Abstract
Human centromeres contain multi-megabase-sized arrays of alpha satellite DNA, a family of satellite DNA repeats based on a tandemly arranged 171bp monomer. The centromere-specific histone protein CENP-A is assembled on alpha satellite DNA within the primary constriction, but does not extend along its entire length. CENP-A domains are estimated to extend over 1500-2000kb of alpha satellite DNA. However, these estimates do not take into account inter-individual variation in alpha satellite array sizes on homologous chromosomes and among different chromosomes. We defined the genomic distance of CENP-A chromatin on human chromosomes X and Y from different individuals. CENP-A chromatin occupied different genomic intervals on different chromosomes, but despite inter-chromosomal and inter-individual array size variation, the ratio of CENP-A to total alpha satellite DNA size remained consistent. Changes in the ratio of alpha satellite array size to CENP-A domain size were observed when CENP-A was over-expressed and when primary cells were transformed by disrupting interactions between the tumor suppressor protein Rb and chromatin. Our data support a model for centromeric domain organization in which the genomic limits of CENP-A chromatin varies on different human chromosomes, and imply that alpha satellite array size is a more prominent predictor of CENP-A incorporation than chromosome size. In addition, our results also suggest that cancer transformation and amounts of centromeric heterochromatin have notable effects on the amount of alpha satellite that is associated with CENP-A chromatin.
Keywords: chromatin, chromosome, heterochromatin, histone, kinetochore, retinoblastoma
Introduction
Centromeres are specialized regions essential for chromosome segregation in meiosis and mitosis. Normal human centromeres are located at regions of alpha satellite DNA, a ~171bp repeat subunit, that is tandemly organized as either homogenous arrays or heterogenous monomers (Waye and Willard, 1987, Willard and Waye, 1987, Rudd and Willard, 2004). On endogenous human chromosomes, alpha satellite DNA is organized into higher-order repeat units (HOR) that contain a specific number of monomers. Hundreds of copies of the HORs give rise to homogenous arrays at each primary constriction. Alpha satellite DNA arrays show both inter- and intra-chromosomal size variation that ranges from 200kb to 5Mbs (Wevrick and Willard, 1989, Mahtani and Willard, 1990, Lo et al., 1999). Higher order alpha satellite DNA is the site of centromere protein binding, including CENP-A, the centromere-specific histone H3 variant. CENP-A is the structural foundation for the three-dimensional kinetochore, linking centromeric DNA and protein components of the kinetochore. Centromeric chromatin appears on linear chromatin fibers as CENP-A nucleosomes interrupted by blocks of nucleosomes containing H3.
Alpha satellite DNA genetically and functionally defines the human centromere and is an efficient substrate for new centromere formation (Schueler et al., 2001, Grimes et al., 2002, Harrington et al., 1997). Although the arrays extend several megabases on a given chromosome, centromere proteins are usually positioned on only a fraction of the total repeat array (Spence et al., 2002, Lam et al., 2006, Zeng et al., 2004, Blower et al., 2002). Previous studies that used chromatin fibers to view organization of centromeric chromatin domains in various organisms were unable to offer single nucleosome resolution so that CENP-A foci were thought to represent groups of nucleosomes. In fact, the number of CENP-A foci per fiber varied, from a few to more than 50, implying that the sizes of CENP-A chromatin domains and/or the number of CENP-A nucleosomes within subdomains may normally vary at individual centromeres. Several fluorescence-based electron microscopy studies of three-dimensional metaphase chromosomes have also suggested that kinetochore size varies two- to three-fold (Irvine et al., 2004, Cherry et al., 1989, Tomkiel et al., 1994). Such variation implies that the extent of centromeric chromatin, as defined as chromatin containing interspersed CENP-A/H3, correlates with the physical size of kinetochores. It has been estimated from biochemical analysis that CENP-A nucleosomes occupy 1500-3000kb of alpha satellite DNA (Black et al., 2007), but these studies assumed similarly sized or an equal number of CENP-A and H3 subdomains. Furthermore, while these estimates may reflect the average ratio of CENP-A to H3 nucleosomes within the higher order structure of the primary constriction, they did not take into account the extensive range of human alpha satellite DNA array sizes present within single individuals and among different people. In this study, we optically measured the genomic extent of CENP-A chromatin assembly on alpha satellite DNA arrays of defined size on individual Homo sapiens chromosomes X (HSAX) and Y (HSAY). Our results indicate that like alpha satellite array length, the genomic size of the CENP-A domain is heterogeneous but is formed proportionally on alpha satellite DNA, regardless of chromosome origin.
Materials and Methods
Cell lines and culture
Human male cell lines and rodent somatic cell hybrid lines containing a single human Y chromosome were grown in MEM alpha supplemented with 10% fetal bovine serum (FBS), L-glutamine, and 1X antibiotic-antimycotic solution (Invitrogen). Rodent-human somatic cell hybrids containing a single human X were grown in MEM alpha containing 10% FBS, 1X antibiotic antimycotic and 1X HAT (hypoxanthine/aminopterin/thymidine; Invitrogen). Male lymphoblast line DIP1 was grown in RPMI 1640 supplemented with 10-15% FBS, L-glutamine, 1X antibiotic-antimycotic and HAT. Lines DIP4 and DIP5 were grown in HAM’s media containing 10% FBS and 1X antibiotic-antimycotic solution. DIP2 cells were transfected with a FLAG-tagged CENP-A expression vector, and blasticidin-resistant clones were isolated and expanded. Clone 1c1 (named DIP2-OE in this study) in which the total CENP-A (endogenous + FLAG-CENP-A) was determined to be 50% above endogenous levels (Lam et al., 2006) was selected for fiber IF-FISH experiments in this study. Line DIP3-E7 was created by infecting a primary human dermal fibroblast line with an E7 adenoviral expression vector.
Western blot analysis
Nuclear fractions from DIP3 and DIP3-E7 cells were isolated using cytoskeleton buffer (CSK buffer: 10mM Pipes pH 6.8, 100mM NaCl, 300mM sucrose, 3mM MgCl2, 1mM EGTA, 1mM DTT, 0.1mM ATP, 0.1% Triton X-100) supplemented with phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor cocktail (Roche). Chromatin and soluble fractions were separated by sonication and centrifugation (Song et al., 2010). Laemli lysis buffer was added to fractions that were then subjected to SDS-PAGE electrophoresis on 12% acrylamide gels. Proteins were transferred onto PVDF membranes by semi-dry blotting, and Rb was detected by Western blotting using polyclonal antibodies (ab6075, Abcam). Blots were incubated with enhanced chemiluminescence (ECL) reagents and exposed to autoradiographic film (Kodak Biomax XAR).
