Abstract
MicroRNAs (miRNAs) affect gene regulation by base pairing with mRNA and contribute to the control of cellular homeostasis. The first step in miRNA maturation is conducted in the nucleus by the “microprocessor” complex made up of an RNase III enzyme, Drosha, that contains one dsRNA binding domain (dsRBD), and DGCR8, that contains two dsRBDs in tandem. The crystal structure of DGCR8-Core (493–720), containing both dsRBDs, and the NMR solution structure of Drosha-dsRBD (1259–1337) have been reported, but the solution dynamics have not been explored for any of these dsRBDs. To better define the mechanism of dsRNA binding and thus the nuclear maturation step of miRNA processing, we report NMR spin relaxation and MD simulations of Drosha-dsRBD (1259–1337) and DGCR8-dsRBD1 (505–583). The study was motivated by electrophoretic mobility shift assays (EMSAs) of the two dsRBDs, which showed that Drosha-dsRBD does not bind a representative miRNA but isolated DGCR8-dsRBD1 does (Kd = 9.4±0.4 µM). Our results show that loop 2 in both dsRBDs is highly dynamic but the pattern of the correlations observed in MD is different for the two proteins. Additionally, the extended loop 1 of Drosha-dsRBD is more flexible than the corresponding loop in DGCR8-dsRBD1 but shows no correlation with loop 2, which potentially explains the lack of dsRNA binding by Drosha-dsRBD in the absence of the RNase III domains. The results presented in this study provide key structural and dynamic features of dsRBDs that contribute to the binding mechanism of these domains to dsRNA.
MicroRNAs (miRNAs)1 are a large class of gene-regulating RNAs that have been shown to contribute to the control of cellular homeostasis in multicellular organisms by regulating such biologically important processes as apoptosis, cell cycle progression, and cell-type differentiation (1, 2). Mature miRNAs, roughly 22 nucleotides, regulate gene translation by base pairing with mRNA, and it has been reported that a majority of human genes (> 60%) are regulated post transcriptionally by at least one miRNA (3). Disease states leading to cancer, Alzheimer’s disease, heart disease, and autoimmune diseases have been directly linked to aberrant cellular levels of specific miRNAs (4–8). Maturation of miRNA is a two-step process, with each step being independent and spatially separate. The first step involves cleavage of the single-stranded RNA (ssRNA) tail of primary miRNA (pri-miRNA) in the nucleus by the “microprocessor” complex, which contains the RNase III enzyme Drosha and its cofactor, a double-stranded RNA (dsRNA) binding protein DGCR8 (known as Pasha in Drosophila) (9). After pre-miRNA is transported to the cytosol by exportin-5, the RNase III enzyme Dicer cleaves the terminal loop, thus forming mature miRNA (9).
Throughout the maturation of miRNA, dsRNA binding domains (dsRBDs) play vital roles in recruitment and positioning for cleavage (9). dsRBDs are one of the most common RNA-binding motifs, having been reported in viral-encoded proteins and in both prokaryotic and eukaryotic organisms (10). Also, dsRBDs are found in proteins in both the cytoplasm and the nucleus of eukaryotes, as evidenced by the compartmentalization of the first and second stages of miRNA processing in the nucleus and cytosol, respectively (9, 11, 12). The dsRBD is an ~65–68 amino acid motif that forms an αβββα topology, where the two α-helices lie on one face of three antiparallel β-sheets with the other surface of the sheet being solvent-exposed (13–15).
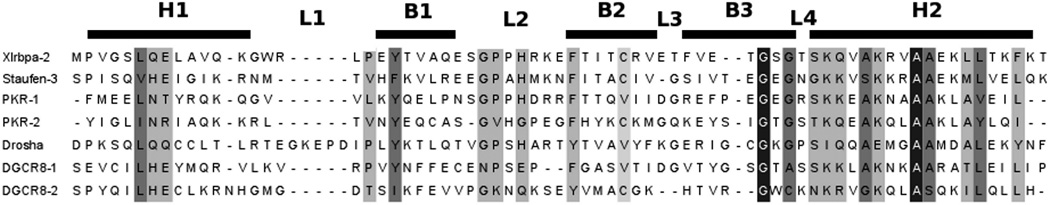
The preferred binding partner of dsRBDs is A-form double-helix RNA, while dsRBDs show little or no affinity for ssRNA, dsDNA, and DNA-RNA hybrids (16, 17). As few as 11 base pairs of dsRNA (coinciding with one turn of canonical A-form helix) have been shown to interact with a single dsRBD (18). Nonsequence specific recognition of dsRNA by dsRBDs is mediated by conserved positively charged residues and hydrogen bond acceptors and donors in loop 2 (L2) and loop 4 (L4), the N-terminal region of helix 2 (H2), and additional contacts from helix 1 (H1) (13, 19–21). These positions of sequence conservation are highlighted in Figure 1 for each dsRBD to be discussed in depth. The dsRBD binding mode differs from the mode employed by the structurally similar RNA recognition motifs (RRMs), exemplified by the two RRMs of U1A, that bind ssRNA by utilizing conserved residues in the central β-strands (10, 22). In dsRBDs, the distance between loop 2 and loop 4 is hypothesized to be a critical determinant of the ability to bind dsRNA (20). The hypothesis originates from analysis of the cocrystal structure of Drosophila Staufen-dsRBD3, in which the spacing of these two loops matches the spacing between the minor and major groove of A-form RNA. The observed spacing is facilitated by the stacking of each loop on opposing faces of a conserved phenylalanine (Figure 1) (19).
Figure 1.
Alignment using Clustal W2 for each of the dsRBDs discussed in this paper showing conserved residues in varying shades of gray, with black corresponding to complete conservation. Approximate location of the secondary elements is shown above the alignment with H representing an α-helix, B representing a β-sheet, and L representing a loop/turn.
In addition to the value of static structure determination, measurement of protein dynamics through NMR spin relaxation experiments has proven to be a powerful step in defining the molecular mechanism of RNA binding by both the RRM (23, 24) and the dsRBD (19, 25). NMR dynamics measurements have been reported for the two dsRBDs of the antiviral response protein, PKR (26), and the dsRBD3 of Staufen free and bound to dsRNA (heteronuclear NOE only) (19). Both showed increased flexibility in loop 2, which in the case of Staufen-dsRBD3 is retained in the bound state (19, 26). In addition, the PKR dynamics indicate that decreased dynamics in loop 2 correlate with decreased dsRNA binding since PKR-dsRBD2, which has a higher order parameter (S2) in loop 2, shows a significantly lower affinity for dsRNA (26).
One of the first dsRBD-containing proteins to be purified and studied was the RNase III enzyme from Escherichia coli (14, 15, 27). Generally, dsRBDs coexist with the RNase III domain in RNase III proteins to the broadest evolutionary extent, except in two mycoplasma (14) and Dicer from Giardia intestinalis (9). RNase III enzymes can still function without dsRBDs, which is illustrated by the naturally occurring RNase III enzymes lacking a dsRBD and data from Sun et al. where the dsRBDs of E. coli RNase III were removed and function was still retained (28). RNase III function requires that two RNase III domains dimerize to form an active catalytic site, where each domain is responsible for hydrolysis of one strand of dsRNA (29), leaving a two-nucleotide 3′ overhang, a notable feature of products of RNase III enzymes (9). At present, the structural biology of dsRBDs derived from RNase III enzymes is limited to solution structures of human Drosha (30) and E. coli RNase III (27), a crystal structure derived from mouse Dicer (31), and two structures of yeast Rnt1p determined by NMR and crystallography, respectively (32). Only one crystal structure has been reported of an RNase III dsRBD bound to RNA (33). No reports of NMR-measured dynamics for RNase III-derived dsRBDs appear in the literature.
