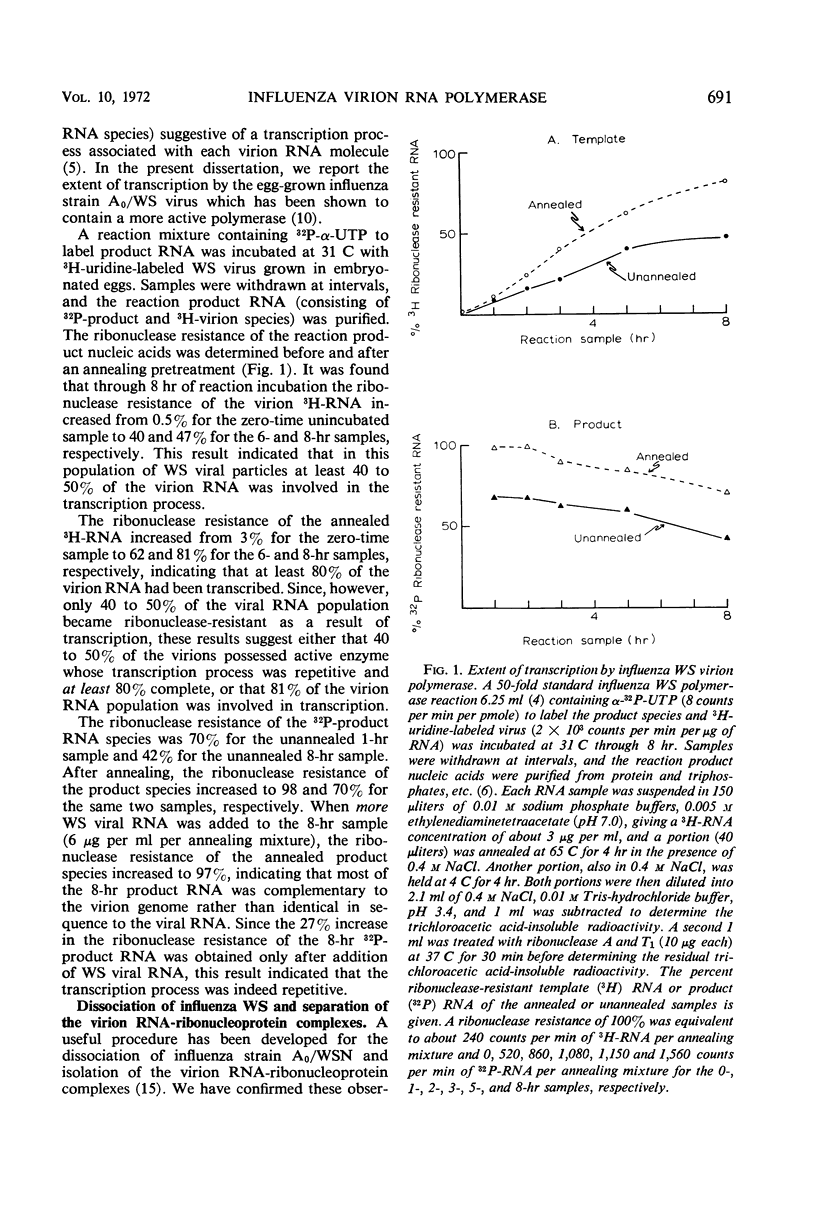
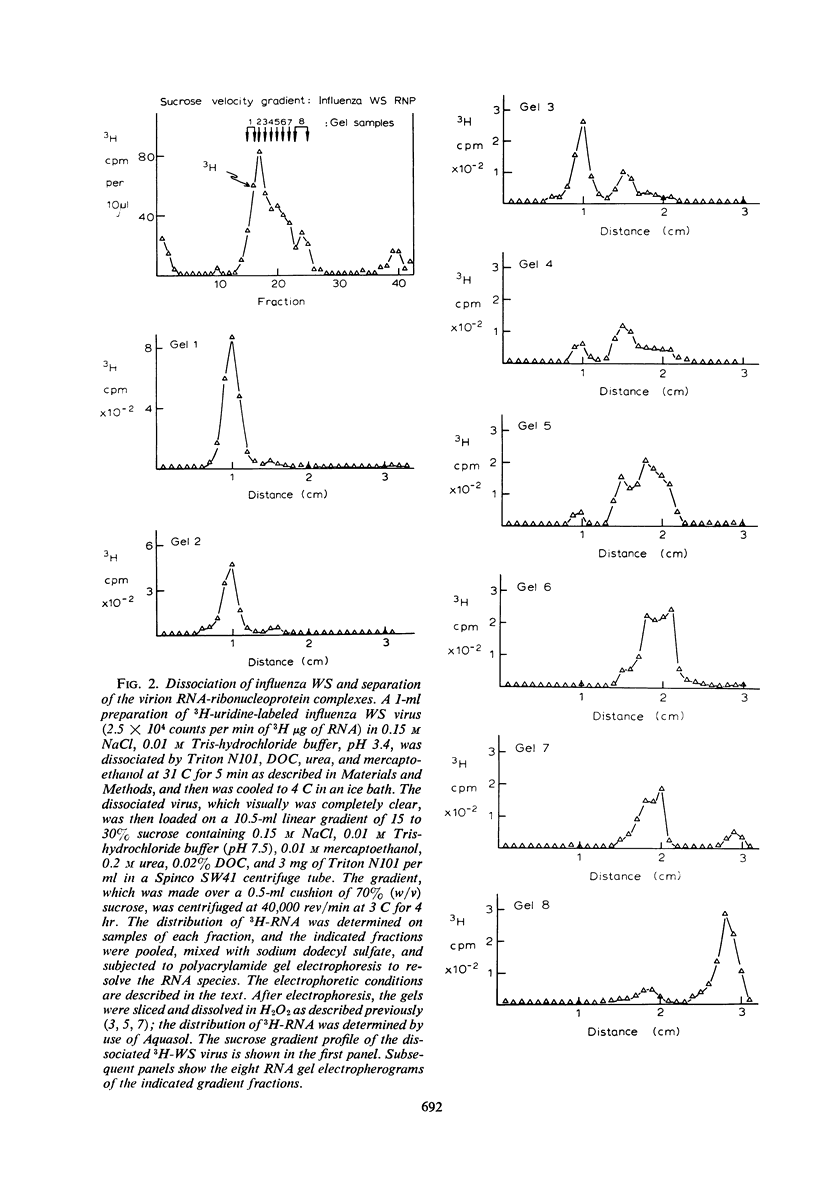
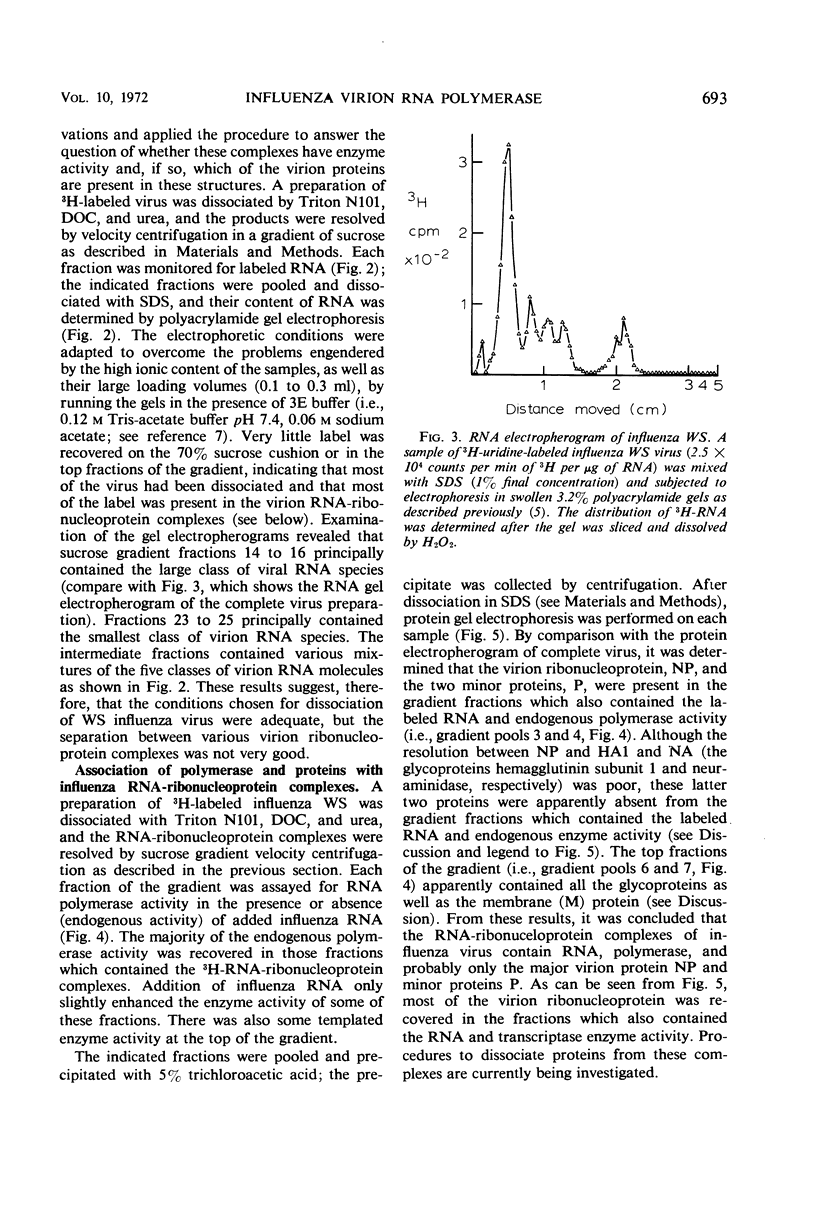
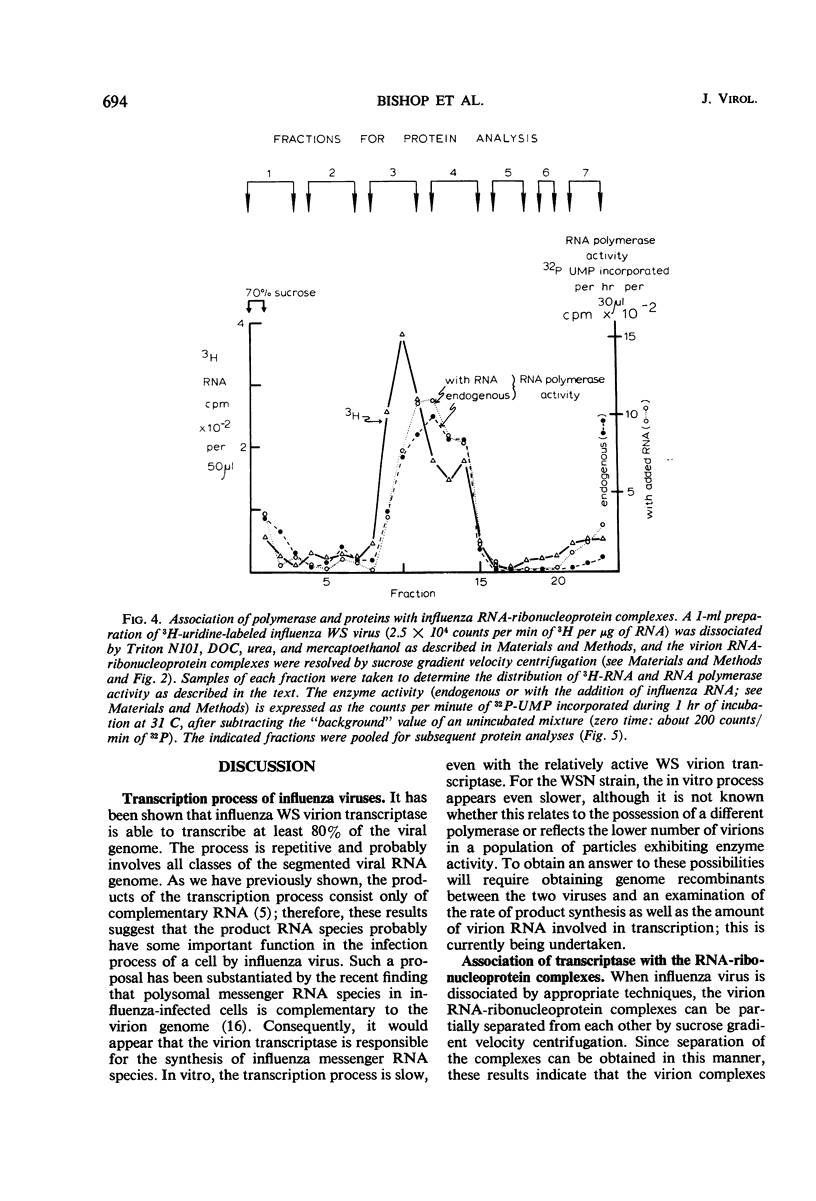
Abstract
The virion ribonucleic acid (RNA) polymerase of influenza strain A0/WS transcribed at least 81% of the viral genome in vitro. The polymerase is shown to be associated with each of the major size classes of the virion RNA-ribonucleoprotein complexes. Under optimal in vitro conditions, at least 45% of the RNA contained in a population of influenza virions was involved in a repetitive transcription process. The detectable proteins associated with enzymatically active complexes containing RNA, ribonucleoprotein, and polymerase have been identified by polyacrylamide gel electrophoresis.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERNKOPF H. Cultivation of influenza virus in the chorio-allantoic membrane of deembryonated eggs. Proc Soc Exp Biol Med. 1949 Dec;72(3):680–682. doi: 10.3181/00379727-72-17541. [DOI] [PubMed] [Google Scholar]
- Bikel I., Duesberg P. H. Proteins of Newcastle disease virus and of the viral nucleocapsid. J Virol. 1969 Oct;4(4):388–393. doi: 10.1128/jvi.4.4.388-393.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. H., Claybrook J. R., Spiegelman S. Electrophoretic separation of viral nucleic acids on polyacrylamide gels. J Mol Biol. 1967 Jun 28;26(3):373–387. doi: 10.1016/0022-2836(67)90310-5. [DOI] [PubMed] [Google Scholar]
