Abstract
RNA interference (RNAi) is a mechanism that regulates genes by either transcriptional (TGS) or posttranscriptional gene silencing (PTGS), required for genome maintenance and proper development of an organism. Small non-coding RNAs are the key players in RNAi and have been intensively studied in eukaryotes. In plants, several classes of small RNAs with specific sizes and dedicated functions have evolved. The major classes of small RNAs include microRNAs (miRNAs) and small interfering RNAs (siRNAs), which differ in their biogenesis. miRNAs are synthesized from a short hairpin structure while siRNAs are derived from long double-stranded RNAs (dsRNA). Both miRNA and siRNAs control the expression of cognate target RNAs by binding to reverse complementary sequences mediating cleavage or translational inhibition of the target RNA. They also act on the DNA and cause epigenetic changes such as DNA methylation and histone modifications. In the last years, the analysis of plant RNAi pathways was extended to the bryophyte Physcomitrella patens, a non-flowering, non-vascular ancient land plant that diverged from the lineage of seed plants approximately 450 million years ago. Based on a number of characteristic features and its phylogenetic key position in land plant evolution P. patens emerged as a plant model species to address basic as well as applied topics in plant biology. Here we summarize the current knowledge on the role of RNAi in P. patens that shows functional overlap with RNAi pathways from seed plants, and also unique features specific to this species.
Keywords: RNAi, non-coding RNAs, miRNA, siRNA, gene silencing, Physcomitrella patens
1. Introduction
Small non-coding RNAs have been increasingly investigated as important regulators of gene expression. These small RNAs of 20–24 nucleotides (nt) function by causing either TGS or PTGS [1–5]. They were first discovered in the nematode Caenorhabditis elegans [6] and are responsible for the phenomenon known as RNAi, co-suppression, gene silencing, or quelling [7–10]. Shortly after these reports were published, it was shown that PTGS in plants is correlated with the activity of small RNAs [11]. These small RNAs regulate various biological processes in animals and plants by interfering with mRNA translation or directing target RNA cleavage, which is the predominant mode of action in plants. In plants, several classes of small RNAs with specific sizes and dedicated functions have evolved through a series of pathways, namely miRNAs, repeat-associated small interfering RNAs (ra-siRNAs), natural antisense transcript-derived small interfering RNAs (nat-siRNAs), and trans-acting small interfering RNAs (ta-siRNAs) [12–16].
In recent years, the analysis of plant RNAi pathway has been extended to the bryophyte (moss) P. patens, a non-vascular and a non-flowering ancient land plant, that diverged from the higher plants approximately 450 million years ago [17]. P. patens occupies an important phylogenetic position to study the development of higher plants and the adaptation to the land environment. In terms of evolutionary distance of P. patens to flowering plants, it equals the evolutionary distance from fish to humans. P. patens has emerged as a model plant species to address basic as well as applied topics in plant biology. P. patens exhibits a high frequency of homologous recombination which makes it an ideal model system for reverse genetics approaches by the simple generation of targeted gene knockout mutants [18]. Furthermore, the P. patens genome is available that makes it a valuable tool for reconstructing the evolution of plant genomes and functional genomics approaches [19].
Recently, a small RNA database has been established in P. patens [5,12,20–23]. Besides the analysis of P. patens small RNA pathways, molecular tools were developed exploiting the mode of action of small RNAs for the down-regulation of genes in reverse genetics applications. These approaches include the use of conventional inverted RNAi constructs [24,25] as well as the expression of highly specific artificial miRNAs [26]. Even though the major RNAi pathways are evolutionarily conserved in P. patens, there are particular differences in the functional components of small RNA pathways and the biological function of small RNAs. These include a specific amplification of initial miRNA and ta-siRNA signals by the generation of transitive siRNAs, deviating functions and specificities of DICER-LIKE proteins and an epigenetic gene silencing pathway that is triggered by miRNAs. These findings underline that P. patens serves as a valuable model system to study the evolution, diversity, and complexity of plant RNAi pathways.
2. Physcomitrella patens Small RNAs
High throughput sequencing approaches led to the identification of small RNAs from diverse plant species with specific origins, sizes and functions [2]. Several classes of small RNA have been identified in P. patens which can be distinguished based on their specific biogenesis: miRNAs, ta-siRNAs, ra-siRNAs and secondary siRNAs (Figure 1A–D) [2,20,21,23,27]. In general, small RNAs are generated from complete or partially dsRNA precursors by the action of DICER-LIKE (DCL) proteins [1,28]. The small RNA duplexes generated by Dicer activity have a characteristic 2-nucleotide overhang at the 3′ end due to an offset slicing activity of the DCL proteins. In plants these 3′ overhangs are stabilized by 2′-O-methylation [29–32]. Only one strand of the processed small RNA duplex subsequently associates with an RNA-induced silencing complex (RISC) that scans for nucleic acids complementary to the loaded small RNA to execute its function [33–36]. In plants, small RNAs act in gene silencing by different ways, namely by mediating RNA slicing [37–40], translational repression [41–43], and histone modification and DNA methylation [5,44,45]. The first two mechanisms control gene expression posttranscriptionally, whereas the latter affects gene expression at the transcriptional level.
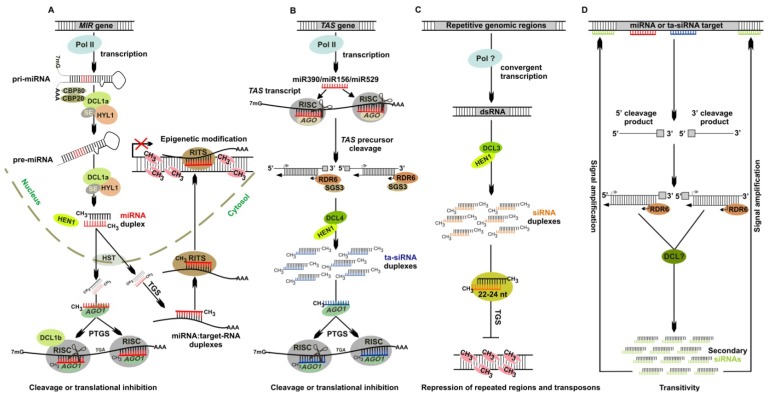
Figure 1.
Different endogenous small interfering RNA (siRNA) pathways of P. patens. Only PpDCLs and PpRDR6 have been functionally characterized in P. patens; evidence for proteins shown in figure comes from Arabidopsis and their homologous exist in P. patens. (A) P. patens miRNA pathway. MIR genes are transcribed by RNA polymerase II into pri-miRNA transcripts that are further processed into pre-miRNAs harboring a characteristic hairpin structure. From the stem of the pre-miRNA the miRNA/miRNA* duplex is excised by PpDCL1a and can be assisted by HYL and SE proteins. These are then methylated by HUA ENHANCER 1 (HEN1) and transported to the cytoplasm by HASTY (HST). The miRNA guide strand is selected, incorporated, and stabilized in dedicated AGO1 protein. miRNA-guided AGO1-containing RNA-induced silencing complex (RISC) directs mRNA cleavage or translation inhibition of the target transcript. Highly abundant miRNAs are either loaded into a RITS complex and subsequently interact with their target to form a duplex, or these duplexes are formed at first and then loaded into RITS. The miRNA:RNA duplexes bound by RNAi-induced transcriptional silencing complex (RITS) initiate DNA methylation at complementary genomic loci. (B) P. patens ta-siRNA pathway. TAS genes are transcribed by RNA polymerase II into TAS precursors harbouring miR390, miR156 and miR529 binding sites. After TAS precursor cleavage at these miRNA sites the middle cleavage product is converted into double-stranded RNAs (dsRNA) by PpRDR6 and subsequently processed into phased ta-siRNAs by PpDCL4. ta-siRNAs are loaded into RISC where they act like miRNAs. (C) P. patens siRNA pathway from repetitive genomic regions primarily LTR-retrotransposons and helitron DNA transposons. dsRNA processed into siRNAs by PpDCL3 and HEN1-mediated siRNA stabilization, the PpDCL3-dependent 22–24 nt siRNAs caused a de-repression of LTR retrotransposon-associated reverse transcriptases pointing to an epigenetic control of these elements. (D) Secondary siRNAs in P. patens. dsRNA is synthesised from cleaved miRNA or ta-siRNA targets by RdRP and processed into secondary siRNAs that mediate cleavage of the target RNA upstream and downstream of the miRNA/ta-siRNA recognition motif resulting in an amplification of the initial small RNA trigger.
