Abstract
Ribonucleotide reductase 1 (RRM1) is a determinant of gemcitabine efficacy in non-small cell lung cancer and pancreatic cancer. We investigated the protein levels of RRM1 and two other DNA repair enzymes, ERCC1 and BRCA1, in 55 metastatic breast cancer (MBC) patients undergoing gemcitabine-based chemotherapy. With automated in situ protein quantification (AQUA v1.6), the average scores for RRM1, ERCC1 and BRCA1 ranged from 245.6–2,774.1, 74.0–410.3 and 54.4–1,833.1, respectively. They were significantly associated with each other (Spearman’s rho ≥ .36; P ≤ 0.007). Given their pattern of distribution, RRM1 and BRCA1 are potentially suitable markers for clinical decision making in MBC.
Keywords: BRCA1, ERCC1, gemcitabine, metastatic breast cancer, RRM1
INTRODUCTION
Breast cancer is the most frequently diagnosed cancer in women in western countries, and it is the second most frequent cause of cancer-related deaths [1, 2]. Despite recent improvements in the diagnosis and management of early disease, approximately 50% of women with breast cancer will develop distant metastases [3]. Metastatic breast cancer (MBC) is a molecularly heterogeneous disease. While therapeutic response has a significant impact on survival, it cannot be precisely predicted [4, 5]. The identification of women that will benefit from palliative chemotherapy would allow physicians to deliver effective treatments to sensitive patients, while preventing others from suffering the side effects of inactive drugs. To date, estrogen receptor (ER)-α, progesterone receptor (PgR) and HER2 represent the only biomarkers used in clinical practice to aid treatment decisions in both early and metastatic disease. Although several other markers are being investigated, none have been sufficiently validated for inclusion into routine clinical practice [6–7].
Gemcitabine (2',2'-difluorodeoxycytidine [dFdC]) is a cell-cycle specific antimetabolite recently approved in combination with taxanes for the treatment of MBC patients pretreated with anthracyclines [8]. Its mechanism of action is well characterized [9]. Intracellularly, gemcitabine is converted into two active metabolites, gemcitabine diphosphate (dFdCDP) and gemcitabine triphosphate (dFdCTP). As gemcitabine triphosphate competes with deoxycytidine triphosphate (dCTP) for incorporation into DNA during replication, which leads to termination of chain elongation, gemcitabine diphosphate inhibits subunit 1 of ribonucleotide reductase (RRM1), the key enzyme for the production of deoxynucleotides. Preclinical studies in multiple model systems have shown that increased tumoral expression of RRM1 is the major determinant of resistance to gemcitabine [10–13]. In addition, low RRM1 levels have been reported to correlate significantly with tumor response in non-small cell lung cancer (NSCLC) patients treated with gemcitabine-based chemotherapy and pancreatic cancer patients treated with gemcitabine alone [12, 13]. To date, the relationship between RRM1 expression and sensitivity to gemcitabine has not been addressed in breast cancer.
In the present study we evaluated RRM1 protein expression in a population of MBC patients treated with gemcitabine-based chemotherapy and studied its association with clinical outcome. In addition, we measured the levels of the excision repair cross-complementation group 1 (ERCC1) protein, given the statistically significant relationship between RRM1 and ERCC1 in NSCLC [12, 14–18]. We also investigated the expression of breast and ovarian cancer susceptibility gene 1 (BRCA1) in view of its emerging role as a marker of sensitivity to chemotherapy in breast cancer [19].
MATERIALS AND METHODS
All data and tumor specimens were collected at the Regina Elena Cancer Institute, Rome, Italy. Analyses of in situ protein expression of RRM1, ERCC1 and BRCA1 and statistical evaluations were done at the Moffitt Cancer Center, Tampa, Florida. The study was approved by the local Ethics Committee of the Regina Elena Cancer Institute, and all patients gave written informed consent for marker analyses.
Patients
Fifty-five consecutive MBC patients eligible for gemcitabine-based chemotherapy were included and followed prospectively. All patients had histologically confirmed diagnoses of breast cancer pretreated with anthracyclines. Treatment consisted of gemcitabine given in combination with paclitaxel or docetaxel in taxane-naïve patients or pegylated liposomal doxorubicin in patients who had received prior taxanes. In all cases, disease progression following the most recent treatment prior to gemcitabine-based chemotherapy was documented. No more than 1 line of chemotherapy for metastatic disease were allowed. The presence of at least one measurable lesion, performance status ≤ 2, life expectancy > 3 months and adequate hematological, hepatic and renal function were inclusion criteria. For staging evaluations, computed tomography scans of the chest and abdomen and mammography were mandatory. Patients with asymptomatic and/or controlled brain metastases were eligible. Patients with HER2 positive tumors [3+ by immunohistochemistry (IHC) on the HercepTest (Dako A/S) or 2+ by IHC and dual color fluorescence in situ hybridization (FISH; PathVision HER2 DNA Probe Kit, Vysis, Inc.) positivity] were also eligible, provided they had progressed after at least one trastuzumab-based therapy for metastatic disease. Only patients receiving at least 2 cycles of gemcitabine-based chemotherapy were considered in the study.
Tissue microarray construction
Formalin-fixed tissue embedded in paraffin (FFPE) obtained at the time of diagnosis was used for construction of a tissue microarray (TMA). Whole tissue sections were stained with hematoxilin and eosin, and representative tumor areas were marked. Triplicate tissue cores with a diameter of 0.6 mm were punched and randomly arrayed into a recipient block using a tissue arrayer (Beecher Instrument, Silver Spring, MD). Sections of 5 µm thickness were cut, transferred using tape to 4x adhesive-coated slides (Instrumedics, Hackensack, NJ) and exposed to UV light for 30 seconds to enhance adherence.