- Bishop D. H., Obijeski J. F., Simpson R. W. Transcription of the influenza ribonucleic acid genome by a virion polymerase. I. Optimal conditions for in vitro activity of the ribonucleic acid-dependent ribonucleic acid polymerase. J Virol. 1971 Jul;8(1):66–73. doi: 10.1128/jvi.8.1.66-73.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. H., Obijeski J. F., Simpson R. W. Transcription of the influenza ribonucleic acid genome by a virion polymerase. II. Nature of the in vitro polymerase product. J Virol. 1971 Jul;8(1):74–80. doi: 10.1128/jvi.8.1.74-80.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. H., Roy P. Dissociation of vesicular stomatitis virus and relation of the virion proteins to the viral transcriptase. J Virol. 1972 Aug;10(2):234–243. doi: 10.1128/jvi.10.2.234-243.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. H., Roy P. Kinetics of RNA synthesis by vesicular stomatitis virus particles. J Mol Biol. 1971 May 14;57(3):513–527. doi: 10.1016/0022-2836(71)90106-9. [DOI] [PubMed] [Google Scholar]
- Bishop D. H., Roy P. Properties of the product synthesized by vesicular stomatitis virus particles. J Mol Biol. 1971 Jun 28;58(3):799–814. doi: 10.1016/0022-2836(71)90041-6. [DOI] [PubMed] [Google Scholar]
