Abstract
The RE1-silencing transcription factor (REST) represses the expression of neuronal-specific genes in non-neuronal cells by recruiting histone deacetylases (HDACs) and other histone modifying and chromatin remodeling proteins to the DNA. REST regulation of the expression of neuronal genes is required for the orderly developmental transition from a neuroepithelial stem cell to a functional neuron. Here, we examined the expression and function of REST in neonatal rat oligodendrocyte precursor cells (OPCs). OPCs develop from the same neuroepithelial stem cells as neurons, can be reprogrammed to act as neural stem-like cells in vitro, and require HDAC-mediated gene repression to develop into mature oligodendrocytes. We show that OPCs express functional REST protein and that REST interacts with several neuronal-specific genes whose expression is repressed in OPCs. REST transcript and protein expression increased fourfold during the first 48 h of oligodendrocyte differentiation. During this differentiation, the expression of RE1 containing neuronal genes further decreased as the transcription of oligodendrocyte-specific genes was activated. Expression of a dominant-negative form of REST in OPCs prevented the cells from developing into mature MBP-positive oligodendrocytes. Rather, the cells began to develop a neuronal phenotype characterized by increased expression of neuronal proteins, transcription factors, and cell-type-specific marker antigens. REST overexpression promoted the development of O4-positive pre-oligodendrocytes from OPCs. Together, these results show that REST function is required for the differentiation of OPCs into oligodendrocytes. By regulating the expression of neuronal genes, REST may also regulate the phenotypic plasticity of OPCs.
Introduction
Oligodendrocytes, the myelin-forming cells of the CNS, develop from an identified precursor/progenitor cell referred herein as an oligodendrocyte precursor cell (OPC). The differentiation of OPCs occurs in a stepwise or stagewise progression and is regulated by both genetic and epigenetic mechanisms. Marin-Husstege et al. (2002) first showed that class 1 histone deacetylases (HDACs) are required for the differentiation of OPCs. By removing acetyl residues from histone tails, HDACs help place the chromatin in a condensed state such that transcription activating factors cannot access the DNA and gene expression is repressed. HDACs are recruited to the chromatin as multiprotein complexes by sequence-specific DNA binding proteins, and these binding proteins confer cellular specificity to the covalent modifications. Inhibiting HDACs in developing OPCs not only prevents oligodendrocyte differentiation but also confers lineage plasticity on the cells and promotes the development of neuronal properties (Marin-Husstege et al., 2002; Liu et al., 2007; Lyssiotis et al., 2007). Understanding how epigenetic mechanisms regulate this plasticity is important because OPCs can give rise to neurons, astrocytes, and oligodendrocytes throughout the life of the organism (Belachew et al., 2003; Aguirre and Gallo, 2004; Rivers et al., 2008; Zhu et al., 2008; Guo et al., 2009, 2010).
Principal among the proteins that recruit HDACs and other chromatin and DNA modifying enzymes to DNA in the developing nervous system is the transcriptional regulator REST (RE1-silencing transcription factor, also known as neural restricted silencing factor or NRSF) (Ballas and Mandel, 2005). Through interactions with a 21 bp repressor element in DNA (the RE1), REST regulates the expression of a large number of neuronal genes, including those that code for transcription factors, neurotransmitter receptors, cytoskeletal proteins, ion channels, and neurotrophins (Bruce et al., 2004, 2009; Otto et al., 2007). REST recruits the corepressors mSin3 and CoREST to its N- and C-terminal domains, respectively, to mediate gene repression (Grimes et al., 2000). REST can form complexes with other corepressors, and these diverse complexes regulate different genes in different cell types (Johnson et al., 2008; Mulligan et al., 2008). Last, REST can also recruit gene-silencing enzymes, including MeCP2 and methyltransferase G9a to effectively silence large regions of chromatin, including genes that lack an RE1 (Lunyak et al., 2002).
REST regulates the neuronal differentiation of embryonic and neural stem cells (Ballas et al., 2005), but its role in the oligodendrocyte lineage is unknown. Approximately 10% of the genes whose expression is highly reduced during the OPC to oligodendrocyte transition contain RE1s (Bruce et al., 2004; Cahoy et al., 2008), many of which are occupied by either REST or CoREST (Abrajano et al., 2009). These observations suggest that REST function may be required during oligodendrocyte differentiation. Here, we asked whether REST can regulate OPC differentiation. We used loss-of-function (LOF) and gain-of-function approaches to show that REST is required for the repression of neuronal properties in OPCs and their development into oligodendrocytes. These results suggest that REST may play important roles in regulating the development and plasticity of OPCs.
Materials and Methods
Cell culture.
Rat glial cultures were prepared from postnatal day 1 (P1) to P3 rat pups of either sex using a modified shaking method (McCarthy and de Vellis, 1980; Yang et al., 2005). Dissociated cortical cells were grown for 10 d in T-75 flasks and shaken at 37°C at 250 rpm overnight (16–20 h), followed by an additional 30 min at 300 rpm. Nonadherent cells were filtered through a 20 μm nylon mesh filter (Spectrum Laboratories) to remove astrocyte aggregates. The remaining cells underwent two rounds of differential adhesion on tissue culture dishes (Corning) for 30 min each at 37°C. The nonadherent cells (OPCs) were plated on 25 μg/ml poly-l-lysine (PLL) (Sigma) coated tissue culture dishes or 50 μg/ml PLL-coated glass coverslips and grown in proliferation media [Neurobasal A medium supplemented with B27 containing 1% penicillin/streptomycin (P/S), 1 mm l-glutamine (all from Invitrogen), 10 ng/ml platelet-derived growth factor AA (PDGFAA), and 5 ng/ml fibroblast growth factor basic (bFGF) (PreproTech)]. To initiate oligodendrocyte differentiation, OPCs were switched to DMEM supplemented with N2 (Invitrogen), 1% P/S, 1% fetal bovine serum (FBS) (Gemini Bio-Products), 40 ng/ml thyroxin, and 30 ng/ml triidothyronine (T3) (Sigma). For protein analysis, the cells were grown in oligodendrocyte media for 6 d. Type 2 astrocyte (2A) development was initiated by switching the cells to DMEM, 1% P/S, and 10% FBS. In some experiments, the cells were grown in a defined media comprising Neurobasal A medium/B27 supplemented with 1% P/S, 1% FBS, and 1 mm l-glutamine. Type I astrocytes were prepared by removing the adherent cells from the 75 cm2 tissue culture flasks after the overnight shake with trypsin and growing them in DMEM containing 1% P/S and 10% FBS. Primary cultures of rat optic nerve cells and cerebellar granule neurons were grown as described previously (Levine, 1989; Dou and Levine, 1994). Rat embryonic fibroblast (REF) and HEK293 cell lines were grown in DMEM supplemented with 1% P/S and 10% FBS. Rat pheochromocytoma PC12 cells were maintained in DMEM containing 1% P/S, 5% FBS, and 10% horse serum. Plat-E cells were grown in DMEM containing 10% FBS, 1% P/S, 1 μg/ml puromycin dihydrochloride (Sigma), and 10 μg/ml Blasticidin S HCl (Invitrogen). HCN cells were obtained from Dr. F. Gage (Salk Institute, San Diego, CA) and maintained as described (Hsieh et al., 2004). HT22 cells were a gift from Dr. S. Ge (State University of New York, Stony Brook, NY). All cell lines were passed 1:5 every 3–5 d.
Nucleofections.
Transfections were performed using the Nucleofector electroporation system as per the instructions of the manufacturer (Amaxa). Cells were nucleofected with 1–3 μg of DNA, and 3 × 106 cells were used per nucleofection. Before nucleofection, the cells were grown overnight in proliferation media with high levels of growth factors (20 ng/ml PDGFAA and 20 ng/ml bFGF) in uncoated Petri dishes at a concentration of 4–6 × 106 cells per dish. OPCs were harvested by gentle pipetting, centrifuged, and incubated in DMEM containing 1% P/S and 10% FBS (∼3 × 106 cells/ml media) for 1 h at 37°C before nucleofection. The cells were nucleofected with either pcDNA1–DnREST (Chong et al., 1995; Otto et al., 2007), pcDNA3–REST–VP16 (Immaneni et al., 2000) (a gift from Dr. S. Majumder, Ohio State University, Columbus, OH), pcDNA1.1–REST (a gift from Dr. N. Ballas, State University of New York, Stony Brook, NY), or an empty vector (Invitrogen), all mixed with 0.5–1.0 μg of pMAXeGFP (Amaxa).
Immunofluorescence staining.
A list of all primary antibodies can be found at the end of Materials and Methods. Secondary antibodies used for immunocytochemistry (IC) include Alexa Fluor 594 donkey anti-rabbit, Alexa Fluor 488 donkey anti-rabbit, Alexa Fluor 594 donkey anti-mouse (all from Invitrogen). Live-cell staining of cell surface antigens was performed using antibodies against NG2, A2B5, and O4 as described previously (Levine and Stallcup, 1987). Cells (unless being double labeled to visualize REST expression; see below for REST detection) were fixed with 4% buffered paraformaldehyde (PFA) for 10–15 min and washed 3× in PBS. To visualize the expression of MBP, GFAP, green fluorescent protein (GFP), and TUJ1, cells were fixed in PFA, rinsed as above (in some cases, cells were first live stained), and incubated in a permeabilizing solution (PBS containing 5% normal donkey serum, 0.01% sodium deoxycholate, and 0.02% NP-40) for 30 min at room temperature. Primary and secondary antibodies were diluted in permeabilizing solution and staining was performed as above.
For immunofluorescence staining to detect REST protein, cells underwent microwave fixation. Coverslips were rinsed two times in PBS, covered in 4% PFA, and placed on a glass plate over ice for 20 s in a microwave (Pelco 3451 Lab Microwave System with a Pelco 3420 Microwave Load Cooler and a Pelco 3430 Microwave Power Controller). Cells were rinsed three times with PBS, blocked, permeabilized, and stained as above. Cells were examined, counted, and photographed with a Carl Zeiss Axioplan2 microscope.
5-Bromo-2′-deoxyuridine labeling.
Retroviral infected OPCs were grown in proliferation media for 4 d, followed by a media switch to either proliferation, oligodendrocyte, defined media, or 2A growth media for 44 h. 5-Bromo-2′-deoxyuridine (BrdU) (10 μm; Sigma) was added to the culture media, and the cells were incubated for 4 h at 37°C. Cells were rinsed in PBS, fixed in 4% PFA for 10 min, washed and then immunofluorescently stained to detect GFP and BrdU.
Clonal analysis.
