Summary
Genomic imprinting occurs when expression of an allele differs based on the sex of the parent that transmitted the allele. In D. melanogaster, imprinting can occur, but its impact on allelic expression genome-wide is unclear. Here, we search for imprinted genes in D. melanogaster using RNA-seq to compare allele-specific expression between pools of 7–10 day old adult female progeny from reciprocal crosses. 119 genes with allelic expression patterns consistent with imprinting were identified and showed significant clustering within the genome. Surprisingly, additional analysis of several of these genes showed that either genomic heterogeneity or high levels of intrinsic noise caused imprinting-like allelic expression. Consequently, our data provide no convincing evidence of imprinting for D. melanogaster genes in their native genomic context. Elucidating sources of false positive signals for imprinting in allele-specific RNA-seq data, as done here, is critical given the growing popularity of this method for identifying imprinted genes.
Introduction
More than 50 years ago, Helen Crouse coined the term ‘imprinting’ to describe a case inSciard flies where the sex of the parent influenced the inheritance of a chromosome (Crouse, 1960). Since this time, the definition of imprinting has been expanded to include any parent-of-origin dependent chromosome marking, especially those causing differential gene activity or expression (Ferguson-Smith, 2011). Recently, genomic scans for imprinting at the level of RNA abundance in plants and mammals have shown that only a small percentage of genes (typically on the order of 100 genes) appear to be imprinted (Babak et al., 2008; Gehring et al., 2011; Hsieh et al., 2011; Luo et al., 2011; Wang et al., 2011; Wang et al., 2008; Waters et al., 2011; Wolff et al., 2011); these genes are sometimes found in clusters within the genome (Ferguson-Smith, 2011; Wood and Oakley, 2006); and their imprinting is often required for normal development (McGrath and Solter, 1984; Surani et al., 1984) and physiology (Buiting et al., 1995; Weksberg et al., 1993).
In Drosophila melanogaster, studies of imprinting have yielded conflicting results. Euchromatic genes inserted onto the heterochromatic Y chromosome and genes located on chromosomes with deletions, duplications, rearrangements, and/or translocations can show differences in their activity depending on the parent from which they are inherited, demonstrating that D. melanogaster is capable of imprinting (Anaka et al., 2009; Golic et al., 1998; Haller and Woodruff, 2000; Joanis and Lloyd, 2002; Lloyd et al., 1999; MacDonald et al., 2010; Maggert and Golic, 2002; Menon and Meller, 2009). However, when Wittkopp et al. (2006) tested for evidence of imprinting by analyzing allele-specific expression of eight genes that showed strong parent-of-origin effects on total gene expression in a genomic survey of D. melanogaster (Gibson et al., 2004), no evidence of imprinting was observed. Furthermore, gynogenetic and androgenetic D. melanogaster, which inherit all of their genetic information from a single parent, are viable, suggesting that imprinting is not essential in this species (Fuyama, 1984; Komma and Endow, 1995). Consequently, even though it is clear that D. melanogaster can form parent-of-origin specific imprints that affect gene activity, the prevalence of imprinted genes in their native genomic context within the D. melanogaster genome remains unclear (Menon and Meller, 2010).
Results and Discussion
To search for imprinting genome-wide, we used Illumina sequencing in conjunction with a novel bioinformatics pipeline to infer allele-specific RNA transcript abundance in progeny from reciprocal crosses. This method uses transcribed sequence polymorphisms to distinguish sequencing reads derived from each of the two parental alleles in F1 offspring. To maximize the proportion of sequencing reads informative for allele-specific expression, we used a cosmopolitan (M-type) and an African (Z-type) line of D. melanogaster (Hollocher et al., 1997). The M-type line used was the zygotic hybrid rescue line (zhr) first described by Sawamura and colleagues (1993) and the Z-type line was a Zimbabwean isofemale line (z30) isolated in 1990 (Begun and Aquadro, 1993; Wu et al., 1995). To improve the accuracy of allele assignments, we sequenced the M-type (zhr) and Z-type (z30) genomes to 23.2X and 21.5X coverage (Table S1) and used these data to assemble line-specific genomic sequences (see Supplemental Experimental Procedures).
M-type females were crossed to Z-type males, producing F1 hybrids hereafter referred to as MZ. Likewise, Z-type females were crossed to M-type males, producing F1 hybrids hereafter referred to as ZM (Figure S1A). MZ and ZM hybrid flies were collected 7–10 days after eclosion, and total RNA was extracted from a pool of 20 hybrid females for each genotype. MZ and ZM RNA samples were used to make cDNA sequencing libraries, which were sequenced using an Illumina GAIIx machine. The resultant paired end (2X76bp) sequencing reads (Table S1) were aligned to the strain-specific M-type and Z-type genomes. Using two strain-specific genome sequences for mapping avoids mapping biases introduced by using only a single reference genome (Degner et al., 2009; Graze et al., 2012). 86% and 87% of the reads from the MZ and ZM samples, respectively, were aligned without mismatches to unique genomic loci (Table S1). In each case, 21% of the uniquely mapping reads aligned perfectly to only one genome and were used to infer allele-specific expression (Figure S1B, Table S1).