2.1. miRNAs (microRNAs)
miRNAs are small RNAs of 20–22 nt that are encoded by endogenous MIR genes. In the miRNA biogenesis pathway, primary miRNAs (pri-miRNAs) are transcribed from MIR genes by RNA polymerase II (Pol II) into transcripts harboring a characteristic hairpin [46] (Figure 1A). In Arabidopsis processing of pri-miRNAs into pre-miRNAs is catalyzed by DCL1 and assisted by HYPONASTIC LEAVES 1 (HYL1) and SERRATE (SE) [36]. The pre-miRNA hairpin precursor is further processed into a 20–22 nt miRNA/miRNA* duplex by DCL1. This duplex is then methylated at the 3′ terminus by HUA ENHANCER 1 (HEN1) and exported into the cytoplasm by HASTY (HST) protein [47,48]. Within the cytoplasm, one strand of the duplex (the miRNA) is incorporated into RISC and guides binding of the complex to cognate target transcripts by sequence complementarity. In P. patens PpDCL1a is responsible for production of miRNAs [5]. In addition to the control of targets at the posttranscriptional level, P. patens miRNAs regulate gene expression by causing epigenetic changes such as DNA methylation (Figure 1A) [5,49–51].
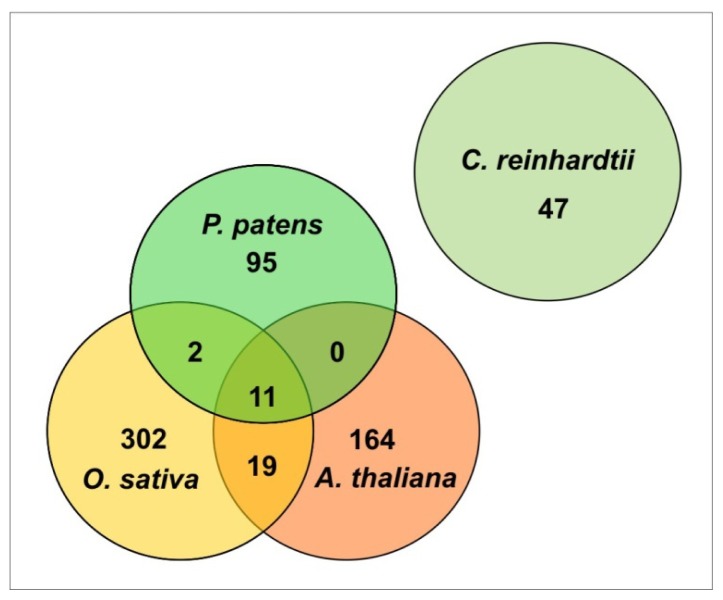
Independent studies on small RNAs in P. patens revealed the existence of a diverse miRNA repertoire including highly conserved miRNA families [2,12,20,23,27]. Conserved miRNAs families between P. patens and other land plants were discovered using the microHARVESTER algorithm that analyszes and predicts conserved MIR genes in genomic sequences [20]. The reported P. patens miRNAs were identified from wild type plants covering major developmental stages (protonema, young gametophores, gametophores, and sporophytes). Further, the identification of miRNAs was restricted to plants that were cultivated under standard growth conditions, and thus miRNAs which respond to certain physiological conditions such as abiotic stress may have escaped identification. These analyses led to the identification of 108 miRNA families in P. patens. Interestingly, 11 miRNA families are conserved between P. patens, A. thaliana and O. sativa, while the 47 miRNAs families identified in the green alga C. reinhardtii are species-specific and are not related to land plant miRNAs (Figure 2).
Figure 2.
Venn diagram comparing miRNA families from the seed plants A. thaliana (dicot) and O. sativa (monocot), the moss P. patens and the unicellular alga C. reinhardtii based on miRBase database (Release 19.0, http://www.mirbase.org/). C. reinhardtii miRNA families are species-specific since they do not show sequence similarity to miRNA families from land plants. 11 miRNA families (miR156, miR160, miR166, miR167, miR171, miR319, miR390, miR395, miR408, miR414 and miR419) are conserved between A. thaliana, O. sativa, and P. patens. Two additional miRNA families, (miR529 and miR535) are conserved between O. sativa and P. patens.
Certain miRNAs regulate various key developmental programs including auxin signaling, lateral root formation, leaf development and polarity, meristem boundary formation and organ separation, transition from the juvenile-to-adult vegetative phase and vegetative-to-flowering phase, floral organ identity, and reproduction, and also plant responses to different stress conditions [50]. Most conserved miRNAs target mRNAs of conserved transcription factor families, for example miR156, miR159/319, miR160, miR166 andmiR171 target transcripts encoding SBPs, MYBs/TCPs, ARFs, HD-ZIPs andGRAS domain proteins, respectively [50]. Some conserved miRNAs and their targets have been identified in P. patens, but functional analysis in P. patens is still required to address this interesting topic of the evolution of miRNAs in plants. Some moss-specific miRNAs that target mRNAs encoding transcription factors that are associated with developmental control have been functionally characterized. For example, miR534 controls mRNAs encoding ankyrin repeat containing proteins homologous to the A. thaliana BLADE ON PETIOLE 1 and 2, miR538 cleaves mRNAs of the MADS box transcription factor family and miR902 controls several mRNAs coding for basic helix-loop-helix transcription factors [2,27,52]. Some miRNAs are expressed in a restricted spatiotemporal manner, so they control target genes only in specific expression domains to establish boundaries for developmental decisions. One example is the cell-type specific and cytokinin-dependent repression of the P. patens specific miR534a that acts in the juvenile-to-adult transition and controls bud formation through an accompanying derepression of PpBOP expression within the same cells [53].
2.2. ta-siRNAs (Trans-Acting Small Interfering RNAs)
Ta-siRNAs are plant specific small RNAs that are generated from genome encoded TAS transcripts; initially the TAS precursor is cleaved by a miRNA, subsequently converted into double stranded RNA and processed into phased siRNA duplexes by DCL proteins (Figure 1B). In A. thaliana main components of ta-siRNA pathway are DCL4, RDR6, SGS3, and DRB4 proteins [13,14,16,54–57]. In A. thaliana four TAS gene families (TAS1-4) have been identified. miR173-mediated cleavage generates ta-siRNA from TAS1 and TAS2 precursors, miR390 is required for TAS3 cleavage while miR828 is assigned to TAS4. TAS2 and TAS4 are encoded by single genes while TAS1 and TAS3 each harbour three members [13–16,57]. In A. thaliana most miRNAs are incorporated into ARGONAUTE1 (AGO1)-containing RISC to direct cleavage of their targets, whereas miR390 specifically interacts with AGO7. The miR390-AGO7 complex has a specific role in the phased processing of ta-siRNAs from TAS3 precursors [58]. Like A. thaliana the rice genome also encodes three TAS3 precursors (TAS3a-c) and all TAS3 precursors from both species have dual miR390 binding sites but miR390-directed cleavage only occurs at the 3′ miR390 binding site. Consequently, ta-siRNAs are only generated from the cleavage product located 5′ to the cleaved miR390 site [4,23]. Recently a new TAS family, TAS5, has been identified in tomato, which required miR482-directed initial slicing [59]. The P. patens genome encodes TAS3 and TAS6 families. The P. patens TAS3 family comprises six members (TAS3a–f) each harbouring dual miR390 sites. The TAS3 precursors are cleaved at both miR390 binding sites and subsequently the middle cleavage product is converted into dsRNA by PpRDR6 (Figure 1B) [23]. The P. patens TAS6 family is encoded by three loci (TAS6a–c) that are in close proximity to TAS3 loci and are typified by sliced miR156 and miR529 target sites [60]. P. patens ΔPpDCL1a, ΔPpRDR6 and ΔPpDCL4 mutants lack ta-siRNA production based on specific functions of the respective proteins in ta-siRNA biogenesis: the absence of ta-siRNAs in the ΔPpDCL1a mutant is due to the lack of the specific miRNAs (miR390, miR156 and miR529) that are required to initiate the ta-siRNA pathway by the cleavage of the respective TAS precursors, PpRDR6 the essential enzyme catalysing the conversion of cleaved precursors into dsRNA while PpDCL4 is the DCL protein that processes ta-siRNAs from the dsRNA precursors in a phased manner [5,12,60].
Like for miRNAs, ta-siRNAs targets can be predicted on the basis of sequence complementarity between a ta-siRNA and its target RNA. In A. thaliana TAS3 ta-siRNAs regulate several AUXIN RESPONSE FACTOR (ARF) mRNAs including ARF3 and ARF4 which regulate leaf polarity and proper timing of vegetative shoot development [13,54,56,61–63]. Likewise, rice TAS3-derived ta-siRNAs control ARF2/3 [4]. Strikingly, P. patens TAS3 ta-siRNAs, also target ARF transcripts (Phypa_203442, Phypa_224167) even though the sequences of the respective ta-siRNAs are not conserved between moss and seed plants [2]. These observations indicate that at least ARF targeting TAS3 ta-siRNA function is conserved between A. thaliana, rice and P. patens irrespective of the varying ta-siRNA sequences which indicate a strong evolutionary conservation of TAS3 ta-siRNAs and ARF regulation in mosses and seed plants.