OPCs were purified and plated in 25 μg /ml PLL-coated 100 mm tissue culture dishes in proliferation media for 1–2 d and then infected with either pMXsIG or pMXsIG–DnREST (see below). OPCs were grown for 5 d after infection in proliferation media and then passed onto coated 18 mm glass coverslips (50 μg/ml PLL) at a density of ∼1000 cells per coverslip. The cells were then grown for 2 additional days in proliferation media, washed with L15, and fed proliferation, oligodendrocyte, defined, or 2A differentiation media. Nucleofected OPCs were plated directly at clonal density without the intervening proliferation period.
Cell extracts and immunoblot analysis.
To isolate optic nerve protein, optic nerves were homogenized in a solution containing 2% SDS, 10 mm Tris, pH 7.5, 5 mm EDTA, pH 8.0, 200 μm phenylmethanesulfonyl fluoride (PMSF), 1 μg/ml aprotinin, and leupeptin using a Wheaton Instruments overhead stirrer and glass on glass tissue grinder (Duall 20; Kontes Glass Co). Samples were centrifuged at 30,000 rpm for 15 min at 20°C in an Optima TLX ultracentrifuge (Beckman Instruments). The supernatant was collected and protein concentration determined using a Pierce BCA Protein Assay (Micro BCA Protein Assay kit; Thermo Fisher Scientific). For primary cells and cell lines, whole-cell lysates were prepared by rinsing the cells two times in cold PBS and incubating them on ice in NP-40 lysis buffer (25 mm Tris, pH 7.5, 125 mm NaCl, 1.0% NP-40, 1 mm EDTA, 200 μm PMSF, 1 μg/ml aprotinin, and leupeptin) for 30 min. Samples were centrifuged at 14,000 rpm for 10 min at 4°C, and the supernatant was collected. Nuclear extracts were prepared using a modified Dignam method (Grimes et al., 2000). Protein concentrations were measured using a Bradford assay (Bio-Rad). All samples were boiled for 3 min in 6× SDS-loading buffer. SDS-PAGE and immunoblotting were performed using standard techniques. Proteins were electrophoresed on 6–8% polyacrylamide gels, electrophoretically transferred to nitrocellulose (HybondTM-ECL; GE Healthcare), and blocked in Tris-buffered saline (TBS) or TBS + 0.5% Triton X-100 containing 5% nonfat dry milk. Primary antibodies used are listed at the end of Materials and Methods, and detection was performed with ECL anti-rabbit IgG horseradish-peroxidase-linked whole antibody (from donkey) or ECL anti-mouse Ig horseradish-peroxidase-linked whole antibody (from sheep) secondary antibodies (GE Healthcare).
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (Ballas et al., 2001). The chromatin was immunoprecipitated after diluting the cleared chromatin 1:10 in an immunoprecipitation buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.0, 167 mm NaCl, and complete mini protease inhibitor cocktail) with 4 μg/ml of either rabbit anti-REST–P73 (Chong et al., 1995), rabbit anti-REST-C (Ballas et al., 2001), or a nonspecific rabbit IgG antibody (Santa Cruz Biotechnology) for 12–16 h at 4°C. The following primers were used to amplify DNA regions containing an RE-1 sequence: Scn2a1 left, CCAAGATTATATGTCAGCTCGCAG; Scn2a1 right, CAACTTCGTCAAGCAGGGTCAGG; neurofilament-M (NF-M) left, GTTCATTGTGCCAACTCTGCAGTGC; NF-M right, GGACATCTGAAGCTTCTAGCAGGTAAGG; SCG10 left, ACAGTCCGAAGTGCAAATGCCAG; SCG10 right, CTGCCGTTCAGCAAATCATCCAG; NeuroD2 left, GACTGGAGGCATAGGTCACCTTTCTAC; NeuroD2 right, GGAAGTCACCACTAGCACATTTCGCG; Grik3 left, CCTGTTAGGAGGGTCGCCCAG; and Grik3 right, CCGCCACAGAAGGCAGAAGACC.
Primers were built around a randomly chosen region of the NG2 gene (CSPG4) that does not contain an RE1 (left, CTTCTCTGGACCCCACTCAC; right, TGGCCCTCAAATATGTCCAC).
A 50 μl PCR reaction contained 2 μl of eluted chromatin, 5 μl of 10× PCR buffer (Roche), 5 μl of 10× PCR Enhancer (Invitrogen), 200 mm dNTPs (Roche), 200 nm forward and reverse primers, and 0.5 μl of Taq polymerase (Roche). DNA was amplified using a Gene AMP PCR System 9700 (Applied Biosystems) with an initial denaturation at 94°C for 4 min, 40 cycles of 94°C for 30 s, 56–58°C for 30 s, and 72°C for 1 min, ending with a final 7 min incubation at 72°C. Samples were analyzed on 1.5% agarose gels.
Quantitative real-time PCR.
Total RNA from primary cells, cell lines, and minced whole optic nerve was isolated by TRIZOL RNA extraction (Invitrogen) and treated with RNase-free DNase (TURBO DNA-free; Ambion) following both of the protocols of the manufacturers. RNA concentrations were measured using a NanoDrop 1000 spectrophotometer, and 250–300 ng of total RNA was used for reverse transcription with Superscript II Reverse Transcriptase (Invitrogen) via the protocol of the manufacturer. Quantitative real-time (qRT)-PCR was performed using FastStart Universal SYBR Green Master Mix (Roche) in an Applied Biosystems 7300 Real-Time PCR System under the cycling conditions described previously (Otto et al., 2007). The relative abundance of the specific mRNA being examined was normalized to glyceraldehydes 3-phosphate dehydrogenase (GAPDH). All primers were designed using Massachusetts Institute of Technology's Primer3 software: Calb1 left, CCGAACAGATCTTGCCCTTA; Calb1 right, GCGCACAGTTATGGTTTTAGA; CelsR3 left, CCATCCTTGCCTCTTTCAAC; CelsR3 right, TGGAAGGTCCAAGTGCAGA; CNP left, AGTGCAAGACGCTGTTCATC; CNP right, AAGCATCAGCAGACACCATCT; DCX left, ACAGAACCAGAACCTTGCAG; DCX right, AGCGTACACAATCCCCTTGA; GFAP left, AGTTACCAGGAGGCACTCG; GFAP right, CGATGTCCAGGGCTAGCTTA; GluR2 left, GGGCGCTGATCAAGAATACA; GluR2 right, GCGAAACTGTTGGCTACCTC; Hes1 left, CTGACGGCCAATTTGCTT; Hes1 right, GTTAGGACCCACCGAGGTC; Hes5 left, GAGATGCTCAGTCCCAAGGA; Hes5 right, GCAGTTTCAGCTGCTCAATG; Hes5 left, AAGAGCCTGCACCAGGACTA; Hes5 right, CGCTGGAAGTGGTAAAGCAG; Id2 left, CTCCAAGCTCAAGGAACTGG; Id2 right, CGATCTGCAGGTCCAAGATA; Id4 left, GTGCGATATGAACGACTGCT; Id4 right, CTGCAGGTCCAGGATGTAGTC; MAG left, GTGGGCTCCCTTTCTCTTG; MAG right, ATGTTGGCAGAGAGGAGCAG; Mash1/acl1 left, GCCCAGCACTCTCTCACTTC; Mash1/acl1 right, TGGGATTATTTGGCTGAACC; MBP left, CACAAGAACTACCCACTACGG; MBP right GGGTGTACGAGGTGTCACAA; MOBP left, CCCACCCTTCACCTTCCT; MOBP right, CCAGTCCTCCTCCTTCTTCTG; MYT1 LIKE left, CGTGACTACTTTGACGGAAATG; MYT1 LIKE right, ATTCCTCTCACAGCCTGCTT; Myt1 left, TCATTGCAACTTCCCTCCTAA; Myt1 right, GGTCCTTTTCACCCTCATCA; Nav1.2 left, GATCCTCATCTGCCTCAACA; Nav1.2 right, TGAACAGGACAATGAACACCA; NF-M left, GAGAGCAGCCTCGACTTCAG; NF-M right, CCCTGCAGCTGCTCTTTCT, NG2 left, ACCCAGCAGGATGGCTTC; NG2 right, CTCATTCACGTCCCTCACAG; NeuroD1 left, CCAAATCATACAGCGAGAGC; NeuroD1 right, CTCCTTCTTGTCTGCCTCGT; NeuroD1 left, AGGAACACGAGGCAGACAAG; NeuroD1 right TCTTGGGCTTTTGATCATCC; Nkx2.2 left, GTGATCATCGTTGCCAAATG; Nkx2.2 right, AAACCGATGCAACTCAAACA; Olig1 left, AACTCCTTGGACCAGTGCAT; Olig1 right, CGCCAGTTAAATTCGGCTAC; Olig2 left, GTGCATCACCCCATCCTC; Olig2 right, CCGATGGAGACTTGAGCAG; PDGFαR left, CACACCGGATGGTACACTTG; PDGFαR right, GCAGAATCGTCCTCTTCCAC; PLP left, GCACTGTTCTGTGGATGTGG; PLP right, GAAGAAGAAAGAGGCAGTTCCA; PLP left, GCTGTGCTAGATGTCTGGTAGG; PLP right, GCTTCATGTCCACATCCACA; POU4 F3 left, GCTCAAGCCAGTCCTCCA; POU4F3 right, GTTTGCGCTTACGCTCACT; POU4F3 left, AGAGAAGCGCTCACTCGAAG; POU4F3 right, GGACAGCAGAGTACTTCATTCG; SCG10 left, GGCATTTGTGCCACTGTAAG; SCG10 right, CAGCAACCTTCAGGAGTTGG; SoxII left, ATGGGCTCTGTGGTCAAGTC; SoxII right, CTGATCATGTCCCGGAGGT; Sox10 left, AACCTCATCCCTCACCTAACTG; Sox10 right, GCAGTGCTAACTGAGGCTGA; SYN1 left, GAAGCCAGACTTTGTGCTGA; SYN1 right, ACTGGGGATCCCAGCATAC; SYT II left, GATCTTCGTAGGCAGCAACG; SYT II right, CCTCAGGCTTCAGAGAGTGC; TrkC left, ACTATGTGGGCTCCGTGCT; TrkC right, CCAGGAGGGGAAAGAGGTT; Tubb3; left, CATGAACGACCTGGTGTCTG; Tubb3 right, CTGGGCTTCCGACTCCTC; Yin Yang1 (YY1) left, ATGAGAAAGCATCTGCACACC; YY1 right, CCAGCTGGTGGTCGTTTTAGC.