- Bishop D. H., Ruprecht R., Simpson R. W., Spiegelman S. Deoxyribonucleic acid polymerase of Rous sarcoma virus: reaction conditions and analysis of the reaction product nucleic acids. J Virol. 1971 Nov;8(5):730–741. doi: 10.1128/jvi.8.5.730-741.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow N. L., Simpson R. W. RNA-dependent RNA polymerase activity associated with virions and subviral particles of myxoviruses. Proc Natl Acad Sci U S A. 1971 Apr;68(4):752–756. doi: 10.1073/pnas.68.4.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compans R. W., Klenk H. D., Caliguiri L. A., Choppin P. W. Influenza virus proteins. I. Analysis of polypeptides of the virion and identification of spike glycoproteins. Virology. 1970 Dec;42(4):880–889. doi: 10.1016/0042-6822(70)90337-5. [DOI] [PubMed] [Google Scholar]
- Content J., Duesberg P. H. Electrophoretic distribution of the proteins and glycoproteins of influenza virus and Sendai virus. J Virol. 1970 Dec;6(6):707–716. doi: 10.1128/jvi.6.6.707-716.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joss A., Gandhi S. S., Hay A. J., Burke D. C. Ribonucleic Acid and protein synthesis in chick embryo cells infected with fowl plague virus. J Virol. 1969 Dec;4(6):816–822. doi: 10.1128/jvi.4.6.816-822.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penhoet E., Miller H., Doyle M., Blatti S. RNA-dependent RNA polymerase activity in influenza virions. Proc Natl Acad Sci U S A. 1971 Jun;68(6):1369–1371. doi: 10.1073/pnas.68.