Abstract
Ducks can survive infection with highly pathogenic avian influenza viruses that are lethal to chickens. We showed that the influenza detector, RIG-I, can initiate antiviral responses in ducks, but this gene is absent in chickens. We can reconstitute this pathway by transfecting chicken DF-1 embryonic fibroblast cells with duck RIG-I, which augments their antiviral response to influenza and decreases viral titre. However, the genes downstream of duck RIG-I that mediate this antiviral response to influenza are not known. Using microarrays, we compared the transcriptional profile of chicken embryonic fibroblasts transfected with duck RIG-I or empty vector, and infected with low or highly pathogenic avian influenza viruses. Transfected duck RIG-I expressed in chicken cells was associated with the marked induction of many antiviral innate immune genes upon infection with both viruses. We used real-time PCR to confirm upregulation of a subset of these antiviral genes including MX1, PKR, IFIT5, OASL, IFNB, and downregulation of the influenza matrix gene. These results provide some insight into the genes induced by duck RIG-I upon influenza infection, and provide evidence that duck RIG-I can function to elicit an interferon-driven, antiviral response against influenza in chicken embryonic fibroblasts.
1. Introduction
Influenza A viruses cause largely asymptomatic infection in ducks, the natural reservoir (Webster et al., 1992). Most strains replicate in the duck, including 16 hemagglutinin (HA) subtypes and 9 neuraminidase (NA) subtypes (Webster et al., 1992). Because there is seldom disease in ducks, the classification of influenza as highly or low pathogenic refers to infections in chickens (Suarez and Schultz-Cherry, 2000). Highly pathogenic avian influenza can be deadly in poultry in 1–3 days, while low pathogenic influenza causes only mild disease signs (Pantin-Jackwood and Swayne, 2009). Influenza strains of H5 and H7 HA subtypes can replicate in chickens, and can evolve from low to highly pathogenic avian influenza (HPAI) (Pasick et al., 2005; Swayne and Suarez, 2000). Since these changes typically happen under immune pressure in alternate hosts, chickens are an important intermediate in the genesis of novel influenza strains.
Circulating H5N1 HPAI strains remain of concern because of their pronounced pathogenicity and transmissibility among chickens, as well as for their ability to occasionally infect humans. Of the 600 human cases in the last 10 years, mortality exceeds 50% http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_arch ives/en/. Because these individuals were infected from direct contact with birds, controlling H5N1 strains in poultry is key to prevention of human infection. Recently it was demonstrated that as few as 4 substitutions in H5 from A/Vietnam1203/2004 (H5N1), together with reassortment, can generate a mammalian transmissible virus (Imai et al., 2012). Also, given the fact that some strains circulating in nature already have some of these changes, the possibility exists that a respiratory droplet transmissible strain could evolve in a mammalian host (Russell et al., 2012), as experimentally demonstrated by passaging of A/Indonesia5/2005 (H5N1) in ferrets (Herfst et al., 2012). These recent papers highlight the possibility of a pandemic virus evolving in nature.
The highly pathogenic H5N1 strains are universally lethal to chickens but only certain strains can kill ducks. Typically, ducks are able to limit H5N1-induced pathology (Sturm-Ramirez et al., 2005; Swayne and Pantin-Jackwood, 2006). Because of the acute onset of the infection, we predict that innate immune mechanisms are crucial in this response. The rapid detection of viruses by pattern recognition receptors initiates signalling cascades leading to production of type I interferons (IFN-α/β). Toll-like receptor 7 (TLR7) and retinoic acid-like receptor-I (RIG-I) are the main detectors for influenza viruses. While TLR7 is responsible for IFN-α production by leukocytes, cytosolic detection in the cells initially infected by influenza involves RIG-I. The cells initially infected with H5N1 strains are respiratory tract epithelial cells (Swayne and Pantin-Jackwood, 2006) (Swayne, 2007). We have previously shown that ducks have an intact and functional RIG-I, while chickens appear to lack the gene for the receptor (Barber et al., 2010). Chickens do have the MDA5 receptor, which uses the same signalling pathway as RIG-I, downstream of IPS-1/MAVS/CARDIF. Chicken DF-1 cells cannot respond to the RIG-I ligand, 5' triphosphate RNA (Barber et al., 2010; Karpala et al., 2011b), but transferring the duck receptor to chicken cells reconstitutes recognition (Barber et al., 2010). Chickens do respond to influenza, and cells can respond to in vitro transcribed-RNA in a 5' triphophosphate independent manner, involving MDA5 (Liniger et al., 2012). Thus the pathway downstream of IPS-1 is intact in chickens. Using duck RIG-I and the signalling downstream of MDA5, we showed that induction of the immune genes MX1, IFNB and PKR upon influenza infection was significantly higher in chicken DF-1 cells transfected with duck RIG-I (Barber et al., 2010). Additionally, the presence of duck RIG-I in DF-1 cells reduces the titre of both A/British Columbia 500/2005 (H5N2) and A/Vietnam 1203/2004 (H5N1) influenza strains (Barber et al., 2010). However, the full complement of genes that contribute to this antiviral effect are not known.
