Abstract
The cloning of DNA directly from environmental samples provides a means to functionally access biosynthetic gene clusters present in the genomes of the large fraction of bacteria that remains recalcitrant to growth in the laboratory. Herein we demonstrate a method by which complementation of phosphopantetheine transferase deletion mutants can be used to restore siderophore biosynthesis and to therefore selectively enrich eDNA libraries for nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) gene sequences to unprecedented levels. The common use of NRPS/PKS-derived siderophores across bacterial taxa makes this method generalizable and should allow for the facile selective enrichment of NRPS/PKS-containing biosynthetic gene clusters from large environmental DNA libraries using a wide variety of phylogenetically diverse bacterial hosts.
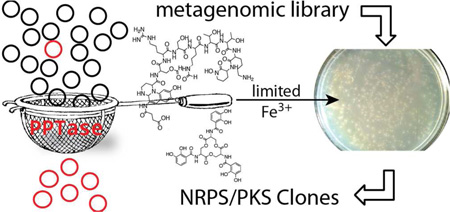
Difficulties associated with culturing bacteria from the environment have prevented the vast majority of microbes from being examined for the production of bioactive small molecules.1–3 The cloning of DNA extracted directly from environmental samples (eDNA) provides a means of accessing secondary metabolite gene clusters found in the genomes of environmental bacteria without the requirement of initially culturing these organisms.4 In most sequenced bacteria less than 2% of the genome is devoted to secondary metabolism.5 By extension, only a small fraction of the clones found in an eDNA library is expected to contain genes involved in small molecule biosynthesis. Many studies have circumvented this problem using homology-based screening methods, which rely on the PCR amplification of conserved natural product biosynthetic gene sequence motifs to identify and then recover gene clusters from large environmental DNA libraries.4, 6–8 The development of metagenomics-driven drug discovery platforms would benefit greatly from methods that permit the selective enrichment of large eDNA libraries for clones containing secondary metabolite biosynthetic genes. Here we show that complementation of phosphopantetheine transferase (PPTase) deletion mutants by eDNA clones encoding PPTase enzymes permits growth under iron limiting conditions through restoration of siderophore biosynthesis, resulting in the facile selective enrichment of large eDNA libraries for clones containing secondary metabolite biosynthesis genes.
Nonribosomal peptides and polyketides comprise the majority of pharmacologically relevant microbial secondary metabolites. These two classes of molecules are assembled by distinct but organizationally similar enzymatic assembly lines.9, 10 In each case, the transfer of a growing metabolite from one assembly line module to the next is achieved by tethering the intermediate to a flexible phosphopantetheine prosthetic group. Phosphopantetheine is covalently appended to a conserved serine on PKS-associated acyl-carrier-domains (ACP) and NRPS-associated peptidyl-carrier-domains (PCP) by enzymes known as PPTases.11 In the absence of this posttranslational modification neither biosynthetic system is functional. The dependence of NRPS/PKS biosynthesis on phosphopantetheine has in the past formed the basis of functional screens designed to identify biosynthetic gene sequences.12, 13 As PPTase genes frequently appear in bacterial NRPS/PKS gene clusters, we reasoned that the development of a method for the selective enrichment of eDNA libraries for PPTase genes would simultaneously provide a robust strategy for enriching large eDNA libraries with biosynthetic gene sequences.