The power to infer allele-specific expression using RNA-seq data (which is necessary to test for imprinting with this method) depends upon the expression level of a gene as well as the density of transcribed polymorphisms within it (Fontanillas et al., 2010). Prior work has shown that obtaining at least 20 allele-specific reads for a gene results in reproducible measures of relative allelic expression (McManus et al., 2010). Retaining only genes with 20 or more allele-specific reads (allele 1 + allele 2 ≥ 20) in both the MZ and ZM samples, 7206 genes were tested for allelic-expression patterns consistent with imprinting (Table S2). This includes 3% of the 4875 genes with an FPKM (number of fragments per kilobase per million mapped reads) less than one, 51% of the 1706 genes with an FPKM between one and five, and 83% of the 7430 genes with an FPKM greater than five (Figure S2). The modENCODE consortium used a threshold of FPKM = 1 to classify D. melanogaster genes as ‘expressed’ or ‘not expressed’ (Graveley et al., 2011) and according to this definition we tested 77% of the 9136 genes expressed (in the 7–10 day old adult females we examined) for imprinting.
To assess the accuracy of our allele-specific expression measurements, we compared the allelic expression ratios determined by RNA-seq to estimates from pyrosequencing (Ahmadian et al., 2000) of individual genes. Ten genes selected at random were used for pyrosequencing of the same MZ and ZM samples used for RNA-seq (Table S3). Pyrosequencing measurements were highly correlated (R2 = 0.88) with estimates from RNA-seq (Table S3, Figure S3A), suggesting that RNA-seq produces reliable measures of relative allelic expression. This is consistent with previous comparisons of RNA-seq and pyrosequencing measures of allelic expression that used distinct bioinformatic pipelines (McManus et al., 2010; Emerson et al., 2010).
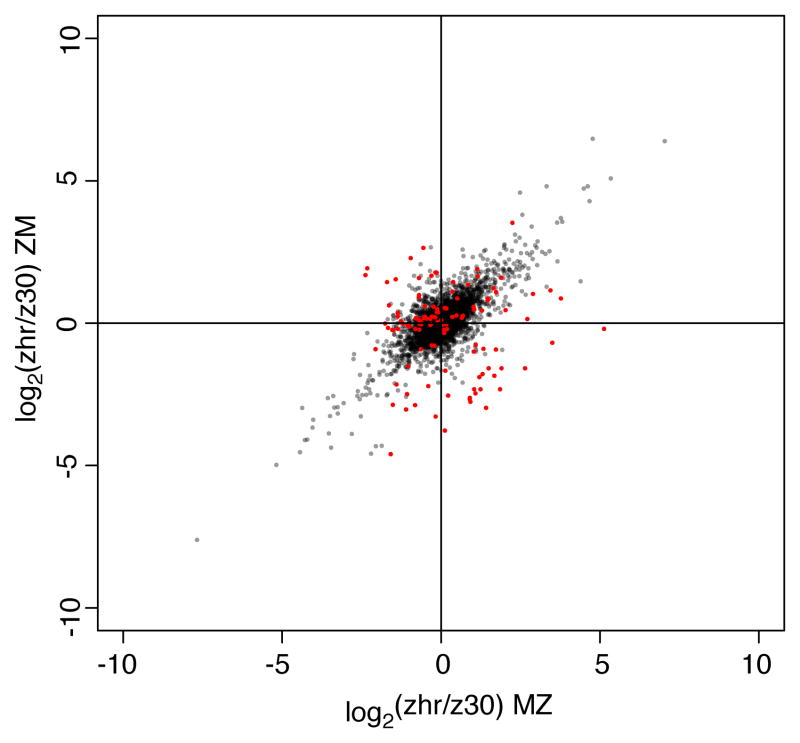
To identify genes that might be imprinted, we tested for differences in relative allele-specific expression between MZ and ZM using Fisher’s Exact Tests (FET). This test evaluates whether differential allelic expression (when present) is equal in magnitude and direction in the two genotypes. At a false discovery rate (FDR) of 5%, 119 (1.65%) of the 7206 genes analyzed had significant differences (FET, q < 0.05) in relative allelic expression between the two types of F1 hybrid progeny (Figure 1, Table S2). To evaluate the accuracy of RNA-seq measurements of allele-specific expression specifically for putatively imprinted genes (PIGs), we used pyrosequencing to independently measure allele-specific expression for four genes in this class using the same ZM and MZ samples as those used for RNA-seq. We again observed strong concordance (R2 = 0.85, Figure S3B) between pyrosequencing and RNA-seq measures of allele-specific expression, suggesting that inaccurate quantification of expression levels in cDNA pools by RNA-seq is unlikely to explain the observed differences in relative allelic expression between hybrid genotypes.
Figure 1. Allelic expression from reciprocal crosses suggests < 2% of genes in the genome might be imprinted.
Log2 transformed allelic expression ratios (zhr/z30) from MZ on the x-axis and log2(zhr/z30) allelic expression ratio from ZM on the y-axis. Each point represents one gene. Points are color-coded by significance in false discovery rate corrected Fisher’s exact tests where red points indicate q < 0.05. Note that the power to detect differences in allelic expression between ZM and MZ differs from gene-to-gene and is dependent upon the number of Illumina sequencing reads obtained that map to that gene. See also Figures S1–S3, Tables S1–S2.