Immunofluorescence and FISH (IF-FISH) on extended chromatin fibers
Immunofluorescence with CENP-A antibodies followed by FISH on metaphase chromosomes was performed as previously described (Sullivan and Warburton, 1999). Extended chromatin fibers were produced using published methods (Blower et al., 2002, Lam et al., 2006). X alpha satellite (DXZ1) and Y alpha satellite (DYZ3) probes for in situ hybridization were generated by cloning PCR products (Warburton et al., 1991). Probes were labeled with biotin-16-dUTP (Roche), digoxygenin-11-dUTP (Roche), or dUTPs conjugated to Alexa Fluor 488 or Alexa Fluor 568 (Molecular Probes). Commerical directly-labeled (red or green) alpha satellite FISH probes were also used (Abbott Laboratories). Fibers were prepared from at least two independent experiments and combined CENP-A immunostaining and FISH was performed using two different schemes to account for potential hybridization or detection bias with fluorescent secondary antibodies. For every cell line, four slides were hybridized. On two, CENP-A was detected with FITC or Alexa Fluor 488 secondary antibodies and alpha satellite probes were directly labeled with Alexa Fluor 568-dUTP or biotin-labeled and detected with Cy3 avidin (Jackson ImmunoResearch). On the other two slides, CENP-A was detected with Cy3 donkey anti-mouse antibodies (Jackson ImmunoResearch) and FISH probes were directly labeled with Alexa Fluor 488-dUTP or biotin-labled and detected with Alexa Fluor 488 streptavidin (Molecular Probes/Invitrogen). The specificity and quality of hybridization was confirmed by analyzing unlysed nuclei contained within the preparations on each slide. Only hybridizations showing multiple CENP-A foci but only one signal for each alpha satellite probe were scored. At least 20 fibers were analyzed for each cell line.
Microscopy and Image Analysis
All images were acquired using an inverted Olympus IX-71 microscope connected to the Deltavision Spectris or Deltavision RT Restoration Imaging System (Applied Precision) equipped with a Photometric CoolSNAP HQ CCD camera. Fibers extending through multiple fields of view were captured using the Panels option in the softWoRx Acquire 3D program and merged into single images using the “Stitch” function. The ‘measure distances’ tool was used to calculate fluorescent signals representing euchromatic probe, alpha satellite probes or CENP-A immunostaining. CENP-A domain size was measured by comparing the length of CENP-A antibody staining (in micrometers) and the length of overlapping alpha satellite FISH probe. Alpha satellite FISH probe signal length represented total satellite array size that had been determined by PFGE (see below). CENP-A domain size was then calculated from the ratio of the length of CENP-A antibody signal over the total length of alpha satellite FISH signal (Lam et al., 2006). Significant differences in the sizes of CENP-A domains in DIP2/DIP2-OE and DIP3/DIP3-E7 comparisons were determined using a Student’s t-test. P values less than 0.05 were considered statistically significant. In control experiments that evaluated extent of fiber stretching in CENP-A versus non-CENP-A chromatin, probe lengths (μm) or resolution (kb/μm) were graphed as dot plots, and linear regression analyses were performed using Kalediagraph graphing and data analysis software (Synergy Software).
Pulsed Field Gel Electrophoresis and Southern Blotting
Alpha satellite array sizes were estimated by pulsed field gel electrophoresis (PFGE) and Southern blotting following established protocols (Mahtani and Willard, 1990, Mahtani and Willard, 1998, Floridia et al., 2000). High molecular weight genomic DNA was embedded in agarose plugs and digested with restriction endonucleases that release the entire alpha satellite array (Wevrick and Willard, 1989, Mahtani and Willard, 1990). S. cerevisiae, and H. wingei chromosomes embedded into agarose were used as size standards (BioRad). Pulsed field gel electrophoresis was performed on a CHEF-DR II apparatus (Bio-Rad) in either 1X TAE (for cell lines containing HSAX) or 0.5X TAE (for cell lines containing HSAY) at 14°C. Plasmid or PCR-generated probes specific for DXZ1 and DYZ3 (Warburton et al., 1991) were labeled with biotin or digoxygenin and hybridized to membranes using the NEBlot Phototope kit (New England Biolabs). Hybridizations were carried out at 68°C overnight using ~20ng/mL of denatured probe, followed by stringent post-hybridization washing in 0.1X SSC/0.1% SDS at 68°C. Probes were detected using the NEB Phototope-Star Detection Kit and exposed to x-ray film (Kodak BioMax XAR). DXZ1 array sizes had been previously determined for cell lines DIP1 and SCX2 by Mahtani and Willard (1990), but were re-confirmed to show that alpha satellite array sizes were as stable as originally reported two decades ago.
Chromatin immunoprecipitation (ChIP) and Polymerase Chain Reaction (PCR)
ChIP was carried out on oliognucleosomes isolated from native chromatin as previously described (Mravinac et al., 2009). Antibodies recognizing lysine-specific histone modifications included: H3K4me2 (ab7766; Abcam), H3K9me2 (ab1220 or ab730; Abcam), H3K9me3 (ab8898; Abcam), H4K20me3 (ab9053; Abcam) or H4K20me3 (generous gift of J.C. Rice, Univ. of Southern California) (Sims et al., 2006). To control for non-specific binding, a mock control with no antibody was included in each ChIP experiment. At least three independent ChIP experiments for each antibody were performed. Immunoprecipitated DNA (IP DNA) was amplified with DXZ1 and DYZ3-specific primers for semi-quantitative PCR using an iCycler (Biorad, Burlingame CA). Primers used for amplifying higher-order alpha satellite have been published previously (Bashamboo et al., 2005, Warburton et al., 1991). 0.5ul of IP DNA was used in each PCR reaction, and amplifications were done in duplicate. PCR products were subjected to 2% agarose gel electrophoresis and bands were quantified using Image J software (http://rsb.info.nih.gov/). Levels of modified histones on DXZ1 or DYZ3 (antibody or ab) were calculated as a percentage of input [(IP-Mock)/(Input-Mock)Query]. Two genic sites GAPDH and AFM were used as non-centromeric controls for enrichment of histone modifications. Significant differences in levels of modified histones at DXZ1 in DIP3 versus DIP3-E7 were determined using a Student’s t-test. P values less than 0.05 were considered statistically significant.