In situ detection and quantification of RRM1, ERCC1 and BRCA1 protein expression
Immunofluorescence combined with automated quantitative analysis (AQUA) was used to assess in situ expression of the target molecules [20]. Antigens were retrieved by microwave oven treatment for 15 minutes. The slides were blocked for 30 minutes with 0.3% BSA and then incubated overnight at room temperature with the primary antibodies. For RRM1 analysis a custom antiserum was used (R1-AS6b, 1:400) [14], while commercially available antibodies were used for the detection of ERCC1 (mouse clone SPM243, 1:200, Santa Cruz Biotech) and BRCA1 (mouse clone MS110, 1:30, Calbiochem). For identification of breast cancer cells, pancytokeratin antibodies were used (anti-human pancytokeratin AE1/AE3, 1:200, #M3515 and #Z0622, Dako Cytomation). Slides were washed and incubated with 2 different secondary antibodies for 1 hour (Envision labeled polymer-HRP anti-mouse #K4007 and anti-rabbit antibody #K4011; Alexa 555 goat anti-rabbit, #A21429 and Alexa 555 mouse anti-rabbit #A21424, 1:200, Dako Cytomation). For fluorescence amplification, slides were exposed to Cy5-Tyramide (1:50) for 10 minutes at room temperature. They were mounted with Prolong Gold antifade reagent with DAPI (4´-6-diamino-2-phenylindole) mound solution. The final slides were scanned with SpotGrabber and image data were analyzed with AQUA (PM-2000, version 1.6, HistoRx, New Haven, Conn). The maximal range of AQUA scores with version 1.6 is 0–33,333; the observed range for the proteins evaluated was 0–4,096.
Statistical methods
This study was prospectively designed to find a correlation between RRM1 expression and response to treatment. Assuming a response rate of 50%, 55 pts give a power of 80% to identify a difference of about 40% between patients with high and low tumoral RRM1 levels. The expression of ERCC1 and BRCA1 were assessed retrospectively.
The distributions of the expression data were examined using descriptive graphical and numerical statistics and the Anderson-Darling statistic. Due to the departure from normality, non-parametric methods were used to analyze the data. Spearman’s correlation coefficient was used to assess the correlation between RRM1, ERCC1 and BRCA1. For two group comparisons, the Wilcoxon rank sum test was used and for multi-group comparisons the Kruskal-Wallis test was used. Survival curves were estimated by the Kaplan-Meier method, and the log-rank test was used to assess differences among curves.
Disease response was categorized as progressive disease (PD), stable disease (SD), and partial remission (PR) according to RECIST [21]. Progression-free survival (PFS) was the time elapsed from the date of first gemcitabine-based treatment to the date of first evidence of disease progression or death. Overall survival (OS) was the time elapsed from the date of first gemcitabine-based treatment to the date of death. Patients without an event were censored at the last date of follow-up (April 2008).
RESULTS
Patients characteristics
Fifty-five patients were enrolled from a single institution from September 2004 to December 2007 (table 1). The median age was 54 years (range 35–78). All patients had received prior anthracyclines, and 9 (16.5%) had received prior taxanes. Twenty-two patients (40%) were chemonaïve for metastatic disease. Treatment consisted of gemcitabine plus a taxane in 46 (83.5%) patients and gemcitabine plus pegylated liposomal doxorubicin in 9 (16.5%) patients.
Table 1.
Patients characteristics
| Characteristics | Patients (%) |
RRM1 mean |
ERCC1 mean |
BRCA1 mean |
|---|---|---|---|---|
| All patients | ||||
| (median age 54 y, range 35–78) | 55(100%) | 1,237.00 | 229.6 | 794.1 |
| Stage at diagnosis | ||||
| Localized | 47 (85.5%) | 1.271.7 | 235.8 | 849.9 |
| Advanced | 8 (14.5%) | 1,126.10 | 198.2 | 560.1 |
| Histotype | ||||
| Ductal carcinoma | 43 (78%) | 1,294.50 | 227.2 | 833 |
| Other | 12 (22%) | 1,079.20 | 251.1 | 697 |
| Grading | ||||
| 1 | 3 (5.5%) | 1,182.80 | 179.3 | 644.9 |
| 2 | 15 (27%) | 1,252.10 | 235.7 | 808.1 |
| 3 | 25 (45.5%) | 1,265.10 | 228.2 | 806.3 |
| Not available | 12 (22%) | 1,173.70 | 233 | 777.6 |
| Hormone receptor | ||||
| ER and/or PgR positive | 40 (73%) | 1,236.90 | 227.5 | 818.1 |
| ER and PgR negative | 13 (23.5%) | 1,355.40 | 251.8 | 816.7 |
| Not available | 2 (3.5%) | 468.3 | 126.9 | 167.9 |
| HER2 status | ||||
| HER2 positive | 6 (11%) | 1,248.60 | 208.1 | 975.7 |
| HER2 negative | 47 (85.5%) | 1,236.20 | 232.9 | 784.2 |
| Not available | 2 (3.5%) | 1,220.60 | 216.2 | 482.1 |
| Prior anthracyclines* | 55 (100%) | 1,237.00 | 229.6 | 794.1 |
| Prior taxanes* | 9 (16.5%) | 928.5 | 219.5 | 542.5 |
| Visceral disease | ||||
| Yes | 43 (78%) | 1,172.80 | 230.4 | 765.7 |
| No | 12 (22%) | 1,466.80 | 226.9 | 895.9 |
| Prior chemotherapies for MBC | ||||
| 0 | 22 (40%) | 1,395.00 | 243.3 | 873.8 |
| 1 | 33 (60%) | 1,188.80 | 226 | 830.8 |
| Type of gemcitabine-based chemotherapy | ||||
| Gemcitabine plus a taxane† | 46 (83.5%) | 1,297.30 | 231.6 | 843.3 |
| Gemcitabine plus PLD § | 9 (16.5%) | 928.5 | 219.5 | 542.5 |
MBC, metastatic breast cancer; PLD, pegylated liposomal doxorubicin
Either in the adjuvant or metastatic setting
Gemcitabine 1250 mg/m2 dd 1,8 – paclitaxel 175 mg/m2 d 1 q 21 (35 patients, 63.5%); gemcitabine 1000 mg/m2 dd 1,8 – docetaxel 80 mg/m2 d 8 q 21 (11 patients, 20%)
Gemcitabine 800 mg/m2 dd 1,8 – pegylated liposomal doxorubicin 25 mg/m2 d 8 q 21
In situ RRM1, ERCC1 and BRCA1 protein expression
In situ RRM1, ERCC1 and BRCA1 protein expression was determined in a TMA containing triplicates of 55 tumor samples for a total of 165 spots on the same array. Table 2 lists the breast cancer specimens used to construct the TMA.