Putatively imprinted genes (PIGs) are clustered in the genome
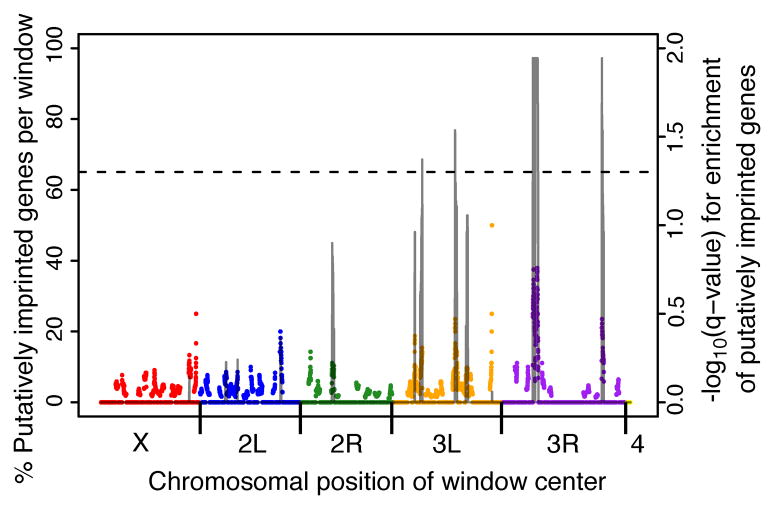
In mammals, imprinted genes are often found in clusters throughout the genome (Ferguson-Smith, 2011; Wood and Oakley, 2006) and this clustering might relate to the mechanism by which they are regulated (Caspary et al., 1998; DiNardo-Mancini et al., 2006; Lewis et al., 2004; Lopes et al., 2003). To determine if this was also true for the PIGs in the D. melanogaster genome, we used a sliding window Monte Carlo sampling approach with FDR corrected approximate permutation tests to investigate potential clustering. We found that there were four regions in the D. melanogaster genome that showed significant clustering (permutation test, q < 0.05) of PIGs (Figure 2). Interestingly, all four significant clusters were found on chromosome 3, with two on the left arm (3L) and two on the right arm (3R) of the chromosome. Together, these four regions contain 27% (32/119) of the PIGs, with one cluster located on chromosome arm 3R (6550000 – 8280000) containing 17% (20/119) of all PIGs (Figure 2). Clustering of PIGs in the genome is consistent with previously described mechanisms of imprinting, but could also be caused by other factors.
Figure 2. Putatively imprinted genes clustered significantly on chromosomes.
Using a sliding window analysis, the proportion of genes within each 500kb window that were identified as putatively imprinted is indicated for positions across the genome. Each chromosome arm is indicated on the x-axis, with one point representing each window. Using a Monte Carlo sampling approach and approximate permutation tests that control for differences in the number of genes within each window, and followed by a multiple testing correction we identified regions of the genome that were significantly enriched for PIGs. FDR corrected p-values are indicated by the solid line and the dotted line indicates the threshold used to identify significant clusters (q < 0.05).
Low frequency deletion(s) responsible for some cases of apparent imprinting
To further test for evidence of imprinting, we more closely examined twelve genes within the largest and most significant cluster of PIGs (3R 6.5–8.3MB region, Figure 2). Seven of these genes were PIGs and five were genes that showed no significant differences in relative allelic expression between ZM and MZ. Pyrosequencing was again used to obtain an independent measure of relative allelic expression, except instead of testing the same biological sample used for RNA-seq (as described above), we analyzed four independent biological replicate pools of ZM and MZ flies, each containing twenty 7–10 day old adult females (Table S3). From each pool, we sequentially extracted genomic DNA (gDNA) and RNA.
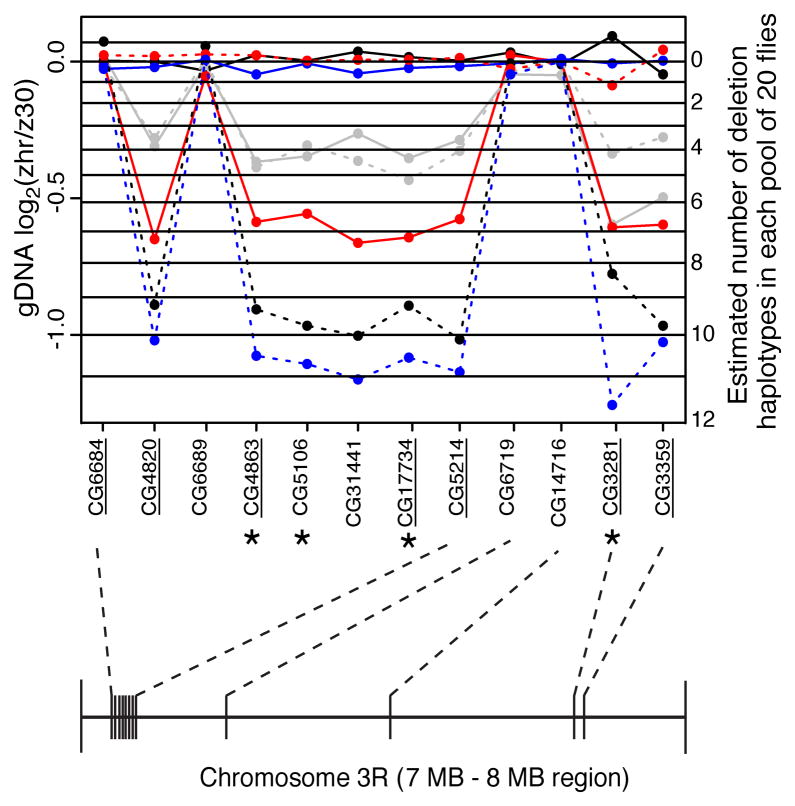
F1 flies produced by crossing two highly inbred lines are expected to be genetically identical, thus analysis of gDNA serves as a control for differential amplification of the two alleles during PCR prior to pyrosequencing (Landry et al., 2005; Wittkopp, 2011; Wittkopp et al., 2004; Wittkopp et al., 2008). Surprisingly, and unlike for the 34 genes located outside of clustered PIGs that we analyzed (data not shown), relative allelic abundance differed greatly for the gDNA samples among the biological replicates -- between the MZ and ZM genotypes as well as among replicate MZ or ZM samples (Figure 3). When present, deviations from equal allelic abundance in gDNA were similar for genes throughout the cluster within a replicate pool, but differed among pools. The M-type (zhr) allele was always the allele underrepresented (Figure 3).