Results
Using stretched chromatin fibers to optically measure genomic lengths of CENP-A domains
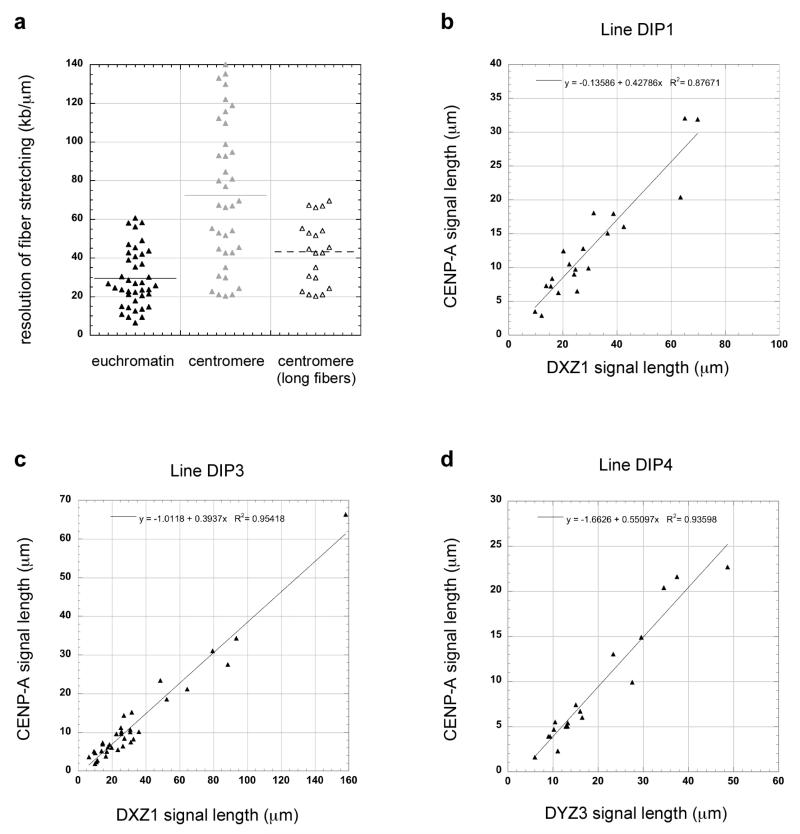
In this study, we examined how much of a multi-megabased array of alpha satellite DNA was occupied by CENP-A. Previous studies, including our own, had shown that CENP-A is usually assembled on only a portion of alpha satellite DNA (Mravinac et al., 2009, Lam et al., 2006, Spence et al., 2002). However, it was not clear what proportion of alpha satellite is associated with CENP-A and if CENP-A domain sizes are similar or different among chromosomes. Due to the repetitive nature of alpha satellite DNA and the absence of contiguous genomic assemblies spanning entire arrays, commonly used methods, such as chromatin immunoprecipitation, cannot assign CENP-A or other proteins to exact genomic coordinates or positions within alpha satellite arrays. To circumvent these challenges, we used stretched chromatin fibers to simultaneously visualize alpha satellite DNA and CENP-A immunostaining, thereby obtaining a long-range view of an entire centromere region. This approach presumes that CENP-A regions (centromeric) and non-CENP-A chromatin (euchromatin) stretch to the same extent. To test this assumption, we hybridized chromatin fibers from the same experiment and cell line with FISH probes specific for either alpha satellite DNA or euchromatic regions of the genome. The mean length of the FISH signal for a euchromatic BAC probe specific for a 155kb region of chromosome 6 was 7.12 +/−4.59 μm (n=41), equating to resolution of 29.42 +/− 14.86 kb/μm (Figure 1a). Centromeric fibers exhibited a mean length of 57.9 +/− 38.2 μm (n= 38), representing genomic resolution of 72.5 +/− 37.2 kb/μm (Figure 1a). Our experiments indicated that the extent of fiber stretching varied within the preparation. Quantification of stretching depended on the fibers that were analyzed, since ~50% of the centromere regions were stretched to the same extent (kb/μm) as euchromatic portions of the genome (Figure 1a). In our analyses, we randomly captured chromatin fibers without deliberately selecting the shortest or longest fibers. However, all preparations generated many extremely long, alpha satellite fibers that spanned multiple fields of view. If we exclusively included these long fibers in our comparisons, centromeric stretching was 43.1 +/− 17 kb/μm (n=20), indicating that centromeric and euchromatic fibers can stretch to similar extents.
Figure 1. Range of centromeric fiber stretching and relationship between CENP-A immunofluorescence and centromeric fiber length.
(a) Comparison of chromatin fiber resolution (kb/μm) in a representative euchromatic region (HSA6) versus a centromere (DXZ1) showed that there was more variability in stretching within centromeric regions. When only long centromeric fibers (i.e. those that spanned multiple fields of view) were exclusively analyzed, euchromatic and centromeric regions were stretched to similar extents. Graphs show the linear relationship between increasing fiber length and fluorescence for centromeres. CENP-A immunofluorescence (y-axis) and alpha satellite fluorescence (x-axis) was plotted for DXZ1 in two representative cell lines (b) DIP1 and (c) DIP3 and for (d) DYZ3 in a third cell line DIP4. Linear regression was performed using Kaleidagraph software. The R2 values and equation for the line are indicated on each graph.
We also tested if CENP-A antibodies could be equivalently detected on short versus long fibers. We plotted the lengths of fluorescent signals representing CENP-A immunostaining and alpha satellite DNA probes on the same fiber. If the ability to detect CENP-A immunofluorescence signals on longer chromatin fibers decreased because the signals became less intense, a non-linear relationship between CENP-A and alpha satellite DNA linear signal length was expected. When we plotted linear lengths of CENP-A and alpha satellite DNA fluorescence among fibers generated from the same experiment, we observed a clear linear relationship, with the length of CENP-A immunostaining increasing with longer alpha satellite FISH signal length (Figure 1b-d). Such results were consistently observed among different cell lines for centromeres of chromosome X (DXZ1) (Figure 1b, c), chromosome Y (DYZ3) (Figure 1d), and chromosome 17 (D17Z1; data not shown). These experiments confirmed that stretched chromatin fibers could be confidently used to study both CENP-A and non-CENP-A chromatin and to estimate the genomic extent of CENP-A domain size among different centromeres.
CENP-A chromatin is assembled across a similar proportion of alpha satellite DNA regardless of array size
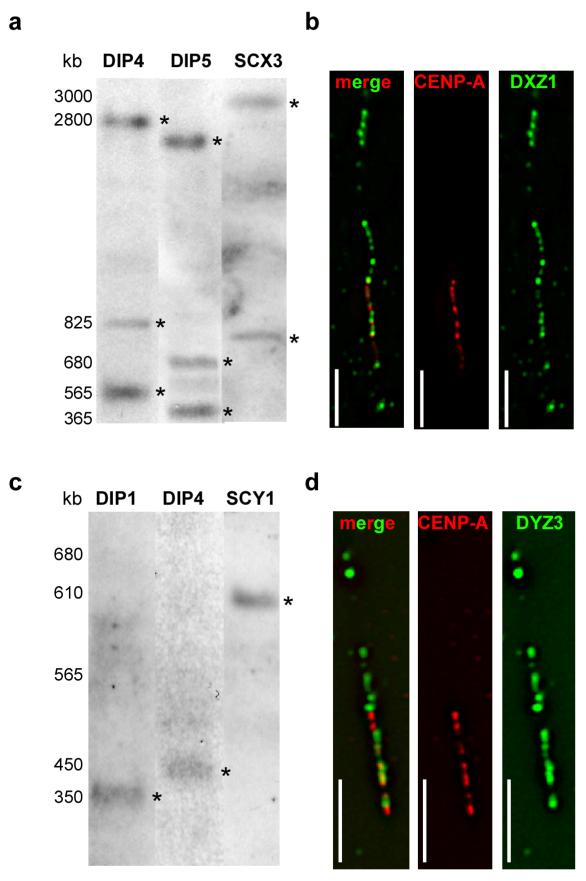
To investigate the correlation between alpha satellite array size and CENP-A domain size among different chromosomes, we measured total array sizes of DXZ1 on HSAX and DYZ3 on HSAY in different male cell lines as well as rodent-human somatic cell hybrids (SC) containing single HSAX or HSAY (Table 1). These chromosomes were chosen since they are haploid and allow unequivocal assignment of an array size to a specific chromosome. High molecular weight genomic DNA was digested with restriction enzymes that do not cut, or cut infrequently, within DXZ1 and DYZ3. Such enzymes released entire DXZ1 or DYZ3 arrays as single or a few fragments that were resolved by PFGE (Mahtani and Willard, 1990, Floridia et al., 2000) (Figure 2a, 2c). Prior analyses of DXZ1 indicated that array sizes vary 2-3 fold among individuals (Mahtani and Willard, 1990). Among the eight X chromosomes studied here, DXZ1 array length also varied, supporting these previous findings (Figure 2a, Table 1). DYZ3, alpha satellite array on HSAY is also polymorphic in size. DYZ3 arrays cluster into two size groups, 100-600kb and 700-1100kb, within the human male population (Abruzzo et al., 1996, Oakey and Tyler-Smith, 1990). In the 5 cell lines we analyzed, we also observed a similar distribution (Table 1). DYZ3 was ≤600kb in three males, but ~1Mb in the two other males (Figure 2c).