Table 2.
Breast cancer specimens used for construction of tissue microarray
| Total number of samples | All samples no. = 55 |
RRM1 mean |
ERCC1 mean |
BRCA1 mean |
|---|---|---|---|---|
| Primary breast cancer | 31 (56.5%) | 1,310.6 | 243.8 | 827.1 |
| Metastases | ||||
| Soft tissue | 6 (11%) | |||
| Lymph nodes | 5 (9%) | |||
| Lung | 5 (9%) | 1,108.20 | 04.82 | 736.3 |
| Skin | 4 (7%) | |||
| Liver | 2 (3.5%) | |||
| Bone | 1 (2%) | |||
| Brain | 1 (2%) | |||
Figure 1 shows the distribution of RRM1, ERCC1 and BRCA1 values. Replicate sample scores were averaged to produce a single score for each patient and marker. For RRM1, these scores ranged from 245.6 to 2,774.1 with a median of 1,290.3 (mean 1,237.0; standard deviation 500.5). For ERCC1, the scores ranged from 74.0 to 410.3 with a median of 226.4 (mean 229.7; standard deviation 66.1). For BRCA1, the scores ranged from 54.4 to 1,833.1 with a median of 771.6 (mean 794.2; standard deviation 385.0). In all cases, the pattern of marker expression was nuclear (figure 2–4).
Figure 1.
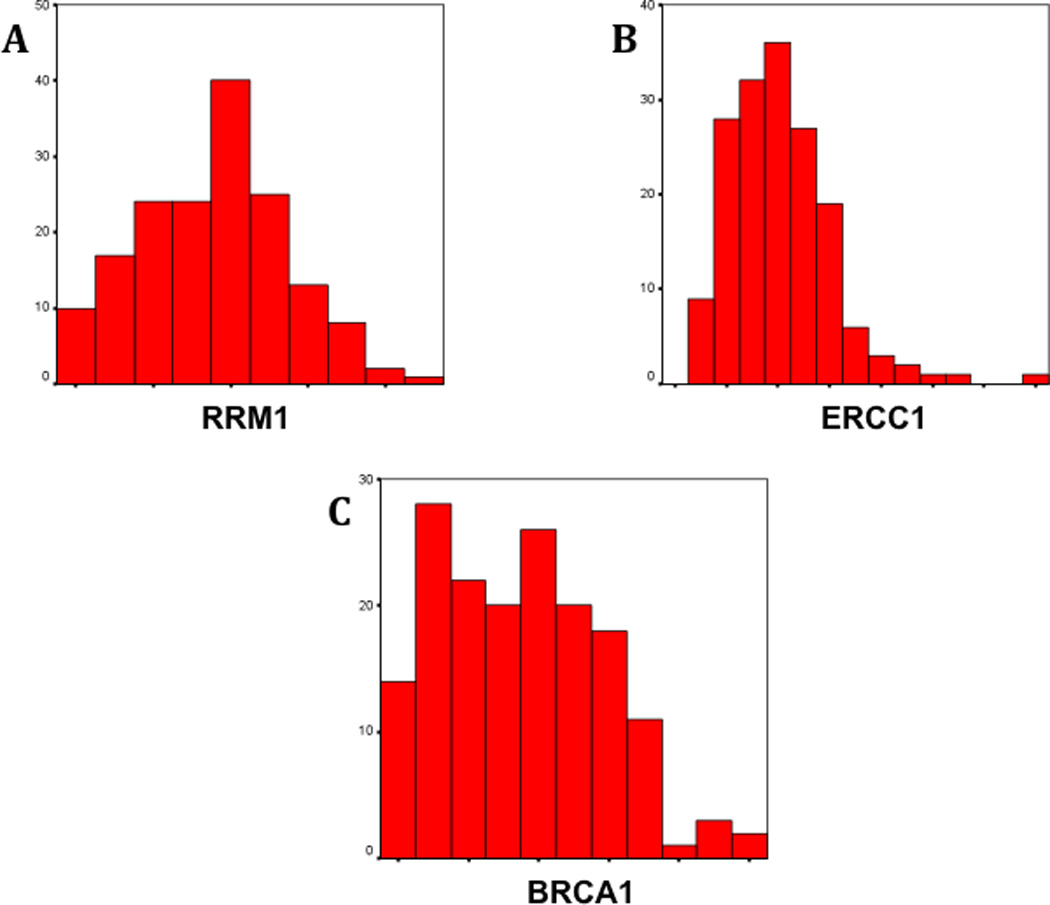
Distribution of AQUA scores for RRM1 (A), ERCC1 (B) and BRCA1 (C). RRM1 values were near normally distributed. ERCC1 and BRCA1 histograms show a skewness to the left, which is more pronounced for ERCC1 (ERCC1 skewness = 1.31; BRCA1 skewness = 0.34).
Figure 2.