Figure 3. Replicate pools of flies showed different allele frequencies in genomic DNA for putatively imprinted genes located in a cluster.
For 12 genes in the region containing the largest cluster of putatively imprinted loci (7,000,000 – 8,000,000 on chromosome 3R), 7 that were identified as putatively imprinted (underlined) and 5 that were not, we used pyrosequencing to determine the relative abundance of zhr and z30 alleles in genomic DNA in additional biological replicate pools containing 20 F1 heterozygous flies each. The log2(zhr/z30) ratio is plotted for gDNA from each biological replicate pool, with the four ZM pools indicated by solid lines and the four MZ pools indicated by dotted lines. Replicates are arbitrarily colored blue, grey, red and black. The genomic arrangement of these genes is shown below the plot. Genes labeled with an asterisk were also genotyped in individual flies (Table S4). Note that CG6684 is underlined because it showed significant evidence of allelic expression differences between MZ and ZM in the RNA-seq data; however this gene does not appear to be included within the deleted region(s). Pyrosequencing analysis of CG6684 showed no evidence of differential allelic expression between MZ and ZM and normal variance among replicate biological samples, suggesting that it was a false positive in the RNA-seq data. CG5106 and CG31441 appear to be included within the deleted region, but showed no significant evidence of an imprinting-like pattern of allelic expression in the RNA-seq data, probably due to lack of power as these two genes had the lowest read counts for those tested. See also Tables S3–S4.
A polymorphic deletion(s) in the M-type (zhr) strain or a polymorphic duplication(s) in the Z-type (z30) strain could account for the differences in gDNA content observed among replicate pools of F1 flies. To directly test for evidence of a deletion or duplication, we used pyrosequencing to genotype 48 individual F1 progeny (24 MZ and 24 ZM) at four loci within the 3R 6.5–8MB region that showed a cluster of PIGs (indicated with asterisks in Figure 3) as well as at two loci on other chromosomes. 46 of the 48 F1 hybrid flies showed evidence of one M-type and one Z-type allele at all six loci tested, as expected. The remaining two hybrids showed evidence of only the Z-type (z30) allele at the four loci within the cluster, but both flies showed both alleles at the two loci tested on other chromosomes (Table S4); the presence of these heterozygous sites demonstrates that these two flies are in fact F1 hybrids and not contaminating flies with parental genotypes. Based on these data, we conclude that the M-type (zhr) strain contains one or more deletion(s) in this region on 3R that remains heterozygous despite years of inbreeding followed by 10 generations of pair mating immediately prior to the start of this experiment. Residual heterozygosity such as this has also been reported in D. melanogaster following extensive inbreeding in lines used for genomic sequencing (Mackay et al., 2012).
The presence of this deletion haplotype at low frequency in the zhr line used to produce MZ and ZM hybrids, suggests that differences in its frequency in the pools of 20 MZ and 20 ZM hybrid flies used for RNA-seq are more likely to be responsible for the observed difference in relative allelic expression than imprinting. Indeed, after controlling for differences in the alleles present in gDNA among the replicate pools analyzed by pyrosequencing (see Experimental Procedures), relative allelic expression in cDNA samples was not significantly different (p > 0.05 for all tests). It remains to be seen whether genotypic differences between the MZ and ZM pools of flies used for RNA-seq are also responsible for differences in relative allelic expression observed for other clustered PIGs, but we believe it is likely.
Non-clustered PIGs have higher than normal intrinsic noise
Our initial RNA-seq survey for imprinting identified all genes with significant differences in relative allelic expression between F1 hybrid progeny from reciprocal crosses as PIGs; however, imprinting is often defined in a more limited way in which only one allele of a gene (either the maternally- or paternally-inherited allele) accounts for the majority (or all) of the expression of the imprinted gene. Among the original set of 119 PIGs, only 18 showed patterns of allelic expression consistent with this more strict definition (Table S2, Figure S4), none of which were located in the clusters described above (Figure 2). To further test these 18 genes for evidence of imprinting, we analyzed allelic expression for each gene in the MZ and ZM biological replicates described in the preceding section (Table S3). Unlike for clustered PIGs examined in these samples, no significant differences in allele frequency were found among replicate gDNA samples for any of these 18 genes.