Luciferase assay.
Luciferase reporter assays were performed as described previously (Conaco et al., 2006; Otto et al., 2007).
Adenoviral vectors and infection.
Adenoviral vectors (pAdTrackCMV vector containing GFP) expressing dominant-negative REST (DnREST, lacks the sequences coding for the N- and C-terminal repressor domains), REST–VP16 (which contains the same region of the REST gene as DnREST fused to the activation domain of the herpes simplex viral protein VP16), or expressing GFP were prepared and used as described previously (Chong et al., 1995; Ballas et al., 2005). OPCs were purified and infected after 2–3 d in proliferation media. For oligodendrocyte and 2A infection, OPCs were purified, grown in proliferation media for 2–3 d, and switched to either oligodendrocyte or 2A differentiation media for 3 d before infection. Type I astrocytes were isolated as described previously and were infected 3–6 d after purification. All cells were infected for 8–16 h with adenovirus at a multiplicity of infection of 10–25.
Retroviral vectors and infection.
DnREST was cloned into the pMXs–IRES–GFP (pMXsIG) vector (Kitamura et al., 2003) between the EcoRI and XhoI sites. PMXsIG and pMXsIG–DnREST retrovirus were assembled in Plat-E cells (Morita et al., 2000). Puromycin dihydrochloride, Blasticidin S HCl, and P/S were removed from Plat-E cell media 24 h before transfection, and retroviral vectors were transfected into Plat-E cells using TransIT-293 transfection reagent (MirusBio) following the protocol of the manufacturer. After 24, 36, and 48 h, the supernatant was collected and stored at −80°C. The retrovirus-containing supernatant was concentrated by ultracentrifugation for 90 min at 4°C at 25,000 rpm and resuspended in a small volume of PBS. OPCs were purified and infected after 1–2 d in proliferation media. Cells were infected to 60–80% efficiency by incubating the cells with virus for 5–8 h with 3 μg/ml Polybrene (Sigma) (final retroviral concentrations were at least 106 colony forming units per milliliters).
Tissue processing and immunofluorescence staining of optic nerve.
Cryostat sections of developing and adult optic nerve were processed and stained as described previously (Tan et al., 2006).
Primary antibodies.
Antibodies listed were used for immunofluorescence staining, Western Blot analysis (WB), cell isolation/purification (CP), and ChIP. Mouse anti-NG2 (Millipore Bioscience Research Reagents) IC, mouse anti-A2B5 (American Type Culture Collection) IC, CP, rabbit anti-REST-C (Gail Mandel, Oregon Health and Science University, Portland, OR) IC, immunohistochemistry, WB, ChIP, rabbit anti-REST P73 (Gail Mandel) ChIP, mouse anti-TUJ1 (Covance) IC, WB, mouse anti-O4 (a gift from R. Bansi, University of Connecticut Health Science Center) IC, mouse anti-MBP (Millipore) IC, mouse anti-GFAP (Sigma) IC, rabbit anti-synapsin I (Millipore) WB, mouse anti-neurofilament (Developmental Studies Hybridoma Bank) WB, mouse anti-β-actin (Sigma) WB, rabbit anti-histone H3 (Cell Signaling Technology) WB, rabbit anti-CoREST (Gail Mandel) IC, WB, rabbit anti-Snap25 (Abcam) WB, rabbit anti-GFP (Invitrogen) IC, rabbit anti-SCG10/Stathmin-2 (University of California, Davis/National Institutes of Health Neuromab Facility) WB, mouse anti-CNPase (Sigma) WB, and mouse anti-BrdU (Sigma) IC.
Statistical methods.
The significance of all pairwise comparisons was determined using Student's t test. For multiple comparisons, we used a one-way ANOVA, followed by the Holm–Sidak post hoc test.
Results
REST is expressed in glial cells
To examine REST expression and function in primary rat glia, we dissociated newborn rat cortices and isolated different types of glia using a modification of the shaking method (Yang et al., 2005). OPCs, oligodendrocytes, type I astrocytes, and 2As were grown in different media as described in Materials and Methods. In the astrocyte and OPC cultures, ∼95% of the cells expressed the appropriate cell-type-specific marker antigens (data not shown). When grown under conditions that foster oligodendrocyte differentiation for 5 d, 90% of the cells expressed the O4 antigen and 40% expressed MBP (data not shown). As shown in Figure 1A, all glial cells examined expressed REST protein in their nuclei. Immunoblot analysis of nuclear protein extracts confirmed that the different glial types expressed both REST and its corepressor protein CoREST (Fig. 1B). In these biochemical experiments, REFs served as positive controls and undifferentiated PC12 cells, which express trace amounts of REST mRNA, served as negative controls. The different types of glia expressed REST transcripts (Fig. 1C) and protein (Fig. 1B) at comparable levels. Glial cells also express mSin3a (Ballas et al., 2009), suggesting that a functional repressor complex exists in these cells. Because these glial cells were grown in culture for 10–12 d before shaking, we wanted to rule out the possibility that REST expression was induced by the relatively long cell culture period. We therefore dissociated cells from postnatal day 7 rat optic nerve, plated the cells onto coated coverslips for 3 h, and immunofluorescently stained the cells with anti-REST antibodies. REST protein was detected in >90% of OPCs, preoligodendrocytes, and astrocytes (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). To confirm REST expression in developing glia in vivo, we stained sections of postnatal day 12 rat optic nerve with anti-REST antibodies. As shown in Figure 1D, the antibodies detected linear arrays of nuclei, a staining pattern typical for developing glia of the oligodendrocyte lineage. Both REST protein and mRNA were present in newborn and P7 optic nerves, and levels of both the protein and mRNA declined with increasing age (Fig. 1E,F). Together, these data show that developing glia, including cells of the oligodendrocyte lineage, express REST. The low levels of REST in adult glia suggest that REST function may be required principally during development.
Figure 1.
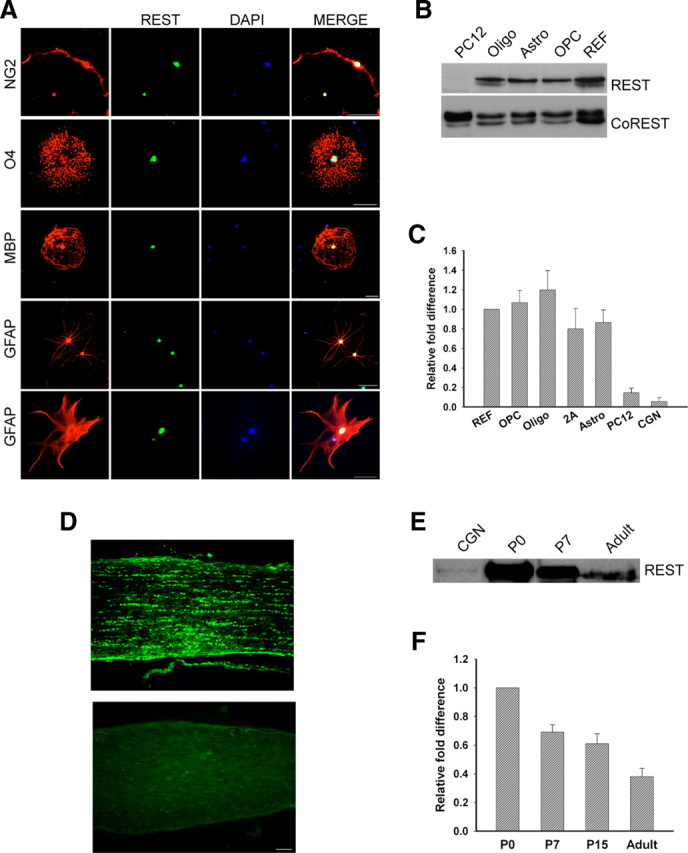
REST is expressed in the nuclear compartment of primary rat glia and in rat optic nerve. Purified OPCs, astrocytes, and oligodendrocytes were grown as described in Material and Methods for 5–6 d, and REST protein and transcript levels were measured. A, Immunofluorescence staining detects REST in the nuclei of cultured glial cells. Scale bar, 50 μm. B, Immunoblot showing the detection of REST and CoREST protein in nuclear extracts prepared from oligodendrocytes (Oligo), astrocytes (Astro), OPCs, and REFs. REST was not present in extracts of PC12 cells. C, Quantitative real-time PCR analysis showing the fold changes in REST mRNA in rat glia relative to REFs after normalization to GAPDH. REST expression is lower in PC12 cells and CGNs. Error bars represent the SD; n = 3. D, Immunofluorescence detection of REST in postnatal day 12 rat optic nerve (top). The bottom is control staining with secondary antibody alone. Scale bar, 50 μm. E, Immunoblot analysis of REST protein in P0, P7, and adult rat optic nerve. CGNs serve as a negative control. F, Real-time PCR analysis showing the fold change in REST expression in P7, P15, and adult rat optic nerve relative to P0 after normalization to GAPDH. Error bars represent the SD from six PCR runs; three runs each from two separate experiments.
