is split into three sections. In each section, the order and numbers of domains is listed in N- to C-terminal order with presence of predicted transmembrane segment indicated.
A. C-type-lectin/FN3 receptors.
B. EGF/FN3 receptors.
C. Novel domain organizations – except for SPU_027145 which encodes an Eph receptor.
Gene predictions including LRR and, in most cases, seven transmembrane segments. The order and numbers of domains are listed in N- to C-terminal order with presence of predicted seven transmembrane segments indicated. Also indicated is whether or not the domain composition/organization has been reported in protostomes and/or deuterostomes or whether it is novel.
Gene predictions including adhesion domains and, in most cases, seven transmembrane segments. The order and numbers of domains is listed in N- to C-terminal order with presence of predicted seven transmembrane segment indicated. Also indicated is whether or not the domain composition/organization has been reported in protostomes and/or deuterostomes or whether it is novel. Two examples of sea urchin homologues of (a) stn/fmi in Drosophila – CELSR in vertebrates and (b) VLGR-1 in vertebrates, are indicated and further discussed in the text. The later entries are included because of their GPS/7TM2 domain structure even in the absence of predited adhesion domains – these are less firmly implicated in cell adhesion but because this gene set is poorly understood in any species, they are included here for reference, pending further study.
The sea urchin genome encodes six complete laminins (three α chains, two β chains and one γ chain). One laminin α chain (Sp_LAMAdrome) is located on two different scaffolds and cannot be assembled with the data currently available. In addition, there are two gene fragments that are likely to be laminins based on BLAST analysis but their type is unclear. The common name, gene prediction identifier, representative sequence identifier and domain architecture of the predicted proteins are displayed in the table.
The sea urchin genome contains at least seven fibrillar collagen genes, two type IV collagens in a head-to-head arrangement in the genome and a single homolog of collagen 15/18, a homolog of an uncharacterized collagen-olfactomedin domain protein and col26a1(EMID2) homolog. There is also a non-fibrillar, non-VWA-containing TSPN domain-containing collagen (Sp_col91). With the exception of a honeybee collagen, this type of molecule is only found in chordates. The domain architecture suggests that Sp_col91 may be a homolog of human col9a1 but the phylogenetic analysis is inconclusive (Suppl. Figure 11). There are also six gene models that contain at least one collagen repeat but further characterization is difficult due to incomplete information.
The TSPN domains from a selection of known collagens and one sea urchin collagen were subjected to phylogenetic analysis using clustalX. The urchin TSPN domain clusters with vertebrate non-fibrillar TSPN collagens (col9a1, col19a1), a honeybee protein (col9a1_Bee) and a group of vertebrate VWA domain-containingTSPN collagens (col12a1, col14a1, coll20a1, col21a2 and col22a1). The urchin protein is in red, branches with >95% bootstrap support are indicated by red stars. The portion of the proteins used in the alignment is indicated by the inset domain diagram.
The sea urchin genome contains about 30 genes that encode proteins with Sema domains. These include two semaphorins, two semaphorin-like fragments and 27 plexin-like proteins and one homolog of the tyrosine kinase receptor Met/Ron.
The sea urchin genome contains four thrombospondin genes. Sp-TSPA1 contains TSP1 domains and, therefore, is a subgroup A TSP. This represents the first non-chordate example of this TSP subtype. TSPB1 is a chordate-type subgroup B TSP due to N-terminal TSPN domain and is the first non-chordate example of this type of TSP. Sp-TSP-like-1 resembles vertebrate COMP and the protostome TSP homologs in that it lacks the TSPN and TSP1 domains. Sp-TSP-like-2 is likely to be an incomplete gene model and phylogenetic analysis (not shown) suggests that it is related to Sp-TSP-like-1. The common name, gene prediction identifier, representative sequence identifier and domain architecture of the predicted proteins are displayed in the table.
The two sea urchin fibillins (see Table 1) were combined with fibrillins from Ciona intestinalis, human and honeybee and phylogenetic analysis was performed using clustalX. There are two sea urchin fibrillin genes and both are of the type found in Ciona and honey bee. Homologs of the vertebrate latent TGF β binding proteins were not detected. The sea urchin proteins are in red, branches with > 95% bootstrap support are indicated by red stars.
The sea urchin genome encodes five membrane-associated proteoglycans, four basement membrane proteoglycans and a variety of enzymes involved in GAG synthesis and modification. This collection of proteoglycans more closely resembles the set found in protostomes than that of chordates.
All the domains presented in the manuscript figures as well as hyperlinks to the domain annotation pages are shown in tabular format.
Sequences for substantially edited gene models are available in this file. The information in the FASTA definition line is equivalent to the Common Name field in the Supplemental Tables.
The embryonic expression levels of the 850 adhesome genes identified in this study as reported by the genomic tiling experiment of Samanta et al., 2006. Genes with an average expression level greater than 4 are likely to be expressed during embryonic development.