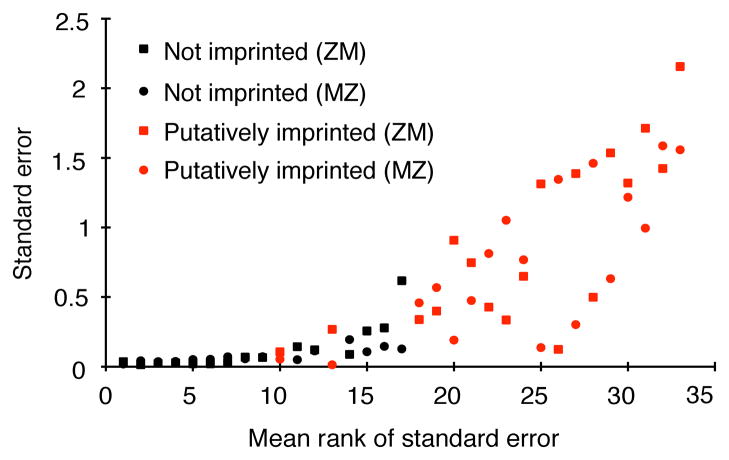
The relative allelic expression for these genes in the four MZ and four ZM biological replicates was still not typical, however: these 18 genes showed greater variance in relative allelic expression among the biological replicate pools than most genes that we have analyzed with pyrosequencing. Indeed, a Wilcoxon rank-sum test showed that the standard errors of log2-transformed allelic expression ratios were significantly greater for the 18 PIGs than for 16 genes selected at random (W = 260, p = 2.68E-7; Figure 4). Additional statistical tests showed no evidence for imprinting of these genes (q > 0.05 for all tests). Given (i) the high degree of variability we observed among replicate pools with the same genotype (MZ or ZM) for these genes, (ii) the lack of evidence for imprinting by pyrosequencing, and (iii) that we only analyzed one pool of flies for each genotype by RNA-seq, we conclude that significant differences observed between MZ and ZM for relative allelic expression in the RNA-seq data are most likely caused by sampling error.
Figure 4. Putatively imprinted genes have high intrinsic noise.
For each gene for each sample type (ZM or MZ) the standard error for log2 transformed allelic expression ratios from biological replicate pools of flies is shown, with black points representing genes selected at random from the genome (all of which showed no significant evidence of imprinting) and red points representing PIGs. Square marks represent the ZM sample and circles represent the MZ sample with one rank for each gene tested. The x-axis is rank of standard error for the two samples for each gene. See also Figure S4 and Table S3.
What role does imprinting play in regulating D. melanogaster gene expression?
As described above, RNA-seq analysis (validated by pyrosequencing) identified 119 of 7206 genes as having differences in relative allele-specific expression in reciprocal hybrids; however, analysis of gDNA and cDNA from additional replicate biological samples identified other factors (the presence of a polymorphic deletion(s) and using a single measurement to represent a highly variable phenotype) that are more likely than imprinting to be responsible for the differences in allelic expression observed in our RNA-seq data. Consequently, we conclude that these data provide no convincing evidence of imprinting affecting expression of endogenous D. melanogaster genes in their native genomic contexts - at least in the 7–10 day old adult females we examined.
Given the evidence of imprinting in other studies of D. melanogaster, why do we fail to find evidence of it in our genomic analysis? We cannot rule out the possibility that imprinting affects allelic activity in males, at other developmental stages, in limited tissues (with the signal masked by the absence of imprinting in the majority of cells sampled), or for genes with expression and/or polymorphism levels that cause them to be below our detection threshold, but there is also no evidence suggesting that imprinting is occurring under any of these conditions. In addition, as described by Menon and Meller (2010), evidence of imprinting in D. melanogaster comes from studying particular genotypes and imprinting might not impact gene expression in all genotypes: “In Drosophila, imprints are detected by alteration in expression of genes on rearranged chromosomes, but there is little to suggest that expression of any gene in karyotypically normally (sic) flies is governed by imprinting.” We tested 77% of the expressed genes in the D. melanogaster genome for imprinting in this study, and evidence that imprinting affects the expression of genes in their native genomic context is still lacking.
Genomic surveys for imprinting using RNA-seq: proceed with caution
In addition to providing insight into imprinting in D. melanogaster, this study identifies important considerations for using RNA-seq to test for imprinting in any species. RNA-seq has been used to search for imprinted loci in both plants and animals, including mouse (Babak et al., 2008; Wang et al., 2008; Wang et al., 2011), Arabidopsis (Gehring et al., 2011; Hsieh et al., 2011; Wolff et al., 2011) maize (Waters et al., 2011), and rice (Luo et al., 2011); but this approach is not without its pitfalls. For example, a study using RNA-seq to identify imprinted genes in various mouse tissues reported over 1000 imprinted loci (Gregg et al., 2010a; Gregg et al., 2010b), but most of these loci were subsequently shown to be false positives caused by biased sequencing and the failure to measure and account for technical and biological variability (DeVeale et al., 2012).
Data presented here and in DeVeale et al. (2012) clearly show the importance of validating putatively imprinted genes identified by RNA-seq with independent techniques (and, ideally, independent biological samples) prior to concluding that they are imprinted. To focus validation efforts on the loci most likely to be imprinted, RNA-seq experiments should include both biological and technical replicates, as well as, whenever possible, the analysis of gDNA extracted from the same tissue homogenate as the RNA. This final control is particularly important when working with small organisms (e.g., flies) for which multiple inbred individuals (that could have residual heterozygosity) are typically pooled prior to RNA extraction and cDNA sequencing, but it can also detect and control for differences in genomic content that might exist among cells from the same individual due to somatic mutations. For example, Shibata et al. (2012) have recently shown that microdeletions can cause genomic heterogeneity among mouse and human cells. Sequencing gDNA and cDNA derived from the same tissue sample can also allow corrections for bias introduced during the library preparation and sequencing. With more and more researchers turning to RNA-seq to study genomic imprinting, it is important to keep these caveats in mind.