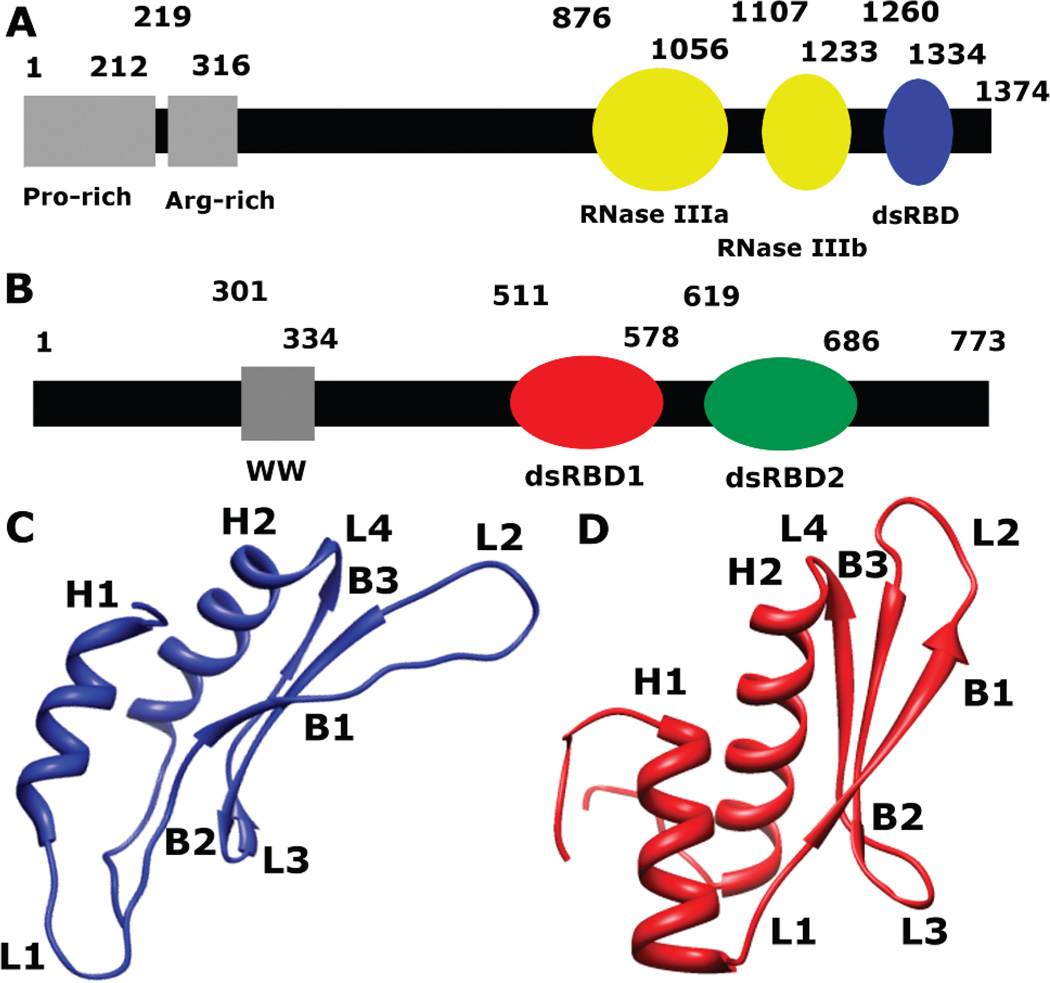
Drosha, a class II RNase III enzyme, is a 1374 amino acid residue protein in its primary human isoform that contains a proline-rich region and an arginine/serine-rich region at the N-terminus, with two RNase III domains and a dsRNA binding domain at the C-terminus (Figure 2A) (9). Association with the dsRNA binding protein DGCR8 is required for Drosha to specifically cleave pri-miRNA roughly 11 base pairs from the ss-ds RNA junction (34). To date, only the solution structure of the dsRBD of Drosha (1259–1337) has been determined (Figure 2C) (30). Drosha-dsRBD adopts the αβββα secondary structure seen in all dsRBDs but also features a unique extension of loop 1 that is negatively charged (Figures 1 and 2C) (30). Although unconfirmed experimentally, sequence features important for RNA recognition are conserved in Drosha-dsRBD, suggesting that Drosha retains the ability to bind dsRNA and is in fact necessary for pri-miRNA processing in vitro (35). Zheng and Cullen demonstrated that the arginine-rich region of Drosha is responsible for ssRNA recognition (36) and that Drosha is also capable of recognizing terminal loops (37).
Figure 2.
Schematic representation of the primary sequence of (A) Drosha and (B) DGCR8. (C) A ribbon diagram representing the solution structure of Drosha-dsRBD (PDB 2KHX, residues 1259–1337) shows the extended loops 1 (L1) and 2 (L2). (D) A ribbon diagram representing the crystal structure of DGCR8-dsRBD1 (PDB 2YT4, residues 505–583) shows a less elongated fold of the dsRBD than Drosha-dsRBD.
In order for Drosha to specifically cleave pri-miRNA, it must be associated with another dsRNA binding protein, DGCR8(38), which is a 773 amino acid residue protein that contains two dsRBDs in tandem at its C-terminus (Figure 2B) (39). The rest of the protein is intrinsically disordered and without known function, except for a WW motif that is proposed to mediate heme-activated dimerization (40). The crystal structure of the Core region of DGCR8 (residues 492–720 of the human sequence) revealed that two dsRBDs are arranged in a pseudo-2-fold symmetry and packed against a well-defined secondary structure formed from the linker and the C-terminal tail of the construct (39). This well-organized arrangement of the dsRBDs is in contrast to the NMR structure of PKR, which shows a flexible linker and structural independence of the two dsRBDs in the apo state (41). The two dsRBDs of DGCR8 contribute to high-affinity binding of pri-miRNA, Kd = 2.1 ± 1.1 µM for pri-miR-16-1 (38, 39). The binding affinities for the individual dsRBDs of DGCR8 have not been reported. Like Drosha-dsRBD, no NMR dynamics measurements have been reported for DGCR8-Core or its individual dsRBDs; however, we have recently reported molecular dynamics simulations of the Core and its dsRBDs that suggest a functional role for conformational dynamics in pri-miRNA binding (42).
Here we present both NMR and MD studies of Drosha-dsRBD, as well as NMR dynamics studies of DGCR8-dsRBD1, in order to provide a dynamic profile of these dsRBDs. Combined with binding assays utilizing the same dsRBDs, this work presents a unified picture for the role of dynamics in binding. The present study aims to incorporate qualitative and quantitative data on structural and dynamic contributions to dsRNA binding by dsRBDs, resulting in a predictive model transferrable to other dsRBDs.
MATERIALS AND METHODS
Protein Preparation
Drosha-dsRBD (1259–1337) was amplified by PCR from an ATCC plasmid (I.M.A.G.E Clone ID 5114643). The PCR product was then cloned into pET47b (Novagen), which encodes a 6× His tag and a 3C protease recognition site upstream of the cloning site, followed by transformation into BL21(DE3) competent cells. A liter growth in M9 minimal media with [15N] ammonium chloride as the only nitrogen source and either [12C]- or [13C] glucose as the only carbon source produced uniformly 15N or uniformly 15N, 13C-labeled protein. Expression was induced using 500 µL of 1.0 M IPTG at OD600 ≈ 0.5, and the cells were harvested after 3.5 h. The cells were lysed by sonication at 4 °C. The suspension was scentrifuged for 30 min at 11500 rpm in a Beckman Coulter Allegra 25R using a TA-14-50 rotor. The clear supernatant was then passed over a Ni-NTA (Novagen) column, and the protein was eluted with imidazole (200 mM). The His tag was cleaved using 3C protease at 4 °C overnight while also dialyzing away the imidazole. The content of the dialysis bag was then passed over the same Ni-NTA column, and the flow-through was collected. The protein was concentrated and buffer exchanged using an Amicon Ultra centrifugal filter device (Millipore) that contained a PES 3000 MWCO membrane. Drosha was buffer exchanged into 100 mM cacodylate, pH 7.0, 100 mM potassium chloride, 1 mM dithiothreitol, 0.02% sodium azide, and 10% deuterium oxide. Final concentration of the 15N NMR sample used for spin relaxation measurements was determined by UV absorption using ε = 5600 M−1 cm−1 at 278 nm to be 380 µM.
A synthetic DGCR8 (493–720) gene was purchased from Geneart, and DGCR8-DSRBD1 (505–720) was amplified by PCR. The PCR product was then cloned into pET49b (Novagen), which encodes a 6× His tag, a GST fusion tag, and a 3C protease recognition site upstream of the cloning site, and then transformed into BL21(DE3) competent cells. The protein expression and purification were the same as for Drosha-dsRBD, except that DGCR8-dsRBD1 was buffer exchanged into 20 mM HEPES, pH 7.0, 100 mM sodium chloride, 1 mM dithiothreitol, 0.02% sodium azide, 5 mM β-mercaptoethanol, and 10% deuterium oxide. Final concentration of the 15N NMR sample used for spin relaxation measurements was determined by UV absorption using ε = 4200 M−1 cm−1 at 278 nm to be 350 µM.