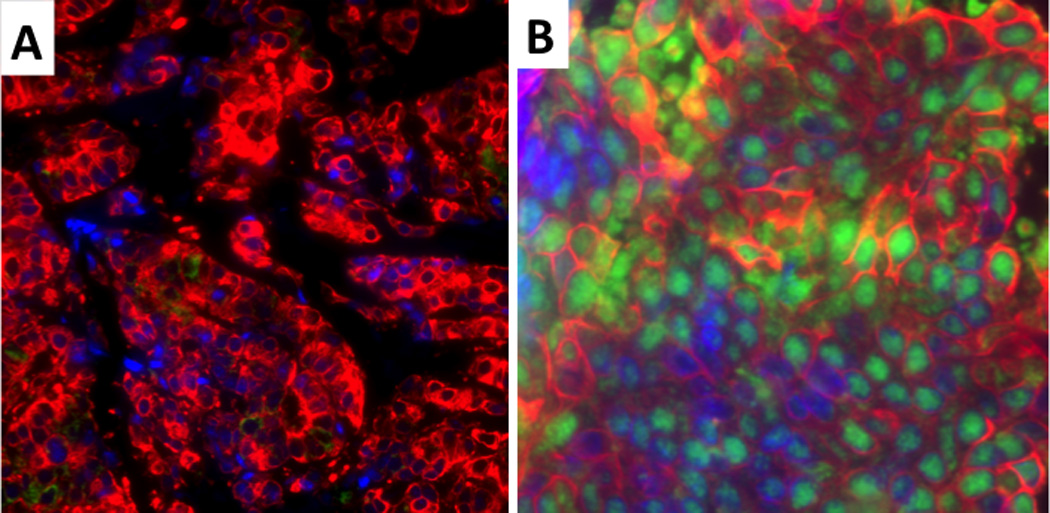
Low (A) and high (B) RRM1 protein expression. The nuclei are blue, RRM1 is green and the tumor cytoplasm is red. RRM1 is located in the nucleus and shows a granular pattern.
Figure 4.
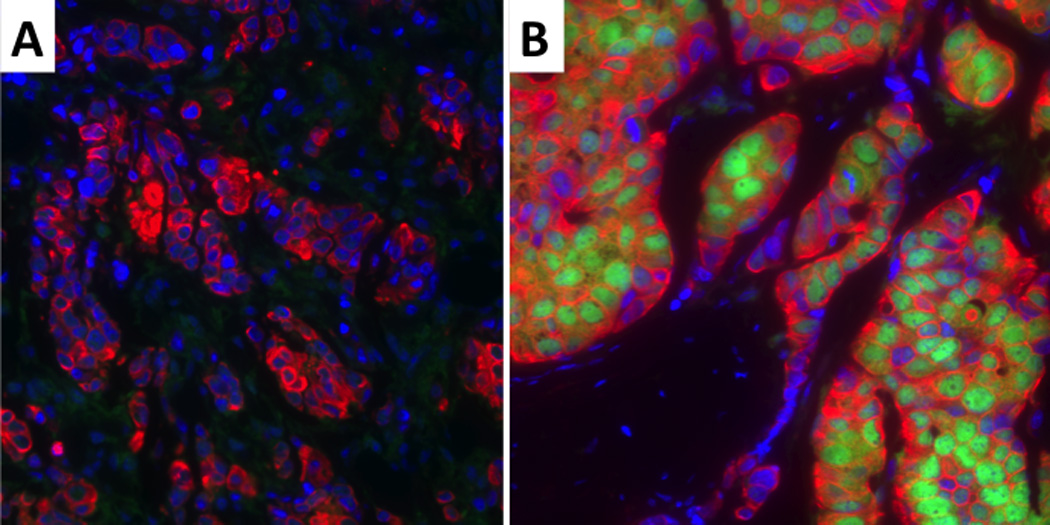
Low (A) and high (B) BRCA1 protein expression. The nuclei are blue, BRCA1 is green and the tumor cytoplasm is red. BRCA1 shows nuclear localization.
A statistically significant correlation was found between RRM1 and ERCC1 protein levels (rho = .4030, p = 0.0024). Similarly, BRCA1 levels were significantly correlated with RRM1 (rho = .6580, p < 0.0001) and ERCC1 levels (rho = .3600, p = 0.0071).
No statistically significant associations were observed between each marker and the following variables: age (≥ vs < 50 years), tumor stage at diagnosis (localized vs advanced), histology (ductal vs other), hormone receptor status (positive vs negative), HER2 status (positive vs negative), presence of visceral disease (yes vs no) and pretreatment for metastatic disease (yes vs no) (table 1).
No significant association was found for the expression of each marker in primary breast cancers and metastases (table 2). Also, no difference in expression was observed within metastatic sites classified as visceral vs non-visceral metastases (data not shown).
Efficacy of treatment and correlative studies
All 55 patients received at least 2 cycles of gemcitabine-based chemotherapy (median cycles 5, range 2–10). The best treatment response was a PR in 21 patients (38%; 95% CI 25.4–52.3%), and it was SD in 19 (34.5%) and PD in 15 (27.5%) patients. As of April 2008, 48 patients had progressed and 32 had died. Seven patients were alive without disease progression (3.5–29 months). The median PFS was 5.8 months (95% CI 4.0–7.6 months), and the 12- and 24-month PFS rates were 16.2% and 5.8% respectively. The median OS was 16.8 months (95% CI 6.6–27.1 months), and the 12- and 24-month OS rates were 64.7% and 42.7% respectively.
No statistically significant associations were observed between RRM1, ERCC1 or BRCA1 and response to treatment (p = 0.49 for RRM1, p = 0.13 for ERCC1, p = 0.41 for BRCA1). Likewise, no significant associations were noted between each marker and PFS or OS (figure 5).
Figure 5.