Experimental Procedures
Fly strains, rearing, and collections
The D. melanogaster strain zhr carrying the hybrid rescuing Zhr1 chromosome (full genotype: XYS.YL.Df(1)Zhr, (Ferree and Barbash, 2009; Sawamura et al., 1993) and the Zimbabwean isofemale line z30 (Begun and Aquadro, 1993; Wu et al., 1995) were used for this study. All flies were reared on cornmeal medium on 16:8 light:dark cycle at 20° C. Prior to crossing, both strains were subjected to 10 generations of sibling pair matings to reduce genome-wide heterozygosity, and this was followed by 3 generations of population expansion to generate the quantity of flies needed for crosses. For each reciprocal cross performed, 10 vials were set up with 3 female and 3 male flies. Virgin female progeny were allowed to mate from the time of eclosion to 3 days post eclosion, then males and females were separated and females aged to 7–10 days post eclosion. All flies were collected during the same time of day to minimize the effects of circadian rhythm and flies were snap frozen in liquid N2.
Library preparation and Illumina sequencing
Pools of 20 female flies were used for total RNA extraction with TRIzol reagent according to manufacturer instructions (Invitrogen). Illumina sequencing libraries were prepared (see Supplemental Experimental Procedures) as previously reported (McManus et al., 2010). Two lanes of paired end (2X76 bp) Illumina GAIIx sequencing were performed. The sequencing data from this study have been submitted to the National Center for Biotechnology Information Sequence Read Archive under accession number SRA052065.
Quantifying total and allele-specific expression from sequencing reads
We developed a bioinformatics pipeline to quantify gene expression from the Illumina sequencing output (Figure S1B, Supplemental Experimental Procedures). Briefly, we aligned each mate of the paired-end RNA-seq reads separately to the newly built D. melanogaster genomes (zhr and z30, Supplemental Experimental Procedures) keeping only those reads that aligned to one genomic location. Reads that did not map were trimmed by 13 bases and realigned in three iterations. Reads that did not align were then discarded. We then converted zhr and z30 genomic coordinates of aligned reads to sequenced D. melanogaster genomic coordinates using the liftOver utility from the UCSC Genome Browser (Kent et al., 2002). Aligned sequence reads were then filtered based on their alignment to a previously identified set of overlap filtered constitutively expressed exons within the D. melanogaster genome (McManus et al., 2010) using intersectBed module of BedTools (Quinlan and Hall, 2010) (Version 2.12.0).
Remaining sequencing reads that aligned to only one of the two line specific genomes were used for quantification of allele-specific gene expression. Down-sampling followed by rounding to the nearest integer was used to account for differences in overall sequencing output between MZ and ZM and differences in mappability between zhr and z30 alleles. For each gene, allele-specific expression levels are reported (Table S2). To reduce the effect of sampling error (Fontanillas et al., 2010; McManus et al., 2010), we analyzed only genes that had more than 20 allele-specific reads (allele 1 + allele 2 ≥ 20) in both ZM and MZ. To test for unequal allelic expression between ZM and MZ, we performed Fisher’s exact tests using zhr and z30 allelic counts. Due to the multitude of tests performed, a false discovery rate (FDR) significance threshold of 5% was used to determine significance (Benjamini and Hochberg, 1995). Statistical analyses were performed in R (version 2.12.2, CRAN).
FPKM values reflecting total expression levels for individual genes were calculated by dividing the total number of paired-end reads mapped to a gene (including reads that were and were not informative for allele-specific expression) by the length of the sequence representing that gene in kilobases and then dividing this value by the number of millions of mapped reads from that sample.
Sliding window analyses with Monte Carlo sampling and approximate permutation tests
Genomic clustering of putatively imprinted genes was analyzed using a sliding window approach where we divided the genome into 11,726 overlapping 500kb windows and moved stepwise, offsetting by 10kb with each step. For each window, we counted the number of total genes and PIGs within each region. To test whether the observed clustering was significant, we used a Monte Carlo sampling approach to approximate the null distribution of imprinted genes randomly scattered along the genome. A Monte Carlo sampling approach was used to approximate the null distribution because the number of permutations required for an exact test in this case was exceedingly large (7206 choose 119). From the total set of 7,206 genes, we randomly sampled 119 genes without replacement, assigned them imprinting status, and aggregated new imprinting counts for each window. This was done 10,000 times, resulting in an approximate null distribution of the number of imprinted genes expected by chance in each window. To calculate an approximate p-value for each window, we summed the number of occurrences where the permuted value exceeded the observed value. Due to the multitude of tests performed, an FDR corrected significance threshold of 5% was used to determine significance (q < 0.05). Significant windows were collapsed to four regions based on overlap (Figure 2).
Pyrosequencing
To evaluate the accuracy of allelic expression measurements derived from our RNA-seq data and analysis, new cDNA pools were synthesized from the same RNA samples used for Illumina sequencing and used for pyrosequencing. cDNA was synthesized from total RNA using T(18)VN primers and Superscript II (Invitrogen) according to manufacturer recommendations. Both cDNA and gDNA were analyzed using pyrosequencing. For each gene assayed, PCR was performed in triplicate on both the cDNA and gDNA samples (separately) and followed by pyrosequencing (Qiagen). The genomic DNA was extracted from an independent pool of F1 flies and was used to normalize cDNA measurements (Wittkopp, 2011). Log2 transformed cDNA allelic expression ratios from Illumina and pyrosequencing were compared after normalization using type 2 regressions in R.