Rest is a functional gene repressor in developing glia
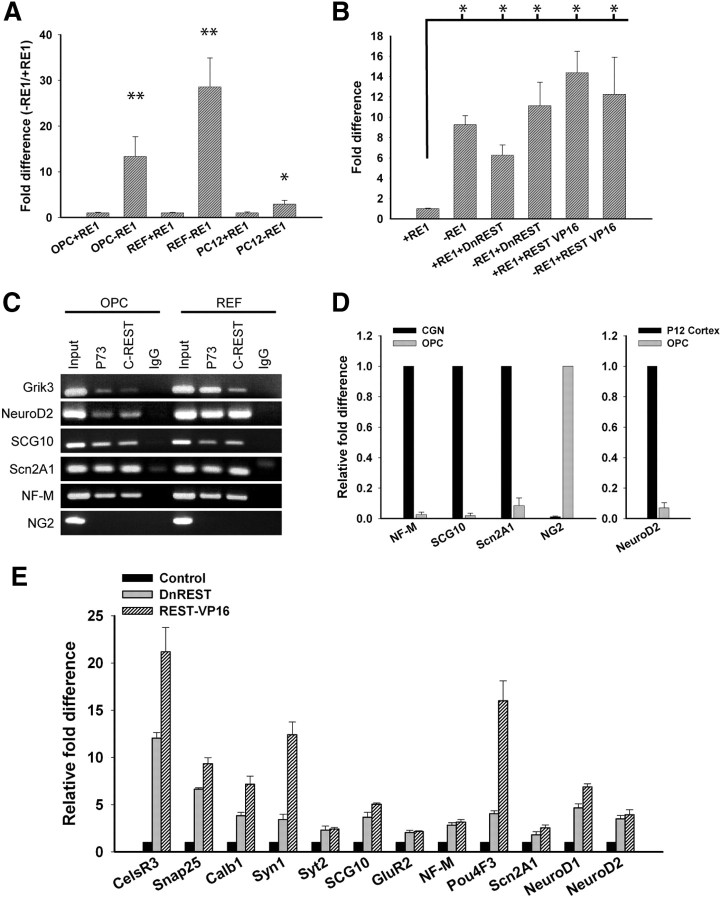
We used a luciferase reporter assay (Otto et al., 2007) to determine whether REST functions as a repressor in oligodendrocyte lineage cells. OPCs, REFs, and PC12 cells were nucleofected with a plasmid containing a region of the GAD1 gene promoter with or without an RE1 site upstream of a minimal TK promoter capable of driving Photalis pyralis luciferase expression. If cells express functional REST protein, then luciferase activity is reduced when the RE1 is present. As shown in Figure 2A, luciferase activity was 13.4-fold higher in OPCs expressing the RE1-negative construct compared with cells expressing the RE1-containing construct. To confirm that a REST/RE1 interaction was responsible for the decreased luciferase activity, cells were conucleofected with a plasmid expressing DnREST or REST–VP16. The DnREST construct contains the DNA binding domain but not the N- and C-terminal corepressor binding domains (Chong et al., 1995). It competes with endogenous REST for DNA binding but does not repress gene transcription. The REST–VP16 construct has both corepressor domains deleted, and the C-terminal domain is replaced by the activator domain of VP16 (Immaneni et al., 2000). When transfected into cells, this construct activates the transcription of RE1-containing genes and initiates neuronal differentiation (Su et al., 2004). DnREST derepressed luciferase gene expression only when the RE1 was present: REST–VP16 further activated luciferase expression, also in an RE1-dependent manner, although the difference between REST–VP16 and DnREST was not statistically significant (Fig. 2B). These results demonstrate that REST can act as a functional transcriptional repressor in OPCs.
Figure 2.
REST is a functional repressor protein in OPCs. A, Luciferase reporter assay showing that RE1-containing reporters are repressed in OPCs but not PC12 cells. Error bars represent the SD from three separate experiments. *p < 0.03, **p < 0.01. B, Luciferase reporter assay showing that DnREST and REST–VP16 derepressed luciferase gene expression in the presence of an RE1. Error bars represent the SD; n = 3. *p < 0.01. C, ChiP assays showing that REST binds to an RE1 element in Grik3, NF-M, SCG10, Scn2a1, and NeuroD2 but not to a randomly chosen site upstream of the NG2 (CSPG4) gene in both OPCs and REFs. D, Quantitative real-time PCR showing the fold difference in NF-M, SCG10, and Scn2a1 relative to CGNs, NG2 relative to OPCs, and NeuroD2 relative to P12 rat cortex after normalization to GAPDH. Error bars represent the SD; n = 3. E, DnREST and REST–VP16 induced changes in the expression of RE1-containing genes. Purified OPCs were infected with DnREST, REST–VP16, or GFP control adenoviruses and grown in proliferation media for 72 h. Transcript levels of REST-regulated neuronal genes were analyzed by real-time PCR. The relative fold change in gene expression is shown relative to control-infected cells after normalization to GAPDH. Error bars represent the SD from six PCR runs; all gene expression changes had p < 0.01.
We used chromatin immunoprecipitation assays to determine whether REST interacts with the RE1 element in known REST-regulated genes. REST protein/DNA complexes were immunoprecipitated with polyclonal antibodies against the DNA binding domain of REST (P73) and the C-terminal CoREST binding domain (anti-REST-C). As shown in Figure 2C, in OPCs, REST bound to the RE1 elements in Grik3, NeuroD2, SCG10, Scn2A1, and NF-M but not to a randomly chosen site upstream of the NG2 gene that does not contain an RE1. qRT-PCR demonstrated that these neuronal genes are transcriptionally repressed in OPCs compared with cerebellar granular neurons (CGNs) or postnatal day 12 rat cortex. Conversely, OPCs contain high levels of transcripts for NG2 that were barely detectable in the CGN cultures (Fig. 2D). If REST/RE1 interactions are responsible for transcriptional repression in glia, then perturbing REST function should alter the expression of RE1-containing genes. To examine this, we infected OPCs with adenoviruses expressing DnREST, REST–VP16, or GFP alone and measured gene expression by qRT-PCR 72 h later (Fig. 2E). Expression of either DnREST or REST–VP16 was sufficient to derepress or activate the transcription of RE1-containing genes in OPCs (Fig. 2E). Expression of these and other genes was also derepressed when differentiated astrocytes and oligodendrocytes were infected with the REST–VP16 adenovirus (supplemental Table S1, available at www.jneurosci.org as supplemental material). These data show that REST is a functional gene repressor in glia. The variations in the levels of expression of individual genes among different cell types suggest that REST function can vary in a cell-type-specific manner.
Loss of REST repression inhibits oligodendrocyte differentiation and induces neuronal gene expression
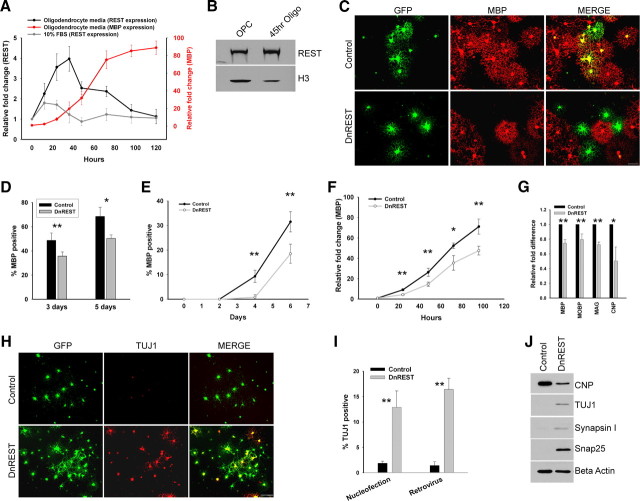
Given that OPCs express functional REST, we next asked whether REST regulates the development of oligodendrocyte lineage cells. We first examined REST expression during the differentiation of OPCs into oligodendrocytes. OPC cultures were expanded in proliferation media for 3 d and then induced to differentiate into oligodendrocytes by plating in serum-free media containing thyroid hormones. As shown in Figure 3A, REST mRNA expression increased 4.0-fold (±0.6, n = 3) relative to OPCs 36 h after inducing differentiation and then returned to baseline as the cells matured into MBP-expressing oligodendrocytes. This increase in mRNA was accompanied by an approximate fourfold increase in REST protein (Fig. 3B). Protein levels remained elevated after 4 d in oligodendrocyte media (data not shown). REST mRNA levels increased modestly but significantly when the cells were induced to develop into 2As by the addition of fetal calf serum to the media. There was no additional increase in REST when T3 was added to the serum-containing media. Adding T3 (30 ng/ml) did not increase REST transcription in either REFs or HCN neural stem cells but did cause a 2.3-fold increase in the HT22 neuronal progenitor cell line (data not shown). The expression of myelin-specific genes increased and that of RE1-containing genes decreased during oligodendrocyte differentiation (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). These data suggest that REST is active as a gene repressor during the initiation of oligodendrocyte differentiation.
Figure 3.
REST repression is necessary for proper oligodendrocyte maturation and to repress neuronal gene and protein expression in OPCs. A, Real-time PCR of REST mRNA expression after the initiation of oligodendrocyte and 2A differentiation. Purified OPCs were switched to oligodendrocyte or 2A differentiation media at time 0, and total RNA was extracted at the indicated time points. mRNA expression is normalized to GAPDH, and fold difference at each time point is relative to time 0. Error bars represent the SD; n = 3. B, Immunoblot analysis shows that REST protein levels increase during the onset of oligodendrocyte differentiation. Nuclear protein was isolated from OPCs in proliferation media and OPCs in oligodendrocyte media for 45 h. Histone H3 is used as a loading control. C, D, Purified OPCs were conucleofected with a plasmid expressing GFP and either a plasmid expressing DnREST or an empty vector. C, Immunofluorescence staining for MBP (red) and GFP (green) 5 d after nucleofection. D, The percentage of the nucleofected cells expressing MBP. Error bars represent the SD; n = 3. *p < 0.02; **p < 0.007. E–J, OPCs were infected with control GFP or DnREST retroviruses as described in Materials and Methods and analyzed for the expression of properties associated with either mature oligodendrocyte (E–G) or immature neurons (H–J). E, Quantification of the percentage of infected cells expressing MBP at the indicated time points subsequent to the initiation of oligodendrocyte differentiation. Error bars represent the SD; n = 3. **p < 0.001. F, Real-time PCR of MBP expression subsequent to the initiation of oligodendrocyte differentiation. Fold change is shown relative to time 0. n = 3. *p < 0.01, **p < 0.005. G, Real-time PCR analysis of myelin gene expression after normalization to GAPDH. RNA was harvested 3 d after initiating oligodendrocyte differentiation. Error bars represent the SD; n = 6. *p < 0.05, **p < 0.001. H, Immunofluorescence staining for TUJ1 5 d after induction. I, The percentage of nucleofected or retroviral infected cells that express TUJ1. Error bars represent the SD; n = 3 for the nucleofection experiments, n = 4 for the retroviral infection experiments. **p < 0.0001. J, Immunoblot analysis of the expression of the indicated neuronal proteins 5 d after infection for cells grown in oligodendrocyte media.