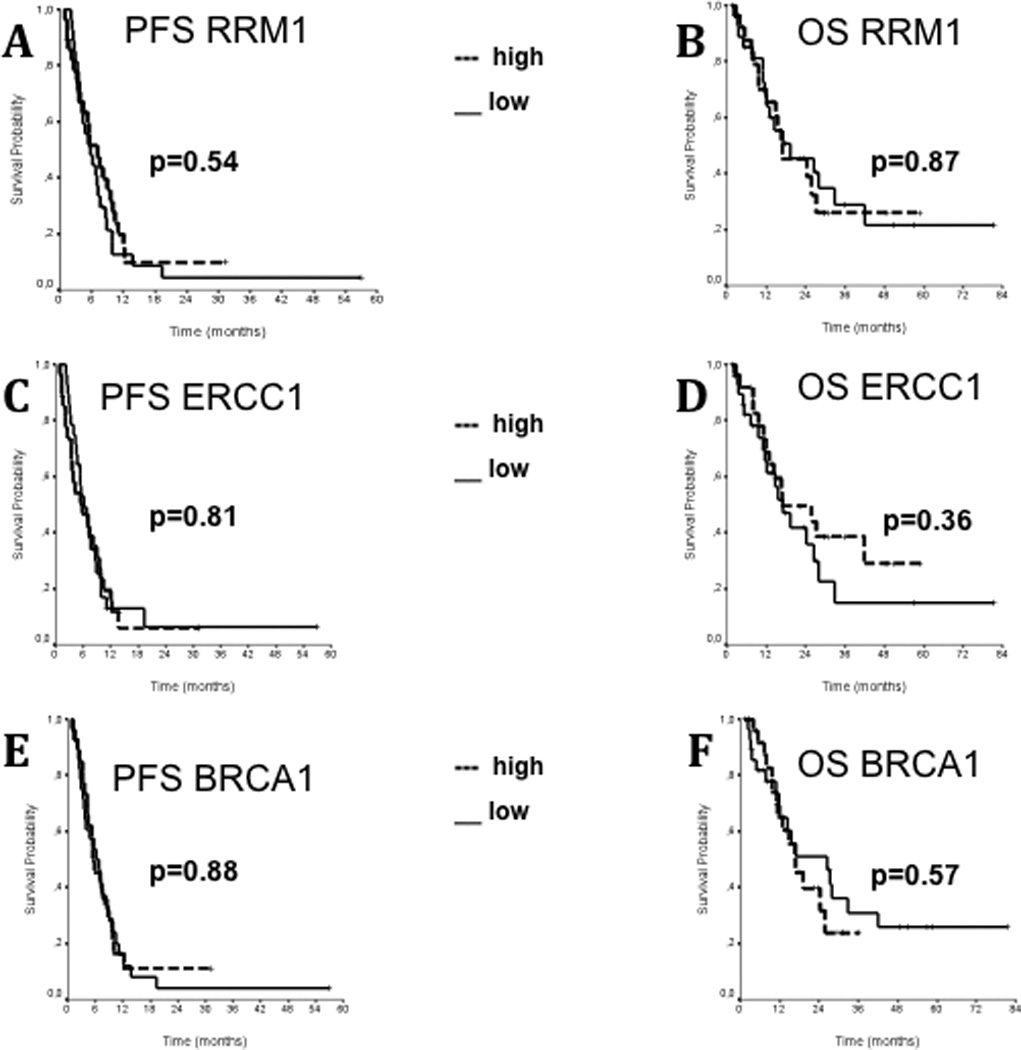
Progression free survival (A,C,E) and overall survival (B,D,F) of patients according to the expression of RRM1, ERCC1 and BRCA1 proteins, respectively. For each marker, median values of AQUA scores were chosen to divide patients into a high- and low-expression group. No statistically significant differences were reported.
DISCUSSION AND CONCLUSIONS
Gemcitabine has become the mainstay of treatment for several cancers, such as metastatic NSCLC, bladder cancer and pancreatic cancer [22]. Its recent approval for treatment of MBC [8] made gemcitabine a useful addition to the therapeutic armamentarium already available for this disease. Pharmacogenomic studies conducted in NSCLC have demonstrated that RRM1 is a reliable marker of sensitivity to gemcitabine [12, 16, 17]. In fact, in NSCLC patients treated with gemcitabine-based therapy, low tumoral RRM1 levels are not only associated with response [12], but also with improved survival as compared to patients with high tumoral RRM1 levels [16, 17]. In light of these results, we undertook a prospective study in MBC patients to investigate the spectrum of RRM1 expression and the relationship between RRM1 and efficacy of gemcitabine-based regimens. Two additional molecules involved in DNA repair, namely ERCC1 and BRCA1, were studied.
To assess the protein levels of RRM1, ERCC1 and BRCA1 we used a fully automated and quantitative immunofluorescence technique (AQUA) that has the advantage to render protein expression analyses on paraffin-embedded tumor tissue objective, reliable and reproducible [20].
Consistent with their function in multiple steps throughout DNA synthesis and repair, we found nuclear expression for each marker (figure 2–4). The distribution of values was near normal for RRM1, skewed to low values for BRCA1, and markedly skewed to low values for ERCC1 (figure 1). RRM1 expression levels had an approximately 10-fold range, and they were higher than in NSCLC [14]. ERCC1 values had a 5-fold range, compared to near 100-fold range in NSCLC, with comparable median values in both diseases [14]. Consistent with what has been shown in NSCLC [12, 14–18], RRM1 and ERCC1 levels were significantly correlated. This finding suggests that the coexpression of both DNA repair proteins may be universal to epithelial malignancies. However, the relatively narrow range of ERCC1 expression in the mid-range of the maximal expression spectrum suggests that MBC is only moderately platinum sensitive, and that ERCC1 expression levels may not be useful for therapeutic decisions on platinum use for MBC [23]. In contrast, BRCA1 expression levels had a wide range in MBC, which may provide an opportunity for its use as a predictive marker for platinum efficacy [24]. However, in contrast to RRM1 and ERCC1 where activating or inactivating mutations have not been described, assessment of the mutational status of BRCA1 may be crucial to the interpretation of the prognostic and predictive utility of its protein levels [25–27]. Interestingly, we found that BRCA1 levels were significantly correlated with both RRM1 and ERCC1 levels, similarly to what has been described in NSCLC [18]. Therefore, we conclude, that RRM1 and BRCA1 levels, as determined by AQUA, display characteristics that make them suitable as biomarkers for both NSCLC and MBC (table 3).