Besides ARF transcripts, P. patens TAS3 ta-siRNAs also regulate an mRNA encoding an AP2/EREPB transcription factor (Phypa_65352) [12]. AtTAS1 and AtTAS2 family ta-siRNAs target PPR (pentatricopeptide repeat) transcripts whereas MYB transcription factor transcripts are regulated by AtTAS4 ta-siRNAs [13–16]. Tomato TAS5 ta-siRNA target the resistance gene Bs4, and P. patens TAS6 ta-siRNA targets an mRNA encoding a zinc finger protein (Pp1s286_43v6.1) [59,60]. In addition to their role in mediating RNA target cleavage A. thaliana ta-siRNAs may also function in the nucleus to control mRNA splicing. A binding site of a TAS1a-derived ta-siRNA was found in an intron of a pre-mRNA that encodes a FAD binding domain-containing protein (At2g46740) and elevated levels of unspliced At2g46740 mRNA was detected in ta-siRNA deficient mutants [16]. However, evidence for a role of P. patens ta-siRNAs in the control of RNA splicing is still missing.
2.3. ra-siRNAs (Repeat Associated Small Interfering RNAs)
dsRNAs are sources of siRNAs biogenesis and they can have different origins such as RNA transcribed from inverted repeats, pairing of natural cis-antisense transcripts, transcript from retroelement-rich genome regions, synthesis by the action of RNA-dependent RNA polymerases (RDRs) or the replication of RNA viruses [64,65]. dsRNAs are cleaved into 21–24 nt siRNAs by DCL proteins whereby the size of the siRNAs depends on the specific DCL protein that catalyzes the processing of siRNAs from these precursors (Figure 1C). Like miRNAs, siRNAs are loaded into AGO protein containing RISC that guide recognition of target genes by base pairing and silence them [39,66]. In addition, siRNAs can be incorporated into RNAi-induced transcriptional silencing complex (RITS) and transcriptionally silence the loci mainly by DNA methylation. For most of the ra-siRNAs this is the predominant mode of action in particular for all those that are derived from repetitive genomic regions. The first proof of siRNA generation and subsequent gene silencing in P. patens was obtained from the expression of inverted GUS and GFP RNAi constructs that caused silencing of GUS and GFP signals in P. patens lines constitutively expressing GUS and GFP [24,25]. The total endogenous small RNA population of flowering plants is characterized by two distinct peaks at 21 nt and 24 nt [15,67,68]. The 21nt size constitutes mainly miRNAs and ta-siRNAs while 24 nt small RNAs are predominantly generated from intergenic and repetitive genomic regions [15,68]. In A. thaliana 24 nt small RNAs are primarily generated by AtDCL3 [69].
The P. patens top 100 non-miRNA and non-ta-siRNA siRNA producing regions fall into two distinct categories and were classified on the basis of generated siRNA size and abundance [21]. One class is dominated by 21 nt RNAs whereas the second class comprises a mixture of 21–24 nt RNAs in a strikingly consistent ratio. The loci generating these two types of small RNAs were annotated as Pp21SR (21 nt small RNA) and Pp23SR (21, 23, and 24 nt small RNA) loci, respectively [21]. The Pp23SR loci constitute larger genomic regions between 5 and 50 kb in length (median 11.9 kb) whereas the Pp21SR loci cover small regions that range from 100 to 1000 nt in length (median 247.5 nt). Pp21SR loci mainly generate single stranded small RNA precursors for siRNA production while Pp23SR loci are templates for the production of long dsRNA precursors which are processed into siRNAs as inferred from the sense and antisense polarity of siRNAs derived from these loci. Most of the Pp23SR loci overlap with LTR-retrotransposons and helitron elements, and PpDCL3 generates 22–24 nt siRNA from these loci [21]. Pp23SR loci are characterized by dense cytosine-methylation and the depletion of the 22–24 nt siRNAs that originate from these loci in ΔPpDCL3 mutants caused a de-repression of LTR retrotransposon-associated reverse transcriptases indicating an epigenetic control of these elements by the specific set of 22–24 nt siRNAs (Figure 1C). Thus, the 22–24 nt small RNAs from Pp23SR loci are functionally similar to AtDCL3 generated 24 nt siRNAs and are involved in the repression of transposons [21]. The biogenesis pathways and functions of siRNAs that derive from Pp21SR loci and of the 21 nt siRNA fraction originating from Pp23SR loci are not clear yet.
2.4. Secondary siRNAs
Recent studies involving plants and the nematode C. elegans reported the amplification of silencing related RNA and explain how strong, persistent silencing can be initiated with small amounts of initiator dsRNA [23,70,71]. The amplification process has implications for application of RNAi to control gene expression in biotechnology and for understanding the effects of silencing RNAs on cell function and organism development. The initiator of transitivity is a dsRNA that is first processed by Dicer into siRNA or a related type of RNA referred to as miRNA, these 21–25 nucleotide single stranded RNAs are the primary silencing RNAs in the transitive process [72,73]. These siRNAs can not only trigger their cleavage and subsequent degradation of target RNAs, but also serve as primers for RdRP activity. With the help of RdRP target RNAs are converted into double strands RNAs which serves as templates for the production of secondary siRNAs by Dicer action, in this way siRNA population is generated which is distinct from the initiator siRNA [73,74]. In plants, the transitivity occurs in both directions of the initial siRNA trigger whereas in animals spreading of the initial signal occurs only upstream of the trigger [23,73–78].
In P. patens amplification of miRNA-mediated RNA cleavage sites by 5′RACE yielded several products including the expected miRNA-directed RNA cleavage products. These additional degradation products are the consequence of the action of secondary siRNAs that guide cleavage of the miRNA targets at additional sites [5]. These secondary siRNAs biogenesis involves RDR6 activity and secondary siRNAs were detected both in sense and antisense orientation [5]. Furthermore, secondary siRNAs were generated from upstream and downstream regions relative to the miRNA binding site (Figure 1D) [5]. Secondary siRNAs were also generated after ta-siRNA-mediated cleavage of the ta-siRNA target PpEREBP/AP2. The biogenesis of transitive siRNAs in P. patens is different from biogenesis of secondary siRNAs in C. elegans where transitivity only occurs in antisense polarity due to an unprimed de novo synthesis of dsRNA by RdRP [70,71]. Furthermore, transitivity in P. patens only occurs after miRNA or ta-siRNA mediated cleavage of target RNAs, since ΔPpDCL1b mutants defective in target cleavage do not generate transitive siRNAs [5]. Recently it is shown that secondary siRNA biogenesis in A. thaliana is triggered by 22 nt rather than by the more typical 21 nt miRNAs and ta-siRNAs. So for eight A. thaliana miRNAs (miR168, miR173, miR393, miR447, miR472, miR473, miR828 and miR856) and one ta-siRNA (ta-siR2140) are known triggers of siRNA production [79]. Since secondary siRNAs in P. patens are generated from RNAs that are targeted by 21nt miRNAs (miR160, miR166) and a 21nt ta-siRNA (ta-siRNA 6(+)) further studies are required to address the differences and specificities in secondary siRNA production in mosses and seed plants.
3. Physcomitrella patens Homologues of RNAi Pathway Components
In P. patens the functional analysis of RNAi pathway components is limited to PpDCL1a, PpDCL1b, PpDCL3, PpDCL4, and PpRDR6. To obtain a comprehensive view of the genes that are present in P. patens, we used A. thaliana proteins that are involved in different small RNA pathways as queries for BLASTP searches in P. patens V1.6 protein database (available online: http://www.cosmoss.org (accessed on 10 December 2012)). This in silico analysis identified presence of A. thaliana homologues of all known protein families involved in small RNA pathways in P. patens (Table 1) that indicate a wide conservation over evolutionary time and point to large functional overlaps in different plant species. However, the size of certain RNAi protein families varies between P. patens and seed plants. For example, six AGO family members were found in P. patens whereas ten members are present in A. thaliana [2,37]. These ten AGO proteins are clustered into three clades: first clade comprises AtAGO1, AtAGO5 and AtAGO10, second clade has AtAGO2, AtAGO3 and AtAGO7, and third clade includes AtAGO4, AtAGO6, AtAGO8 and AtAGO9 proteins [80]. P. patens encodes three homologues of AtAGO1 which is the core component of RISC, and three homologous of AtAGO4, AtAGO6 and AtAGO9 proteins, respectively (Table 1). Thus, beside a large overlap of small RNA-related proteins, there are particular differences in the protein repertoire that may cause deviating functions of small RNA pathways in P. patens and seed plants.