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons M. W. Isolation of influenza virus ribonucleoprotein from infected cells. Demonstration of the presence of negative-stranded RNA in viral RNP. Virology. 1971 Oct;46(1):149–160. doi: 10.1016/0042-6822(71)90014-6. [DOI] [PubMed] [Google Scholar]
- Pons M. W., Schulze I. T., Hirst G. K., Hauser R. Isolation and characterization of the ribonucleoprotein of influenza virus. Virology. 1969 Oct;39(2):250–259. doi: 10.1016/0042-6822(69)90045-2. [DOI] [PubMed] [Google Scholar]
- Pons M. W. Studies on the replication of influenza virus RNA. Virology. 1972 Mar;47(3):823–832. doi: 10.1016/0042-6822(72)90574-0. [DOI] [PubMed] [Google Scholar]
- Schulze I. T. The structure of influenza virus. I. The polypeptides of the virion. Virology. 1970 Dec;42(4):890–904. doi: 10.1016/0042-6822(70)90338-7. [DOI] [PubMed] [Google Scholar]
- Schulze I. T. The structure of influenza virus. II. A model based on the morphology and composition of subviral particles. Virology. 1972 Jan;47(1):181–196. doi: 10.1016/0042-6822(72)90251-6. [DOI] [PubMed] [Google Scholar]
- Skehel J. J. RNA-dependent RNA polymerase activity of the influenza virus. Virology. 1971 Sep;45(3):793–796. doi: 10.1016/0042-6822(71)90197-8. [DOI] [PubMed] [Google Scholar]
- Skehel J. J., Schild G. C. The polypeptide composition of influenza A viruses. Virology. 1971 May;44(2):396–408. doi: 10.1016/0042-6822(71)90270-4. [DOI] [PubMed] [Google Scholar]