Figure 2. Alpha satellite array size and CENP-A domain size at centromeres of chromosomes X and Y.
Molecular sizes of alpha satellite arrays were determined by pulsed field gel electrophoresis (PFGE) of high molecular weight DNA embedded in agarose, followed by non-radioactive Southern blotting. (a) Sizes of X alpha satellite (DXZ1) arrays in two diploid lines (DIP4, DIP5) and a mouse-human somatic cell hybrid containing a single human X chromosome (SCX3) ranged from 3.7-4.2Mb. The total array size for DXZ1 in each cell line was estimated by adding the molecular weights of all bands that appeared in the Southern blot. Digestion of DNA embedded within agarose plugs with BglI (shown) or BstEII (not shown) released DXZ1 as two or three bands that were detected by Southern blotting. High molecular weight bands are denoted with asterisks (*). PFGE conditions for DXZ1 were 3 V/cm, initial switch time of 250s, and a final switch time of 900s, for 50 hours. The figure shows three cell lines that were run on separate but identically sized gels under identical conditions using the same high molecular weight standards. The gel for each cell line was cropped identically and aligned according to the molecular weight standards. (b) CENP-A domains sizes were determined using CENP-A immunostaining on extended chromatin fibers followed by FISH with a DNA probe specific for DXZ1. Domain sizes were calculated by assigning molecular sizes as determined by PFGE to the length of the alpha satellite signals (in micrometers). CENP-A domains were calculated by comparing the length of the immunostaining on the fiber to the length of the alpha satellite fluorescent signal (see Materials and Methods for additional details). The representative image from cell line DIP3 shows CENP-A (red) immunostaining overlapping with only a portion of DXZ1 (green). The merged image is shown on the left, followed by individual image channels for CENP-A (middle) and DXZ1 (right). Scale bar is 10 micrometers. (c) Molecular sizing of Y alpha satellite (DYZ3) arrays in two diploid lines (DIP1, DIP4) and a mouse-human somatic cell hybrid containing a single human Y chromosome (SCY1) showed arrays that ranged from 350-600kb. Digestion of DNA embedded within agarose plugs with BamHI released the entire DYZ3 array that was detected by Southern blotting as a single band. High molecular weight bands are denoted with asterisks (*). PFGE parameters were 6 V/cm, 10s initial switch time, 80s final switch time, for 23 hours. The figure shows three cell lines that were run on separate but identically sized gels under identical conditions and using the same high molecular weight standards. The gel for each cell line was cropped identically and aligned according to the molecular weight standards. (d) Representative image from cell line DIP4 showing CENP-A (red) immunostaining overlapping with a portion of DYZ3 (green). The merged image is shown on the left, followed by individual image channels for CENP-A (middle) and DYZ3 (right). Scale bar is 5 micrometers.
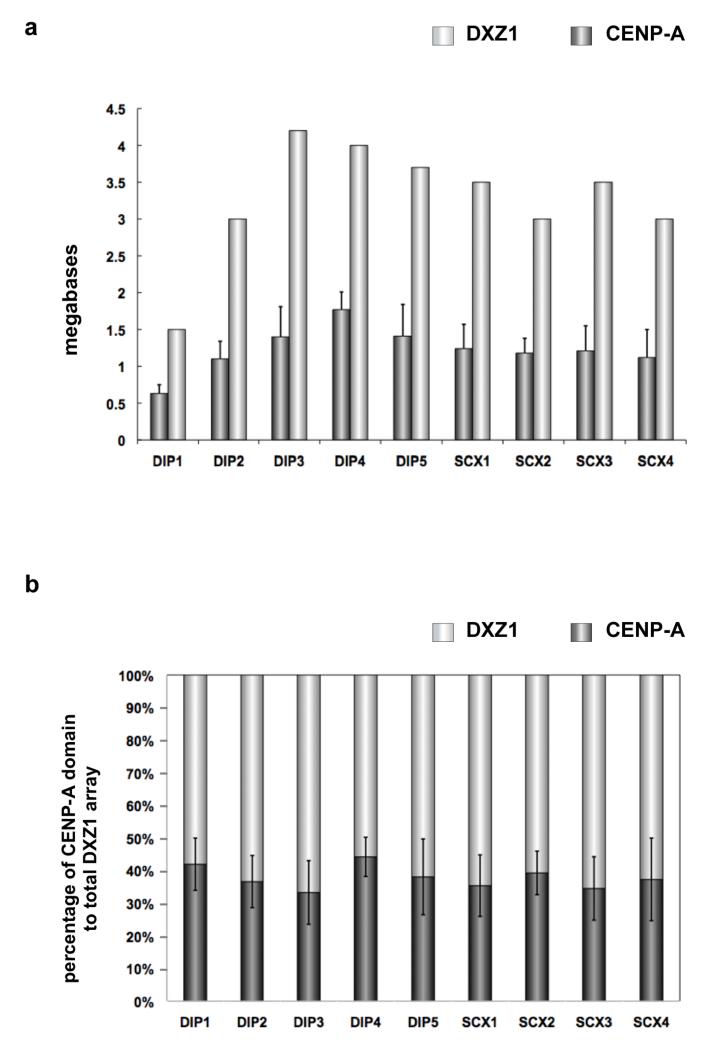
To define genomic sizes of CENP-A chromatin domains, stretched chromatin fibers were prepared from each cell line and immunostained for CENP-A, followed by FISH with probes specific to DXZ1 or DYZ3 (Figure 2b, 2d) (Blower et al., 2002, Lam et al., 2006, Mravinac et al., 2009). Within nine cell lines, the CENP-A domains on DXZ1 (CENP-ADXZ1) varied from 630kb to 1.8Mb (Figure 3). Line DIP1 had the smallest CENP-ADXZ1domain, occupying 630kb (+/− 120kb), or ~42% of the array (Figure 3a, 3b). DXZ1 in DIP1 had been previously reported to be very small (1.5Mb) (Mahtani and Willard, 1990). The other HSAX centromeres exhibited larger alpha satellite arrays (3-4Mb) and also much larger CENP-ADXZ1 domains that spanned 1.1-1.8Mb. A general size trend was observed in that equivalently sized alpha satellite arrays had similarly sized CENP-ADXZ1 domains (Table 1). For instance, the DXZ1 array in lines DIP2, SCX2 and SCX4 was ~3Mb, and the CENP-ADXZ1 domains were 1.1-1.2Mb. Likewise, DXZ1 was ~3.5Mb in lines SCX1 and SCX3, and the CENP-ADXZ1 domains were each 1.2Mb. Our results indicate that like alpha satellite array size, CENP-ADXZ1 domain size varies in different individuals. Overall, CENP-ADXZ1 domains were assembled on 35-45% of the array, regardless of total alpha satellite array size.