To determine whether REST function is required for oligodendrocyte differentiation, we used an LOF approach. OPCs were nucleofected with plasmids expressing either DnREST and GFP or GFP alone and grown in oligodendrocyte media for 3 and 5 d, and oligodendrocyte differentiation was assayed by staining with antibodies against MBP to identify mature oligodendrocytes. The percentage of MBP-positive cells decreased when DnREST was expressed (Fig. 3C,D). Because protein expression after nucleofection of plasmids declines after 3–4 d, we infected OPCs with retroviruses expressing either DnREST (pMXsIG–DnREST) or control GFP (pMXsIG). The cells were infected, grown in proliferation media for 5–6 d to allow for expression of the virally encoded proteins, plated onto coverslips for 2 d, and then switched to oligodendrocyte media. As shown in Figure 3E, the development of MBP-positive oligodendrocytes was retarded after infection with DnREST viruses. MBP-positive cells were slower to appear, and, after 6 d in oligodendrocyte-inducing media, the percentage of MBP-positive cells was reduced significantly among the DnREST-infected cells relative to control-infected cells. The number of cells expressing the O4 antigen, a marker for preoligodendrocytes, was also reduced in the REST LOF cells (data not shown; see also Fig. 7H). MBP gene transcription lagged behind that of control cells and never reached the levels of control cells (Fig. 3F). The transcription of several other myelin genes was also reduced (Fig. 3G), and the DnREST-infected cells also had lower levels of CNP protein (Fig. 3J). REST LOF led to multiple changes in gene expression during differentiation (Table 1). Genes known to be highly regulated by REST, such as Celr3 and Snap25, increased 5-fold to 10-fold, whereas other genes (with and without identified RE1s) increased more modestly. Several genes involved in the regulation of oligodendrocyte differentiation, including Id2, Id4, olig2, Nkx2.2, and YY1, did not change. The magnitude of any changes in gene transcription is likely underestimated in Table 1 because the infection efficiency was only between 60 and 80%. These data suggest that REST function is required for oligodendrocyte differentiation.
Figure 7.
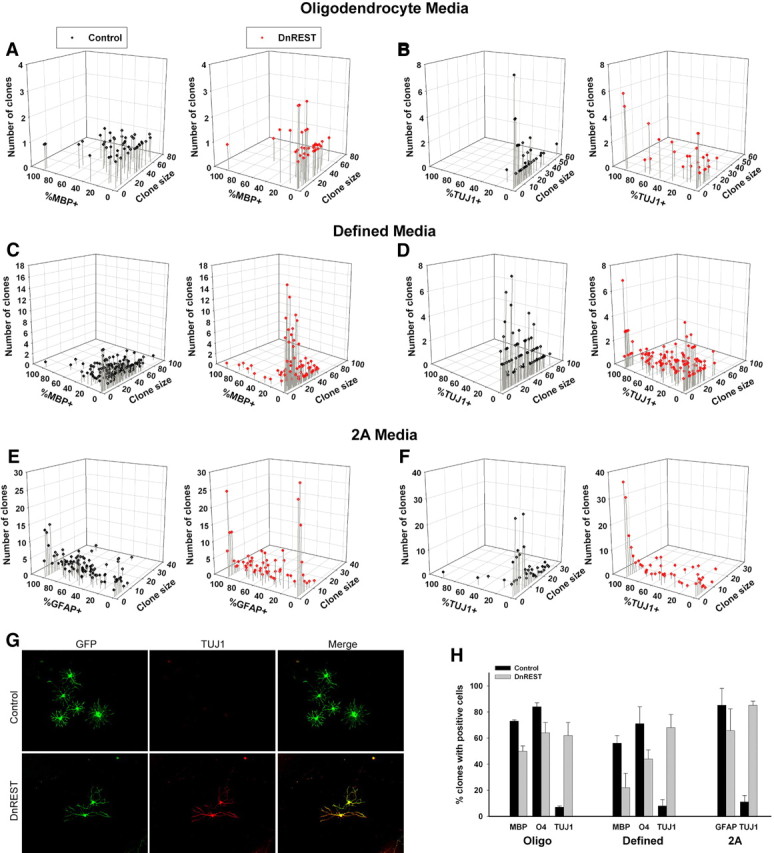
Clonal analysis of OPC differentiation. OPCs were infected and grown at clonal density as described in Materials and Methods and analyzed for the expression of MBP, TUJ1, or GFAP after 5 d or O4 after 3 d in their indicated media. A–F, The scatter plots show the size and composition of all individual clones for the indicated growth conditions. G, Immunofluorescence staining for TUJ1 in a control- and DnREST-infected clone grown in 2A differentiation media for 5 d. H, The percentage of clones containing cells positive for the indicated marker antigens. Total number of clones analyzed is given in the legend to Figure 6. p < 0.05 for all pairwise comparisons.
Table 1.
REST loss-of-function causes multiple changes in gene expression during OPC differentiation
| Oligodendrocyte |
2A |
||
|---|---|---|---|
| Gene name | Relative fold difference | Gene name | Relative fold difference |
| CelsR3 | 10.48 ± 4.16** | SCG10 | 13.73 ± 2.24** |
| SCG10 | 6.85 ± 0.25** | CelsR3 | 11.33 ± 3.92** |
| Snap25 | 5.25 ± 0.35** | Snap25 | 4.88 ± 1.09** |
| NFM | 4.89 ± 0.98** | Nestin | 3.65 ± 2.05* |
| Myt1 | 3.61 ± 0.12** | Myt1L | 2.95 ± 0.78** |
| βIII tubulin | 2.08 ± 0.28** | βIII tubulin | 2.93 ± 0.27** |
| Syn1 | 1.96 ± 0.24** | ATOH | 2.50 ± 0.08** |
| Nestin | 1.83 ± 0.03** | DCX | 2.42 ± 1.17 |
| Hes5 | 1.82 ± 0.05** | NGN3 | 2.36 ± 0.07** |
| Hes1 | 1.76 ± 0.75 | NeuroD1 | 2.29 ± 1.13 |
| Syt2 | 1.75 ± 0.95 | NF-M | 2.03 ± 0.02** |
| Calb1 | 1.71 ± 0.08** | Myt1 | 1.98 ± 0.15** |
| Myt1L | 1.58 ± 0.07** | Hes1 | 1.71 ± 0.13** |
| Olig2 | 1.49 ± 0.74 | Syn1 | 1.55 ± 0.29* |
| ASCL1 | 1.40 ± 0.04** | Nkx2.2 | 1.48 ± 0.20** |
| DCX | 1.35 ± 0.06** | NGN2 | 1.48 ± 0.02** |
| PLP | NC | Calb1 | 1.47 ± 0.09** |
| Sox2 | NC | GluR2 | 1.40 ± 0.07** |
| PDGFαR | NC | Olig1 | 1.39 ± 0.03** |
| GluR2 | NC | Olig2 | 1.25 ± 0.11* |
| NG2 | NC | β-actin | NC |
| Olig1 | NC | Sox2 | NC |
| Sox10 | NC | ID4 | NC |
| YY1 | NC | YY1 | NC |
| GFAP | NC | ASCL1 | NC |
| CTNNb1 | NC | Hes5 | NC |
| Id2 | NC | PLP | NC |
| Nkx2.2 | NC | PDGFαR | NC |
| β-actin | NC | ID2 | NC |
| Id4 | NC | CNP | 0.74 ± 0.22 |
| MOBP | 0.79 ± 0.07** | NG2 | 0.68 ± 0.36 |
| MBP | 0.74 ± 0.05** | MOBP | 0.66 ± 0.38 |
| MAG | 0.72 ± 0.03** | MBP | 0.65 ± 0.05** |
| CNP | 0.50 ± 0.31* | MAG | 0.58 ± 0.18** |
| GFAP | 0.41 ± 0.11** | ||
| Brca1 | 0.39 ± 0.02** | ||
Gene changes during differentiation. OPCs were infected with control- or DnREST-expressing retroviruses and grown in proliferation media for 5 d. Cells were switched to either oligodendrocyte media or 2A differentiation media for 3–4 d. Total RNA was collected, and transcript levels were analyzed by real-time reverse transcription-PCR. Relative fold difference in gene expression in DnREST-infected cells is shown relative to control-infected cells. The SD was taken from four to six total PCR runs from two separate experiments. NC, No change.
*p < 0.05,
**p < 0.005. When not indicated, the change was not statistically significant.
It is possible that the OPCs that did not develop into oligodendrocytes were differentiating along a neuronal lineage (Kondo and Raff, 2000). We therefore stained the nucleofected and infected cells with antibodies that identify young neurons (TUJ1). A total of 12.9 ± 3.2% of the DnREST-nucleofected and 16.4 ± 2.2% of the pMSxIG–DnREST-infected cells were βIII tubulin (TuJ1) positive, whereas only 1.89 ± 0.42 and 1.42 ± 0.75% of the control cells were positive (Fig. 3H,I). Consistent with the changes in gene expression shown in Table 1, βIII tubulin and other REST-regulated neuronal proteins (Snap25, Synapsin1) were expressed in the DnREST-infected cells but not the control-infected cells (Fig. 3J). These data show that REST function normally represses the expression of neuronal genes and proteins in differentiating OPCs.
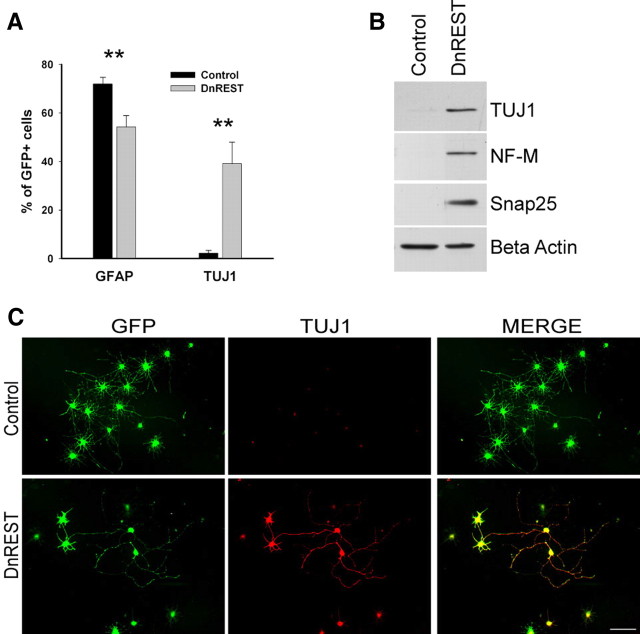
OPCs are highly plastic cells, and their development in vitro is responsive to different media conditions (Raff et al., 1983). To rule out the possibility that the expression of neuronal antigens after REST LOF was attributable to factors found in the oligodendrocyte media used here, we examined the effects of REST LOF on OPCs grown in media containing 10% FBS to promote 2A differentiation. A total of 71.9 ± 2.8 of the control-infected cells expressed GFAP compared with 54.2 ± 7.2% of the DnREST-infected cells (n = 4) (Fig. 4A). The percentage of cells expressing TUJ1 increased (2.2 ± 1.2% in control-infected cells, 39.1 ± 8.9% in the DnREST-infected cells) (Fig. 4A,C). Immunoblot analysis detected βIII tubulin, NF-M, and Snap25 protein in the DnREST-infected cells (Fig. 4B). As was the case with cells grown in oligodendrocyte media, there were complex changes in the levels of gene expression with the transcription of RE1-containing genes increasing 3-fold to 14-fold, whereas myelin and astrocyte-specific genes decreased (Table 1). Thus, when grow under conditions that foster the appearance of astrocytes, REST LOF reduces astrocytic differentiation and drives the cells toward a neuronal phenotype.