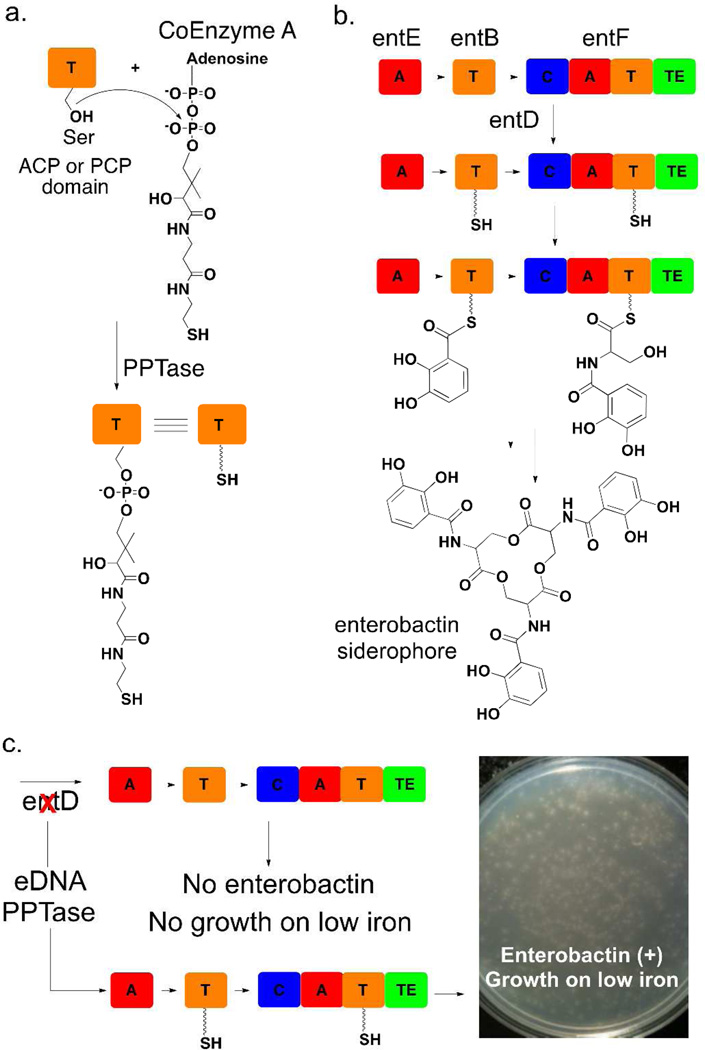
To obtain sufficient iron from the environment to support growth, bacteria commonly produce NRPS-derived siderophores that transport iron into the bacterial cytosol.14 As with all NRPSs, the function of NRPS siderophore biosynthetic machinery is dependent on PPTase mediated posttranslational modification. In fact, PPTase gene deletion mutants cannot produce NRPS derived siderophores and are often unable to grow under iron limited conditions. This is the case in E. coli where the NRPS derived siderophore enterobactin is obligatory for growth under low iron conditions. In addition to core NRPS enzymes directly involved in the biosynthesis of enterobactin, the enterobactin gene cluster also encodes the PPTase EntD that appends phosphopantetheine onto enterobactin biosynthesis PCP domains (Figure 1, 2a).14, 15 E. coli entD deletion mutants do not produce enterobactin and as a result cannot grow under iron limited conditions. As the largest eDNA libraries ever constructed are hosted in E. coli, our initial efforts to selectively enrich eDNA libraries for clones containing secondary metabolite biosynthesis genes relied on restoring the ability of E. coli entD mutants to grow on low iron medium. For this selection, an eDNA cosmid library constructed in an enterobactin proficient E. coli background was electroporated into an entD mutant and plated on iron depleted medium. Approximately 1 in every 2,000–2,500 library members grew on these selection plates. In the initial plating, each large colony was surrounded by a halo of smaller colonies that appeared to be scavenging the enterobactin produced by the central complementation colony. Upon re-plating to rid the library of satellite colonies approximately 1 in every 20 library members complemented the entD deletion, representing a 125-fold enrichment of the original library (Figure 1c). After a second round of selection up to 90% of the library was able to complement the entD deletion mutant. As this is a selection and not a screen, even the largest eDNA libraries currently available can be enriched with this strategy using minimal effort (Figure 1c).
Figure 1. Enterobactin Complementation Strategy.