Table 1.
P. patens and A. thaliana homologues of proteins involved in RNAi pathways (Updated after [81]).
| Protein family | P. patens homologues | NCBI/Gene model number | E-value, % Identity | Molecular function | References |
|---|---|---|---|---|---|
| AtDCL1 | PpDCL1a | EF670436 | 0.0, 68% | miRNA biogenesis Indispensable for target cleavage | [5,29,82,83] |
| PpDCL1b | DQ675601 | 0.0, 65% | |||
|
| |||||
| AtDCL2 | n.i 1 | - | - | Generates endogenous siRNAs from a convergently transcribed and overlapping gene pairs Transitive silencing of transgenes Produces viral siRNAs |
[74,84,85] |
|
| |||||
| AtDCL3 | PpDCL3 | EF670437 | 1e−116, 32% | Generates siRNAs that guide chromatin modification in P. patens and A. thaliana | [21,69] |
|
| |||||
| AtDCL4 | PpDCL4 | EF670438 | 1e−124, 33% | Generates trans-acting siRNAs (ta-siRNAs) | [54,55,57,60,86,87] |
|
| |||||
| AtAGO1 | PpAGO1a 2 | Phypa_205541 | 0.0, 78% | Associates with the majority of miRNAs to guide the cleavage of their targets | [2,39,40,88,89] |
| PpAGO1b 2 | Phypa_158832 | 0.0, 77% | |||
| PpAGO1c 2 | Phypa_141045 | 0.0, 75% | |||
|
| |||||
| AtAGO2 | n.i | - | -- | Known to be function in antiviral defense and ta-siRNAs biogenesis | [90,91] |
|
| |||||
| AtAGO3 | n.i | - | - | Not analyzed | |
|
| |||||
| AtAGO4 | PpAGO42 | Phypa_200513 | 1e−164, 38% | Involved in 24nt siRNA mediated gene silencing | [92–97] |
|
| |||||
| AtAGO5 | n.i | - | - | Not analyzed | |
|
| |||||
| AtAGO6 | PpAGO62 | Phypa_117253 | 1e−152, 39% | Involved in 24nt siRNA mediated DNA methylation | [93,98] |
|
| |||||
| AtAGO7 | n.i | - | - | Associates specifically with miR390 and directs cleavage of the AtTAS3 precursor | [13–16,56,58] |
|
| |||||
| AtAGO8 | n.i | - | - | Not analyzed | |
|
| |||||
| AtAGO9 | PpAGO92 | Phypa_134255 | 6.1e−160, 40% | Preferentially interacts with 24nt siRNAs derived from transposable elements (TEs), required to silence TEs in female gametes and their accessory cells. Cell fate determination in the ovule. | [93,99] |
|
| |||||
| AtAGO10 | n.i | - | - | Implicated in miRNA-directed translational inhibition and repression of miR165/166 levels | [42,100,101] |
|
| |||||
| AtRDR1 | PpRDR12 | Phypa_219654 | 1.3e−204, 48% | Synthesis of long dsRNA from transgenes that can initiate different RNAi pathways Biogenesis of secondary siRNAs from RNA viruses |
[102–104] |
|
| |||||
| AtRDR2 | n.i | - | - | Biogenesis of 24nt siRNAs from repeat loci involved in DNA methylation | [105,106] |
|
| |||||
| AtRDR3a | n.i. | - | - | Not analyzed | [107,108] |
| AtRDR3b | PpRDR3b 2 | Phypa_169723 | 1.8e−96, 33% | ||
| AtRDR3c | PpRDR3c 2 | Phypa_172848 | 6.1e−89, 31% | ||
|
| |||||
| AtRDR6 | PpRDR6 | Phypa_379 | 2.8e−226, 42% | Initiation and maintenance of dsRNA-induced RNAi in A. thaliana Conversion of TAS precursors into dsRNA in P. patens and A. thaliana |
[12,14,109,110] |
|
| |||||
| AtHEN1 | PpHEN12 | Phypa_148777 | 3e−56, 33% | Methylates miRNA and siRNA duplexes at the 3′ end | [29–31,111] |
|
| |||||
| AtHYL | PpHYL12 | Phypa_34761 | 7e−31, 50% | Interacts with AtDCL1 and confers stability to miRNA precursors | [69,112–114] |
|
| |||||
| AtHASTY | PpHASTY12 | Phypa_137344 | 1.3e−228, 40% | Exports miRNA-miRNA * duplex to the cytoplasm | [14,48] |
| PpHASTY22 | Phypa_151199 | 2.1e−173, 41% | |||
|
| |||||
| AtSE | PpSE12 | Phypa_133793 | 1.7e−92, 41% | Interacts with AtDCL1 and confers stability to miRNA precursors | [69,82,112,113] |
| PpSE22 | Phypa_124567 | 1.8e−70, 41% | |||
| PpSE32 | Phypa_99415 | 3.3e−53, 35% | |||
|
| |||||
| AtCPL1 | PpCPL12 | Phypa_432395 | 1e−126, 49% | Required for HYL1 dephosphorylation, which in turn is essential for accurate miRNA processing and strand selection. | [115] |
| PpCPL22 | Phypa_429817 | 1e−126, 51% | |||
|
| |||||
| AtCBP20 | PpCBP20a2 | Phypa_442048 | 5e−42, 53% | Involved in pre-miRNA splicing and miRNA processing | [116,117] |
| PpCBP20b2 | Phypa_442049 | 7e−71, 58% | |||
| PpCBP20c2 | Phypa_442050 | 7e−69,76% | |||
|
| |||||
| AtCBP80 | PpCBP80.12 | Phypa_425787 | 0.0, 47% | Involved in pre-miRNA splicing and miRNA processing | [116,117] |
| PpCBP80.22 | Phypa_432264 | 0.0, 47% | |||
|
| |||||
| AtSQN/CYP40 | PpSQNa2 | Phypa_433182 | 1e−136, 66% | Required for miRNA activity by promoting the activity of AGO1. Plays a unique and important role in plant RISC assembly |
[118–120] |
| PpSQNb2 | Phypa_433181 | 1e−136, 66% | |||
|
| |||||
| AtHSP90 | PpHsp90.12 | Phypa_456075 | 0.0, 80% | Plays a unique and important role in plant RISC assembly | [119,120] |
| PpHsp90.22 | Phypa_454408 | 0.0, 80% | |||
| PpHsp90.32 | Phypa_452062 | 0.0, 79% | |||
| PpHsp90.42 | Phypa_452093 | 0.0, 80% | |||
|
| |||||
| AtSGS3 | PpSGS32 | Phypa_448213 | 3.0e−71, 37% | Involved in the production of ta-siRNAs, through direct or indirect stabilisation of TAS cleavage products | [14,110] |
|
| |||||
| AtPol IV | PpPol IV2 | Phypa_132119 | 1.3e−72, 49% | Required for the biogenesis of 24nt siRNAs (with RDR2 and DCL3) that associate with AGO4 and direct DNA and histone modifications | [94–96] |
|
| |||||
| AtPol V | PpPol V2 | Phypa_129844 | 1e−132, 70% | Generates transcripts from heterochromatic regions (with DRD1) that are discussed to bind siRNA-AGO4 complexes directing DNA and histone modifications | [94–96] |
|
| |||||
| AtDRM1 | PpDRM12 | Phypa_148057 | 5e−92, 51% | Involved in the siRNA-directed de novo DNA methylation and maintenance of DNA methylation at CHH sites | [105,121,122] |
| AtDRM2 | PpDRM22 | Phypa_133529 | 6.3e−87, 47% | ||
|
| |||||
| AtDRD1 | PpDRD12 | Phypa_113504 | 1e−109, 35% | Cooperates with Pol V | [94,96,122–127] |
|
| |||||
| AtSNF2 | PpSNF22 | Phypa_211797 | 1.3e−187, 46% | Involved in the spreading of transgene silencing (with AtRDR2 and AtPol IV) and in the production of endogenous 24 nt siRNAs | [128–131] |
|
| |||||
| AtRDM12 | PpRDM122 | Phypa_98999 | 1e−46, 26% | Involved in the de novo DNA methylation and siRNA-mediated maintenance of DNA methylation | [132,133] |
Protein sequences from A. thaliana (TAIR; available online: http://www.arabidopsis.org (accessed on 10 December 2012)) were used for reciprocal BLASTP searches against the P. patens V1.6_proteins database (available online http://www.cosmoss.org (accessed on 10 December 2012)).
Not identified.
Unknown function in P. patens.