Our reconstitution of the RIG-I pathway in chicken cells allows us to exploit the genomic resources available for chickens to identify the avian genes that are altered downstream of RIG-I upon influenza infection. We used 44k Agilent chicken microarrays to compare RIG-I or empty-vector transfectants in a chicken embryonic fibroblast (DF-1) cell line following infection with BC500 or VN1203 viruses. These microarrays showed significantly increased transcripts of innate antiviral genes in chicken cells carrying duck RIG-I, as compared to empty vector following influenza infection. Thus, duck RIG-I is functional in chicken cells and elicits a robust immune response to influenza. The global transcriptional profile reveals important mediators of the RIG-I dependent antiviral response.
2. Methods
2.1. Cloning, transfections and infections
UMNSAH/DF-1 cells, a spontaneously immortalized chicken embryonic fibroblast cell line derived from East Lansing strain eggs (Schaefer-Klein et al., 1998) was obtained from ATCC, and maintained in DMEM supplemented with 10% FBS. Cloning of duck RIG-I into pcDNA3.1, cell transfections and infections were performed as previously described (Barber et al., 2010). Briefly, DF-1 cells were seeded overnight at 1.25 × 105 per well in 24-well plates. Cells were transfected with 1 µg of pcDNA-RIG or empty pcDNA3.1. Twenty-four hours after transfection, cells were infected at a multiplicity of infection (MOI) of 1 with A/mallard/BC/500/05 (H5N2) or reverse genetics A/Vietnam/1203/04 (H5N1) virus. L-(tosylamido-2-phenyl) ethyl chloromethyl ketone–treated trypsin (Worthington Biochemicals) was used for infection with BC500 at a low concentration (0.1 µg/mL). Exogenous trypsin was not added for infection with VN1203. Fifteen hours after infection, RNA was extracted from cells for quantitative RT-PCR (qRT-PCR) and microarrays. This timepoint was selected to identify downstream effectors of the interferon response. Control transfections carried out with 0.5 µg pDsRed-N1 consistently yielded transfection efficiencies of ~ 40–60% for DF-1 cells. Infection with BC500 and VN1203 at an MOI of 1 is expected to infect all cells, and most cells appeared infected by cytopathic effect. We previously observed signalling through RIG-I to our IFNβ luciferase reporter, and a decrease in viral titre in presence of RIG-I by 15 hours post-infection (Barber et al., 2010) in the supernatant of the same experimental samples used for RNA extraction. Influenza virus titer was determined by plaque assay on MDCK cells. Viral titre for BC500 was 5 × 105 in RIG-I transfected cells, and 1.5 × 106 in empty-vector transfected cells, while VN1203 replicated to 2×106 in DF-1 cells transfected with empty vector, or 1×106 in RIG-I transfected cells (Barber et al., 2010). VN1203 was handled in biosafety level 3+ facilities approved by the United States Department of Agriculture and Center for Disease Control and Prevention.
2.2 Microarray hybridization and analysis
Chicken microarrays from Agilent-015068, Chicken Gene Expression 4×44K (Agilent, Santa Clara, CA) consisting of 42,034 60-mer in situ synthesized oligonucleotides were used. Labelled cRNA was prepared from 500 ng of total RNA using the Agilent labelling protocol, and microarray hybridization was performed at 65°C for 18 hours in Agilent's microarray hybridization chambers, followed by wash procedures according to the manufacturer’s recommended protocols. Microarrays were carried out using the reference design, whereby RNA from each experimental sample is compared to a single reference RNA sample. Our reference RNA sample was prepared from uninfected, untransfected DF-1 cells. The testing samples (Cy5) were co-hybridized with uninfected, untransfected reference DF-1 samples (Cy3). Three independent experiments were performed with duck RIG-I transfected cells along with empty vector transfected cells, followed by BC500 infection; while two replicate experiments of transfections followed by VN1203 infection were performed. The microarrays were scanned in an Agilent scanner at 3 µm resolution, and the array data was extracted with Agilent feature extraction software (version 10.5.1.1) using the GE2_105_Jan09 protocol. Reproducibility and reliability of each single microarray was assessed using Quality Control report data. Gene expression ratios compared with the reference control (uninfected, untransfected DF-1 cells) were calculated and log2 transformed. Lowess normalization on background-subtracted signal intensity was performed to correct the intensity bias, and additional filtering steps, including valid measurement in at least 50% of the tested samples, were applied to further improve the quality of the data set. The relative differential gene expression between RIG-I transfected and empty vector transfected samples was reported as the difference of log2 ratios between these two groups, and a p-value using t-test was calculated for each gene as statistical measurement of the replicates. The selection of the genes for Gene Ontology term analysis (DAVID, http://david.abcc.ncifcrf.gov (Huang et al., 2009) was based on a change of at least twofold and a probability of >95% (P < 0.05) of differential expression in at least two experiments. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE29596 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSEGSE29596).