(a) Apo ACP and PCP (T) domains are phosphopantetheinylated by a PPTase to create holo domains. (b) The biosynthesis of the siderophore enterobactin by E. coli requires the activity of the PPTase EntD [A=adenylation, C=condensation, T=thiolation (ACP or PCP), TE=thioesterase]. (c) entD deletion mutants do not grow on low iron due to the absence of enterobactin; however, growth on low iron media can be restored by complementing this deletion with eDNA clones expressing PPTases. Approximately 5,000,000 eDNA clones hosted in an E. coli entD deletion mutant were plated on low iron media for this image.
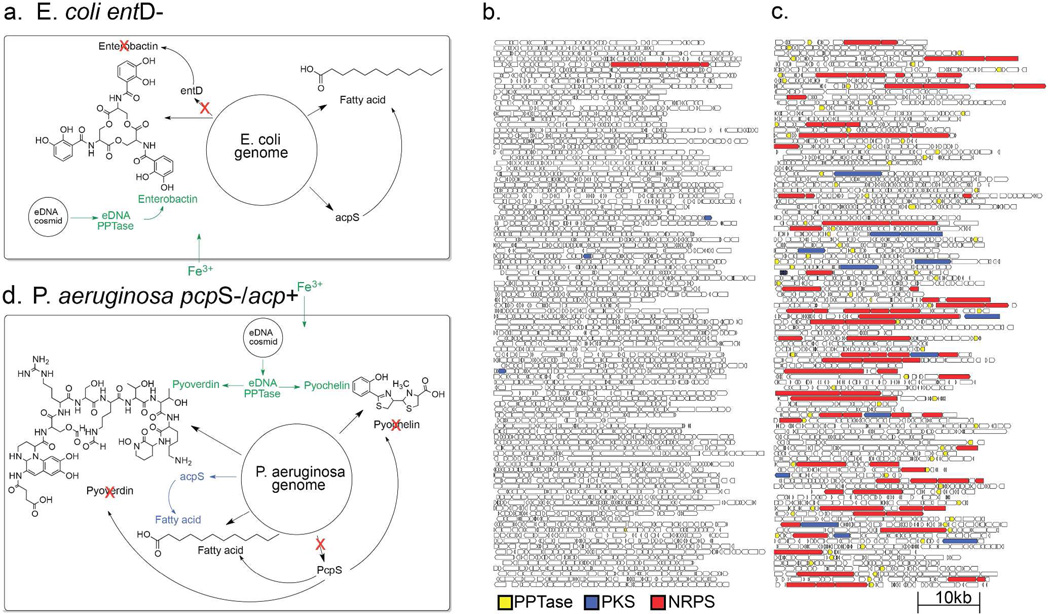
Figure 2. Library enrichment schemes and sequencing.
(a) E. coli PPTase selection scheme. One hundred cosmid clones from un-enriched (b) and E. coli PPTase enriched (c) eDNA libraries. PPTase (yellow), NRPS (red) and PKS (blue) genes are colored. (d) P. aeruginosa PPTase selection scheme. Genome positions in (a) and (d) are not drawn to scale.