DCL, Dicer Like; AGO, ARGONAUTE; RDR, RNA-Dependent RNA Polymerase; HEN1, Hua Enhancer 1, HYL1, Hyponastic Leaves 1; SE, Serrate; CPL1, C-Terminal Domain Phosphatase-like 1; CBP20, Cap-binding Protein 20; CBP80, Cap-binding Protein 80; SQN/CYP40, Squint/Cyclophilin 40; HSP90, Heat Shock Protein 90; SGS3, Suppressor of Gene Silencing 3; DRM, Domains Rearranged Methylase; DRD1, Defective in RNA-directed DNA Methylation 1; RDM12, RNA- directed DNA Methylation 12.
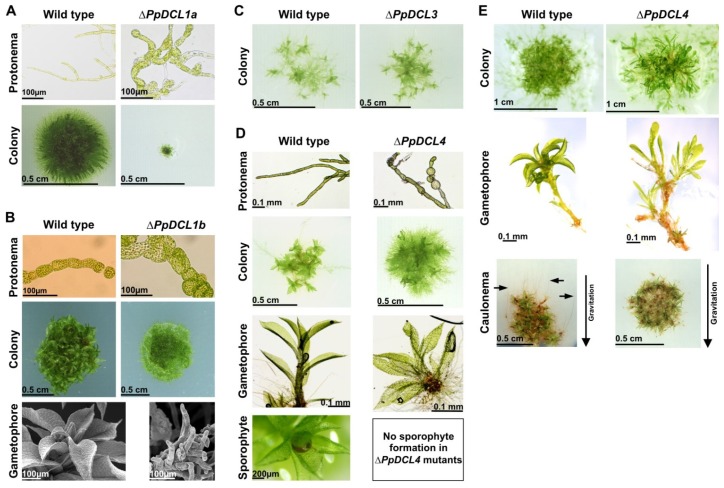
Like A. thaliana, there are four DCL proteins present in P. patens (Table 1) [2], but the DCL repertoire differs. P. patens encodes two proteins similar to AtDCL1 and two DCL proteins homologous to AtDCL3 and AtDCL4 respectively, whereas no AtDCL2 homologue is present in P. patens (Table 1). Consequently, the P. patens proteins were named PpDCL1a, PpDCL1b, PpDCL3 and PpDCL4. Although PpDCL1a and PpDCL1b are highly similar to the A. thaliana AtDCL1 protein but experimental evidences indicate distinct functions [5]. Strongly reduced expression levels or complete lack of miRNAs and elevated steady-state transcript levels of cognate miRNA targets were detected in ΔPpDCL1a null mutants [5]. This is similar to A. thaliana where dcl1 mutants that lack miRNAs accompanied with increased miRNA target expression levels in addition, Atdcl1 mutants are embryo lethal [29,134]. Similarly, ΔPpDCL1a mutants displayed severe developmental disorders affecting cell size and shape, retarded growth that was partially complemented by growth on medium supplemented with vitamins, and developmental arrest at the filamentous protonema stage since these mutants failed to developed leafy gametophores (Figure 3A). The lack of gametophores also causes sterility of ΔPpDCL1a mutants since the gametophores bear the male and female sex organs (antheridia and archegonia). So, it was concluded that PpDCL1a is the functional equivalent to the AtDCL1 protein from A. thaliana. Similar to ΔPpDCL1a mutants, ΔPpDCL1b mutants showed developmental disorders (Figure 3B) including abnormalities in cell division, cell size, cell shape and growth polarity, and they developed only a small number of gametophores which in addition were malformed [5]. PpDCL1b is involved in miRNA directed target genes cleavage, this is a novel mechanism not known so far in other organism.
Figure 3.
P. patens DCL mutant phenotypes. (A) Phenotypic comparison of P. patens wild type and a ΔPpDCL1a mutant line: in mutants cell size and shape is affected, they have retarded growth, and developmental arrest at the filamentous protonema stage is obserbed. (B) Phenotypic comparison of P. patens wild type and a ΔPpDCL1b mutant line: mutants show deviating cell division, cell size, cell shape and growth polarity, and they developed only a small number of gametophores which in addition are malformed (C) Phenotypic comparison of P. patens wild type and a ΔPpDCL3 mutant line: mutants show accelerated gametophore development. (D) Phenotypic comparison of P. patens wild type and ΔPpDCL4 mutant lines under standard growth conditions: mutants show several brachycytes in the protonema, colony produces relative more protonema and less leafy gametophores, gametophores are stunted in growth, and mutants are sterile (E) Phenotypic comparison of P. patens wild type and ΔPpDCL4 mutant lines under short day conditions: mutants generates malformed gametophores, and fail to produce caulonema under dark conditions.
The P. patens PpDCL3 generates 22–24 nt small RNAs mainly from intergenic regions repetitive regions. Repetitive siRNAs control P. patens development since ΔPpDCL3 mutants show an accelerated gametophore formation (Figure 3C) [21]. PpDCL3 generated 22–24 nt are analogous in function to AtDCL3 generated 24 nt siRNAs and are involved in the repression of transposons [21]. The ΔPpDCL4 loss-of-function mutants have dramatic developmental abnormalities [60], mutants display phenotypic aberration throughout all the developmental stages and in addition are sterile (Figure 3D,E). In Arabidopsis dcl4 mutants, the lack of ta-siRNAs only has minor developmental effects, namely the formation of slightly elongated and down-ward curled rosette leaf margins and accelerated juvenile-to-adult vegetative phase changes [54,55]. P. patens ΔPpRDR6 mutants show mild phenotypic deviation, only an accelerated transition from juvenile to adult gametophyte stage was observed [12]. Similarly A. thaliana rdr6 mutants exhibit mild phenotypic deviations, rosette leaves are elongated and slightly downwards curled, and an accelerated transition to the adult phase [14]. Like Arabidopsis and P. patens rice, rdr6 (OsSHL2) is required for ta-siRNA production but in contrast, strong rdr6 mutants display severe phenotypes and they lack the shoot apical meristem [135,136] whereas the weak mutations leads to defects in the adaxial-abaxial patterning of floral organs [137]. These severe phenotypes of rice hint to a broader activity of rdr6 protein, it could be also an indication of the stronger ta-siRNA directed regulation on developmental programs of rice.
4. Physcomitrella patens and Epigenetic Modification
In some cases, endogenous siRNAs trigger epigenetic effects at target loci and are associated with RNA-directed DNA methylation (RdDM) and chromatin remodeling [69,92,138]. In plants, dsRNAs which contain sequences that are homologous to promoter regions can trigger promoter methylation and transcriptional gene silencing [139,140]. RdDM leads to de novo methylation of cytosine that occurs in all sequence contexts, not just in symmetrical CG dinucleotides, which are considered the usual targets for methylation. Methylation is largely restricted to the region of RNA-DNA sequence homology [141,142]. So, unlike RNAi based heterochromatin, which can spread over several kilobases from the RNA-targeted nucleation site, RdDM does not usually infiltrate substantially into adjacent sequences [143]. RdDM is a stepwise process that is initiated by RNA signals and site-specific DNA methyltransferases [121,144]. The methylation of cytosine residues can result in transcriptional silencing. Recently in Arabidopsis thaliana a nucleolar complex involved in the siRNA-directed silencing of endogenous repeat regions has been identified. This complex constitutes several proteins which are linked to RdDM including RDR2, DCL3 and AGO4. By the action of this nucleolar complex 24 nt siRNAs generated from repetitive regions methylate DNA and do a role in silencing of repetitive regions. Homologues of silencing nucleolar complex are also present in P. patens (Table 1), functional analyses is required to obtain further mechanistic insight into P. patens RdDM pathways.
In yeast Schizosaccharomyces pombe, an RITS contains AGO1, a chromodomain protein Chp1, and TAS3 [145]. RITS and Rdrp1 are recruited to silenced loci [102] and it is proposed that it is involved in a self-reinforcing loop, in which DNA-interacting proteins and the siRNAs loaded RITS guide methylation of lysine 9 or histone 3 that result in heterochromatin formation. If a transcript is formed from these loci it will be converted to siRNA precursors by Rdrp1 and will keep loci in heterochromatin state [34]. However, it is not clear whether siRNAs alone target the complex directly to the genomic DNA or if the interaction requires an additional RNA molecule closely associated with the genomic DNA.