Figure 3. Human chromosome X (HSAX) alpha satellite (DXZ1) array size compared to CENP-A chromatin domain size.
(a) DXZ1 arrays (light gray bars) were measured on different HSAXs present in human diploid (DIP) male lines or rodent-human somatic cell hybrids (SCX) in which HSAX was the only human chromosome. CENP-A chromatin domain size in megabases (dark gray bars) was determined using CENP-A immunostaining and FISH with a DXZ1 probe. DXZ1 arrays and CENP-A domain sizes varied up to three-fold among individual HSAXs. Error bars represent standard deviations in the genomic length (in megabases). (b) Calculation of the percentage of DXZ1 occupied by the CENP-A domain (dark gray bars) showed that the CENP-A domain was proportionally assembled on DXZ1 arrays (light gray bars) that varied in size among different HSAXs. Thirty-five to forty-five percent of DXZ1 was occupied by CENP-A chromatin. Error bars represent standard deviations in the percentage of DXZ1 occupied by the CENP-A domain.
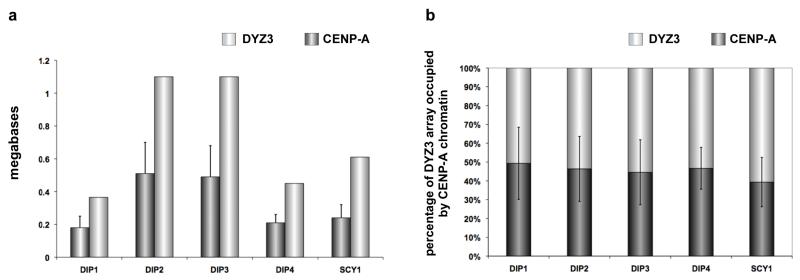
CENP-A domain sizes on DYZ3 (CENP-ADYZ3) were determined in 8 cell lines. Like CENP-A domains on HSAX, CENP-ADYZ3 domains were also heterogeneous in size among individuals. In general, they extended over slightly larger proportions (40-50%) of DYZ3 as compared to CENP-ADXZ1 domains (Figure 4a, 4b). However, like HSAX, we observed the same correlation between total array size and CENP-A domain size. For instance, lines DIP1 and DIP4, which had the smallest DYZ3 arrays (370-450kb), also had the smallest CENP-A domains (180-200kb). Likewise, larger DYZ3 arrays had larger CENP-A domains. Taken together, the DXZ1 and DYZ3 experiments indicate that CENP-A chromatin domains are proportionally assembled on at least one-third to one-half of alpha satellite arrays, regardless of total size.
Figure 4. Human chromosome Y (HSAY) alpha satellite (DYZ3) array size compared to CENP-A chromatin domain size.
(a) DYZ3 arrays (light gray bars) were measured on different HSAYs present in human diploid (DIP) male lines or rodent-human somatic cell hybrids (SCY) in which HSAY was the only human chromosome. CENP-A chromatin domain size in megabases (dark gray bars) was determined using CENP-A immunostaining and FISH with a DYZ3 probe. As previously reported for DYZ3, arrays were distributed into two size groups: those <600kb and those >1Mb (Abruzzo et al., 1996). Accordingly, CENP-A domains clustered into groups that were either <200kb or >500kb. Error bars represent standard deviations in the genomic length (in megabases). (b) Calculation of the percentage of DYZ3 occupied by the CENP-A domain (dark gray bars) showed that the CENP-A domain was proportionally assembled on DYZ3 (light gray bars) that varied in size among different HSAYs. Typically, 45-50% of DYZ3 was occupied by CENP-A chromatin. Error bars represent standard deviations in the percentage of DYZ3 occupied by the CENP-A domain.
CENP-A domain size expands in transformed cells
Many genomic and epigenetic changes accompany cellular transformation, including changes in DNA methylation and histone modification as well as aberrant gene expression. Increased CENP-A expression and mislocalization have been described in some cancers and have been linked with mitotic defects and aneuploidy (Amato et al., 2009, Tomonaga et al., 2003). Increased CENP-A domain size may be a feature of tumorigenesis, with chromosomal defects reflecting altered proportions of CENP-A and/or heterochromatin at endogenous centromeres. To test this hypothesis, we mimicked cancer cells by transforming primary human cell line DIP3 via over-expression of human papillomavirus E7 oncoprotein (HPV-E7), creating line DIP3-E7 (Table 1). HPV-E7 mediates cellular transformation by binding to retinoblastoma tumor susceptibility protein Rb and inhibiting its association with E2F transcription factors (Dyson et al., 1989, Munger et al., 1989). In the presence of HPV-E7, cells prematurely enter S phase and become tumorigenic. In addition to suppressing transcription of cell cycle genes regulated by E2F, Rb interacts with DNA and histone methyltransferases to maintain heterochromatin assembly and centromere stability (Gonzalo and Blasco, 2005, Gonzalo et al., 2005, Siddiqui et al., 2007, Manning et al., 2010).