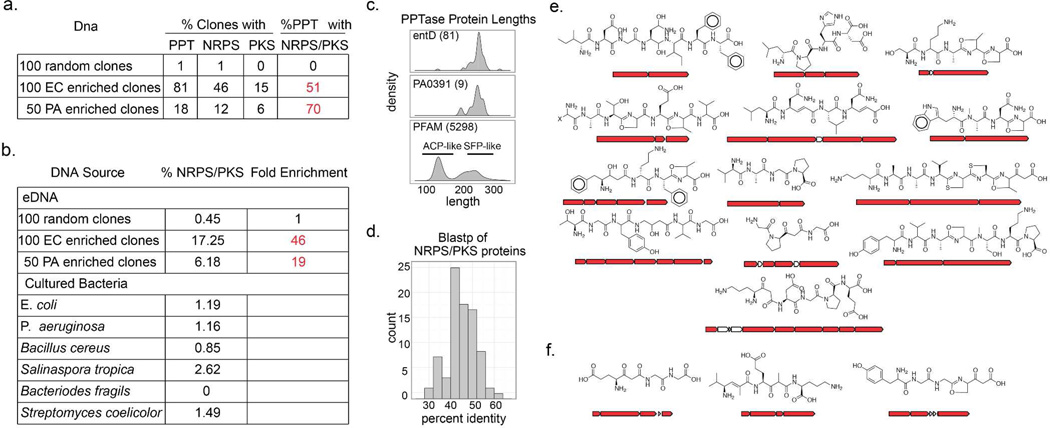
One hundred randomly selected clones from the enriched library, the equivalent of approximately one bacterial genome of DNA (~3–4 Mb), were sequenced and compared to sequences derived from an equivalent population of clones randomly selected from the un-enriched parent eDNA library. A simple qualitative analysis of these datasets (Figures 2b and 2c) indicates that not only was library successfully enriched for PPTase genes, but it was also enriched for NRPS/PKS biosynthetic genes. To quantitatively assess the success of this enrichment strategy the percentage of nucleotides contained within predicted NRPS/PKS genes was calculated for each of the above datasets, as well as for several phylogenetically diverse sequenced bacterial genomes (Figure 3b). The fraction of sequence space devoted to NRPS/PKS genes in most genomes is on the order of 1%, therefore it was no surprise to find a similarly low level of NRPS/PKS representation (0.45%) in randomly selected eDNA clones. In the case of the selectively enriched library the fraction of sequence space devoted to NRPS/PKS genes jumped to 17%, an almost 50 fold increase over the un-enriched library. In fact, the DNA captured in the PPTase enriched library is richer in secondary metabolism than any pool of genomic DNA sequenced to date.5 While Enterobacteriaceae, like E. coli, have been of limited utility as sources of bioactive natural products, selective enrichment of metagenomic libraries provides a strategy for rendering even these organisms potentially rich sources of secondary metabolites.
Figure 3. Library enrichment statistics.
(a) E. coli and P. aeruginosa library enrichment statistics (EC=E. coli, PA=P. aeruginosa, PPT=PPTase). Similar statistics from the analysis of a phylogentically diverse collection of sequenced bacterial species are also shown. (c) PPTase length comparisons can distinguish sfp- from acpS-like PPTases. Frequency histograms are plotted as density to allow co-plotting of samples with different sizes (in parentheses). PFAM proteins greater than 350 amino acids were excluded from the analysis. (d) Frequency histogram of percent identity for the top BlastP hit of each NRPS/PKS protein in the combined enriched dataset. Substructure predictions for the subset of large canonical NRPS/PKS operons found in the enriched libraries (e) E. coli (f) P. aeruginosa.
This selective enrichment strategy relies on the host’s recognition of foreign promoters located upstream of eDNA derived PPTase genes. As bacterial species are known to differ in their ability to utilize foreign genetic material, we wanted to test the feasibility of expanding this method to bacterial hosts with different expression capabilities. Pseudomonas aeruginosa produces two well-studied fluorescent NRPS derived siderophores, pyoverdine and pyochelin.16, 17 Unlike E. coli, which utilizes separate PPTases for primary and secondary metabolism, P. aeruginosa relies on a single PPTase, PcpS, to install phosphopantetheine onto all PCP/ACP domains of its proteome (Figure 2d). For a PPTase selection strategy to work in P. aeruginosa, the promiscuous pcpS gene would first need to be replaced with a PPTase that is selective for primary metabolic ACPs. Fortuitously, this had already been done in the previously described P. aeruginosa strain PAO391, in which pcpS was replaced with the E. coli fatty acid specific PPTase, acpS.18 To test the utility of PAO391 for the selective enrichment of eDNA libraries, a soil library constructed in the broad host range cosmid vector pJWC119, 20 was conjugated from E. coli into PAO391, and the resulting library was then passed through two rounds of low iron selection, as previously described for E. coli based libraries. Eighteen percent of the clones picked from the final selection plates encoded recognizable PPTase homologs, and 70% of the PPTase-containing clones also contained genes encoding NRPS/PKS enzymes. This corresponds to an almost 20-fold enrichment in NRPS/PKS genes compared to the un-enriched library, and suggests that PPTase selection should be a generalizable strategy for the enrichment of PKS/NRPS genes. As with E. coli, P. aeruginosa almost exclusivley enriched for secondary metabolism associated PPTases like sfp over fatty acid associated PPTases like acpS (Figure 3c). The reduced enrichment efficiency observed with P. aeruginosa is likely due to the fact that P. aeruginosa colonies are larger, leading to increased satellite colony cross contamination.