In P. patens PpDCL3-dependent 22–24 nt siRNAs are involved in epigenetic silencing of LTR retrotransposons [21]. Additional evidence for the existence of small RNA-mediated epigenetic gene silencing was obtained from the analysis of ΔPpDCL1b mutants [5]. In ΔPpDCL1b mutants miRNA-triggered cleavage of target RNAs was abolished, but it is unlikely that PpDCL1b is directly involved in the cleavage of target RNAs since AGO proteins residing in RISC are the catalytic enzymes responsible for RNA-directed target cleavage [66]. Thus, similar to animal dicers which were shown to be components of RISC-loading complexes (RLC) PpDCL1b was proposed to play a role in loading miRNAs into RISC [66,146–148] (Figure 1A). Even though miRNA-directed cleavage of target RNAs was abolished in the ΔPpDCL1b mutants and elevated steady-state transcript levels of miRNA targets were expected, all analyzed miRNA targets had strongly reduced expression levels in ΔPpDCL1b mutants. Subsequent analysis of revealed cytosine methylation at CpG residues within the cognate miRNA target loci causing transcriptional silencing of these loci. Furthermore, DNA methylation was not restricted to the region of the encoded miRNA binding site, but spread into upstream and downstream regions including introns and promoter regions [5]. In two miRNA target genes, PpHB10 and PpC3HDZIP1, the miRNA binding site is disrupted by intron making it unlikely that DNA methlyation is initiated by the formation of an miRNA:DNA hybrid. In addition, stable miRNA:mRNA duplexes were observed in the ΔPpDCL1b mutants leading to the hypothesis that these duplexes interact with RITS to guide it to cognate genomic regions to initiate DNA methylation (Figure 1A). Moreover, it was hypothesized that DNA methylation of miRNA target genes in the ΔPpDCL1b mutants is triggered by a high miRNA:target RNA ratio due to the abolished target cleavage [5]. The influence of the miRNA:target RNA ratio on DNA methylation and transcriptional silencing was also proved by the analysis of miR1026 and its target gene PpbHLH in P. patens wild type. Upon ABA treatment miR1026 accumulates to elevated steady-state levels which results in a higher miR1026:PpbHLH. Consequently, reduced PpbHLH steady-state levels were observed which, at least in part, are caused by transcriptional silencing of the PpbHLH locus due to DNA methylation at CpG sites within the PpbHLH genomic locus.
5. Physcomitrella patens and Autoregulation of miRNA Biogenesis
The miRNA pathway controls many important genes, and hence requires proper regulation of its own pathway. One of the regulations might be regulation of transcripts encoding catalytic small RNA pathway components and indeed miRNAs are involved in the negative feedback control of important miRNA pathway genes. For example, in A. thaliana miR162 targets the AtDCL1 transcript which is the key gene of miRNA biogenesis [149]. Another feedback control may affect the maturation of the AtDCL1 pre-mRNA. Intron 14 of the AtDCL1 gene harbours a miR828 precursor sequence [15]. Processing of the miR828 by AtDCL1 could compete with the splicing of the AtDCL1 pre-mRNA to control functional AtDCL1 mRNA levels. So far, no miRNA has been identified that targets PpDCL1a or PpDCL1b. However, miR1047 precursor is present in intron 7 of PpDCL1a that is essential for miRNA biogenesis, suggesting a similar role of miR828 in AtDCL1 [2]. Another conserved miRNA-mediated feedback was reported for AGO1 mRNAs in A. thaliana and P. patens, AGO1 is a key protein in RISC and is required for miRNA directed cleavage of target transcripts [89,150]. In A. thaliana the single AtAGO1 mRNA is targeted by miR168, whereas miR904 targets three PpAGO1 homologues (PpAGO1a–c) [2,88,89]. This present a negative feedback loop, perturbation of this control loop by the expression of a miR168-resistant AtAGO1 mRNA led to elevated AtAGO1 transcript levels and an increased abundance of miRNA target RNAs suggesting that elevated AtAGO1 levels interfere with proper miRNA-RISC activity. In addition, miR168-resistant AtAGO1 lines show developmental defects indicating the biological relevance of this negative control loop [89]. Further studies are required to know whether the miR904-mediated control of the P. patens PpAGO1a–c homologues has a similar function in the maintenance of miRNA-RISC activity. In addition, functional analysis will reveal if the three PpAGO1 homologues act redundantly and are functionally equivalent to the single A. thaliana AtAGO1 or they exhibit diverse functions. The intronic mature miRNAs or its precursor sequences of ath-miR838 and ppt-miR1047 regulating DCL1 or and ath-miR168 and ppt-miR904 targeting AGO1 transcripts are not conserved between both species pointing to a convergent evolution of these control pathways.
6. Physcomitrella patens and Artificial miRNAs
The successful use of artificial miRNAs (amiRNAs) for the specific down-regulation of genes was shown for the dicotyledonous plants Arabidopsis, tomato and tobacco, and for the monocot rice [150–156]. In most of the cases the amiRNA was expressed from endogenous miRNA precursors. However, high expression rates of amiRNAs were achieved in tobacco and tomato using the Arabidopsis miR164b precursor sequence indicating correct processing of conserved pre-miRNAs within seed plants [150]. The Arabidopsis miR319a precursor was used for the expression of amiRNAs in P. patens targeting PpFtsZ2-1 and PpGNT1 genes [26]. The constitutive expression of the modified precursor for both amiRNAs resulted in precisely processing of mature miRNA and effective highly specific knockdown of the corresponding transcripts. The amiRNA-mediated knockdown of PpFtsZ2-1 that is indispensable for plastid division caused the formation of macrochloroplasts which is the exact phenotypic deviation observed in ΔPpFtsZ2-1 null mutants [157]. In addition, similar to natural P. patens miRNAs the overexpression of amiRNAs caused transitivity by the generation of secondary siRNAs [26]. The expression of amiRNAs can be beneficial to target several related genes or they can be expressed by tissue specific or inducible manner. The use of amiRNA may replace conventional inverted repeat-based RNAi constructs because of off-targeting effects [25].
7. Conclusions and Future Prospects
Understanding of P. patens RNAi pathways was made by high-throughput and “degradome” sequencing as well as the functional analysis of essential components of RNAi. In future, a combination of these techniques and the inclusion of gene expression profiling using RNA-seq technique can be applied to the available P. patens mutants with perturbed small RNA pathways. The information obtained from such analyses will add to a comprehensive understanding of small RNA pathways on a genome-wide scale.
The functional analysis of RNAi components in P. patens is currently limited to ΔPpDCL1a, ΔPpDCL1b, ΔPpDCL3, ΔPpDCL4 and ΔRDR6 mutants. However, it is now evident that P. patens RNAi pathway have specific features that differ from seed plants. Since all identified components of seed plant RNAi pathway have homologues in P. patens, their detailed analysis may reveal further conserved or deviating functions. In addition, functional studies of P. patens miRNAs by miRNA overexpression, miRNA target mimicry or the generation of miRNA-resistant lines by altering miRNA binding sites may reveal interesting findings. The analysis of conserved miRNAs together with their conserved targets between seed plants and P. patens will clarify whether these miRNAs control homologous and/or analogous processes.
The adaptation to adverse environmental conditions is a major prerequisite to assure survival of plants in nature, analyzing the involvement of small RNAs in the control of stress-induced gene regulatory networks in P. patens will greatly increase our understanding of plant tolerance to biotic and abiotic stresses.