Figure 4.
REST loss of function increases neuronal protein expression during astrocytic differentiation. Purified OPCs were infected with retroviruses expressing either DnREST and/or GFP alone and grown for 5 d in 2A differentiation media. A, The percentage of the infected cells that were positively stained with antibodies against GFAP and TUJ1. Error bars represent the SD; n = 3. **p < 0.001. B, Immunoblot analysis shows the expression of neuronal proteins in the DnREST-infected cells after 5 d in 2A differentiation media. C, Immunofluorescence staining showing the expression of TUJ1 (red) in the infected cells (green). Scale bar, 50 μm.
Last, we grew the OPCs in a defined media lacking known growth and trophic factors but that is permissive for neural stem cell differentiation along oligodendrocytic, astrocytic, and neuronal pathways (Belachew et al., 2003). In the control cultures, the percentage of NG2-positive cells decreased and the MBP-positive population increased slowly and moderately over a 6 d time course (Fig. 5A,C). A small number of the control cells (2.1%) expressed TUJ1 antigens, and this did not change over the time course of the experiment (Fig. 5A). The percentage of cells categorized as “other” (i.e., not identifiable with the antibodies used) in the control cultures also increased by day 6. This pattern of development was altered by REST LOF: 22.1 ± 4.4% of DnREST-infected cells became TUJ1 positive, but only 2.4 ± 0.6% were MBP positive (Fig. 5A,C,D). Immunoblot analysis detected TUJ1, Synapsin1, and Snap25 protein in DnREST-infected cells but not the control cells (Fig. 5B). In both control- and DnREST-infected cells, the number of GFAP-positive cells remained low (data not shown). These results show that preventing REST function induces neuronal protein and mRNA expression in OPCs under a variety of media conditions and support the hypothesis that REST repression of neuronal genes is required for the differentiation of oligodendrocyte progenitor cells into either oligodendrocytes or astrocytes.
Figure 5.
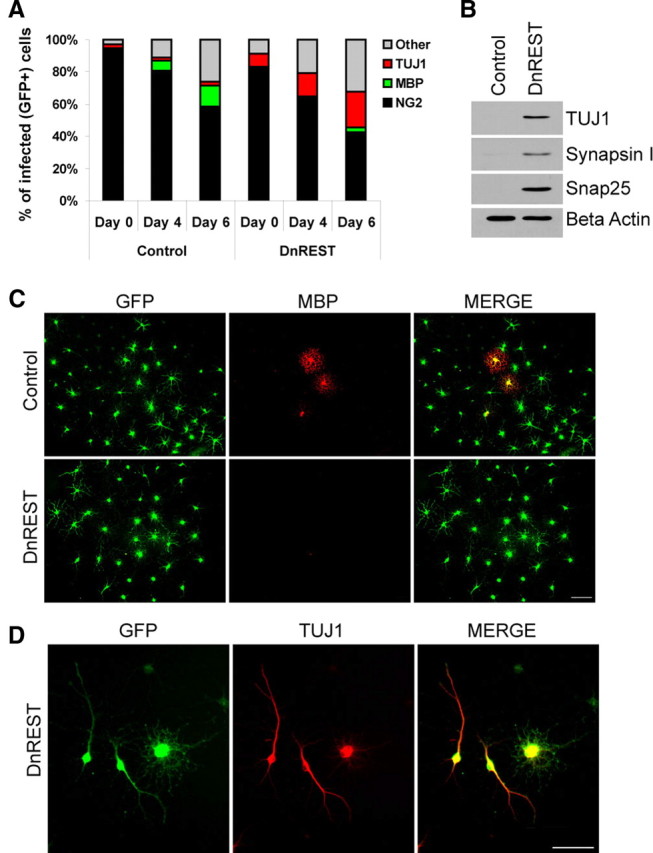
DnREST-infected OPCs grown in a defined media express neuronal proteins. Purified OPCs were infected with control GFP or DnREST retroviruses and grown in a defined media as described in Materials and Methods. A, The stacked bar graph shows the percentage of the cells expressing the indicated marker antigens as determined by immunofluorescence staining. n = 4. B, Immunoblot analysis of the expression of neuronal proteins by the infected OPCs. C, D, DnREST-infected cells fail to express MBP but express TUJ1. Scale bars, 50 μm.
Clonal analysis
One question that arises in the experiments described above is whether all the cells or only a subpopulation of the OPCs are responding to REST LOF. It is possible that the cultures contain undifferentiated neural stem-like cells and that these are the cells that respond to REST LOF by expressing neuronal proteins (Su et al., 2004). To answer this question, we performed a clonal analysis. If all or most OPCs can respond to REST LOF, we predicted that a large number of clones would express TUJ1, whereas if only a subpopulation of stem-like cells responded, the number of clones expressing TUJ1 might be small. Purified OPCs were infected with control or DnREST retrovirus and grown in proliferation media for 5 d. The cells were plated onto coverslips at clonal density, grown in proliferation media for 2 d, and then switched to either oligodendrocyte media, 2A media, or defined media for 3 or 5 d. GFP-expressing clones were identified, and cell number and phenotype for each clone were determined.
Individual clones were scored for the presence of at least one cell expressing MBP, GFAP, or TUJ1. As shown in Figure 7H, regardless of media, between 60 and 80% of the DnREST clones contained TUJ1-positive cells. This rules out the possibility that REST LOF is affecting only a small subpopulation of neural stem-like cells.
Clonal size varied according to the media conditions, but the mean clonal size of the DnREST-infected cells always was smaller than that of control cells when analyzed after 7 d (Fig. 6D). This reduction was attributable to an increased number of small-sized clones and a reduced number of large-sized clones (Fig. 6A–C). To determine whether REST LOF altered cell proliferation, we pulse labeled the cells with BrdU as described in Materials and Methods. As shown in Figure 6E, there was a small but significant reduction in BrdU incorporation in the REST LOF cells. This effect on cell proliferation may lead to the smaller clone size, but we cannot rule out a contribution from either increased cell death or accelerated cell differentiation.
Figure 6.
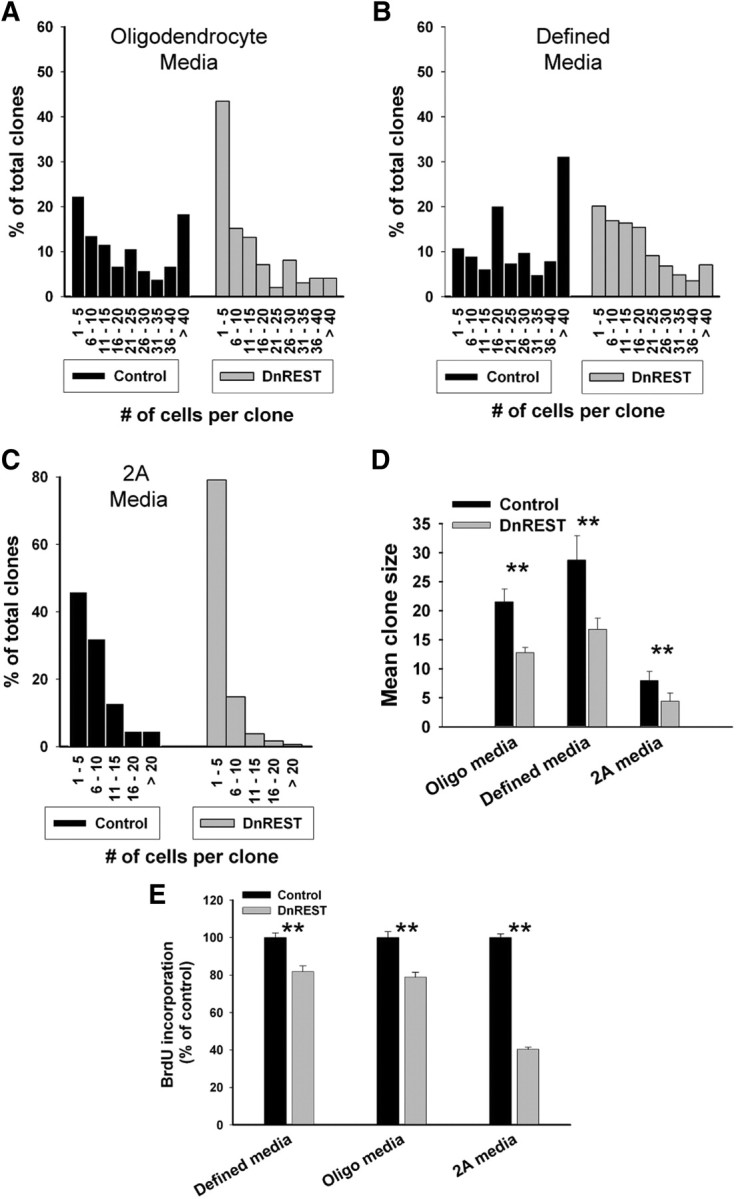
REST LOF decreases clone size and proliferation rate. OPCs were infected and grown at clonal density as described in Materials and Methods. A–C, The histograms show clonal size distribution after growth for 5 d in the indicated media. Total clone number was as follows: oligodendrocyte media, control, n = 103 and DnRest, n = 99; defined media, control, n = 387 and DnREST n = 397; 2A media, control, n = 475 and DnREST, n = 473. D, Mean clonal size in the indicated media. Error bars represent the SD; n = 4. **p < 0.001. E, Control- and DnREST-infected cells were labeled with BrdU as described in Materials and Methods and quantified using immunofluorescence staining. Error bars represent the SD; n = 3. **p < 0.001.
Because OPCs are heterogeneous in terms of their capacity for cell division and because oligodendrocyte differentiation is obligate once an OPC stops dividing (Casaccia-Bonnefil and Liu, 2003), we further analyzed the composition and size of individual clones. The three-dimensional scatter plots in Figure 7 show clearly that the effects of REST LOF on phenotypic differentiation were greater for the smaller-sized clones. In all media, the smaller REST LOF clones contained a lower percentage of MBP-positive (or GFAP-positive) cells and a higher percentage of TUJ1-positive cells (supplemental Fig. S3A–D, available at www.jneurosci.org as supplemental material). For example, in defined media, which had the largest mean clonal size, only 13.5% of the REST LOF clones with 20 or fewer cells contained MBP-positive cells compared with 34.9% of the control-infected clones. Conversely, 4.6% of the smaller control clones contained at least 1 TUJ1-positive cell as opposed to 61% of the LOF clones. LOF lead to a smaller percentage of the cells within any single clone expressing either MBP or GFAP (Fig. 7) and to more clones in which all the cells were 100% positive for the neuronal marker TUJ1 (Fig. 7G) (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). This was especially prominent for the small-sized clones: between 60 and 80% of the one and two cell clones were 100% positive for TJU1 expression (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). There was also an increase in the number of O4-negative clones (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). These data suggest that REST functions are necessary for the differentiation of OPCs into oligodendrocytes. In its absence, the progeny of both rapidly dividing (i.e., founder cells for large clones) and more slowly dividing (i.e., founder cells of smaller clones) develop neuronal properties.