2.3 Analysis of immune gene expression by quantitative RT-PCR (qRT-PCR)
RNA was extracted using TRIzol (Invitrogen), followed by purification from the final aqueous phase using the RNeasy Mini Kit (Qiagen). RNA was DNAse treated and first-strand cDNA synthesized using a mix of random primers (Invitrogen) and Oligo(dT)20 primer (50 µM) (Invitrogen) using the SuperScript III reverse transcriptase kit (Invitrogen). Gene-specific primers and probe sets (Table 1) were designed using the Primer Express Version 3.0 software and IDT real-time primer design program Sirtool. Primers were obtained from Integrated DNA Technologies and validated for linear amplification, and relative amplification efficiency matched to the GAPDH endogenous control. All qRT-PCR assays were performed using Fast Start TaqMan Probe Master Mix (Roche) in the Applied Biosystems 7500 real-time PCR System machine (Applied Biosystems). All experimental samples were assayed in triplicate. Target gene expression was normalized to a constitutively expressed endogenous control gene GAPDH. Quantification was carried out using relative quantitation of gene expression (ΔΔCT) and the analysis was performed with the 7500 Fast System software version 1.4 (Applied Biosystems).
Table 1.
Primer and probe sequences for qRT-PCR.
| Gene | Primer and Probe Sequence (5’→3’) |
|---|---|
| InfA M | Primer 1: GACCRATCCTGTCACCTCTGAC |
| Primer 2: AGGGCATTYTGGACAAAKCGTCTA | |
| Probe:/56-FAM/ TGCAGTCCT/Zen/CGCTCACTGGGCACG/3IABkFQ | |
| IFIT5 | Primer 1: CAGAATTTAATGCCGGCTATGC |
| Primer 2: TGCAAGTAAAGCCAAAAGATAAGTGT | |
| Probe:/56- FAM-TCTGAAGCG/Zen/TGCACTGAAACTGAATCCAA-3IABkFQ | |
| MX1 | Primer 1: GGACTTCTGCAACGAATTG |
| Primer 2: TCCCACAAGTTCATCTGTAAG | |
| Probe:/56-FAM/ CTTCACCTC/Zen/CGCAATCCAGCAAGA/3IABkFQ | |
| OASL | Primer 1:ACATCCTCGCCATCATCGA |
| Primer 2: GCGGACTGGTGATGCTGACT | |
| Probe:/56-FAM/ AGTGCCTCC/Zen/CGACGCTGTCCTTC/3IABkFQ | |
| PKR | Primer 1: CATTGAGGCACATTTCACAGATTATA |
| Primer 2: GGTCAATCAGTTCATACCTTATGTTGA | |
| Probe:/56-FAM/ CTGAACACC/Zen/TCTGCTGGCCTTACTGTCA/3IABkFQ | |
| IFNB | Primer 1: TCCAACACCTCTTCAACATGCT |
| Primer 2: TGGCGTGTGCGGTCAAT | |
| Probe:/56-FAM/AGCAGCCCA/Zen/CACACTCCAAAACACTG/3IABkFQ | |
| GAPDH | Primer 1:GGTGCTAAGCGTGTTATCATCTCA |
| Primer 2: CATGGTTGACACCCATCACAA | |
| Probe:/56-FAM/ CTCCCTCAG/Zen/CTGATGCCCCCATG/3IABkFQ |
3. Results
3.1. Chicken cells expressing duck RIG-I induce immune genes in response to BC500
To define the role of RIG-I in antiviral gene expression during influenza infections, we conducted microarray analyses. Using chicken DF-1 embryonic fibroblast cells, which lack RIG-I, we compared cells transfected with duck RIG-I or empty vector at 15 hours post infection (hpi) with a low pathogenic avian influenza A/mallard/British Columbia/500/2005 (H5N2) (BC500) at a multiplicity of infection (MOI) of 1. Our data show increased expression of a number of genes in the presence of RIG-I, compared to empty vector, following infection with BC500. Of genes that were altered in expression by more than twofold in the presence of RIG-I, the fold-change of 206 of them was statistically significant (P < 0.05) across three independent experiments.