References
- 1.Vazquez F. Arabidopsis endogenous small RNAs: Highways and byways. Trends Plant Sci. 2006;11:460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Axtell M.J., Snyder J.A., Bartel D.P. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 4.Lu C., Jeong D.H., Kulkarni K., Pillay M., Nobuta K., German R., Thatcher S.R., Maher C., Zhang L., Ware D., et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs) Proc. Natl. Acad. Sci. USA. 2008;105:4951–4956. doi: 10.1073/pnas.0708743105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khraiwesh B., Arif M.A., Seumel G.I., Ossowski S., Weigel D., Reski R., Frank W. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.De Carvalho F., Gheysen G., Kushnir S., van Montagu M., Inze D., Castresana C. Suppression of beta-1,3-glucanase transgene expression in homozygous plants. EMBO J. 1992;11:2595–2602. doi: 10.1002/j.1460-2075.1992.tb05324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 9.Napoli C., Lemieux C., Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romano N., Macino G. Quelling: Transient inactivation of gene expression in neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton A.J., Baulcombe D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 12.Talmor-Neiman M., Stav R., Klipcan L., Buxdorf K., Baulcombe D.C., Arazi T. Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 2006;48:511–521. doi: 10.1111/j.1365-313X.2006.02895.x. [DOI] [PubMed] [Google Scholar]
- 13.Allen E., Xie Z., Gustafson A.M., Carrington J.C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 17.Kenrick P., Crane P. The origin and early evolution of plants on land. Nature. 1997;389:33–39. [Google Scholar]
- 18.Schaefer D.G., Zryd J.P. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- 19.Rensing S.A., Lang D., Zimmer A.D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P.F., Lindquist E.A., Kamisugi Y., et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 20.Fattash I., Voss B., Reski R., Hess W.R., Frank W. Evidence for the rapid expansion of microRNA-mediated regulation in early land plant evolution. BMC Plant Biol. 2007;7:13. doi: 10.1186/1471-2229-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho S.H., Addo-Quaye C., Coruh C., Arif M.A., Ma Z., Frank W., Axtell M.J. Physcomitrella patens DCL3 is required for 22–24 nt siRNA accumulation, suppression of retrotransposon-derived transcripts, and normal development. PLoS Genet. 2008;4:e1000314. doi: 10.1371/journal.pgen.1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Axtell M.J., Bartel D.P. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 24.Bezanilla M., Pan A., Quatrano R.S. RNA interference in the moss Physcomitrella patens. Plant Physiol. 2003;133:470–474. doi: 10.1104/pp.103.024901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bezanilla M., Perroud P.F., Pan A., Klueh P., Quatrano R.S. An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol. 2005;7:251–257. doi: 10.1055/s-2005-837597. [DOI] [PubMed] [Google Scholar]
- 26.Khraiwesh B., Ossowski S., Weigel D., Reski R., Frank W. Specific gene silencing by artificial microRNAs in Physcomitrella patens: An alternative to targeted gene knockouts. Plant Physiol. 2008;148:684–693. doi: 10.1104/pp.108.128025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arazi T., Talmor-Neiman M., Stav R., Riese M., Huijser P., Baulcombe D.C. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 29.Park W., Li J., Song R., Messing J., Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z., Ebright Y.W., Yu B., Chen X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;2:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan J., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 33.Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 34.Noma K., Sugiyama T., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S.I. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 35.Hutvagner G., Simard M.J. ARGONAUTE proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 36.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 37.Morel J.B., Godon C., Mourrain P., Beclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 39.Baumberger N., Baulcombe D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi Y., Denli A.M., Hannon G.J. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 41.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 43.Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crete P., Voinnet O., Robaglia C. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell. 2009;21:1762–1768. doi: 10.1105/tpc.108.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matzke M.A., Matzke A.J. Planting the seeds of a new paradigm. PLoS Biol. 2004;2:E133. doi: 10.1371/journal.pbio.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schramke V., Allshire R. Those interfering little RNAs! Silencing and eliminating chromatin. Curr. Opin. Genet. Dev. 2004;14:174–180. doi: 10.1016/j.gde.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim V.N. MicroRNA precursors in motion: Exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Park M.Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R.S. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao N., Lye K.W., Barton M.K. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Khraiwesh B., Zhu J.K., Zhu J.H. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta. 2012;1819:137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., Qi Y. DNA methylation mediated by a microRNA pathway. Mol. Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Addo-Quaye C., Snyder J.A., Park Y.B., Li Y.F., Sunkar R., Axtell M.J. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA. 2009;15:2112–2121. doi: 10.1261/rna.1774909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saleh O., Issman N., Seumel G.I., Stav R., Samach A., Reski R., Frank W., Arazi T. MicroRNA534a control of BLADE-ON-PETIOLE 1 and 2 mediates juvenile-to-adult gametophyte transition in Physcomitrella patens. Plant J. 2011;65:661–674. doi: 10.1111/j.1365-313X.2010.04451.x. [DOI] [PubMed] [Google Scholar]
- 54.Xie Z., Allen E., Wilken A., Carrington J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 56.Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouche N., Gasciolli V., Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 57.Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 59.Li F., Orban R., Baker B. SoMART: A web server for plant miRNA, tasiRNA and target gene analysis. Plant J. 2012;70:891–901. doi: 10.1111/j.1365-313X.2012.04922.x. [DOI] [PubMed] [Google Scholar]
- 60.Arif M.A., Fattash I., Ma Z., Cho S.H., Beike A.K., Reski R., Axtell M.J., Frank W. DICER-LIKE3 activity in physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol. Plant. 2012;5:1281–1294. doi: 10.1093/mp/sss036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams L., Carles C.C., Osmont K.S., Fletcher J.C. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA. 2005;102:9703–9708. doi: 10.1073/pnas.0504029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia D., Collier S.A., Byrne M.E., Martienssen R.A. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 63.Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 64.Dunoyer P., Brosnan C.A., Schott G., Wang Y., Jay F., Alioua A., Himber C., Voinnet O. An endogenous, systemic RNAi pathway in plants. EMBO J. 2010;29:1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Axtell M.J. The small RNAs of Physcomitrella patens: Expression, function and evolution. Ann. Plant Rev. 2009;36:113–142. [Google Scholar]
- 66.MacRae I.J., Ma E., Zhou M., Robinson C.V., Doudna J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. USA. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nobuta K., McCormick K., Nakano M., Meyers B.C. Bioinformatics analysis of small RNAs in plants using next generation sequencing technologies. Methods Mol. Biol. 2010;592:89–106. doi: 10.1007/978-1-60327-005-2_7. [DOI] [PubMed] [Google Scholar]
- 68.Morin R.D., Aksay G., Dolgosheina E., Ebhardt H.A., Magrini V., Mardis E.R., Sahinalp S.C., Unrau P.J. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 2008;18:571–584. doi: 10.1101/gr.6897308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pak J., Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 71.Sijen T., Steiner F.A., Thijssen K.L., Plasterk R.H. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 72.Baulcombe D.C. Molecular biology. Amplified silencing. Science. 2007;315:199–200. doi: 10.1126/science.1138030. [DOI] [PubMed] [Google Scholar]
- 73.Moissiard G., Parizotto E.A., Himber C., Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007;13:1268–1278. doi: 10.1261/rna.541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mlotshwa S., Pruss G.J., Peragine A., Endres M.W., Li J., Chen X., Poethig R.S., Bowman L.H., Vance V. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One. 2008;3:e1755. doi: 10.1371/journal.pone.0001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luo Q.J., Samanta M. P., Koksal F., Janda J., Galbraith D.W., Richardson C.R., Ou-Yang F., Rock C.D. Evidence for antisense transcription associated with microRNA target mRNAs in Arabidopsis. PLoS Genet. 2009;5:e1000457. doi: 10.1371/journal.pgen.1000457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alder M.N., Dames S., Gaudet J., Mango S.E. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA. 2003;9:25–32. doi: 10.1261/rna.2650903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nishikura K. A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell. 2001;107:415–418. doi: 10.1016/s0092-8674(01)00581-5. [DOI] [PubMed] [Google Scholar]
- 79.Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. From the cover: 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA. 2010;107:15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 81.Asif M.A., Fattash I., Khraiwesh B., Frank W. Physcomitrella patens Small RNA Pathways. In: Erdmann V.A., Barciszewski J., editors. Non Coding RNAs in Plants. Springer; Berlin/Heidelberg, Germany: 2011. pp. 139–173. [Google Scholar]