To investigate allelic expression within a cluster of genes on chromosome 3R, we constructed four new replicate pools of 20 individuals each for both ZM and MZ samples and co-extracted RNA and gDNA from a single tissue homogenate of each pool of flies using the Promega SV total RNA extraction system with modified protocol (Wittkopp, 2011). cDNA was made from total RNA as above and both gDNA and cDNA were used for PCR followed by pyrosequencing. To account for differences in gDNA allelic abundance among replicate pools of flies, the log2 allelic expression ratio for gDNA from a particular pool was subtracted from the log2 allelic expression ratios for cDNA samples derived from the same pool of flies (Wittkopp et al., 2004; Landry et al., 2005; Wittkopp et al., 2006; Wittkopp et al., 2008; Wittkopp et al., 2011).
The four biological replicates were used to investigate variation in allelic expression for a set of randomly chosen genes and this was compared to a set of putatively imprinted genes. The standard error for the log2 allelic expression ratio was calculated for each assay-sample combination for the randomly chosen genes and non-clustered PIGs and these two sets were compared using a Wilcoxon rank-sum test in R.
Supplementary Material
Highlights.
RNA-seq identified 119 potentially imprinted genes in D. melanogaster
These potentially imprinted genes were significantly clustered in the genome
Polymorphic deletions and intrinsic noise caused imprinting-like RNA-seq data
We conclude that D. melanogaster genes in their native context are not imprinted
Acknowledgments
We would like to thank all members of the Wittkopp lab, especially Brian Metzger and Bing Yang, for helpful comments on experimental design, data analysis, and manuscript presentation, Sebastian Zoellner for computational resources and Chung-I Wu for the z30 fly line. This work was supported by a National Science Foundation grant to P.J.W (MCB-1021398) and a National Institutes of Health grant to B.R.G. (5R01GM095296) as well as a National Research Service Award from the National Institutes of Health to J.C.D. (1F32GM089009-02). P.J.W. is a Research Fellow of the Alfred P. Sloan Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmadian A, Gharizadeh B, Gustafsson A, Sterky F, Nyren P, Uhlen M, Lundeberg J. Single-nucleotide polymorphism analysis by pyrosequencing. Anal Biochem. 2000;280:103–110. doi: 10.1006/abio.2000.4493. [DOI] [PubMed] [Google Scholar]
- Anaka M, Lynn A, McGinn P, Lloyd VK. Genomic Imprinting in Drosophila has properties of both mammalian and insect imprinting. Dev Genes Evol. 2009;219:59–66. doi: 10.1007/s00427-008-0267-3. [DOI] [PubMed] [Google Scholar]
- Babak T, DeVeale B, Armour C, Raymond C, Cleary MA, van der Kooy D, Johnson JM, Lim LP. Global Survey of Genomic Imprinting by Transcriptome Sequencing. Curr Biol. 2008;18:1735–1741. doi: 10.1016/j.cub.2008.09.044. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature. 1993;365:548– 550. doi: 10.1038/365548a0. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Buiting K, Saltoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B. Inherited microdeletions in the Angelman and Prader Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- Caspary T, Cleary MA, Baker CC, Guan X, Tilghman SM. Multiple Mechanisms Regulate Imprinting of the Mouse Distal Chromosome 7 Gene Cluster. Mol Cell Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse HV. The controlling element in sex chromosome behavior in Sciara. Genetics. 1960;45:1429–1443. doi: 10.1093/genetics/45.10.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner JF, Marioni JC, Pai AA, Pickrell JK, Mkadori E, Gilad Y, Pritchard JK. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveale B, van der Kooy D, Babak T. Critical Evaluation of Imprinted Gene Expression by RNA-Seq: A New Perspective. PLoS Genet. 2012;8:e1002600. doi: 10.1371/journal.pgen.1002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo-Mancini D, Steele SJS, Levorse JM, Ingram JS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Hsieh L, Sung H, Wang T, Huang C, Lu HH, Lu MJ, Wu S, Li W. Natural selection on cis and trans regulation in yeasts. Geneome research. 2010;20:826–836. doi: 10.1101/gr.101576.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- Ferree PM, Barbash DA. Species-Specific Heterochromatin Prevents Mitotic Chromosome Segregation to Cause Hybrid Lethality in Drosophila. PLoS Biol. 2009;7:e1000234. doi: 10.1371/journal.pbio.1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanillas P, Landry CR, Wittkopp PJ, Russ C, Gruber JD, Nusbaum C, Hartl DL. Key considerations for measuring allelic expression on a genomic scale using high-throughput sequencing. Mol Ecol. 2010;19:212–227. doi: 10.1111/j.1365-294X.2010.04472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuyama Y. Gynogenesis in Drosophila melanogaster. Jpn J Genet. 1984;59:91–96. [Google Scholar]
- Gehring M, Missirian V, Henikoff S. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One. 2011;6:e23687. doi: 10.1371/journal.pone.0023687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G, Riley-Berger R, Harshman LG, Kopp A, Vacha S, Nuzhdin SV, Wayne M. Extensive Sex-Specific Nonadditivity of Gene Expression in Drosophila melanogaster. Genetics. 2004;167:1791–1799. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic KG, Golic MM, Pimpinelli S. Imprinted control of gene activity in Drosophila. Curr Biol. 1998;8:1273–1276. doi: 10.1016/s0960-9822(07)00537-4. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graze RM, Novelo LL, Fear JM, Casella G, Nuzhdin SV, McIntyre LM. Allelic imbalance in Drosophila hybrid heads: exons, isoforms and evolution. Mol Biol Evol. 2012 doi: 10.1093/molbev/msr318. Published online January 7, 2012, 2010.1093/molbev/msr2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010a;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010b;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller BS, Woodruff RC. Varied expression of a Y-linked P[w+} insert due to imprinting in Drosophila melanogaster. Genome. 2000;43:285–292. doi: 10.1139/g99-125. [DOI] [PubMed] [Google Scholar]