Table 3.
Comparison of RRM1, ERCC1 and BRCA1 protein expression levels between NSCLC and breast cancer
| Minimum | Median | Maximum | |
|---|---|---|---|
| RRM1 NSCLC [14] | 8.3 | 40.5 | 96.2 |
| RRM1 breast cancer*† | 12.3 | 64.5 | 138.7 |
| ERCC1 NSCLC [14] | 1.9 | 65.9 | 178.7 |
| ERCC1 breast cancer*† | 18.5 | 56.6 | 102.6 |
| BRCA1 NSCLC§ | 19.6 | 65.2 | 164.0 |
| BRCA1 breast cancer*† | 4.1 | 57.9 | 137.5 |
NSCLC, non-small cell lung cancer
Comparisons for RRM1, ERCC1 and BRCA1 AQUA scores are based on version 1.2 scores [1.2 AQUA score = 1.6 AQUA score × (255 × target exposure time / 100.000)]
Current study
Monteiro, Zhong, Bepler, unpublished data for version 1.2 scores
However, we did not find significant associations between RRM1 levels and clinico-pathological parameters, such as age, histology, stage, and hormone receptor status, which is consistent with prior data in NSCLC [15, 17, 28]. Also, we did not find a significant association between RRM1 levels and therapeutic efficacy of gemcitabine-containing chemotherapy in our cohort of 55 MBC patients in terms of disease response, PFS or OS. In prior clinical trials, low RRM1 expression was reported to be significantly associated with benefit from such therapy [12, 16, 17]. There arw several potential explanations for this discrepancy. A plausible explanation may be the choice of chemotherapy combinations. Prior studies had demonstrated an interaction between RRM1 and efficacy of gemcitabine alone or gemcitabine combined with platinum. In our present study, gemcitabine was combined with a taxane in 46 patients or pegylated liposomal doxorubicin in 9 patients. An earlier report by Rosell et al. had demonstrated a significantly different survival in patients with high versus low RRM1 levels when treated with gemcitabine and cisplatin; however, when vinorelbine was added to the combination or when gemcitabine was combined with vinorelbine only, no impact of RRM1 expression levels on survival was observed [16]. It is thus conceivable that the addition of a taxane to gemcitabine may have a similar effect as the addition of vinorelbine to gemcitabine on RRM1’s impact on efficacy.
Another potential explanation may be linked to the coexpression of RRM1 and BRCA1. In fact, recent data suggest that low BRCA1 levels are associated with resistance to taxane therapy [29–32]. We had found a significant correlation between RRM1 and BRCA1 levels in MBC specimens. Thus, patients with low RRM1 levels and sensitivity to gemcitabine are likely to have low BRCA1 levels and thus resistance to taxanes. In contrast, patients with high RRM1 levels and resistance to gemcitabine are likely to have high BRCA1 levels and thus sensitivity to taxanes. As a result, the clinical specimens available to us for the present investigation would not be suitable to provide results demonstrating an interaction between RRM1 levels and gemcitabine efficacy in MBC.
Finally, emerging data suggest that the expression levels of tumoral RRM1 might not be the only indicators of resistance to gemcitabine. A decrease in the level of the equilibration-sensitive nucleoside transporter of gemcitabine and the genotype of RRM1 gene promoter polymorphisms may also play a role in gemcitabine resistance [33, 34].
In conclusion, we found that RRM1 and BRCA1 have a range of expression and pattern of distribution potentially useful for development as clinical markers in MBC (figure 1). Although our results suggest that no association exists between gemcitabine efficacy and RRM1 expression levels in MBC, which is in contrast to studies in lung and pancreatic cancers, this is likely explained by the specific regimens used.
Figure 3.
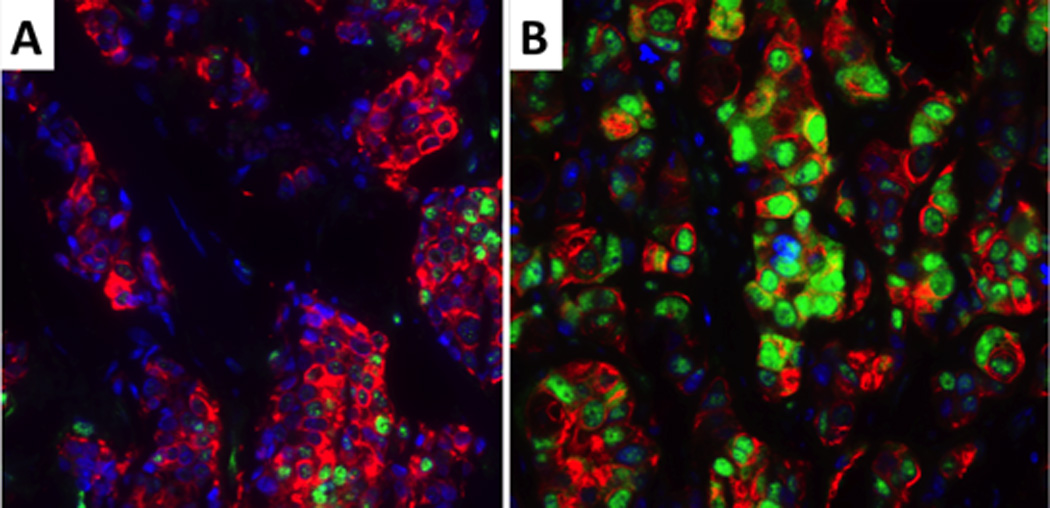
Low (A) and high (B) ERCC1 protein expression. The nuclei are blue, ERCC1 is green and the tumor cytoplasm is red. ERCC1 shows nuclear localization.