The superfamily of PPTases is divided into two sub-groups (ACP and SFP) that are named after the founding genes of their respective classes.21, 22 The ACP-type proteins are generally responsible for activating fatty acid biosynthetic machinery, while the SFP class of enzymes has pan-specificity, activating non-ribosomal peptides synthetases, polyketide synthases and fatty acid synthases. One potential drawback of PPTase enrichment strategies is the possibility that clones containing PPTases associated exclusively with primary metabolism would overwhelm the final enriched library, resulting in the failure to enrich for biosynthetic genes. SFP- and ACP-type PPTases differ in size22 (Figure 3c) and can easily be distinguished from each other based on this size discrepancy. Nearly all of the PPTases found in the enriched libraries resemble the SFP-type PPTase family members (~230 amino acids) that are commonly associated with secondary metabolism. Assuming that barriers to functional expression are equivalent amongst SFP- and ACP-type PPTases, this finding suggests that, as a general rule, metagenome derived ACP-type PPTases do not efficiently complement siderophore PPTase deletion mutants as well as SFP-type PPTases.
The usefulness of enriched eDNA libraries as sources of chemical novelty depends on the ability of library enrichment strategies to produce clones with gene clusters that are functionally distinct from previously characterized biosynthetic systems. Consistent with the fact that many NRPS and PKS gene clusters are larger than the 25–35 Kb of sequence that is typically captured on a single cosmid clone, many of the cosmids obtained using this enrichment strategy contained truncated biosynthetic gene clusters. As partial gene clusters are generally insufficient for facilitating the discovery of novel metabolites, the ultimate utility of this method will likely be realized when it is coupled with larger insert eDNA cloning strategies that are still in development. To assess the novelty of the enriched NRPS/PKS genes themselves, each predicted eDNA-derived megasynth(et)ase protein was compared with megasynth(et)ases from known biosynthetic gene clusters. When NRPS/PKS proteins from the enriched library were submitted for BLAST analysis against the NCBI NR dataset no protein showed greater than 60% identity (at 90% alignment) to any sequence deposited in GenBank, and the average identity was below 45% (Figure 3d), suggesting an enrichment for gene sequences that are only distantly related to previously characterized sequences. As even distantly related gene sequences can encode for the production of similar metabolites, we also assessed the functional novelty of the enriched NRPS/PKS megasynth(et)ases by comparing substructures predicted to arise from these enzymes with known natural products. Although gene-based natural product structure prediction algorithms still fail to predict all of the structural features encoded by complex biosynthetic systems, they are often quite good at predicting substructures from individual NRPS/PKS operons.23, 24 Each large NRPS/PKS operon sequenced from the enriched library was submitted for chemical structure prediction using NP.searcher23 (Figure 3e,3f), and the resulting substructures were searched against the Sci-Finder Scholar database. Several predicted peptides were found to be present as substructures within synthetic peptides but none matched a substructure within any known natural product. Taken together these analyses suggest that the gene clusters arising from PPTase enriched clones are likely to encode metabolites not previously reported in culture based studies.