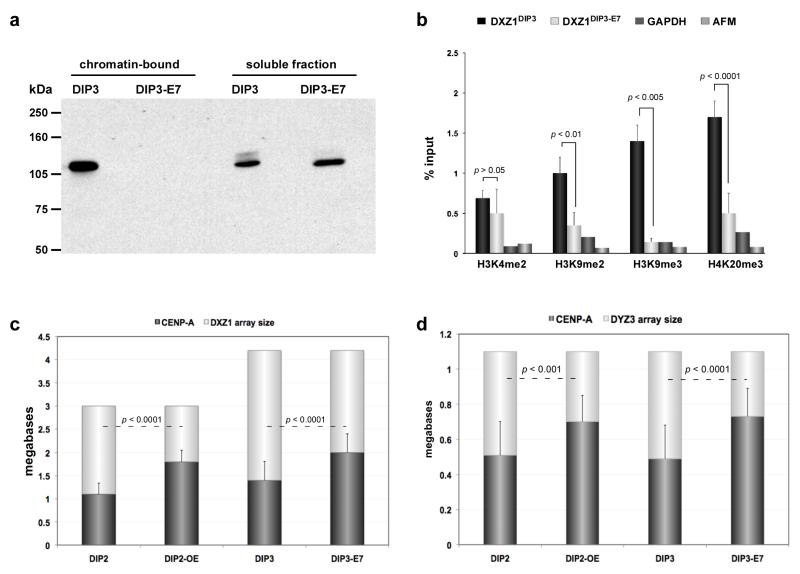
As expected, after HPV-E7 transformation, the amount of chromatin-bound Rb was decreased in DIP3-E7 compared to the parental, untransformed line DIP3 (Figure 5a). We also observed that methylation of H3K9 and H4K20 decreased at DXZ1 in DIP3-E7 (Figure 5b), suggesting that loss of chromatin-bound Rb reduced enrichment of heterochromatin at centromeres. H3K4me2, a euchromatic modification, was not significantly different on DXZ1 in DIP3-E7 compared to DIP3. However, Rb depletion has been correlated with increased amounts of CENP-A (Amato et al., 2009). When CENP-A arrays sizes on HSAX and HSAY were measured in DIP3-E7, we observed that the CENP-A domain had enlarged by an additional two-thirds of its original genomic length. CENP-ADXZ1 was originally 1.3Mb and increased to 2Mb (Figure 5c). CENP-ADYZ3 expanded from 0.5Mb to 0.75Mb (Figure 5d). The increase in the extent of CENP-A chromatin was similar to what was observed in DIP2-OE, a cancer cell line over-expressing FLAG-tagged CENP-A. We previously showed that DIP2-OE had decreased amounts of heterochromatic histone modifications at DXZ1, but the euchromatic modification H3K4me2 was largely unchanged (Lam et al., 2006). In both instances (E7 and CENP-A over-expression), the size of the CENP-A domain increased to encompass almost 70% of the alpha satellite array (Figure 5c, 5d). Collectively, these results suggest that the total genomic extent of CENP-A chromatin at endogenous centromeres expands when cells undergo transformation. Such expansion is associated with decreased enrichment of heterochromatic nucleosomes at centromeres and/or increased expression of CENP-A.
Figure 5. CENP-A domains size changes in response to protein dosage.
(a) Primary cell line (DIP3) was transformed by virally-expressing HPV E7 oncoprotein (DIP3-E7). E7 binds to retinoblastoma protein (Rb) and inactivates it, preventing it from binding to its nuclear targets. Western blotting with anti-Rb antibodies demonstrated that the amount of chromatin-bound Rb was undetectable upon E7 expression and transformation of DIP3. (b) ChIP-PCR showed that in the absence of chromatin-bound Rb, heterochromatic histone modifications H3K9me2, H3K9me3 and H4K20me3 at DXZ1 were decreased. The poised euchromatic mark H3K4me2 was not significantly different at DXZ1 in DIP3 before and after transformation. Enrichment at DXZ1 was calculated as the ratio of immunoprecipitated DNA to input. GAPDH and AFM were used as genic control sites. Error bars represent standard deviations. (c) CENP-A chromatin domain size at DXZ1 was measured using CENP-A immunostaining and FISH in cells over-expressing CENP-A (DIP2-OE) and after HPV-E7-mediated transformation/heterochromatin depletion of a primary fibroblast line DIP3 to create DIP3-E7. Comparisons in CENP-A domain size were made to the parent line (i.e. DIP2 vs DIP2-OE and DIP3 vs DIP3-E7). In each case, CENP-A domain size on DXZ1 increased by 1.5 fold of its original size. Error bars represent standard deviations in the total number of megabases occupied by the CENP-A chromatin domain. (d) CENP-A chromatin domain size on DYZ3 was measured as in (c) in cell lines DIP2-OE and DIP3-E7. Comparisons were made to the parental line in each case (i.e. DIP2 vs DIP2-OE and DIP3 vs DIP3-E7). The domain increased 1.5 times in length over DYZ3 when CENP-A was over-expressed or heterochromatin was depleted from the centromere. Error bars represent standard deviations in the total number of megabases occupied by the CENP-A chromatin domain. Significant differences in (b), (c) and (d) were calculated using a Student’s t-test.
Discussion
In this work, we provide evidence for epigenomic heterogeneity at centromeres reflected by the differences in the genomic distances that CENP-A chromatin occupy on polymorphic alpha satellite DNA arrays of homologous chromosomes. Our results indicate that size of CENP-A chromatin as a domain varies among non-homologous chromosomes. In all cases, the genomic size of CENP-A chromatin was consistently proportional to overall alpha satellite array size, ranging from 30-50% of the array. Such proportionality of CENP-A domain:alpha satellite array size is not restricted to X or Y chromosomes, as we have also observed similar proportions at centromeres of other human chromosomes (BA Sullivan, unpublished observations). Chromosome size has been proposed to be a predictor of kinetochore size and might explain differences in kinetochore size (Irvine et al., 2004). In such a model, small chromosomes will have small centromeres and CENP-A domains. The human Y chromosome (HSAY) is one of the smallest human chromosomes (~60Mb) and has been reported to contain the lowest amount of CENP-A (Irvine et al., 2004). However, in our study, several HSAYs had CENP-A domains that were similar in size to the CENP-A domain on an HSAX (DIP1). HSAX is nearly three times larger (155Mb) than HSAY, thus we suggest that alpha satellite array size is more strongly correlated to CENP-A domain size than is chromosome size. This conclusion is also supported by our previous data showing that the total genomic extent of the CENP-A domain decreases when the size of an alpha satellite array is reduced (Mravinac et al., 2009).
Depletion of CENP-A by RNAi profoundly affects chromosome stability when more than 75% of CENP-A in the cell is depleted (Bergmann et al., 2010). Although genomic length of CENP-A chromatin has not been studied in CENP-A depleted cells, it is reasonable to assume that under these conditions, the genomic size of the CENP-A domain would fall below the normal ratio of 30-50% of each alpha satellite array. In our experiments, HSAX and HSAY were not unstable in cell lines in which CENP-A domain sizes increased at DXZ1 and DYZ3 due to CENP-A over-expression or cellular transformation. Studies in yeast have found that numbers of of CENP-A nucleosomes are normally in excess to the number of microtubule attachments (Joglekar et al., 2008), suggesting that more kinetochore chromatin is assembled than is actually needed. Our results are consistent with this, in that even when CENP-A chromatin spanned more than 60% of a given alpha satellite array, kinetochore assembly and chromosome stability remained normal. However, expansion of CENP-A domain size in cancer cells did change chromatin organization at centromeres. The long-term effects of these changes remain unclear. Many tumors and cancers are marked by abnormalities aberrant histone modifications at satellites and genic regions, defective chromosome structure and segregation, and whole chromosome gains or losses (Amato et al., 2009, Tomonaga et al., 2003, Manning et al., 2010). CENP-A over-expression has also been reported in cancer cells, leading to mislocalization of CENP-A to non-centromeric regions (Tomonaga et al., 2003). Inappropriate CENP-A incorporation appears to be more detrimental to chromosome stability than expansion of the CENP-A domain at centromere regions (Heun et al., 2006, Van Hooser et al., 2001).