- Hollocher H, Ting CT, Wu ML, Wu CI. Incipient Speciation by Sexual Isolation in Drosophila melanogaster: Extensive Genetic Divergence Without Reinforcement. Genetics. 1997;147:1191–1201. doi: 10.1093/genetics/147.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA. 2011;108:1755–1762. doi: 10.1073/pnas.1019273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joanis V, Lloyd VK. Genomic imprinting in Drosophila is maintained by the products of Supressor of variegation and trithorax group, but not the Polycomb group, genes. Mol Genet Genomics. 2002;268:103–112. doi: 10.1007/s00438-002-0731-0. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komma DJ, Endow SA. Haploidy and andorgenesis in Drosophila. Proc Natl Acad Sci USA. 1995;92:11884–11888. doi: 10.1073/pnas.92.25.11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Lloyd VK, Sinclair DA, Grigliatti TA. Genomic Imprinting and Position-Effect Variegation in Drosophila melanogaster. Genetics. 1999;151:1503–1516. doi: 10.1093/genetics/151.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forne T, Murrell A, Constancia M, Bartolomei M, Walter J, et al. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum Mol Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- Luo M, Taylor JM, Spriggs A, Zhang H, Wu X, Russell S, Singh M, Koltunow A. A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 2011;7:e1002125. doi: 10.1371/journal.pgen.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald WA, Menon D, Bartlett NJ, Sperry GE, Rasheva V, Meller V, Lloyd VK. The Drosophila homolog of the mammalian imprint regulator, CTCF, maintains the maternal genomic imprint in Drosophila melanogaster. BMC Biol. 2010;8:105. doi: 10.1186/1741-7007-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert KA, Golic KG. The Y chromosome of Drosophila melanogaster exhibits chromosome-wide imprinting. Genetics. 2002;162:1245–1258. doi: 10.1093/genetics/162.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 2010;20:816–825. doi: 10.1101/gr.102491.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon DU, Meller VH. Imprinting of the Y Chromosome Influences Dosage Compensation in roX1 and roX2Drosophila melanogaster. Genetics. 2009;183:811–820. doi: 10.1534/genetics.109.107219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon DU, Meller VH. Germ line imprinting in Drosophila Epigenetics in search of function. Fly. 2010;4:48–52. doi: 10.4161/fly.4.1.10881. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K, Yamamoto MT, Wantanabe TK. Hybrid Lethal Systems in the Drosophila Melanogaster Species Complex. II. the Zygotic Hybrid Rescue (Zhr) Gene of D. Melanogaster. Genetics. 1993;133:307–313. doi: 10.1093/genetics/133.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, Dutta A. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012;336:82–86. doi: 10.1126/science.1213307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MAH, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Wang X, Soloway PD, Clark AG. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics. 2011;189:109–122. doi: 10.1534/genetics.111.130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sun Q, McGrath SD, Mardis ER, Soloway PD, Clark AG. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One. 2008;3:e3839. doi: 10.1371/journal.pone.0003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh C, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM. Parent-of-Origin Effects on Gene Expression and DNA Methylation in the Maize Endosperm. Plant Cell. 2011;23:4221–4233. doi: 10.1105/tpc.111.092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksberg R, Shen DR, Fei YL, Song QL, Squire J. Disruption of insulin like growth factor 2 imprinting in Beckwith Wiedemann syndrome. Nat Genet. 1993;5:143–150. doi: 10.1038/ng1093-143. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ. Using pyrosequencing to measure allele-specific mRNA abundance and infer the effects of cis- and trans-regulatory differences. In: Orgogozo V, Rockman M, editors. Molecular Methods for Evolutionary Genetics. 2011. pp. 297–317. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Parent-of-origin effects on mRNA expression in Drosophila melanogaster not caused by genomic imprinting. Genetics. 2006;173:1817–1821. doi: 10.1534/genetics.105.054684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp T, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between species. Nat Genet. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MT, Spillane C, Nordborg M, Rehmsmeier M, Köhler C. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 2011;7:e1002126. doi: 10.1371/journal.pgen.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Oakley RJ. Genomic Imprinting in Mammals: Emerging Themes and Established Theories. PLoS Genet. 2006;2:e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Hollocher H, Begun DJ, Aquadro CF, Xu Y, Wu M. Sexual isolation in Drosophila melanogaster: A possible case of incipient speciation. Proc Natl Acad Sci USA. 1995;92:2519–2523. doi: 10.1073/pnas.92.7.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.