- 82.Kurihara Y., Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bouche N., Lauressergues D., Gasciolli V., Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu B., Chen Z., Song X., Liu C., Cui X., Zhao X., Fang J., Xu W., Zhang H., Wang X., et al. Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell. 2007;19:2705–2718. doi: 10.1105/tpc.107.052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dunoyer P., Himber C., Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 88.Vaucheret H., Mallory A.C., Bartel D.P. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vaucheret H., Vazquez F., Crete P., Bartel D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harvey J.J., Lewsey M.G., Patel K., Westwood J., Heimstadt S., Carr J.P., Baulcombe D.C. An antiviral defense role of AGO2 in plants. PLoS One. 2011;6:e14639. doi: 10.1371/journal.pone.0014639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rajeswaran R., Aregger M., Zvereva A.S., Borah B.K., Gubaeva E.G., Pooggin M.M. Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res. 2012;40:6241–6254. doi: 10.1093/nar/gks242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zilberman D., Cao X., Jacobsen S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 93.Havecker E.R., Wallbridge L.M., Hardcastle T.J., Bush M.S., Kelly K.A., Dunn R.M., Schwach F., Doonan J.H., Baulcombe D.C. The Arabidopsis RNA-directed DNA methylation ARGONAUTE functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pontes O., Costa-Nunes P., Vithayathil P., Pikaard C.S. RNA polymerase V functions in Arabidopsis interphase heterochromatin organization independently of the 24-nt siRNA-directed DNA methylation pathway. Mol. Plant. 2009;2:700–710. doi: 10.1093/mp/ssp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pontes O., Li C.F., Nunes P.C., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 96.Wierzbicki A.T., Haag J.R., Pikaard C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zilberman D., Cao X., Johansen L.K., Xie Z., Carrington J.C., Jacobsen S.E. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 98.Zheng X., Zhu J., Kapoor A., Zhu J.K. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Olmedo-Monfil V., Duran-Figueroa N., Arteaga-Vazquez M., Demesa-Arevalo E., Autran D., Grimanelli D., Slotkin R.K., Martienssen R.A., Vielle-Calzada J.P. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Q., Yao X., Pi L., Wang H., Cui X., Huang H. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and leaf polarity establishment by repressing miR165/166 in Arabidopsis. Plant J. 2009;58:27–40. doi: 10.1111/j.1365-313X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- 101.Mallory A.C., Hinze A., Tucker M.R., Bouche N., Gasciolli V., Elmayan T., Lauressergues D., Jauvion V., Vaucheret H., Laux T. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet. 2009;5:e1000646. doi: 10.1371/journal.pgen.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sugiyama T., Cam H., Verdel A., Moazed D., Grewal S.I. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl. Acad. Sci. USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vaistij F.E., Jones L., Baulcombe D.C. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garcia-Ruiz H., Takeda A., Chapman E.J., Sullivan C.M., Fahlgren N., Brempelis K.J., Carrington J.C. Arabidopsis RNA-Dependent RNA polymerases and Dicer-Like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. Plant Cell. 2010;22:481–496. doi: 10.1105/tpc.109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chan S.W., Zilberman D., Xie Z., Johansen L.K., Carrington J.C., Jacobsen S.E. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 106.Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pandey S.P., Gaquerel E., Gase K., Baldwin I.T. RNA-directed RNA polymerase3 from Nicotiana attenuata is required for competitive growth in natural environments. Plant Physiol. 2008;147:1212–1224. doi: 10.1104/pp.108.121319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zong J., Yao X., Yin J., Zhang D., Ma H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: Duplications and possible losses before and after the divergence of major eukaryotic groups. Gene. 2009;447:29–39. doi: 10.1016/j.gene.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 109.Dalmay T., Hamilton A., Rudd S., Angell S., Baulcombe D.C. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 110.Elmayan T., Adenot X., Gissot L., Lauressergues D., Gy I., Vaucheret H. A neomorphic sgs3 allele stabilizing miRNA cleavage products reveals that SGS3 acts as a homodimer. FEBS J. 2009;276:835–844. doi: 10.1111/j.1742-4658.2008.06828.x. [DOI] [PubMed] [Google Scholar]
- 111.Akbergenov R., Si-Ammour A., Blevins T., Amin I., Kutter C., Vanderschuren H., Zhang P., Gruissem W., Meins F., Hohn T., et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006;34:462–471. doi: 10.1093/nar/gkj447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fang Y., Spector D.L. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 2007;17:818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qin H., Chen F., Huan X., Machida S., Song J., Yuan Y.A. Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein-protein interaction. RNA. 2010;16:474–481. doi: 10.1261/rna.1965310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Manavella P.A., Hagmann J., Ott F., Laubinger S., Franz M., Macek B., Weigel D. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012;151:859–870. doi: 10.1016/j.cell.2012.09.039. [DOI] [PubMed] [Google Scholar]
- 116.Kim S., Yang J.-Y., Xu J., Jang I.-C., Prigge M.J., Chua N.-H. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol. 2008;49:1634–1644. doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Rätsch G., Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing inArabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2008 doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smith M.R., Willmann M.R., Wu G., Berardini T.Z., Möller B., Weijers D., Poethig R.S. Cyclophilin 40 is required for microRNA activity inArabidopsis. Proc. Natl. Acad. Sci. USA. 2009 doi: 10.1073/pnas.0812729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iki T., Yoshikawa M., Meshi T., Ishikawa M. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 2012;31:267–278. doi: 10.1038/emboj.2011.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Earley K.W., Poethig R.S. Binding of the cyclophilin 40 Ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J. Biol. Chem. 2011;286:38184–38189. doi: 10.1074/jbc.M111.290130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cao X., Aufsatz W., Zilberman D., Mette M.F., Huang M.S., Matzke M., Jacobsen S.E. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 122.Cao X., Jacobsen S.E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA. 2002;99:16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Law J.A., Ausin I., Johnson L.M., Vashisht A.A., Zhu J.K., Wohlschlegel J.A., Jacobsen S.E. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr. Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huettel B., Kanno T., Daxinger L., Bucher E., van der Winden J., Matzke A.J., Matzke M. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: A versatile pathway for transcriptional gene silencing in plants. Biochim. Biophys. Acta. 2007;1769:358–374. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 125.Chan S.W., Henderson I.R., Zhang X., Shah G., Chien J.S., Jacobsen S.E. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet. 2006;2:e83. doi: 10.1371/journal.pgen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matzke M., Kanno T., Huettel B., Daxinger L., Matzke A.J. RNA-directed DNA methylation and Pol IVb in Arabidopsis. Cold Spring Harbor Symp. Quant. Biol. 2006;71:449–459. doi: 10.1101/sqb.2006.71.028. [DOI] [PubMed] [Google Scholar]
- 127.Huettel B., Kanno T., Daxinger L., Aufsatz W., Matzke A.J., Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jeddeloh J.A., Stokes T.L., Richards E.J. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 129.Shaked H., Avivi-Ragolsky N., Levy A.A. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006;173:985–994. doi: 10.1534/genetics.105.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Morel J.B., Mourrain P., Beclin C., Vaucheret H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- 131.Smith L.M., Pontes O., Searle I., Yelina N., Yousafzai F.K., Herr A.J., Pikaard C.S., Baulcombe D.C. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ausin I., Mockler T.C., Chory J., Jacobsen S.E. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat. Struct. Mol. Biol. 2009;16:1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zheng Z., Xing Y., He X.J., Li W., Hu Y., Yadav S.K., Oh J., Zhu J.K. An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis. Plant J. 2010;62:92–99. doi: 10.1111/j.1365-313X.2010.04130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Golden T.A., Schauer S.E., Lang J.D., Pien S., Mushegian A.R., Grossniklaus U., Meinke D.W., Ray A. Short integuments1/suspensor1/carpel factory, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 2002;130:808–822. doi: 10.1104/pp.003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Satoh N., Itoh J., Nagato Y. The SHOOTLESS2 and SHOOTLESS1 genes are involved in both initiation and maintenance of the shoot apical meristem through regulating the number of indeterminate cells. Genetics. 2003;164:335–346. doi: 10.1093/genetics/164.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nagasaki H., Itoh J., Hayashi K., Hibara K., Satoh-Nagasawa N., Nosaka M., Mukouhata M., Ashikari M., Kitano H., Matsuoka M., et al. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA. 2007;104:14867–14871. doi: 10.1073/pnas.0704339104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Toriba T., Suzaki T., Yamaguchi T., Ohmori Y., Tsukaya H., Hirano H.Y. Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell. 2010;22:1452–1462. doi: 10.1105/tpc.110.075291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hamilton A., Voinnet O., Chappell L., Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Matzke M.A., Birchler J.A. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 140.Melquist S., Bender J. Transcription from an upstream promoter controls methylation signaling from an inverted repeat of endogenous genes in Arabidopsis. Genes Dev. 2003;17:2036–2047. doi: 10.1101/gad.1081603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Aufsatz W., Mette M.F., van der Winden J., Matzke A.J., Matzke M. RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2002;99:16499–16506. doi: 10.1073/pnas.162371499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pelissier T., Thalmeir S., Kempe D., Sanger H.L., Wassenegger M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 1999;27:1625–1634. doi: 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hall I.M., Shankaranarayana G.D., Noma K., Ayoub N., Cohen A., Grewal S.I. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 144.Aufsatz W., Mette M.F., Matzke A.J., Matzke M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol. Biol. 2004;54:793–804. doi: 10.1007/s11103-004-0179-1. [DOI] [PubMed] [Google Scholar]
- 145.Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu Q., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 147.Tabara H., Yigit E., Siomi H., Mello C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 148.Tomari Y., Zamore P.D. Perspective: Machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 149.Xie Z., Kasschau K.D., Carrington J.C. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 150.Alvarez J.P., Pekker I., Goldshmidt A., Blum E., Amsellem Z., Eshed Y. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell. 2006;18:1134–1151. doi: 10.1105/tpc.105.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Niu Q.W., Lin S.S., Reyes J.L., Chen K.C., Wu H.W., Yeh S.D., Chua N.H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006;24:1420–1428. doi: 10.1038/nbt1255. [DOI] [PubMed] [Google Scholar]
- 152.Ossowski S., Schwab R., Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- 153.Parizotto E.A., Dunoyer P., Rahm N., Himber C., Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18:2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Qu J., Ye J., Fang R. Artificial microRNA-mediated virus resistance in plants. J. Virol. 2007;81:6690–6699. doi: 10.1128/JVI.02457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Warthmann N., Chen H., Ossowski S., Weigel D., Herve P. Highly specific gene silencing by artificial miRNAs in rice. PLoS One. 2008;3:e1829. doi: 10.1371/journal.pone.0001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Strepp R., Scholz S., Kruse S., Speth V., Reski R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA. 1998;95:4368–4373. doi: 10.1073/pnas.95.8.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]