By using PPTase selections to parse the extreme biosynthetic diversity present in large eDNA libraries it should be possible to generate highly enriched orthogonal sub-libraries that will render almost any bacterial species a potentially rich source of novel secondary metabolites. In this study both E. coli and P. aeruginosa based PPTase selection strategies led to the substantial selective enrichment of eDNA libraries for clones containing NRPS and PKS gene sequences. NRPS derived siderophores are common to many phylogenetically diverse bacterial species, and therefore complementation of siderophore PPTase deletion mutants should be a generalizable strategy that will permit the facile enrichment of very large eDNA libraries constructed in diverse bacterial expression hosts.
Methods
eDNA library
Library construction methods have been described in detail elsewhere.25 Briefly: Soil was sifted to remove large particulates, and then heated (70 °C) in lysis buffer (100 mM Tris-HCl, 100 mM EDTA, 1.5 M NaCl, 1% (w/v) CTAB, 2% (w/v) SDS, pH 8.0) for 2 hours. Soil particulates were removed from the crude lysate by centrifugation, and eDNA was precipitated from the resulting supernatant with the addition of 0.7 volumes of isopropanol. Crude eDNA was collected by centrifugation, washed with 70% ethanol and resuspended in TE. Gel purified (1% agarose) high-molecular-weight eDNA was blunt ended (Epicentre, End-It), ligated into of pWEB, pWEB-TNC (Epicentre) or the broad host range vector pJWC1, packaged into lambda phage, and transfected into E. coli EC100. Cosmid DNA was midipreped (Qiagen) from EC100 host libraries and electroporated into either E. coli-EC100ΔentD or E. coli S17.1.
Selection in E. coli
Cosmid DNA from EC100 based eDNA libraries was midipreped (Qiagen) and electroporated into E. coli-EC100ΔentD. After 1 hour recovery in SOC at 37°C transformations were diluted (1:50) into LB containing an appropriate selection antibiotic (30 µg/mL kanamycin, pWEB or 12.5 µg/mL chloramphenicol, pWEB-TNC) and then incubated with shaking overnight at 37°C. The resulting libraries were then stored directly as glycerol stocks. Glycerol stocks were titered and then plated on iron-deficient entD M9 selection media (M9 media supplemented with 1 g/L casamino acids, 10 µM thiamine-HCl, 100 µM 2,2-dipyridyl, antibiotic as needed) at titers as high as 5,000,000 clones per 150 mm plate (Figure S1). DNA clones capable of complementing the entD deletion mutant appeared after 12–16 h at 37°C at which time sterile M9 media was added to the selection plates, the colonies were resuspended en masse and stored as a glycerol stock. Glycerol stocks from the first round of selection were used to inoculate LB cultures containing antibiotic and these were grown to an OD600 of 0.5, at which point the culture was washed three times with M9 and plated onto iron-deficient entD complementation selection media. Colonies used in the analysis here were picked from these second round selection plates, arrayed in 96 well plates, grown as individual cultures, pooled, miniprepped as a pool and sequenced by 454 pyrosequencing.
The entD deletion was created using RedET recombination. Primers carrying homology arms identical to the first and the last 39 bp of the coding region of entD gene were used to amplify the disruption cassette containing apramycin resistance marker from pIJ77326 [Primers: entD-KOF ATG GTC GAT ATG AAA ACT ACG CAT ACC TCC CTC CCC TTT ATT CCG GGG ATC CGT CGA CC, entD-KOR TTA ATC GTG TTG GCA CAG CGT TAT GAC TAT CTT TTC TTT TGT AGG CTG GAG CTG CTT C]. The PCR reaction was digested with EcoRI/HindIII to eliminate any circular plasmids and the amplicon was then gel-purified. The gel purified PCR product was used to transform electrocompetent E. coli EC100 harbouring the temperature sensitive λ recombination plasmid pIJ790. Transformants were grown overnight at 37 °C to eliminate the temperature sensitive λ recombination plasmid and successful deletion mutants identified by selection on LB supplemented with 50 µg/ml apramycin.