Our results show that genes with altered expression in chicken cells expressing duck RIG-I upon infection with BC500 group predominantly within host defense and immune response Gene Ontology (GO) categories (Table 2). Several immune genes appear in multiple hierarchical GO classes. Of the 206 differentially expressed genes (with differences greater than twofold), 21 had known roles in the immune response, with the fold-change of 6 of them being statistically significant across replicates (Table 3). These genes are particularly involved in the innate immune defences to influenza.
Table 2.
Categories of upregulated genes in RIG-I transfected compared to empty vector transfected DF-1 cells, 15 hpi with BC500.
| Gene Ontology Term | P-value | Genes |
|---|---|---|
| GO:0006955~immune response | 1.05E-04 | OASL, TNFSF10, CCL20, TLR15, IRF7, IRF8, TNFSF15, CCL19, TLR3 |
| GO:0006952~defense response | 0.0799 | NFKBIZ, TLR15, TLR3, GAL12 |
| GO:0042107~cytokine metabolic process | 0.0851 | IRF7, TNFSF15 |
Table 3.
A selection of immune genes upregulated more than twofold in RIG-I transfected DF-1 cells, 15 hpi with BC500.
| Gene Name | Accession Number |
Fold Change |
P-value | |
|---|---|---|---|---|
| CCL19 | chemokine (C-C motif) ligand 19 | BX929857 | 9.25 | 0.0899 |
| CCL20 | chemokine (C-C motif) ligand 20 | AB101005 | 3.34 | 0.398 |
| DHX58 (LGP2) | probable ATP-dependent RNA helicase DHX58 | AM070728 | 3.67 | 0.0724 |
| EIF2AK2 (PKR) | eukaryotic translation initiation factor 2-alpha kinase 2 | AB125660 | 3.22 | 0.0269 |
| GAL12 | beta-defensin 12 | AY534898 | 2.48 | 0.0678 |
| IFIH1 (MDA5) | interferon-induced helicase C domain-containing protein 1 | CR385175 | 7.33 | 0.0461 |
| IFIT5 | interferon-induced protein with tetratricopeptide repeats 5 | XM_421662 | 27.5 | 0.0200 |
| IFITM5 | interferon induced transmembrane protein 5 | XM_420924 | 2.31 | 0.0686 |
| IRF1 | interferon regulatory factor 1 | L39766 | 3.98 | 0.0364 |
| IRF7 | interferon regulatory factor 7 | U20338 | 2.81 | 0.264 |
| IRF8 | interferon regulatory factor 8 | L39767 | 2.15 | 0.0805 |
| ISG12-2 | interferon stimulated gene 12-2 protein-like | NM_001001296 | 8.23 | 0.0660 |
| MX1 | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 | AB088533 | 32.6 | 0.0096 |
| NFKBIZ | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta | AJ721113 | 2.36 | 0.406 |
| OASL | 2'–5'-oligoadenylate synthetase-like | NM_205041 | 3.81 | 0.0563 |
| TLR15 | toll-like receptor 15 | DQ267901 | 2.76 | 0.557 |
| TLR3 | toll-like receptor 3 | AY633575 | 2.37 | 0.457 |
| TNFSF10 | tumor necrosis factor (ligand) superfamily, member 10 | AJ720191 | 2.16 | 0.507 |
| TNFSF15 | tumor necrosis factor (ligand) superfamily, member 15 | BX930081 | 2.79 | 0.101 |
| ZC3HAV1 | zinc finger CCCH-type, antiviral 1 | CR524013 | 3.73 | 0.0910 |
| ZNFX1 | zinc finger, NFX1-type containing 1 | XM_417395 | 7.96 | 0.0448 |
Genes with statistically significant (P < 0.05) fold-differences across three replicates are indicated in bold.
3.2. Chicken cells expressing duck RIG-I induce immune genes in response to VN1203
To determine the role of RIG-I in detection and downstream gene expression in response to a highly pathogenic avian influenza, we compared DF-1 cells transfected with duck RIG-I or empty vector, 15 hours post-infection with the highly pathogenic avian influenza A/Vietnam/1203/2004 (H5N1). RNA from two independent experiments were used for microarray hybridization and compared to the RNA reference sample from uninfected, untransfected DF-1 cells. Of the RIG-I responsive genes altered more than twofold by VN1203 infection, 377 genes had fold changes that were statistically significant (P value of < 0.05) across both replicates.