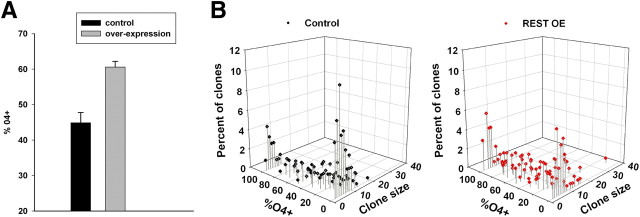
Last, we asked whether increased levels of REST can promote oligodendrocyte differentiation. We nucleofected OPCs with a plasmid containing a full-length REST cDNA under control of the cytomegalovirus (CMV) promoter, grew the cells at clonal density in a serum-free defined media, and determined the expression of the O4 antigen after 3 d. As shown in Figure 8A, REST overexpression resulted in an increased number of clones containing O4-positive cells. When analyzed on a single clone basis, there were fewer clones with no O4-positive cells (Fig. 8B). The average number of O4-positive cells was higher in the overexpressing clones (32.2 ± 3.2 for the control clones vs 43.2 ± 10.4 for the overexpressing clones, n = 3), although this result was not statistically significant (p < 0.09). Nevertheless, these data show that REST overexpression can promote OPC development to the preoligodendrocyte stage.
Figure 8.
REST overexpression induces O4 expression. A, OPCs were nucleofected with either pMAXeGFP or a mixture of pMAXeGFP and full-length REST and grown for 3 d at clonal densities in defined media, and the percentage of clones containing O4-postive cells determined (n = 3; p < 0.005). B, Three-dimensional scatter plots of a typical experiment. The number of O4-negative clones decreased, and there was a general shift to clones containing higher percentages of O4-positive cells after REST overexpression (REST OE).
Discussion
The RE1 binding protein REST was first discovered on the basis of its ability to repress the expression of neuronal genes in non-neuronal cells (Chong et al., 1995; Schoenherr and Anderson, 1995). In addition to regulating the development of neurons from embryonic stem cells, REST regulates heart and pancreatic development and can function as either a tumor suppressor or an oncogene (Lawinger et al., 2000; Kuwahara et al., 2003; Martin et al., 2003; Ballas et al., 2005; Westbrook et al., 2005). Because REST knock-out mice die by embryonic day 11.5 (Chen et al., 1998), little is known about REST function during the gliogenic stages of neuronal development. The data presented here expand the role of REST by demonstrating that REST represses neuronal gene expression during the differentiation of OPCs into oligodendrocytes. Given the unusual mixture of neuronal and glial properties expressed by OPCs (for review, see Bakiri et al., 2009; Bergles et al., 2010), the repression of neuronal genes may be necessary for the initiation of oligodendrocytic development. REST is also required during the bone morphogenetic protein (BMP)-induced astrocytic differentiation of embryonic neural progenitor cells (Kohyama et al., 2010). Together, these studies demonstrate a general requirement for REST function during gliogenesis and suggest that REST is likely to be an important regulator of glial development, differentiation, and phenotypic plasticity.
The oligodendrocyte lineage is one of the most studied cell-specific lineages in the developing CNS (Nishiyama et al., 2009; Trotter et al., 2010). The maturation of OPCs into myelinating oligodendrocytes requires both gene repression, mediated predominantly by class 1 HDACs (Liu and Casaccia, 2010), and gene activation, mediated by transcription factors such as zfp488, MRF, and Nkx2.2 (Qi et al., 2001; Wang et al., 2006; Emery et al., 2009). MicroRNAs also play a role (Lau et al., 2008; Dugas et al., 2010; Zhao et al., 2010). Here, we focused on the functions of REST at the OPC stage of the oligodendrocyte lineage because deleting HDACs prenatally prevents the development of OPCs (He et al., 2007; Ye et al., 2009). In the absence of REST function, OPCs fail to develop into MBP-positive oligodendrocytes. The reduced levels of CNP mRNA and protein (Fig. 3) and the increase in O4-negative clones (supplemental Fig. S3, available at www.jneurosci.org as supplemental material) suggest that differentiation is arrested before the preoligodendrocyte stage. Conversely, REST overexpression was able to promote OPC development to an O4-positive preoligodendrocyte stage under conditions in which oligodendrocyte differentiation is not especially robust (Fig. 8). Our clonal analysis shows that REST LOF reduced the expression of MBP and promoted the expression of TUJ1 in almost all the cells within small-sized clones, whereas the larger-sized clones were more heterogeneous in their cellular composition (Fig. 7). This suggests that REST function is needed at the time when the cells make the decision to differentiate into oligodendrocytes. Because the frequency of clones in which 100% of the cells were positive for either O4, MBP, or GFAP did not change after REST LOF, some OPCs must have already been committed to differentiate. Because of this previous commitment, REST LOF reduced but did not entirely eliminate oligodendrocyte development.
REST acts as a functional gene repressor in OPCs and is physically associated with several neuronal genes whose expression, initially low in OPCs, is further reduced during differentiation but upregulated after LOF (Table 1) (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). This is consistent with the known functions of REST as a repressor of the neuronal phenotype in non-neuronal cells (Ballas and Mandel, 2005). Although the repression of genes encoding the neuronal phenotype may be necessary for oligodendrocyte differentiation and to prevent the development of cells with mixed phenotypic properties, REST LOF also altered the expression of genes that are not commonly considered targets for REST-mediated regulation (Bruce et al., 2004). Among this group are genes associated with neurogenesis (ASCL1, Myt1L, and Olig2) and gliogenesis (GFAP, CNP, MAG, and MBP). Thus, it is unclear whether the failure to develop into oligodendrocytes is attributable to the misregulated expression of either genes that encode neuronal properties, genes that regulate the decision to develop into a neuron or a glial cell, or both. Nevertheless, our findings are consistent with the notion that REST can regulate a wide program of gene expression in cells of the oligodendrocyte lineage (Abrajano et al., 2009) and further show that this regulation is necessary for oligodendrocyte differentiation.
A striking aspect of REST expression during OPCs differentiation is the fourfold increase in mRNA and protein levels after the initiation of differentiation with thyroid hormones. This increase in REST protein is maintained for at least 45 h. Inducing astrocyte development, either from neural progenitor cells with BMPs (Kohyama et al., 2010) or from OPCs with serum (Fig. 3A), also leads to increases in REST, albeit smaller increases than during oligodendrocyte differentiation. Based on the specifications of the manufacturer, we estimate that the concentration of T3 in serum-containing media is 0.1 nm, or ∼460-fold less than that in the oligodendrocyte-inducing media used here. This suggests that the increase in REST is attributable to the initiation of a program of glial differentiation rather than a general response to thyroid hormone. Consistent with this idea, the effects of T3 on REST transcription varied in different neural stem cell lines. This increase in REST mRNA and protein levels is in contrast to the proteasome-mediated downregulation of REST protein that occurs rapidly during retinoic-acid-induced neural differentiation of embryonic stem cells (Ballas et al., 2005; Westbrook et al., 2008). How this increase in REST expression in differentiating OPCs is regulated is unknown. In cortical neurons, REST is transcriptionally repressed (Ballas et al., 2005), but it can be transcriptionally activated in a neuroblastoma cell line (Jiang et al., 2008). Whether REST is regulated by proteasomal degradation, gene transcription, or both is an area for future study.
Most of the effects of REST on gene transcription result from binding to the 21 bp RE1 element. In addition to the canonical RE1, structurally variant RE1 sites have been identified that differ in their affinity for REST (Bruce et al., 2004, 2009; Otto et al., 2007). These low-affinity REST binding sites are often associated with cell-type-specific and cell-tissue-specific genes (Bruce et al., 2009). The transient increase in nuclear REST protein during the initiation of oligodendrocyte differentiation may be necessary to bind to and repress genes with such variant RE1s. For example, the transcription of hes5, which inhibits oligodendrocyte differentiation (Liu et al., 2006), is downregulated normally during OPC differentiation (Kondo and Raff, 2004) (supplemental Fig. S2, available at www.jneurosci.org as supplemental material), and has an expanded RE1 (Otto et al., 2007), was increased ∼3.5-fold to 4-fold when DnREST-infected cells were differentiated into oligodendrocytes. There were no increases when the cells were differentiated into 2As (Table 1). The changes in REST protein levels demonstrated here may provide a novel mechanism by which REST can differentially modify gene expression during cellular differentiation.
HDAC-mediated changes in chromatin structure are required for oligodendrocyte differentiation, and there has been much progress recently in uncovering the mechanisms involved (Shen and Casaccia-Bonnefil, 2008; Liu and Casaccia, 2010). In general, class1 HDACs are recruited to chromatin and repress the transcription of inhibitors of oligodendrocyte differentiation. For example, the transcription of Id2 and Id4, which inhibit oligodendrocyte differentiation, is activated by Wnt signaling during OPC specification (Ye et al., 2009) and then repressed by YY1–HDAC complexes during differentiation (He et al., 2007). The Notch pathway is also regulated by class I HDACs and in turn regulates oligodendrocyte development in complex ways (Kao et al., 1998; Wang et al., 1998; Genoud et al., 2002; Hu et al., 2003). Hes5, a downstream target of activated notch, inhibits myelin gene expression and mice null for hes5 display precocious myelination (Liu et al., 2006; Zhang et al., 2009). It seems possible therefore that there may be a division of labor during OPC development in which factors such as YY1 repress Id expression after Wnt signaling whereas REST counteracts Notch-mediated inhibition by repressing hes5. A delicate balance between the activation and repression of transcription mediated by these and other factors (Li et al., 2009) may be necessary to insure that the proper complement of oligodendrocytes develops on an appropriate time schedule (Rosenberg and Chan, 2009). Disruption of this timing could be a factor in a wide range of cognitive disorders (Fields, 2008).