RNA Preparation
pri-miR-16-1 DNA containing a T7 promoter sequence at the 5′ end and an inverted BsaI cut site at the 3′ end from Geneart was cloned into pUC19 (New England Biolabs) and then transformed into DH5α competent cells. These cells were grown overnight in LB media at 37 °C until an OD600 of approximately 3.75 was reached. The cells were then lysed, and the DNA was purified using a Plasmid Maxi Kit (Omega). The recovered DNA was digested with BsaI overnight at 50 °C. After digestion, calf intestinal alkaline phosphatase was added and incubated another 30 min at 37 °C to prevent self-ligation. Postdigestion, the linearized DNA was extracted with phenolchloroform and precipitated with ethanol. The pri-miR-16-1 was thereafter transcribed by T7 polymerase in a 10 mL reaction mixture of 25 µg/mL linearized DNA, 40 mM Tris, pH 8.0, 25 mM MgCl2, 2 mM dithiothreitol, 1 mM spermidine, and 4 mM each of free NTPs. The transcription incubated at 37 °C for 3 h. The mixture was then purified by polyacrylamide gel electrophoresis in which the desired RNA band was cut out of the gel and soaked overnight at 4 °C in a TEN250 solution. The RNA was then purified from the supernatant by ethanol precipitation and quantified by UV–vis absorption, using ε = 1125400 M−1 cm−1 at 260 nm.
Electrophoretic Mobility Shift Assay
The pri-miR-16-1 RNA prepared by in vitro transcription was dialyzed in 10 mM cacodylate, pH 6.0, and 30 mM KCl. The proteins were dialyzed in 100 mM cacodylate, pH 6.0, 100 mM KCl, and 0.35 µg/mL β-mercaptoethanol. The RNA was radiolabeled using [γ-32P] ATP. Prior to mixing with protein, the RNA was denatured at 95 °C for 1 min and renatured at 1 °C for 1 min. The binding reactions incubated at room temperature for 3 h to ensure full equilibration in the presence of 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 5% glycerol, 100 µg/mL bovine serum albumin, 1 mM dithiothreitol, and 0.1 mg/mL herring sperm DNA to prevent the complex from sticking in the wells. Subsequently, the binding reactions were run on a 0.25× TBE, 10% acrylamide gel at 12 V cm−1 at 4 °C for 3 h, with each lane containing 20 µCi. Signal from free and bound pri-miR-16-1 was quantified by imaging the gel on a Typhoon-9410 imager, and uncertainties were estimated by averaging the intensities in each lane from duplicate gels. The resulting fraction-bound curves were fit to both a general model assuming N-independent and identical binding sites and a fully cooperative model, analogous to that used for Hill analysis. Data fitting was performed using the Levenberg–Marquardt model as implemented in Matlab (MathWorks).
NMR Methods
Standard triple resonance NMR techniques (43, 44) were used to assign the backbone resonances of Drosha-dsRBD on a Bruker Avance III 850 MHz spectrometer (chemical shifts are reported in Supporting Information Table S1). The backbone and side chain resonances of DGCR8-dsRBD1 were assigned using data from Bruker Avance III 500 and 600 MHz spectrometers (chemical shifts are reported in Supporting Information Table S2) (43, 44). Representative 15N-HSQC spectra for both proteins are shown in the Supporting Information. The spin relaxation experiments were performed on Bruker Avance III 500 and 600 MHz spectrometers using standard 15N relaxation methods (45, 46). All spectrometers were equipped with TCI cryoprobes for maximum sensitivity, and the experiments were performed at 25 °C. Spectra were processed by NMR pipe and analyzed with SPARKY (SPARKY3.113; T. D. Goddard and D. G. Kneller, University of California, San Francisco, CA). Data were analyzed in Matlab.
Model-Free Analysis
Lipari–Szabo model-free fitting was performed using the program Model Free 4.20 (47), with diffusion tensor fitting performed using the quadric method (48, 49). The coordinates from the NMR structure of Drosha-dsRBD (2KHX) and the crystal structure of DGCR8 (2YT4) were used as structural references for diffusion tensor determination. T1, T2, and NOE data were fit to a model including the axially symmetric global diffusion parameters with S2 and τint (model 2), except residues 516–518 of Drosha-dsRBD, which were fit with S2 and Rex (model 3) to model internal motions.
Simulations
MD trajectories were run in the AMBER 10.0 software package (50) using the ff99SB (51, 52) force field. Simulations were carried out in explicit solvent represented by the SPC water model (53) under particle mesh Ewald periodic boundary conditions (54). DGCR8-dsRBD1 and Drosha-dsRBD MD simulations were run as previously reported (42). All 10 configurations from the Drosha-dsRBD PDB file (2KHX) were used for initial analysis (30). Chloride counterions sufficient to neutralize the net positive charge on the proteins were added, and the resulting systems were solvated such that no solute atom was within 10 Å of a box edge. The 10 configurations were energy minimized as previously reported (55). Since structure 6 in the PDB file had the lowest energy after minimization, the rest of the simulation was continued with this structure. Following the initial equilibration period, 250 ns of dynamics was run in an isothermal–isobaric (NPT) simulation for each construct. Snapshots from each trajectory were stored to disk every 1.0 ps. The analysis of the trajectories was done in AMBER using the ptraj program (50). Molecular graphic images were created using the UCSF Chimera package (56). Additional analysis and visualization was accomplished in Matlab. MD-derived order parameters were obtained by using iRED analysis of MD trajectories averaged over 5 ns windows, as previously reported (55, 57).
RESULTS
Drosha-dsRBD and DGCR8-dsRBD1 Binding to pri-miR-16-1
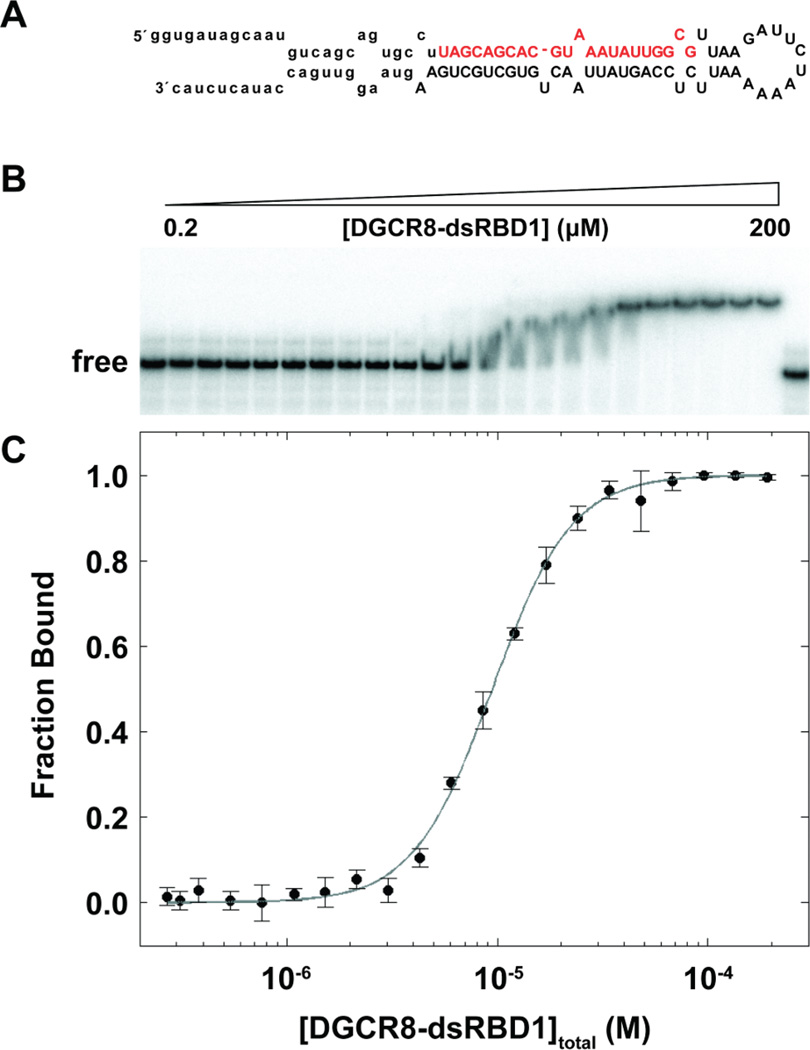
The present study was motivated by electrophoretic mobility shift assays (EMSAs) of pri-miR-16-1 (sequence shown in Figure 3A) binding by Drosha-dsRBD (1259–1337) and DGCR8-dsRBD1 (505–583). Pri-miR-16-1 was chosen as the model pri-miRNA since it has previously been utilized in other studies with DGCR8 (38, 39) and Drosha (34, 37). We observe no binding of pri-miR-16-1 by Drosha-dsRBD, even at 250 µM (see Supporting Information Figure S1). Therefore, we conclude that Drosha-dsRBD does not possess intrinsic binding affinity for pri-miR-16-1 in the absence of the Drosha-RNase III domains and is unlikely to bind other pri-miRNAs as well. In comparison, the EMSAs of DGCR8-dsRBD1 demonstrate that DGCR8-dsRBD1 binds to pri-miR-16-1 with a Kd = 9.4±0.4 µM (Figure 3B,C), which is less than 3-fold weaker binding compared with DGCR8-Core under the same conditions (Kd = 3.7 ± 0.1; Supporting Information Figure S2). The steep slope of the transition contributes to a measured Hill coefficient of n = 2.34± 0.01 that is consistent with cooperative binding of more than one DGCR8-dsRBD1 per pri-miR-16-1. The steep slope could also be explained by loss of free protein during equilibration or dissociation during gel electrophoresis, but gels run with lower incubation times show multiple intermediates reinforcing the assumption that the steep slope is indicative of cooperative binding (Quarles and Showalter, unpublished).