RIG-I expression and detection of influenza induced the expression of a wide array of genes in different functional groups, including those associated with transcriptional and translational regulation, and the immune response (Table 4). Twenty-eight genes were identified as immune genes and their expression was augmented by the presence of RIG-I, with fold-change of 21 of these genes being statistically significant between biological replicates (Table 5). The presence of RIG-I augmented the expression of several key innate immune genes with known or potential antiviral roles in influenza defense.
Table 4.
Categories of upregulated genes in RIG-I transfected compared to empty vector transfected DF-1 cells, 15 hpi with VN1203.
| Gene Ontology Term | P-value | Genes |
|---|---|---|
| GO:0009615~response to virus | 2.14E-05 | IFNB, MYD88, IRF7, IFNG, TLR3 |
| GO:0006955~immune response | 1.90E-04 | OASL, MYD88, CCL20, IRF7, IRF8, IFNG, ZAP70, CCL19, TLR3, IL10 |
| GO:0005125~cytokine activity | 5.48E-04 | IFNB, CCL20, IFNG, CCL19, IL10, THPO |
| GO:0003950~NAD+ ADP-ribosyltransferase activity | 8.11E-04 | PARP9, PARP12, ZC3HAV1, PARP14 |
| GO:0045080~positive regulation of chemokine biosynthetic process | 0.00131 | MYD88, IFNG, TLR3 |
| GO:0006952~defense response | 0.00218 | IFNB, LSP1, LIPA, MYD88, IFNG, TLR3, IL10 |
| GO:0001816~cytokine production | 0.0233 | LIPA, MYD88, IRF7 |
| GO:0045348~positive regulation of MHC class II biosynthetic process | 0.0298 | IFNG, IL10 |
Table 5.
A selection of immune genes upregulated more than twofold in RIG-I transfected DF-1 cells 15 hpi with VN1203 HPAI.
| Gene Name | Accession Number |
Fold Change |
P-value | |
|---|---|---|---|---|
| CCL19 | chemokine (C-C motif) ligand 19 | BX929857 | 7.48 | 0.0201 |
| CCL20 | chemokine (C-C motif) ligand 20 | AB101005 | 4.33 | 0.00970 |
| EIF2AK2 (PKR) | eukaryotic translation initiation factor 2-alpha kinase 2 | AB125660 | 2.44 | 0.0698 |
| EPAS1 | endothelial PAS domain protein 1 | AF129813 | 2.88 | 0.376 |
| GDNF | glial cell derived neurotrophic factor | AF176017 | 2.20 | 0.378 |
| IFIH1 (MDA5) | interferon-induced helicase C domain-containing protein 1 | CR385175 | 18.7 | 0.0159 |
| IFIT5 | interferon-induced protein with tetratricopeptide repeats 5 | XM_421662 | 13.8 | 0.0138 |
| IFITM5 | interferon induced transmembrane protein 5 | XM_420924 | 4.39 | 0.0456 |
| IFNB | interferon beta | AY831397 | 4.21 | 0.00862 |
| IFNG | interferon, gamma | AY705909 | 5.66 | 0.0309 |
| IL10 | interleukin 10 | AJ621254 | 2.38 | 0.245 |
| IRF1 | interferon regulatory factor 1 | L39766 | 2.91 | 0.0474 |
| IRF7 | interferon regulatory factor 7 | U20338 | 3.12 | 0.0314 |
| IRF8 | interferon regulatory factor 8 | L39767 | 3.70 | 0.00949 |
| IRG1 | immunoresponsive 1 homolog (mouse) | AJ720739 | 5.44 | 0.00689 |
| ISG12-2 | ISG12-2 protein-like | NM_001001296 | 14.8 | 0.00209 |
| LIPA | lipase A, lysosomal acid, cholesterol esterase | XM_421661 | 2.73 | 0.0140 |
| LSP1 | lymphocyte-specific protein 1 | AB101641 | 4.36 | 0.171 |
| MAFB | v-maf musculoaponeurotic fibrosarcoma oncogene homolog B | NM_001030852 | 2.81 | 0.0282 |
| MX1 | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 | AB088533 | 20.8 | 0.0209 |
| MYD88 | myeloid differentiation primary response gene (88) | CR389938 | 2.34 | 0.00948 |
| OASL | 2'–5'-oligoadenylate synthetase-like | NM_205041 | 4.43 | 0.0226 |
| THPO | thrombopoietin | AY613434 | 2.06 | 0.124 |
| TLR3 | toll-like receptor 3 | AY633575 | 3.23 | 0.0137 |
| TLX1 | t-cell leukemia homeobox 1 | AF071874 | 2.44 | 0.0156 |
| ZAP70 | zeta-chain-associated protein kinase 70 | XM_418206 | 2.11 | 0.120 |
| ZC3HAV1 | zinc finger CCCH-type, antiviral 1 | CR524013 | 6.43 | 0.0113 |
| ZNFX1 | zinc finger, NFX1-type containing 1 | XM_417395 | 4.51 | 0.0145 |
Genes with fold-changes that are statistically significant (P < 0.05) across two replicates are indicated in bold.