Selection in P. aeruginosa
Midiprepped DNA from soil eDNA libraries constructed using the pJWC1 shuttle cosmid vector were electroporated into S-17.1 an E. coli which is competent for conjugation. The resulting S17.1 based library was then conjugated into a P. aeruginosa strain PAO39118 using established methods.20 In brief, cultures of PAO391 and the S17.1 based library were mixed in a 1:1 ratio and spotted on cellulose discs overlaying LB. After 4 hours the mixture was resuspended from the cellulose disc and plated on LB supplemented with tetracycline (100 µg/ml) and irgasan (25 µg/ml). After 48 h, exconjugants were resuspended en masse from the original selection plates and stored as glycerol stocks. Glycerol stocks were used to inoculate LB cultures (tetracycline, 50 µg/ml; gentamicin, 30 µg/ml), which were grown to an OD600 = 0.6, washed twice in 0.9% NaCl and plated onto Kings B low-iron selection plates (20 g/L Bacto Peptone, 5 g/L glutamine, 1.5 g/L potassium phosphate dibasic, 15 g/L agar, 10 ml/L glycerol, 400 µM 2,2-dipyridyl, 50 µg/ml tetracycline, 30 µg/ml gentamicin). Plates were grown overnight, washed with sterile 0.9% sodium chloride and re-plated onto Kings B low-iron media with an estimated cell density of 20,000 cells/plate. The high density of satellite colonies that appeared after the first round of selection prevented us from determining an accurate initial complementation rate. After the second round of plating, cells were allowed to grow for either two or three days until the fluorescent siderophore pyoverdine was visible. These cells were individually struck out on Kings B low-iron selective plates. Single colonies were grown in deep-well 48 well plates (4 ml) in the presence of BAC autoinduction solution (Epicentre), pooled together, midi prepped and submitted as a pool for 454 pyrosequencing.
Sequence Analysis
454 pyrosequencing data was analyzed by assembling raw data to contigs with the gsAssembler program (Roche) followed by ORF-finding with Metagenemark.27 ORFs were scanned against the PFAM-A database using HMMscan (HMMER 3.0 (March 2010); http://hmmer.org/) using an E value cutoff of 10e−10. PFAM-A domains used for annotation and subsequent analysis including NRPS/PKS calculations are as follows: PPTase ('PF01648'), PKS (‘PF00109'), NRPS ('PF08415', 'PF00668'). Data were then manually curated to remove fatty acid synthesis proteins that were picked up by HMM identification of PKS domains. Genomic plots were made with Biopython28; other plots were generated with R 2.1529, and ggplot2.30 Genomic statistics for previously sequenced genomes were calculated in the same manner. Genome protein datasets were scanned for the presence of the PFAM domains listed above and the genes encoding proteins in these families were included in our NRPS/PKS calculations.
Accession Codes
Genbank accession codes: 100 unselected clones: (JX827880 – JX827979); 100 E. coli-selected clones: (JX827780 – JX827879); 50 P. aeruginosa-selected clones:(JX827730 – JX827779).
Small Molecule Structure Prediction and Analysis
Gene clusters from enriched datasets were picked for small molecule structure prediction if it appeared that NRPS/PKS genes would allow for reliable prediction. Large NRPS/PKS operons were submitted to the NP.searcher webserver to obtain linear substructure predictions.23 Side chain cyclizations were added manually. Each substructure was compared to known metabolites by querying against Sci-Finder scholar using their substructure search algorithm, and displayed using ChemDraw (PerkinElmer).
Acknowledgments
This work was supported by NIH GM077516 and NIH MSTP grant GM07739 (JWC). SFB is an HHMI Early Career Scientist. We thank C. Gleber and D. Kalifidas for their assistance. PAO391 was a gift from H. Schweizer (Colorado State University).