Figure 3.
EMSA of pri-miR-16-1 binding to DGCR8-dsRBD1. (A) Predicted secondary structure of pri-miR-16-1 with the sequence of the mature miRNA shown in red and the region removed by Drosha cleavage indicated through lower case letters. (B) Representative gel showing addition of DGCR8-dsRBD1 (2–200 µM) to 0.25 nM pri-miR-16-1. (C) Fitted EMSA fraction bound as a function of DGCR8-dsRBD1 concentration with data points and uncertainties represented by filled circles and the best fit to the data (see text) represented as a gray line.
NMR Spin Relaxation
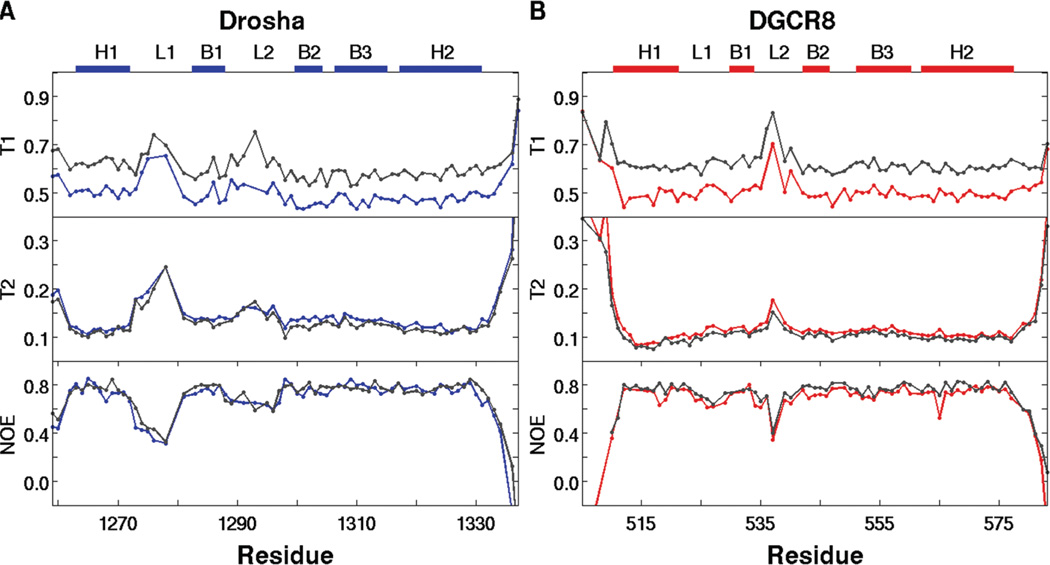
Information on picosecond to nanosecond time scale dynamics can be obtained from backbone 15N NMR spin relaxation data (45, 46, 58, 59). We have measured 15N T1, T2, and {1H}-15N NOE NMR spin relaxation at 500 and 600 MHz field strength for both Drosha-dsRBD and DGCR8-dsRBD1 in the RNA-free state (Figure 4). Analysis of the spin relaxation data using the quadric method (48, 49) indicates that Drosha-dsRBD is more anisotropic in solution having D‖/D⊥= 1.40 and τiso = 6.29 ns compared to D‖/D⊥ = 1.29 and τiso = 7.20 ns for DGCR8-dsRBD1. In both cases, the anisotropy of these domains is significantly higher than previously reported for the isolated dsRBDs of PKR, D‖/D⊥ ≈ 1.06 (τiso was not reported) (26). This trend is qualitatively observed in the ribbon diagrams, where DGCR8-dsRBD1 (Figure 2D) is less elongated than Drosha-dsRBD (Figure 2C).
Figure 4.
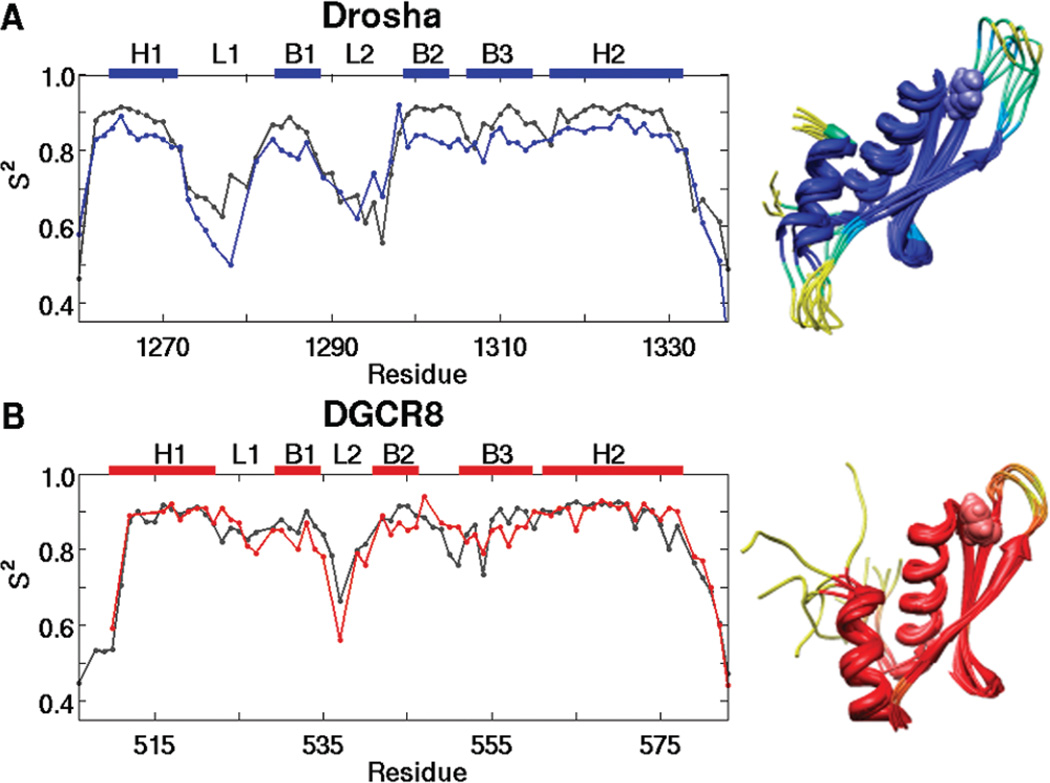
15N spin relaxation data for (A) Drosha-dsRBD and (B) DGCR8-dsRBD1 collected at 500 MHz (blue and red, respectively) and 600MHz (gray). The data show that the extended loop 1 of Drosha-dsRBD is more flexible than that of DGCR8-dsRBD1 on the picosecond to nanosecond time scale. Loop 2 is also seen to be highly dynamic in both proteins. The secondary structure elements for the respective dsRBDs, as well as the positions of loop 1 and loop 2, are represented as colored bars above the plots.