3.3. Innate immune genes show increased expression in chicken cells expressing duck RIG-I
Many immune genes were induced by infection with BC500 and VN1203 and their expression is increased in chicken cells expressing duck RIG-I, relative to empty vector (Figure 1). A heatmap of selected immune genes graphically shows their upregulation in the transfected DF-1 cells carrying RIG-I, compared to empty vector. Of these, several have known roles in influenza defense, including IFIT, MX1, OASL and IFIH1 (MDA5). Some components of the signalling pathway downstream of RIG-I are also upregulated, including IRF7 and IFNB. All of the differentially expressed immune genes were upregulated by RIG-I, consistent with the role of the gene in eliciting a Type I interferon response upon influenza infection. MyD88 was down regulated in all cells, relative to the untransfected, uninfected DF-1 cells.
Figure 1. Upregulation of RIG-I-responsive innate immune genes in influenza infected chicken cells expressing duck RIG-I.
A heat map shows the differential expression of RIG-I-responsive immune genes following influenza infection. Chicken embryonic fibroblasts (DF-1 cells) were transfected with duck RIG-I or empty vector. 24 hours post-transfection, the DF-1 cells were infected with A/mallard/BC/500/2005 (H5N2) (BC500) or A/Vietnam/2004 (H5N1) (VN1203) at a multiplicity of infection (MOI) of 1. At 15 hpi, cellular RNA was extracted for microarray analysis. Micorarray hybridizations were normalized to a reference RNA sample (untransfected, uninfected DF-1 cell RNA). Heatmap is generated from three independent experiments of RIG-I (6 arrays) and empty vector for BC500, or two independent experiments VN1203 (4 arrays).
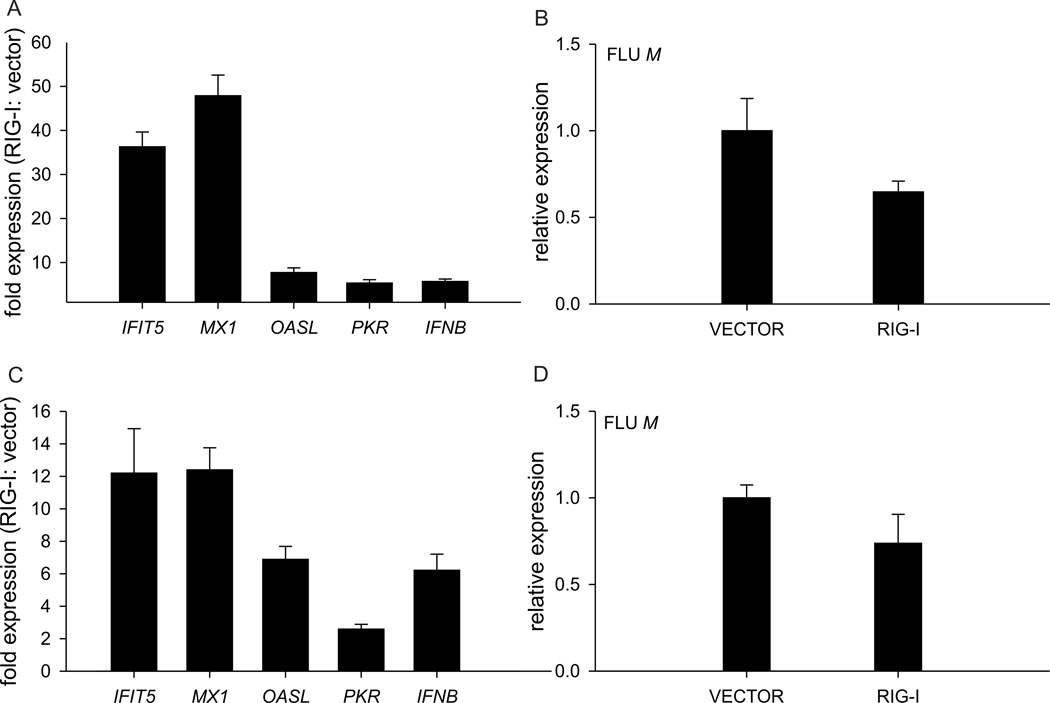
To validate the fold-increases indicated by the microarray analyses, we used qRT-PCR to examine the expression of key innate immune genes including, IFIT5, MX1, OASL, PKR, IFNB and influenza matrix gene (Figure 2). By qRT-PCR, the presence of RIG-I augmented the expression of IFNB following infection with either BC500 and VN1203, by 5-fold and 6-fold respectively. All ISGs showed increased expression in the RIG-I transfected cells, compared to the cells carrying empty vector. IFIT5 was 36-fold or 12-fold higher, while MX1 was 48-fold or 12.4 fold higher in BC500 and VN1203 infected cells, respectively. The presence of RIG-I decreased the influenza matrix gene transcript, consistent with the decrease in viral replication we previously measured by plaque assay. Thus the qRT-PCR results were consistent with the fold-changes observed by microarray.