Cells with the properties of adult OPCs generate new neurons throughout the life of the organism, but little is known about how this process is regulated (Aguirre and Gallo, 2004; Rivers et al., 2008; Guo et al., 2009, 2010). Our data show that REST can regulate genes associated with neuronal differentiation in developing OPCs, suggesting that REST may regulate OPC plasticity. The low levels of REST present in adult glia may be sufficient to repress neuronal and neurogenic genes but allow for their dynamic regulation perhaps in response to environmental stimuli such as depolarization or injury (Calderone et al., 2003; Ballas et al., 2005). Given the central role of HDACs and chromatin remodeling in regulating the developmental plasticity of OPCs (Kondo and Raff, 2004; Liu et al., 2007; Lyssiotis et al., 2007), it will be important to investigate further the functions of REST in adult glia.
Footnotes
This work was supported by National Institutes of Health Grant NS21198 (J.M.L.) and a Multiple Sclerosis Society Collaborative Research Center Award. We thank Dr. H. Sirotkin for his comments on this manuscript and Drs. N. Ballas and G. Mandel for their gifts of reagents.
References
- Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS One. 2009;4:e7665. doi: 10.1371/journal.pone.0007665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre A, Gallo V. Postnatal neurogenesis and gliogenesis in the olfactory bulb from NG2-expressing progenitors of the subventricular zone. J Neurosci. 2004;24:10530–10541. doi: 10.1523/JNEUROSCI.3572-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakiri Y, Attwell D, Káradóttir R. Electrical signalling properties of oligodendrocyte precursor cells. Neuron Glia Biol. 2009;5:3–11. doi: 10.1017/S1740925X09990202. [DOI] [PubMed] [Google Scholar]
- Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Ballas N, Lioy DT, Grunseich C, Mandel G. Non-cell autonomousinfluence of MeCP2-deficient glia on neuronal dendritic morphology. Nat Neurosci. 2009;12:311–317. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belachew S, Chittajallu R, Aguirre AA, Yuan X, Kirby M, Anderson S, Gallo V. Postnatal NG2 proteoglycan-expressing progenitor cells are intrinsically multipotent and generate functional neurons. J Cell Biol. 2003;161:169–186. doi: 10.1083/jcb.200210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergles DE, Jabs R, Steinhäuser C. Neuron-glia synapses in the brain. Brain Res Rev. 2010;63:130–137. doi: 10.1016/j.brainresrev.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Göttgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AW, López-Contreras AJ, Flicek P, Down TA, Dhami P, Dillon SC, Koch CM, Langford CF, Dunham I, Andrews RM, Vetrie D. Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res. 2009;19:994–1005. doi: 10.1101/gr.089086.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone A, Jover T, Noh KM, Tanaka H, Yokota H, Lin Y, Grooms SY, Regis R, Bennett MV, Zukin RS. Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Liu A. Relationship between cell cycle molecules and onset of oligodendrocyte differentiation. J Neurosci Res. 2003;72:1–11. doi: 10.1002/jnr.10565. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramírez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou CL, Levine JM. Inhibition of neurite growth by the NG2 chondroitin-sulfate proteoglycan. J Neurosci. 1994;14:7616–7628. doi: 10.1523/JNEUROSCI.14-12-07616.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields RD. White matter in learning, cognition and psychiatric disorders. Trends Neurosci. 2008;31:361–370. doi: 10.1016/j.tins.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud S, Lappe-Siefke C, Goebbels S, Radtke F, Aguet M, Scherer SS, Suter U, Nave KA, Mantei N. Notch1 control of oligodendrocyte differentiation in the spinal cord. J Cell Biol. 2002;158:709–718. doi: 10.1083/jcb.200202002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes JA, Nielsen SJ, Battaglioli E, Miska EA, Speh JC, Berry DL, Atouf F, Holdener BC, Mandel G, Kouzarides T. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J Biol Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- Guo F, Ma J, McCauley E, Bannerman P, Pleasure D. Early postnatal proteolipid promoter-expressing progenitors produce multilineage cells in vivo. J Neurosci. 2009;29:7256–7270. doi: 10.1523/JNEUROSCI.5653-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Maeda Y, Ma J, Xu J, Horiuchi M, Miers L, Vaccarino F, Pleasure D. Pyramidal neurons are generated from oligodendroglial progenitor cells in adult piriform cortex. J Neurosci. 2010;30:12036–12049. doi: 10.1523/JNEUROSCI.1360-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Aimone JB, Kaspar BK, Kuwabara T, Nakashima K, Gage FH. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J Cell Biol. 2004;164:111–122. doi: 10.1083/jcb.200308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, Takeda Y, Chia W, Sankar N, Ng YK, Ling EA, Maciag T, Small D, Trifonova R, Kopan R, Okano H, Nakafuku M, Chiba S, Hirai H, Aster JC, et al. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115:163–175. doi: 10.1016/s0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- Immaneni A, Lawinger P, Zhao Z, Lu W, Rastelli L, Morris JH, Majumder S. REST-VP16 activates multiple neuronal differentiation genes in human NT2 cells. Nucleic Acids Res. 2000;28:3403–3410. doi: 10.1093/nar/28.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yao M, Shi J, Shen P, Niu G, Fei J. Yin yang 1 directly regulates the transcription of RE-1 silencing transcription factor. J Neurosci Res. 2008;86:1209–1216. doi: 10.1002/jnr.21595. [DOI] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, Cooper ML, Volta M, Chan SS, Lipovich L, Pollard SM, Karuturi RK, Wei CL, Buckley NJ, Stanton LW. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;3:1007–1014. [PubMed] [Google Scholar]
- Kohyama J, Sanosaka T, Tokunaga A, Takatsuka E, Tsujimura K, Okano H, Nakashima K. BMP-induced REST regulates the establishment and maintenance of astrocytic identity. J Cell Biol. 2010;189:159–170. doi: 10.1083/jcb.200908048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M. Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science. 2000;289:1754–1757. doi: 10.1126/science.289.5485.1754. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 2004;18:2963–2972. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara K, Saito Y, Takano M, Arai Y, Yasuno S, Nakagawa Y, Takahashi N, Adachi Y, Takemura G, Horie M, Miyamoto Y, Morisaki T, Kuratomi S, Noma A, Fujiwara H, Yoshimasa Y, Kinoshita H, Kawakami R, Kishimoto I, Nakanishi M, et al. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 2003;22:6310–6321. doi: 10.1093/emboj/cdg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawinger P, Venugopal R, Guo ZS, Immaneni A, Sengupta D, Lu W, Rastelli L, Marin Dias Carneiro A, Levin V, Fuller GN, Echelard Y, Majumder S. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat Med. 2000;6:826–831. doi: 10.1038/77565. [DOI] [PubMed] [Google Scholar]
- Levine JM. Neuronal influences on glial progenitor cell development. Neuron. 1989;3:103–113. doi: 10.1016/0896-6273(89)90119-0. [DOI] [PubMed] [Google Scholar]
- Levine JM, Stallcup WB. Plasticity of developing cerebellar cells in vitro studied with antibodies against the NG2 antigen. J Neurosci. 1987;7:2721–2731. doi: 10.1523/JNEUROSCI.07-09-02721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, He Y, Richardson WD, Casaccia P. Two-tier transcriptional control of oligodendrocyte differentiation. Curr Opin Neurobiol. 2009;19:479–485. doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Han YR, Li J, Sun D, Ouyang M, Plummer MR, Casaccia-Bonnefil P. The glial or neuronal fate choice of oligodendrocyte progenitors is modulated by their ability to acquire an epigenetic memory. J Neurosci. 2007;27:7339–7343. doi: 10.1523/JNEUROSCI.1226-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Lyssiotis CA, Walker J, Wu C, Kondo T, Schultz PG, Wu X. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proc Natl Acad Sci U S A. 2007;104:14982–14987. doi: 10.1073/pnas.0707044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Tawadros T, Meylan L, Abderrahmani A, Condorelli DF, Waeber G, Haefliger JA. Critical role of the transcriptional repressor neuron-restrictive silencer factor in the specific control of connexin36 in insulin-producing cell lines. J Biol Chem. 2003;278:53082–53089. doi: 10.1074/jbc.M306861200. [DOI] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Mulligan P, Westbrook TF, Ottinger M, Pavlova N, Chang B, Macia E, Shi YJ, Barretina J, Liu J, Howley PM, Elledge SJ, Shi Y. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol Cell. 2008;32:718–726. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes(NG2 cells) multifunctional cells with loineage plasticity. Nat Rev Neurosci. 2009;10:9–22. doi: 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- Otto SJ, McCorkle SR, Hover J, Conaco C, Han JJ, Impey S, Yochum GS, Dunn JJ, Goodman RH, Mandel G. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J Neurosci. 2007;27:6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303:390–396. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci. 2008;11:1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SS, Chan JR. Modulating myelination: knowing when to say Wnt. Genes Dev. 2009;23:1487–1493. doi: 10.1101/gad.1824009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Shen S, Casaccia-Bonnefil P. Post-translational modifications of nucleosomal histones in oligodendrocyte lineage cells in development and disease. J Mol Neurosci. 2008;35:13–22. doi: 10.1007/s12031-007-9014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Kameoka S, Lentz S, Majumder S. Activation of REST/NRSF target genes in neural stem cells is sufficient to cause neuronal differentiation. Mol Cell Biol. 2004;24:8018–8025. doi: 10.1128/MCB.24.18.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AM, Colletti M, Rorai AT, Skene JH, Levine JM. Antibodies against the NG2 proteoglycan promote the regeneration of sensory axons within the dorsal columns of the spinal cord. J Neurosci. 2006;26:4729–4739. doi: 10.1523/JNEUROSCI.3900-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter J, Karram K, Nishiyama A. NG2 cells: properties, progeny and origin. Brain Res Rev. 2010;63:72–82. doi: 10.1016/j.brainresrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Dulin J, Wu H, Hurlock E, Lee SE, Jansson K, Lu QR. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133:3389–3398. doi: 10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, Depinho RA, Chin L, Elledge SJ. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, Elledge SJ. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Watanabe M, Nishiyama A. Optimization of oligodendrocyte progenitor cell culture method for enhanced survival. J Neurosci Methods. 2005;149:50–56. doi: 10.1016/j.jneumeth.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, Lu QR. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Argaw AT, Gurfein BT, Zameer A, Snyder BJ, Ge C, Lu QR, Rowitch DH, Raine CS, Brosnan CF, John GR. Notch1 signaling plays a role in regulating precursor differentiation during CNS remyelination. Proc Natl Acad Sci U S A. 2009;106:19162–19167. doi: 10.1073/pnas.0902834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, Wang F, Appel B, Lu QR. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135:145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]