Complete model-free analysis of the spin relaxation data yields an estimation of the amplitude of picosecond to nanosecond time scale backbone conformational dynamics through the generalized order parameter S2 (Figure 5) (58). Loop 2 of both Drosha-dsRBD and DGCR8-dsRBD1 shows increased dynamics compared to the rest of the domain, as expected based on Staufen-dsRBD3 (19) and PKR (41) apo dynamics. Even though loop 2 is highly flexible in both domains under investigation, the C-terminal residue of the region, which corresponds to Tyr-1298 and Phe-542 in Drosha-dsRBD and DGCR8-dsRBD1, respectively, shows a high order parameter compared with its neighbors. These residues correspond with the highly conserved aromatic residue that is hypothesized to be a critical determinant of dsRNA binding by dsRBDs, since it maintains the spacing between loop 2 and loop 4 (20). In addition, the high order parameter indicates that the conserved aromatic residue is very rigid on the fast time scale.
Figure 5.
Order parameter (S2) plots for (A) Drosha-dsRBD and (B)DGCR8-dsRBD1 show that loop 2 in both proteins and loop 1 of Drosha-dsRBD are the most dynamic regions of the domains. Experimental data (blue and red lines for Drosha-dsRBD and DGCR8-dsRBD1, respectively) are plotted against MD predicted order parameters (gray). The secondary structure elements for the respective dsRBDs, as well as the positions of loops 1 and 2, are represented as colored bars above the plots. S2 is represented color imetrically in the MD-derived ribbon bundles for (C) Drosha-dsRBD and (D) DGCR8-dsRBD1, with passage from dark blue and dark red toward yellow indicating increased flexibility, based on the experimental order parameters. The conserved aromatic residues (Tyr-1298 of Drosha-dsRBD and Phe-542 of DGCR8-dsRBD1) are shown in space-filling mode to demonstrate how these residues are orientated to preserve the spacing between loop 2 and loop 4. Both bundles are created by taking the structures from the simulation every 50 ns and superimposing them to remove translation and rotation of the center of mass.
Unlike Staufen-dsRBD3 (19), there is no indication of increased dynamics in loop 4 for the apo state of either dsRBD, but this is consistent with data from PKR (26). In all structures used for alignment, except Staufen-dsRBD3 and DGCR8-dsRBD2 (Figure 1), there is a conserved serine or threonine in loop 4 forming an N-cap for H2 (60) and likely restricting the dynamics of the loop as a result. Instead of a serine or threonine, Staufends-RBD3 has a glycine in this position, which explains the increased flexibility for this region as glycine is a poor N-cap residue (60) and is intrinsically more flexible. The asparagine residue found inDGCR8-dsRBD2 should be capable of forming a strong N-cap and, in this sense, is a conservative mutation from the consensus. Our data indicate that Staufen-dsRBD3 is a unique case among dsRBDs that does not contain a good N-cap residue for helix 2, which causes the loop preceding (loop 4) to have increased flexibility.
The key difference in the order parameter profiles of the two dsRBDs investigated in this study is that the Drosha-dsRBD order parameter profile (Figure 5A) indicates that loop 1 is the most flexible region of the domain, while this same loop in DGCR8-dsRBD1 shows no increased flexibility in comparison to the rest of the domain. This trend is likely due to the addition of five amino acid residues in the loop of Drosha, which is a unique feature of Drosha-dsRBD compared with other dsRBDs (Figure 1) and is likely the cause for the increase in anisotropy of the domain compared to DGCR8-dsRBD1 and the two dsRBDs of PKR. Additionally, the greater flexibility of loop 1 in Drosha-dsRBD is likely disruptive to Drosha-dsRBD’s ability to bind dsRNA unassisted.
Molecular Dynamics Simulations
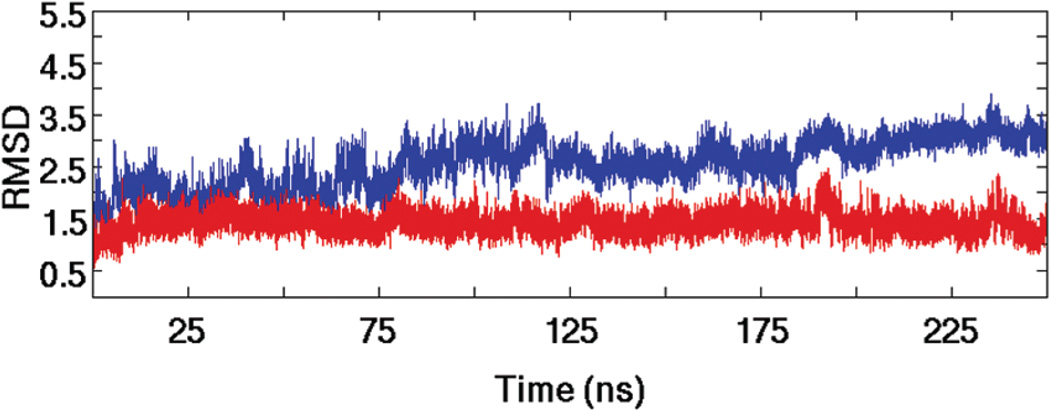
Two MD simulations were run: DGCR8-dsRBD1 (505–583) derived from the crystal structure of DGCR8-Core (PDB 2YT4) and one derived from the lowest energy structure, with respect to the AMBER ff99SB force field, in the solution ensemble of Drosha-dsRBD (PDB 2KHX, 1259–1337). The DGCR8-dsRBD1 trajectory has previously been reported (42), but the analysis reported in this study is novel (except where noted). The Drosha-dsRBD trajectory however has not previously been reported. Protein stability in the 250 ns simulations was verified by analyzing root-mean-square deviation (rmsd) from the starting structure over the course of the trajectories (Figure 6). The rmsd of DGCR8-dsRBD1 (as previously reported) was very low (1.5 Å) for a large majority of the simulation (Figure 6, red line), indicating that this construct is highly stable (42). The rmsd of Drosha-dsRBD is higher than DGCR8-dsRBD1 (Figure 6, blue line) but eventually plateaus at 3.0 Å, demonstrating that this construct is also stable over the simulation time scale. The higher rmsd seen in the Drosha-dsRBD simulation is typical of a simulation initiated from an NMR solution structure rather than a crystal structure, as was used for DGCR8-dsRBD1. Additionally, Drosha-dsRBD loop 1 and loop 2 are larger and more dynamic than the same loops in DGCR8-dsRBD1, which inflates the calculated rmsd by roughly 1.0 Å (data not shown). As final evidence that the high rmsd is not due to local unfolding of secondary elements or tertiary contacts, ribbon bundles for both simulations were generated from the trajectories (Figure 5C,D). The ribbon bundles clearly show that the overall folds are retained throughout both simulations. Additionally, the ribbon diagrams show the increased flexibility of loop 1 and loop 2 in Drosha-dsRBD compared with those in DGCR8-dsRBD1 throughout the simulation.
Figure 6.
The global dynamics of Drosha-dsRBD and DGCR8-dsRBD1 observed in MD simulations indicates qualitative differences between the two domains. The rmsd traces of Drosha-dsRBD (blue line) and DGCR8-dsRBD1 (red line) demonstrate that both proteins are stable over the MD simulations, although Drosha dsRBD is more dynamic overall, as seen in the MD structure bundles found in Figure 5C,D.
MD-Derived Order Parameters
The generalized model-free order parameter S2 was computationally predicted using iRED analysis of molecular dynamics (MD) trajectories averaged over 5 ns windows (Figure 5, gray lines) (55, 57). Qualitatively, the same global trends observed in the experimental data are reproduced computationally in both Drosha-dsRBD and DGCR8-dsRBD1 (Figure 5). The notable exception is that loop 3 of DGCR8-dsRBD1 is computationally predicted to be more dynamic than the adjacent secondary structural elements, which is not observed experimentally. Loop 3 of DGCR8-dsRBD1 has been shown computationally to mediate interfacial interactions with dsRBD2 in the Core structure of DGCR8 (42). The interfacial interactions cause loop 3 of DGCR8-dsRBD1 to be bent toward dsRBD2 in the crystal structure, a conformation enabled by the positive ϕ angle observed for Asp-549 in the crystal structure. This bend of loop 3 is not observed in the solution structure of isolated DGCR8-dsRBD1 (PDB 1X47). Therefore, the disagreement between experimental and computational data for loop 3 of DGCR8-dsRBD1 is likely due to the crystal structure used for the MD simulation and the solution structure of DGCR8-dsRBD1 being different for this region.