Figure 2. Upregulation of key innate immune genes in influenza infected chicken cells expressing duck RIG-I.
RIG-I–transfected DF-1 cells respond to BC500 infection (MOI, 1) (A) or VN1203 infection (C) with increased expression of chicken IFNβ and the IFN-stimulated genes IFIT, MX1, OASL and PKR, and decreased influenza matrix gene expression (B and D), compared to empty vector transfected cells. RNA was extracted from cells for qRT-PCR 15 hpi, and fold difference in gene expression calculated for RIG-I and empty-vector transfected DF-1 cells. A representative of three independent experiments (BC500) or two independent experiments (VN1203) is shown and error bars show RQMin/Max at a 95% confidence level.
4. Discussion
Through a microarray approach, we have characterized the transcriptional response induced by influenza in chicken cells in the absence or presence of duck RIG-I. We demonstrate that the induction of key innate immune genes by both BC500 and VN1203 was augmented by the presence of duck RIG-I in chicken cells. Many of the immune genes that were induced in cells carrying duck RIG-I correspond to those reported as RIG-I responsive in a microarray comparison of wildtype and RIG-I−/− mouse fibroblasts, including IFNB, IRF1, IRF7, MDA5, IFIT and MX1 (Loo et al., 2008). In the mouse, these genes were defined as the RIG-I bioset, i.e. genes that were induced by influenza infection that were dependent on RIG-I, and expression was absent in RIG-I−/− mouse embryonic fibroblasts. RIG-I stimulates the production of IFNβ, which is essential for protection against influenza (Koerner et al., 2007), because it orchestrates the production of antiviral interferon stimulated genes (ISGs). Thus, duck RIG-I regulates a similar subset of genes in the pathway, as well as downstream effectors. This defines a set of candidate avian antiviral effectors, which can be mined for those with direct antiviral function. We expect that some of these effectors are responsible for our previous observation that duck RIG-I reduces influenza virus titre in chicken DF-1 cells (Barber et al., 2010).
Signalling through duck RIG-I, like mouse RIG-I, also upregulates the genes in its own pathway. Both IRF7 and IFNB are induced by influenza infection in chicken cells transfected with duck RIG-I. In chickens, the gene named IRF3 more closely resembles IRF7, and like IRF7 is inducible by viral infection and poly (I-C) (Grant et al., 1995). IRF3 appears to have been selectively lost in avian species (Cormican et al., 2009). It is not known whether IRF7 replaces IRF3 in the signalling pathway downstream of RIG-I, i.e. whether signalling leads to nuclear translocation of chicken IRF7 to drive expression of IFNβ, and the reagents to address this question experimentally in chicken cells do not exist. However, the demonstration of augmentation of expression of downstream ISGs in chicken cells expressing RIG-I implies that this signalling pathway is intact.
DF-1 cells expressing duck RIG-I demonstrated a 33-fold increase in Mx1 expression in response to infection with BC500, and 21-fold by VN1203. The interferon-inducible Mx proteins confer antiviral function in transfected cells and transgenic animals (Engelhardt et al., 2004; Pavlovic et al., 1995; Pavlovic et al., 1990). In mice, Mx1 is interferon-induced and protects against lethal infection by the 1918 pandemic H1N1 strain and VN1203 (Tumpey et al., 2007). Chickens have a single, polymorphic, interferon-stimulated Mx1 gene (Schumacher et al., 1994). Although an original report suggested chicken Mx lacked antiviral activity (Bernasconi et al., 1995), allelic variants have been identified that have antiviral activity (Ko et al., 2002; Ko et al., 2004b) and recent data showed antiviral effects of some alleles against two different HPAI strains (Ewald et al., 2011). However, siRNA knockdown of Mx in chicken embryonic fibroblasts carrying either allelic variant does not impair the antiviral response to low or highly pathogenic avian influenza, and both variants were shown to lack GTPase activity, which may preclude antiviral function (Schusser et al., 2011).