Acknowledgment
The authors are grateful to Diana Giannarelli for assistance with the statistical analysis.
Footnotes
Declaration of interest
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.American Cancer Society. Cancer Facts and Figures 2005. [ http://www.cancer.org/downloads/STT/2008CAFFfinalsecured.pdf.]
- 2.Bosetti C, Bertuccio P, Levi F, Lucchini F, Negri E, La Vecchia C. Cancer mortality in the European Union, 1970–2003, with a joinpoint analysis. Ann. Oncol. 2008;19(4):631–640. doi: 10.1093/annonc/mdm597. [DOI] [PubMed] [Google Scholar]
- 3.De Vita VT, Heliman S, Rosenberg SA. Cancer: principles & practice of oncology. Philadelphia: Lippincott Williams & Wilkins; 2008. [Google Scholar]
- 4.Greenberg PA, Hortobagyi GN, Smith TL, Ziegler LD, Frye DK, Buzdar AU. Long-term follow-up of patients with complete remission following combination chemotherapy for metastatic breast cancer. J Clin. Oncol. 1996;14(8):2197–2205. doi: 10.1200/JCO.1996.14.8.2197. [DOI] [PubMed] [Google Scholar]
- 5.Bruzzi P, Del Mastro L, Sormani MP, Bastholt L, Danova M, Focan C, Fountzilas G, Paul J, Rosso R, Venturini M. Objective response to chemotherapy as a potential surrogate end point of survival in metastatic breast cancer patients. J. Clin. Oncol. 2005;23(22):5117–5125. doi: 10.1200/JCO.2005.02.106. [DOI] [PubMed] [Google Scholar]
- 6.Tanner M, Isola J, Wiklund T, Erikstein B, Kellokumpu-Lehtinen P, Malmström P, Wilking N, Nilsson J, Bergh J Scandinavian Breast Group Trial 9401. Topoisomerase II alpha gene amplification predicts favorable treatment response to tailored and dose-escalated anthracycline-based adjuvant chemotherapy in HER-2/neu-amplified breast cancer: Scandinavian Breast Group Trial 9401. J. Clin. Oncol. 2006;24(16):2428–2436. doi: 10.1200/JCO.2005.02.9264. [DOI] [PubMed] [Google Scholar]
- 7.Galmarini CM, Treilleux I, Cardoso F, Bernard-Marty C, Durbecq V, Gancberg D, Bissery MC, Paesmans M, Larsimont D, Piccart MJ, Di Leo A, Dumontet C. Class III beta-tubulin isotype predicts response in advanced breast cancer patients randomly treated either with single-agent doxorubicin or docetaxel. Clin. Cancer Res. 2008;14(14):4511–4516. doi: 10.1158/1078-0432.CCR-07-4741. [DOI] [PubMed] [Google Scholar]
- 8.Albain KS, Nag SM, Calderillo-Ruiz G, Jordaan JP, Llombart AC, Pluzanska A, Rolski J, Melemed AS, Reyes-Vidal JM, Sekhon JS, Simms L, O'Shaughnessy J. Gemcitabine plus Paclitaxel versus Paclitaxel monotherapy in patients with metastatic breast cancer and prior anthracycline treatment. J. Clin. Oncol. 2008;26(24):3950–3957. doi: 10.1200/JCO.2007.11.9362. [DOI] [PubMed] [Google Scholar]
- 9.Lund B, Kristjansen PE, Hansen HH. Clinical and preclinical activity of 2',2'-difluorodeoxycytidine (gemcitabine) Cancer Treat. Rev. 1993;19(1):45–55. doi: 10.1016/0305-7372(93)90026-n. [DOI] [PubMed] [Google Scholar]
- 10.Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res. 2004;64(11):3761–3766. doi: 10.1158/0008-5472.CAN-03-3363. [DOI] [PubMed] [Google Scholar]
- 11.Bergman AM, Eijk PP, Ruiz van Haperen VW, Smid K, Veerman G, Hubeek I, van den Ijssel P, Ylstra B, Peters GJ. In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as the major determinant. Cancer Res. 2005;65(20):9510–9516. doi: 10.1158/0008-5472.CAN-05-0989. [DOI] [PubMed] [Google Scholar]
- 12.Bepler G, Kusmartseva I, Sharma S, Gautam A, Cantor A, Sharma A, Simon G. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J. Clin. Oncol. 2006;24(29):4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 13.Nakahira S, Nakamori S, Tsujie M, Takahashi Y, Okami J, Yoshioka S, Yamasaki M, Marubashi S, Takemasa I, Miyamoto A, Takeda Y, Nagano H, Dono K, Umeshita K, Sakon M, Monden M. Involvement of ribonucleotide reductase M1 subunit overexpression in gemcitabine resistance of human pancreatic cancer. Int. J. Cancer. 2007;120(6):1355–1363. doi: 10.1002/ijc.22390. [DOI] [PubMed] [Google Scholar]
- 14.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N. Engl. J. Med. 2007;356(8):800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 15.Simon G, Sharma A, Li X, Hazelton T, Walsh F, Williams C, Chiappori A, Haura E, Tanvetyanon T, Antonia S, Cantor A, Bepler G. Feasibility and efficacy of molecular analysis-directed individualized therapy in advanced non-small-cell lung cancer. J. Clin. Oncol. 2007;25(19):2741–2746. doi: 10.1200/JCO.2006.08.2099. [DOI] [PubMed] [Google Scholar]