Another observation to note is that for Drosha-dsRBD there is an offset in the baseline between the experimental and computational order parameter profiles which is not observed in DGCR8-dsRBD1. Intriguingly, it is the experimental order parameters that adopt a low average value compared to the MD, and compared with both sets of order parameters for DGCR8-dsRBD1, and not the other way around. The generally high order parameters seen for the secondary elements in Drosha-dsRBD support the conclusion that the conformation of the protein in the MD simulation was dynamically stable but substantially drifted from the initial configuration during equilibration, producing the high rmsd observed (Figure 6).
Correlated Dynamics in the dsRBDs
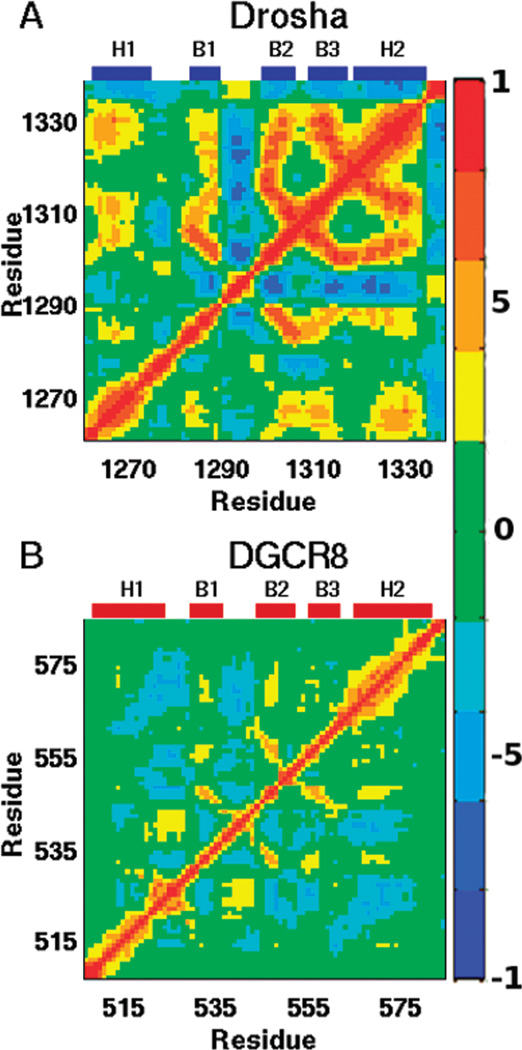
A major advantage of running MD simulations is the retention of dynamic correlations in the data set. Such correlations can be extracted through principal component analysis (61, 62) or a variety of related techniques that have been developed to monitor dynamics that are believed to be functionally relevant (63–65). Analysis of Cα atomic fluctuations in our MD trajectories produces the covariance matrices shown in Figure 7. Note the Cα correlation for DGCR8-dsRBD1 has already been reported (42), but the analysis discussed below is novel. The correlation plots for Drosha-dsRBD and DGCR8-dsRBD1 show similar features, with the features in Drosha-dsRBD indicating a greater absolute magnitude of correlation for a higher percentage of the residues than is the case for DGCR8-dsRBD1. One exception is the positive correlation between loop 1 and loop 2 in DGCR8-dsRBD1, which is absent in Drosha-dsRBD. Previous studies have shown that both loop 2 and helix 1 are involved in binding dsRNA (13, 19, 20), so it makes sense that loop 2 and loop 1 (which is connected to helix 1) show correlated motions in the apo state that would facilitate dsRNA binding. It has already been mentioned that loop 1 of Drosha is structurally and dynamically unique compared to other dsRBDs, and the correlated dynamics further expand on this. The lack of correlations between the two loops in Drosha-dsRBD could further contribute to Drosha-dsRBD not being able to bind dsRNA in the absence of the RNase III domains.
Figure 7.
Cα correlation matrices reveal the collective backbone motions of (A) Drosha-dsRBD and (B)DGCR8-dsRBD1. The color bar on the right shows the scale indicating strong positive correlation (red), strong negative correlation (blue), and noncorrelated motion (green). Labels above each panel indicate the location of secondary structural elements within the sequence.
In both domains under investigation, anticorrelated motions exist between loop 2 and helix 2, with only minor anticorrelations seen between loop 2 and loop 4. Hence, the pivot for the anticorrelated motions observed between loop 2 and helix 2 may be loop 4 itself and the contacts it makes with the C-terminal region of loop 2 and the N-terminal region of sheet 2. Moreover, this yields an alternative explanation for the importance of the conserved aromatic residue (Tyr-1298 and Phe-542 for Drosha-dsRBD and DGCR8-dsRBD1, respectively). The role of a static spacer could easily be filled by a bulky aliphatic side chain, but such a residue would provide a highly flexible stacking surface compared to the rigid ring of a phenylalanine or tyrosine residue. The rigidity of the aromatic moiety from the phenylalanine or tyrosine residue may be preferred evolutionarily because of its enhanced ability to transduce dynamic correlations between its two stacking partners.
DISCUSSION
Both dsRBDs studied are part of the microprocessor complex, which is involved in the nuclear maturation of miRNA, but the binding data presented here point to different roles for the two domains in the complex. The results presented in this study identify key structural and dynamic features of dsRBDs that contribute to the binding mechanism of these domains to dsRNA. Previous studies have shown extensive cooperativity between the dsRBDs of the protein RDE-4 (66), which is involved in the processing of long continuously double-stranded small-interfering RNAs. Especially when considered in light of the correlated conformational dynamics our previous MD studies of DGCR8-Core have revealed (42), this work points to a central role for cooperative complex assembly in the microprocessor as a means of establishing specificity in the pri-miRNA cleavage reaction.
Intrinsic Binding Affinity of the dsRBDs
The slightly weaker binding of DGCR8-dsRBD1 reported here (Kd = 9.4 ± 0.4 µM) compared to DGCR8-Core (Kd = 3.7 ± 0.1 µM; Supporting Information Figure S2) is noteworthy, as it suggests a dominant role for dsRBD1 in RNA binding, with dsRBD2 present to fine-tune the interaction. This is similar to the respective roles of PKR-dsRBD1 and PKR-dsRBD2; albeit PKR-dsRBD1 binds ~30-fold weaker than the two domains in tandem (26, 67, 68). Analysis of the EMSA results with a binding model that assumes N-independent and identical sites on pri-miR-16-1 for DGCR8-dsRBD1 resulted in poor fit quality (see Supporting Information Figure S3) that was completely relieved through the assumption of fully cooperative binding by more than one copy of the protein. The result that more than one dsRBD can be loaded onto a single pri-miRNA is unsurprising. The construct possesses approximately three full turns of (interrupted) A-form helix (Figure 3A), and there are at least three dsRBDs in the microprocessor (assuming a 1:1:1 DGCR8:Drosha:miRNA stoichiometry). Hill analysis of binding data in the Supporting Information of Sohn et al. also points to the possibility of cooperative loading of more than one DGCR8-Core molecule onto the same pri-miRNA (39). DGCR8-dsRBD1 packs against dsRBD2 in the crystal structure of DGCR8-Core, for which we have previously reported dynamic correlations between the two dsRBDs by MD (42). The apparent cooperative loading of multiple DGCR8-dsRBD1 molecules onto pri-miR-16-1 could be a manifestation of the intrinsic capability of the domain to assemble and further supports our hypothesis that collective positioning of the dsRBDs in the microprocessor imparts some or all of the specificity inherent in the endonuclease activity of the complex. Further studies will be needed to quantitatively define the stoichiometry in the complex.