Variation in mitotic kinetochore size has been previously reported (Tomkiel et al., 1994). These differences could be explained by centromere compaction, numbers of centromere proteins, or sizes of centromeric DNA or chromatin. Our data indicating that there are a range of CENP-A domain sizes among human centromeres support the latter possibility that kinetochore size might vary by the genomic extent of CENP-A chromatin. While it is generally accepted that centromeric/kinetochore chromatin as an overall domain is defined by multiple CENP-A subdomains interrupted by blocks of H3-containing nucleosomes (Blower et al., 2002, Brinkley et al., 1992), such interspersion of CENP-A and H3 nucleosomes raises questions as to how many CENP-A nucleosomes are present in subdomains. Our study was not focused on quantifying molecules of CENP-A or numbers of CENP-A nucleosomes at each centromere, and clearly this question remains outstanding. Neocentromeres, i.e. new centromeres that form on unique sequences in euchromatin, have provided the most detailed estimates of the genomic range of CENP-A chromatin as a domain and within subdomains. CENP-A subdomains on neocentromeres can range from 5kb to 90kb (Alonso et al., 2007, Alonso et al., 2010, Lo et al., 2001). As a complete domain, CENP-A chromatin on neocentromeres ranges from 120-330kb (Alonso et al., 2007, Alonso et al., 2003, Chueh et al., 2005, Lo et al., 2001). These domain sizes are similar to CENP-A regions on the smallest HSAYs in our study (180-200kb), and may represent lower genomic limits of human CENP-A chromatin. We did not observe CENP-A domains that were smaller than 180kb or exceeded 2Mb. Broad upper and lower limits for centromere size may protect or de-sensitize cells from becoming aneuploid when there are minor fluctuations in protein availability. Moreover, larger domains might be limited by physical barriers, such as those present at centromeres in fission yeast (Scott et al., 2006) or by epigenetic boundaries (Nakano et al., 2008, Maggert and Karpen, 2001).
Our results highlight several directions for future study, such as how the variation in genomic occupancy of the CENP-A domain as a whole translates into distribution (size and frequency) of CENP-A subdomains, and the size and frequency of CENP-A and H3 subdomains at individual centromeres. CENP-A is loaded in late mitosis/G1 and subsequently diluted by replication in S phase, and it is unclear how replication affects the number and distribution of CENP-A nucleosomes. Alpha satellite is highly homogenous and it is not known if the edges or boundaries of CENP-A chromatin or the CENP-A subdomains themselves are strictly fixed within the large arrays of repeats. The CENP-A domain may expand and contract on an alpha satellite array depending on where and how much new CENP-A is incorporated each cell cycle. The identification of unique markers within alpha satellite to serve as genomic anchors within this expansive repetitive region will be important for testing these models experimentally. Future studies will also be important for determining if especially small or large CENP-A chromatin domain sizes occur in specific types of cancer and/or if aberrant domain sizes contribute to aneuploidy and chromosome instability.
Acknowledgements
We thank Cyrus Vaziri (University of North Carolina, Chapel Hill) for providing cell lines DIP3 and DIP3-E7 and Chris Shaw for technical assistance. This work was supported in part by grants from the American Cancer Society (ACS IRG-72-001-29-IRG), March of Dimes Birth Defects Foundation (6-FY06-377 and 6-FY-10-294), and NIH/NIGMS (R01 GM069514) to B.A.S.
Abbreviations
- CENP-A
centromere protein A
- CCD
charged coupled device
- CHEF
contour-clamped homogenous electric field
- ChIP
chromatin immunprecipitation
- CSK
buffer cytoskeleton buffer
- DNA
deoxyribonucleic acid
- E7
E7 protein subunit of HPV
- FISH
fluorescence in situ hybridization
- FITC
fluorescein isothiocyanate
- FBS
fetal bovine serum
- HAT
hypoxanthine/aminopterin/thymidine
- HPV
human papillomavirus
- HOR
higher order repeat
- HSAX
Homo sapiens chromosome X
- HSAY
Homo sapiens chromosome Y
- Kb
kilobase
- MEM
minimal essential medium
- Mb
megabase
- PCR
polymerase chain reaction
- PVDF
polyvinylidene fluoride
- Rb
retinoblastoma protein
- RPMI
Roswell Park Memorial Institute medium
- PFGE
pulsed field gel electrophoresis
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
References
- ABRUZZO MA, GRIFFIN DK, MILLIE EA, SHEEAN LA, HASSOLD TJ. The effect of Y-chromosome alpha-satellite array length on the rate of sex chromosome disomy in human sperm. Hum Genet. 1996;97:819–23. doi: 10.1007/BF02346196. [DOI] [PubMed] [Google Scholar]
- ALONSO A, FRITZ B, HASSON D, ABRUSAN G, CHEUNG F, YODA K, RADLWIMMER B, LADURNER AG, WARBURTON PE. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 2007;8:R148. doi: 10.1186/gb-2007-8-7-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALONSO A, HASSON D, CHEUNG F, WARBURTON PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALONSO A, MAHMOOD R, LI S, CHEUNG F, YODA K, WARBURTON PE. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum Mol Genet. 2003;12:2711–21. doi: 10.1093/hmg/ddg282. [DOI] [PubMed] [Google Scholar]
- AMATO A, SCHILLACI T, LENTINI L, DI LEONARDO A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer. 2009;8:119. doi: 10.1186/1476-4598-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASHAMBOO A, RAHMAN MM, PRASAD A, CHANDY SP, AHMAD J, ALI S. Fate of SRY, PABY, DYS1, DYZ3 and DYZ1 loci in Indian patients harbouring sex chromosomal anomalies. Mol Hum Reprod. 2005;11:117–27. doi: 10.1093/molehr/gah135. [DOI] [PubMed] [Google Scholar]
- BERGMANN JH, RODRIGUEZ MG, MARTINS NM, KIMURA H, KELLY DA, MASUMOTO H, LARIONOV V, JANSEN LE, EARNSHAW WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2010 doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BLACK BE, JANSEN LE, MADDOX PS, FOLTZ DR, DESAI AB, SHAH JV, CLEVELAND DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–22. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- BLOWER MD, SULLIVAN BA, KARPEN GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;3:1–11. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRINKLEY BR, OUSPENSKI I, ZINKOWSKI RP. Structure and molecular organization of the centromere-kinetochore complex. Trends Cell Biol. 1992;2:15–21. doi: 10.1016/0962-8924(92)90139-e. [DOI] [PubMed] [Google Scholar]
- CHERRY LM, FAULKNER AJ, GROSSBERG LA, BALCZON R. Kinetochore size variation in mammalian chromosomes: an image analysis study with evolutionary implications. J Cell Sci. 1989;92(Pt 2):281–9. doi: 10.1242/jcs.92.2.281. [DOI] [PubMed] [Google Scholar]
- CHUEH AC, WONG LH, WONG N, CHOO KH. Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum Mol Genet. 2005;14:85–93. doi: 10.1093/hmg/ddi008. [DOI] [PubMed] [Google Scholar]
- DYSON N, HOWLEY PM, MUNGER K, HARLOW E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–7. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- FARR CJ, STEVANOVIC M, THOMSON EJ, GOODFELLOW PN, COOKE HJ. Telomere-associated chromosome fragmentation: applications in genome manipulation and analysis. Nat Genet. 1992;2:275–82. doi: 10.1038/ng1292-275. [DOI] [PubMed] [Google Scholar]