References
- 1.Torsvik V, Goksøyr J, Daae FL. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 3.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banik JJ, Brady SF. Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules. Curr. Opin. Microbiol. 2010;13:603–609. doi: 10.1016/j.mib.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia JAL, Fernández-Guerra A, Casamayor EO. A close relationship between primary nucleotides sequence structure and the composition of functional genes in the genome of prokaryotes. Mol. Phylogenet. Evol. 2011;61:650–658. doi: 10.1016/j.ympev.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, Crews P, Piel J. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat. Chem. Biol. 2009;5:494–501. doi: 10.1038/nchembio.176. [DOI] [PubMed] [Google Scholar]
- 7.Brady SF, Simmons L, Kim JH, Schmidt EW. Metagenomic approaches to natural products from free-living and symbiotic organisms. Nat. Prod. Rep. 2009;26:1488–1503. doi: 10.1039/b817078a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donia MS, Ruffner DE, Cao S, Schmidt EW. Accessing the hidden majority of marine natural products through metagenomics. ChemBioChem. 2011;12:1230–1236. doi: 10.1002/cbic.201000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 10.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 11.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem. Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 12.Owen JG, Robins KJ, Parachin NS, Ackerley DF. A functional screen for recovery of 4’-phosphopantetheinyl transferase and associated natural product biosynthesis genes from metagenome libraries. Environ. Microbiol. 2012;14:1198–1209. doi: 10.1111/j.1462-2920.2012.02699.x. [DOI] [PubMed] [Google Scholar]
- 13.Yin J, Straight PD, Hrvatin S, Dorrestein PC, Bumpus SB, Jao C, Kelleher NL, Kolter R, Walsh CT. Genome-wide high-throughput mining of natural-product biosynthetic gene clusters by phage display. Chem. Biol. 2007;14:303–312. doi: 10.1016/j.chembiol.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Raymond KN, Dertz EA, Kim SS. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Z, Lai JR, Walsh CT. Directed evolution of aryl carrier proteins in the enterobactin synthetase. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11621–11626. doi: 10.1073/pnas.0705122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelis P. Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 2010;86:1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 17.Mossialos D, Amoutzias GD. Siderophores in fluorescent pseudomonads: new tricks from an old dog. Future Microbiol. 2007;2:387–395. doi: 10.2217/17460913.2.4.387. [DOI] [PubMed] [Google Scholar]
- 18.Barekzi N, Joshi S, Irwin S, Ontl T, Schweizer HP. Genetic characterization of pcpS, encoding the multifunctional phosphopantetheinyl transferase of Pseudomonas aeruginosa. Microbiology. 2004;150:795–803. doi: 10.1099/mic.0.26823-0. [DOI] [PubMed] [Google Scholar]
- 19.Craig JW, Chang F-Y, Brady SF. Natural products from environmental DNA hosted in Ralstonia metallidurans. ACS Chem. Bio. 2009;4:23–28. doi: 10.1021/cb8002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craig JW, Chang F-Y, Kim JH, Obiajulu SC, Brady SF. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl. Environ. Microbiol. 2010;76:1633–1641. doi: 10.1128/AEM.02169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Copp JN, Roberts AA, Marahiel MA, Neilan BA. Characterization of PPTNs, a cyanobacterial phosphopantetheinyl transferase from Nodularia spumigena NSOR10. J. Bacteriol. 2007;189:3133–3139. doi: 10.1128/JB.01850-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Copp JN, Neilan BA. The phosphopantetheinyl transferase superfamily: phylogenetic analysis and functional implications in cyanobacteria. Appl. Environ. Microbiol. 2006;72:2298–2305. doi: 10.1128/AEM.72.4.2298-2305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li MH, Ung PM, Zajkowski J, Garneau-Tsodikova S, Sherman DH. Automated genome mining for natural products. BMC Bioinformatics. 2009;10:185. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brady SF. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat. Protoc. 2007;2:1297–1305. doi: 10.1038/nprot.2007.195. [DOI] [PubMed] [Google Scholar]
- 26.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2012. [Google Scholar]
- 30.Wickham H. ggplot2: elegant graphics for data analysis. Springer New York: 2009. [Google Scholar]