While our results show DGCR8-dsRBD1 possesses intrinsic RNA binding affinity, we also observe a lack of pri-miRNA binding in vitro for the isolated Drosha-dsRBD that is likely due to both structural and dynamic features that are inconsistent with those of other studied dsRBDs known to bind dsRNA. First, dsRBDs known to bind dsRNA, like PKR-dsRBD1 (17, 26) and Staufen-dsRBD3 (19), generally contain three lysine residues that are on the N-terminal part of helix 2 and positioned between helix 1 and the β-sheet on the protein surface (Figure 1). Even though Drosha-dsRBD lacks the conserved lysine residues, Mueller et al. show a positively charged region on the Drosha-dsRBD surface which could facilitate binding of the negatively charged phosphate backbone in this region, albeit in an atypical manner compared with models from dsRNA bound to dsRBDs in the literature (30). Second, loop 1 of Drosha-dsRBD is five residues longer than other dsRBDs, and this increase in size causes increased flexibility in the loop along with increased anisotropy of the domain (Figures 4 and 5). The increased dynamics of loop 1 in Drosha-dsRBD, which is negatively charged, allows it to sample conformational space that potentially would be occupied by dsRNA, thus hindering binding due to charge repulsion. Also, in contrast to DGCR8-dsRBD1 (Figure 7), loop 1 of Drosha-dsRBD shows no correlated motions with the rest of the protein construct, which would be an effective means of maintaining its distance from the presumptive binding surface if they were present,. However, our data do not rule out the possibility that Drosha-dsRBD contributes to dsRNA binding in more inclusive constructs containing the RNase III domains, and ongoing studies in our laboratory aim to address this possibility.
Dynamic Profile of Drosha-dsRBD and DGCR8-dsRBD1
Dynamically, Drosha-dsRBD is not a total anomaly among dsRBDs: it shows increased flexibility in loop 2 like Staufen-dsRBD3 (19), PKR-dsRBD1 and PKR-dsRBD2 (26), and DGCR8-dsRBD1. The increased flexibility of loop 2 has been hypothesized to be critical for binding, since the loop needs to rotate toward the RNA in order to interact with the 2′-hydroxyl groups from the minor groove of dsRNA (19). In the case of PKR-dsRBD2, the higher order parameter in loop 2 compared to PKR-dsRBD1 correlated with a much lower affinity of dsRNA (26). While being important, increased flexibility of loop 2 is not by itself a sufficient determinant of dsRNA binding, since Drosha-dsRBD which possesses this feature does not bind dsRNA.
Even though loop 2 is dynamic, the C-terminal region of the loop in the protein still remains in close proximity to loop 4. Our data indicate that loop 4 is not dynamic on the fast time scale, which is consistent with data from PKR (26) but differs from Staufen-dsRBD3 (19). Loop 4 is not dynamic in these cases due to a conserved serine or threonine that forms a good N-cap for helix 2, while Staufen-dsRBD3 contains a glycine in this position that causes increased flexibility. However, the added dynamics of loop 4 in Staufen-dsRBD3 is not detrimental to binding since it binds dsRNA with micromolar affinity (19).
Distance between Loop 2 and Loop 4
The overall good agreement between the experimental and computational order parameters for the two dsRBDs led us to further investigate the molecular dynamic simulations for structural and correlated features of the two domains. Chang and Ramos hypothesized that a critical determinant of RNA binding by dsRBDs is the distance separating loop 2 and loop 4, which corresponds with the spacing between the minor and major groove of A-form RNA (20). In the case of Staufen-dsRBD3, the distance between the two loops is maintained by a phenylalanine (13), which based on alignment is strongly conserved (as either phenylalanine or tyrosine) and is present in both DGCR8-dsRBD1 and Drosha-dsRBD (Figure 1A). As previously mentioned, the order parameter for Phe-542 inDGCR8-dsRBD1 and Tyr-1298 in Drosha-dsRBD is high in comparison to its neighbors, indicating that these residues are rigid and could potentially be involved in maintaining the spacing between the aforementioned loops.
As a further investigation, we calculated the distance between α-carbons in loop 2 and loop 4 found in our MD trajectories. For DGCR8-dsRBD1, Glu-540 in loop 2 maintains an average distance from Ala-558 and Ser-559 of 4.7 and 4.4Å, respectively. More than 99% of the time, Glu-540 is within 6Å of both of these residues. In Drosha, the average distance between Thr-1297 and Pro-1315 is 4.7Å, and they are kept within 6Å of each other 99% of the time as well. The order parameters for the residues in loop 2 (Glu-540 and Thr-1297 for DGCR8-dsRBD1 and Drosha-dsRBD, respectively) indicate that these residues are dynamic while the residues in loop 4 are not, but the distances between the residues in loop 2 and loop 4 are maintained on average throughout the simulation. In both cases, no hydrogen bonds or salt bridges are formed between the residues mentioned to preserve the close proximity of the atoms. Instead, for DGCR8-dsRBD1, a salt bridge is formed between Glu-540 in loop 2 and Lys-567 in helix 2. This salt bridge is evident in the crystal structure with the charged moieties being 4.8 Å apart. Throughout the simulation, the average distance of the two charged atoms is 5.8 Å. A similar salt bridge is not seen in Drosha-dsRBD since in the Drosha-dsRBD structure a glycine residue is present instead of a lysine. In fact, the Lys-567 of DGCR8-dsRBD is not a conserved residue in the other dsRBD sequences either (Figure 1); thus this would only help to explain the close proximity of loop 2 and loop 4 observed in DGCR8-dsRBD but not dsRBDs as a whole.
The conserved aromatic residue hypothesized to be responsible for maintaining the spacing between these two loops is observed to potentially be a pivot for anticorrelated motions between loop 2 and helix 2, which caused it to have a high order parameter compared to neighboring residues and to maintain the loop 2 to loop 4 spacing. In the case of DGCR8-dsRBD1, the additional salt bridge observed between Glu-540 and Lys-567 further controls the amount of correlation and dynamics of these two regions. The anticorrelated motions of these regions of the domain are potentially responsible for the ability of dsRBDs in the miRNA maturation pathway to bind a large variety of heterogeneous dsRNA precursors. Further NMR spin relaxation and MD simulations are needed to verify the generality of our conclusions and the predictive quality of our model for the effect of dynamics on dsRNA binding.
Supplementary Material
ACKNOWLEDGMENT
We thank Prof. Phil Bevilacqua for helpful evaluation of the manuscript and members of the Bevilacqua laboratory for assistance in initiating the EMSA assays.
Footnotes
This work was supported by start-up funds provided to S.A.S. by The Pennsylvania State University
Abbreviations: miRNA, microRNA; ss, single stranded; ds, double stranded; dsRBDs, dsRNA binding domains; DGCR8, DiGeorge critical region 8; NMR, nuclear magnetic resonance; MD, molecular dynamics; EMSA, electrophoretic mobility shift assay; RRMs, RNA recognition motifs; NOE, nuclear Overhauser effect; PKR, dsRNA-activated protein kinase R; PCR, polymerase chain reaction; IPTG, isopropyl β-d-1-thiogalactopranoside; Ni-NTA, nickel nitrilotriacetic acid; MWCO, molecular weight cutoff; GST, glutathione S-transferase; Tris, tris(hydroxymethyl)aminomethane; HEPES, N-(2-hydroxyethyl)-piperazine-N′-2-ethanesulfonic acid; PES, polyether sulfone; NTPs, nucleoside triphosphates; TEN250, 10 mM Tris, 1 mM EDTA, and 250 mM NaCl; HSQC, heteronuclear single-quantum coherence; rmsd, root-mean-square deviation.