- FLORIDIA G, ZATTERALE A, ZUFFARDI O, TYLER-SMITH C. Mapping of a human centromere onto the DNA by topoisomerase II cleavage. EMBO Rep. 2000;1:489–93. doi: 10.1093/embo-reports/kvd110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONZALO S, BLASCO MA. Role of Rb family in the epigenetic definition of chromatin. Cell Cycle. 2005;4:752–5. doi: 10.4161/cc.4.6.1720. [DOI] [PubMed] [Google Scholar]
- GONZALO S, GARCIA-CAO M, FRAGA MF, SCHOTTA G, PETERS AH, COTTER SE, EGUIA R, DEAN DC, ESTELLER M, JENUWEIN T, BLASCO MA. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–8. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- GRIMES BR, RHOADES AA, WILLARD HF. DNA and Vector Composition Influence Rates of Human Artificial Chromosome Formation. Mol Ther. 2002;5:798–805. doi: 10.1006/mthe.2002.0612. [DOI] [PubMed] [Google Scholar]
- HARRINGTON JJ, VAN BOKKELEN G, MAYS RW, GUSTASHAW K, WILLARD HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–55. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- HEUN P, ERHARDT S, BLOWER MD, WEISS S, SKORA AD, KARPEN GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–15. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRVINE DV, AMOR DJ, PERRY J, SIRVENT N, PEDEUTOUR F, CHOO KH, SAFFERY R. Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Res. 2004;12:805–15. doi: 10.1007/s10577-005-5377-4. [DOI] [PubMed] [Google Scholar]
- JOGLEKAR AP, BOUCK D, FINLEY K, LIU X, WAN Y, BERMAN J, HE X, SALMON ED, BLOOM KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–94. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAM AL, BOIVIN CD, BONNEY CF, RUDD MK, SULLIVAN BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci U S A. 2006;103:4186–91. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LO AW, LIAO GC, ROCCHI M, CHOO KH. Extreme reduction of chromosome-specific alpha-satellite array is unusually common in human chromosome 21. Genome Res. 1999;9:895–908. doi: 10.1101/gr.9.10.895. [DOI] [PubMed] [Google Scholar]
- LO AWI, MAGLIANO DJ, SIBSON MC, KALITSIS P, CRAIG JM, CHOO KHA. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 2001;11:448–457. doi: 10.1101/gr.167601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAGGERT KA, KARPEN GH. The Activation of a Neocentromere in Drosophila Requires Proximity to an Endogenous Centromere. Genetics. 2001;158:1615–28. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAHTANI MM, WILLARD HF. Pulsed-field gel analysis of alpha-satellite DNA at the human X chromosome centromere: high-frequency polymorphisms and array size estimate. Genomics. 1990;7:607–13. doi: 10.1016/0888-7543(90)90206-a. [DOI] [PubMed] [Google Scholar]
- MAHTANI MM, WILLARD HF. Physical and genetic mapping of the human X chromosome centromere: repression of recombination. Genome Res. 1998;8:100–10. doi: 10.1101/gr.8.2.100. [DOI] [PubMed] [Google Scholar]
- MANNING AL, LONGWORTH MS, DYSON NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010;24:1364–76. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MRAVINAC B, SULLIVAN LL, REEVES JW, YAN CM, KOPF KS, FARR CJ, SCHUELER MG, SULLIVAN BA. Histone modifications within the human X centromere region. PLoS One. 2009;4:e6602. doi: 10.1371/journal.pone.0006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUNGER K, PHELPS WC, BUBB V, HOWLEY PM, SCHLEGEL R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–21. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAKANO M, CARDINALE S, NOSKOV VN, GASSMANN R, VAGNARELLI P, KANDELS-LEWIS S, LARIONOV V, EARNSHAW WC, MASUMOTO H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–22. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OAKEY R, TYLER-SMITH C. Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics. 1990;7:325–30. doi: 10.1016/0888-7543(90)90165-q. [DOI] [PubMed] [Google Scholar]
- RUDD MK, WILLARD HF. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–33. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- SCHUELER MG, HIGGINS AW, RUDD MK, GUSTASHAW K, WILLARD HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–15. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- SCOTT KC, MERRETT SL, WILLARD HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol. 2006;16:119–29. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- SIDDIQUI H, FOX SR, GUNAWARDENA RW, KNUDSEN ES. Loss of RB compromises specific heterochromatin modifications and modulates HP1alpha dynamics. J Cell Physiol. 2007;211:131–7. doi: 10.1002/jcp.20913. [DOI] [PubMed] [Google Scholar]
- SIMS JK, HOUSTON SI, MAGAZINNIK T, RICE JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem. 2006;281:12760–6. doi: 10.1074/jbc.M513462200. [DOI] [PubMed] [Google Scholar]
- SONG IY, PALLE K, GURKAR A, TATEISHI S, KUPFER GM, VAZIRI C. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J Biol Chem. 2010;285:31525–36. doi: 10.1074/jbc.M110.138206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPENCE JM, CRITCHER R, EBERSOLE TA, VALDIVIA MM, EARNSHAW WC, FUKAGAWA T, FARR CJ. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. Embo J. 2002;21:5269–80. doi: 10.1093/emboj/cdf511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SULLIVAN B, WARBURTON P. Studying the progression of vertebrate chromosomes through mitosis by immunofluorescence and FISH. In: BICKMORE W, editor. Chromosome structural analysis: a practical approach. IRL Press; 1999. [Google Scholar]
- TOMKIEL J, COOKE CA, SAITOH H, BERNAT RL, EARNSHAW WC. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J Cell Biol. 1994;125:531–45. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOMONAGA T, MATSUSHITA K, YAMAGUCHI S, OOHASHI T, SHIMADA H, OCHIAI T, YODA K, NOMURA F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–6. [PubMed] [Google Scholar]
- VAN HOOSER AA, OUSPENSKI II, GREGSON HC, STARR DA, YEN TJ, GOLDBERG ML, YOKOMORI K, EARNSHAW WC, SULLIVAN KF, BRINKLEY BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–42. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- WARBURTON PE, GREIG GM, HAAF T, WILLARD HF. PCR amplification of chromosome-specific alpha satellite DNA: definition of centromeric STS markers and polymorphic analysis. Genomics. 1991;11:324–33. doi: 10.1016/0888-7543(91)90139-6. [DOI] [PubMed] [Google Scholar]
- WAYE JS, WILLARD HF. Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: a survey of alphoid sequences from different human chromosomes. Nucleic Acids Res. 1987;15:7549–69. doi: 10.1093/nar/15.18.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEVRICK R, WILLARD HF. Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proc Natl Acad Sci U S A. 1989;86:9394–8. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLARD HF, WAYE JS. Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet. 1987;3:192–98. [Google Scholar]
- ZENG K, DE LAS HERAS JI, ROSS A, YANG J, COOKE H, SHEN MH. Localisation of centromeric proteins to a fraction of mouse minor satellite DNA on a mini-chromosome in human, mouse and chicken cells. Chromosoma. 2004;113:84–91. doi: 10.1007/s00412-004-0299-z. [DOI] [PubMed] [Google Scholar]