- 16.Rosell R, Danenberg KD, Alberola V, Bepler G, Sanchez JJ, Camps C, Provencio M, Isla D, Taron M, Diz P, Artal A. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin. Cancer Res. 2004;10(4):1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 17.Ceppi P, Volante M, Novello S, Rapa I, Danenberg KS, Danenberg PV, Cambieri A, Selvaggi G, Saviozzi S, Calogero R, Papotti M, Scagliotti GV. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann. Oncol. 2006;17(12):1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 18.Rosell R, Skrzypski M, Jassem E, Taron M, Bartolucci R, Sanchez JJ, Mendez P, Chaib I, Perez-Roca L, Szymanowska A, Rzyman W, Puma F, Kobierska-Gulida G, Farabi R, Jassem J. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS ONE. 2007;2(11):e1129. doi: 10.1371/journal.pone.0001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James CR, Quinn JE, Mullan PB, Johnston PG, Harkin DP. BRCA1, a potential predictive biomarker in the treatment of breast cancer. Oncologist. 2007;12(2):142–150. doi: 10.1634/theoncologist.12-2-142. [DOI] [PubMed] [Google Scholar]
- 20.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat. Med. 2002;8(11):1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 21.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 2000;92(3):205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 22.Toschi L, Finocchiaro G, Bartolini S, Gioia V, Cappuzzo F. Role of gemcitabine in cancer therapy. Future Oncol. 2005;1(1):7–17. doi: 10.1517/14796694.1.1.7. [DOI] [PubMed] [Google Scholar]
- 23.Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V, Taranchon E, Filipts M, Pirker R, Popper HH, Stahel R, Sabatier L, Pignon JP, Tursz T, Le Chevalier T, Soria JC. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N. Engl. J. Med. 2006;355(10):983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 24.Taron M, Rosell R, Felip E, Mendez P, Souglakos J, Ronco MS, Queralt C, Majo J, Sanchez JM, Sanchez JJ, Maestre J. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum. Mol. Genet. 2004;13(20):2443–2449. doi: 10.1093/hmg/ddh260. [DOI] [PubMed] [Google Scholar]
- 25.Chappuis PO, Goffin J, Wong N, Perret C, Ghadirian P, Tonin PN, Foulkes WD. A significant response to neoadjuvant chemotherapy in BRCA1/2 related breast cancer. J. Med. Genet. 2002;39(8):608–610. doi: 10.1136/jmg.39.8.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goffin JR, Chappuis PO, Bégin LR, Wong N, Brunet JS, Hamel N, Paradis AJ, Boyd J, Foulkes WD. Impact of germline BRCA1 mutations and overexpression of p53 on prognosis and response to treatment following breast carcinoma: 10-year follow up data. Cancer. 2003;97(3):527–536. doi: 10.1002/cncr.11080. [DOI] [PubMed] [Google Scholar]
- 27.Byrski T, Huzarski T, Dent R, Gronwald J, Zuziak D, Cybulski C, Kladny J, Gorski B, Lubinski J, Narod SA. Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res. Treat. 2009 doi: 10.1007/s10549-008-0128-9. in press. [DOI] [PubMed] [Google Scholar]
- 28.Bepler G, Sharma S, Cantor A, Gautam A, Haura E, Simon G, Sharma A, Sommers E, Robinson L. RRM1 and PTEN as prognostic paramenters for overall and disease-free survival in patients with non-small-cell lung cancer. J. Clin. Oncol. 2004;22(10):1878–1885. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Quinn JE, Kennedy RD, Mullan PB, Gilmore PM, Carty M, Johnston PG, Harkin DP. BRCA1 Functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003;63(19):6221–6228. [PubMed] [Google Scholar]
- 30.Kennedy RD, Quinn JE, Mullan PB, Johnston PG, Harkin DP. The role of BRCA1 in the cellular response to chemotherapy. J. Natl. Cancer Inst. 2004;96(22):1659–1668. doi: 10.1093/jnci/djh312. [DOI] [PubMed] [Google Scholar]
- 31.Chabalier C, Lamare C, Racca C, Privat M, Valette A, Larminat F. BRCA1 downregulation leads to premature inactivation of spindle checkpoint and confers paclitaxel resistance. Cell Cycle. 2006;5(9):1001–1007. doi: 10.4161/cc.5.9.2726. [DOI] [PubMed] [Google Scholar]
- 32.Lafarge S, Sylvain V, Ferrara M, Bignon YJ. Inhibition of BRCA1 leads to increased chemoresistance to microtubule-interfering agents, an effect that involves the JNK pathway. Oncogene. 2001;20(45):6597–6606. doi: 10.1038/sj.onc.1204812. [DOI] [PubMed] [Google Scholar]
- 33.Oguri T, Achiwa H, Muramatsu H, Ozasa H, Sato S, Shimizu S, Yamazaki H, Eimoto T, Ueda R. The absence of human equilibrative nucleoside transporter 1 expression predicts nonresponse to gemcitabine-containing chemotherapy in non-small cell lung cancer. Cancer Lett. 2007;256(1):112–119. doi: 10.1016/j.canlet.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 34.Kim SO, Jeong JY, Kim MR, Cho HJ, Ju JY, Kwon YS, Oh IJ, Kim KS, Kim YI, Lim SC, Kim YC. Efficacy of gemcitabine in patients with non-small cell lung cancer according to promoter polymorphisms of the ribonucleotide reductase M1 gene. Clin. Cancer Res. 2008;14(10):3083–30881. doi: 10.1158/1078-0432.CCR-07-4591. [DOI] [PubMed] [Google Scholar]