SUPPORTING INFORMATION AVAILABLE
The Drosha-dsRBD EMSA (Figure S1), the DGCR8-Core EMSA (Figure S2), the N-independent fitting of DGCR8-dsRBD1 EMSA data (Figure S3), the representative 15N-HSQC spectra for Drosha-dsRBD and DGCR8-RBD1 (Figure S4), and assignments for the Drosha-dsRBD (Table S1) and DGCR8-dsRBD1 (Table S2) spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–230. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao Y, Srivastava D. A developmental view of micro-RNA function. Trends Biochem. Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Liang Y, Lu Q. MicroRNA epigenetic alterations: predicting biomarkers and therapeutic targets in human diseases. Clin. Genet. 2008;74:307–315. doi: 10.1111/j.1399-0004.2008.01075.x. [DOI] [PubMed] [Google Scholar]
- 5.Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J. Cell Sci. 2007;120:1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 6.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang GL, Huang QW, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ. Res. 2008;103:1072–1083. doi: 10.1161/CIRCRESAHA.108.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang ZG, Luo XB, Lu YJ, Yang BF. miRNAs at the heart of the matter. J. Mol. Med. 2008;86:771–783. doi: 10.1007/s00109-008-0341-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 10.Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 11.Nowotny M, Yang W. Structural and functional modules in RNA interference. Curr. Opin. Biol. 2009;19:286–293. doi: 10.1016/j.sbi.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 15.St. Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 17.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 18.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund SMV, Bycroft M, St. Johnston D, Varani G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang KY, Ramos A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2005;272:2109–2117. doi: 10.1111/j.1742-4658.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- 21.Carlson CB, Stephens OM, Beal PA. Recognition of double-stranded RNA by proteins and small molecules. Biopolymers. 2003;70:86–102. doi: 10.1002/bip.10413. [DOI] [PubMed] [Google Scholar]
- 22.Nagai K. RNA-protein complexes. Curr. Opin. Struct. Biol. 1996;6:53–61. doi: 10.1016/s0959-440x(96)80095-9. [DOI] [PubMed] [Google Scholar]
- 23.Showalter SA, Hall KB. A functional role for correlated motion in the N-terminal RNA-binding domain of human U1A protein. J. Mol. Biol. 2002;322:533–542. doi: 10.1016/s0022-2836(02)00804-5. [DOI] [PubMed] [Google Scholar]
- 24.Showalter SA, Hall KB. Altering the RNA-binding mode of the U1A RBD1 protein. J. Mol. Biol. 2004;335:465–480. doi: 10.1016/j.jmb.2003.10.055. [DOI] [PubMed] [Google Scholar]
- 25.Lindhout DA, McKenna SA, Aitken CE, Liu CW, Puglisi JD. PKR: a NMR perspective. Prog. Nucl. Magn. Reson. Spectrosc. 2007;51:199–215. [Google Scholar]
- 26.Nanduri S, Rahman F, Williams BRG, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kharrat A, Macias M, Gibson TJ, Nilges M, Pastore A. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 1995;14:3572–3584. doi: 10.1002/j.1460-2075.1995.tb07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun WM, Jun E, Nicholson AW. Intrinsic double-stranded-RNA processing activity of Escherichia coli ribonuclease III lacking the dsRNA-binding domain. Biochemistry. 2001;40:14976–14984. doi: 10.1021/bi011570u. [DOI] [PubMed] [Google Scholar]
- 29.Zhang HD, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 30.Mueller GA, Miller MT, DeRose EF, Ghosh M, London RE, Hall TMT. Solution structure of the Drosha double-stranded RNA-binding domain. Silence. 2010;1:5. doi: 10.1186/1758-907X-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Z, Lee JK, Tjhen R, Stroud RM, James TL. Structural and biochemical insights into the dicing mechanism of mouse Dicer: a conserved lysine is critical for dsRNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2391–2396. doi: 10.1073/pnas.0711506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leulliot N, Quevillon-Cheruel S, Graille M, van Tilbeurgh H, Leeper TC, Godin KS, Edwards TE, Sigurdsson STL, Rozenkrants N, Nagel RJ, Ares MJ, Varani G. A new α-helical extension promotes RNA binding by the dsRBD of rnt1p RNAse III. EMBO J. 2004;23:2468–2477. doi: 10.1038/sj.emboj.7600260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gan JH, Tropea JE, Austin BP, Court DL, Waugh DS, Ji XH. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 34.Han JJ, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho YJ, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 35.Han JJ, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeom KH, Lee Y, Han JJ, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y. Crystal structure of human DGCR8 core. Nat. Struct. Mol. Biol. 2007;14:847–853. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- 40.Senturia R, Faller M, Yin S, Loo JA, Cascio D, Sawaya MR, Hwang D, Clubb RT, Guo F. Structure of the dimerization domain of DiGeorge critical region 8. Protein Sci. 2010;19:1354–1365. doi: 10.1002/pro.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nanduri S, Carpick BW, Yang YW, Williams BRG, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wostenberg C, Noid WG, Showalter SA. MD simulations of the dsRBP DGCR8 reveal correlated motions that may aid pri-miRNA binding. Biophys. J. 2010;99:248–256. doi: 10.1016/j.bpj.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kay LE. NMR studies of protein structure and dynamics. J Magn. Reson. 2005;173:193–207. doi: 10.1016/j.jmr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 44.Kanelis V, Forman-Kay JD, Kay LE. Multi-dimensional NMR methods for protein structure determination. IUBMB Life. 2001;52:291–302. doi: 10.1080/152165401317291147. [DOI] [PubMed] [Google Scholar]
- 45.Palmer AG. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 46.Jarymowycz VA, Stone MJ. Fast time scale dynamics of protein backbones: NMR relaxation methods, applications, and functional consequences. Chem. Rev. 2006;106:1624–1671. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- 47.Mandel AM, Akke M, Palmer AG. Backbone dynamics in Escherichia coli ribonuclease HI correlations with structure and function in an active enzyme. J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 48.Bruschweiler R, Liao XB, Wright PE. Long-range motional restrictions in a multidomain zinc-finger protein from anisotropic tumbling. Science. 1995;268:886–889. doi: 10.1126/science.7754375. [DOI] [PubMed] [Google Scholar]
- 49.Lee LK, Rance M, Chazin WJ, Palmer AG., III Rotational diffusion anisotropy of proteins from simultaneous analysis of 15N and 13C alpha nuclear spin relaxation. J. Biomol. NMR. 1997;9:287–298. doi: 10.1023/a:1018631009583. [DOI] [PubMed] [Google Scholar]
- 50.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The AMBER biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hornak V, Okur A, Rizzo RC, Simmerling C. HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A. 2006;103:915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins: Struct. Funct. Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 54.Darden T, York D, Pedersen L. Particle mesh Ewald—an N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 55.Showalter SA, Bruschweiler R. Validation of molecular dynamics simulations of biomolecules using NMR spin relaxation as benchmarks: application to the AMBER99SB force field. J. Chem. Theory Comput. 2007;3:961–975. doi: 10.1021/ct7000045. [DOI] [PubMed] [Google Scholar]
- 56.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Prompers JJ, Bruschweiler R. General framework for studying the dynamics of folded and nonfolded proteins by NMR relaxation spectroscopy and MD simulation. J. Am. Chem. soc. 2002;124:4522–4534. doi: 10.1021/ja012750u. [DOI] [PubMed] [Google Scholar]
- 58.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules 1 Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 59.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem. Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 60.Doig AJ, MacArthur MW, Stapley BJ, Thornton JM. Structures of N-termini of helices in proteins. Protein Struct. 1997;6:147–155. doi: 10.1002/pro.5560060117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kitao A, Go N. Investigating protein dynamics in collective coordinate space. Curr. Opin. Struct. Biol. 1999;9:164–169. doi: 10.1016/S0959-440X(99)80023-2. [DOI] [PubMed] [Google Scholar]
- 62.Koyama YM, Kobayashi TJ, Tomoda S, Ueda HR. Perturbational formulation of principal component analysis in molecular dynamics simulation. Phys. Rev. E: Stat. Phys. Plasmas, Fluids. 2008;78:11. doi: 10.1103/PhysRevE.78.046702. [DOI] [PubMed] [Google Scholar]
- 63.Amadei A, Linssen ABM, Berendsen HJC. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 64.Berendsen HJC, Hayward S. Collective protein dynamics in relation to function. Curr. Opin. Struct. Biol. 2000;10:165–169. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 65.Antoniou D, Basner J, Nunez S, Schwartz SD. Computational and theoretical methods to explore the relation between enzyme dynamics and catalysis. Chem. Rev. 2006;106:3170–3187. doi: 10.1021/cr0503052. [DOI] [PubMed] [Google Scholar]
- 66.Parker GS, Maity TS, Bass BL. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J. Mol. Biol. 2008;384:967–979. doi: 10.1016/j.jmb.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cole JL. Activation of PKR: an open and shut case? Trends Biochem. Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ucci JW, Kobayashi Y, Choi G, Alexandrescu AT, Cole JL. Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences. Biochemistry. 2007;46:55–65. doi: 10.1021/bi061531o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.