Protein kinase R (PKR) (EIF2AK2) was induced in chicken cells transfected with duck RIG-I after both BC500 and VN1203 infection, at a P value < 0.05 for the BC500 but not VN1203 infection. Chicken PKR is a polymorphic protein with demonstrated antiviral function against VSV infection (Ko et al., 2004a). PKR is required for IFNα/β induction in response to several viruses, but not influenza (Schulz et al., 2010). Recent data shows a role for PKR in the formation of antiviral stress granules involved in orchestration of RIG-I and signalling components, and is inhibited by NS1 (Khaperskyy et al., ; Onomoto et al., 2012). The function of avian PKR is unknown.
The gene encoding OAS-like protein (OASL) was induced approximately 4-fold by both viruses in the presence of duck RIG-I, compared to empty vector transfected cells. Chicken OASL, encodes a domain typical of 2′–5′ oligoadenylate synthase proteins and also two ubiquitin-like (UbL) domains (Tatsumi et al., 2000; Tatsumi et al., 2003). While human OASL lacks oligoadenylate synthetase (OAS) activity, the UbL domains are necessary for antiviral function (Marques et al., 2008). Human OASL inhibits replication of RNA viruses (Marques et al., 2008) (Ishibashi et al., 2010), and is upregulated by influenza through IRF3 (Melchjorsen et al., 2009). OASL was upregulated by influenza infection in chicken lung tissues, and recovered in a screen to identify genes involved in survival of chickens to lethal influenza strains (Uchida et al., 2012).
In both BC500 and VN1203 infections, interferon-stimulated genes were significantly induced. IFIT5, a member of the IFN-induced proteins with tetratricopeptide repeats (IFIT) family, was induced in cells carrying duck RIG-I by 28-fold upon infection with BC500 or 14-fold by VN1203. IFIT proteins sequester viral RNA transcripts in a multiprotein complex (Pichlmair et al., 2011). ISG12-2 was induced in RIG-I transfected compared to empty-vector transfected DF-1 cells by 8-fold in response to BC500 and 15-fold by VN1203. ISG12-2 is induced in chicken lung and tracheal organ culture after influenza infection and is predicted to have antiviral function through an unknown mechanism (Reemers et al., 2009; Reemers et al., 2010). Human ISG12a has been implicated in sensitizing cells for apoptosis by mitochondrial membrane destabilization (Rosebeck and Leaman, 2008). Apoptotic cell death has been proposed as a mechanism to limit influenza spread, and this effect is much greater in duck than chicken cells (Kuchipudi et al., 2011). RIG-I may be inducing apoptosis upon influenza infection through the pronounced expression of ISG12-2.
CCL19 expression is interferon-dependent (Pietila et al., 2007) and was induced by 9-fold by BC500 and 7-fold by VN1203 infection in chicken cells carrying duck RIG-I compared to empty vector, while CCL20 was induced by 3 to 4-fold. Both CCL19 (Forster et al., 2008; Marsland et al., 2005) and CCL20 (Le Borgne et al., 2006) are involved in recruitment of dendritic cells and naïve lymphocytes to initiate adaptive immune responses. CCL19 was upregulated in VN1203 infected duck lung tissues (Fleming-Canepa et al., 2011).
Interestingly, many of the immune genes that were significantly induced downstream of duck RIG-I in both the BC500 and VN1203 infections were augmented by less in the VN1203-infected samples (IFIT5, IRF1, MX1 and ZNFX1). This contrasts with expression in human primary macrophages, where many innate immune genes were induced more by H5N1, than by H1N1 (Lee et al., 2009). Similarly, our analysis in ducks showed VN1203 infection induced a 200-fold induction of RIG-I, and greater than 1000-fold induction of IFIT and OASL. In comparison, these genes are induced by BC500 infection 80 and 95-fold, relative to mock infected ducks (Vanderven et al., 2012). In chicken cells expressing RIG-I, the difference between innate immune responses to these viruses was less dramatic. VN1203 replicated to a higher titre in DF-1 cells compared to BC500-infected cells (Barber et al., 2010), and this may have resulted in an earlier peak of innate responses. VN1203 infection also induced higher expression of innate immune genes in cells transfected with empty vector, making the difference due to RIG-I appear less. Differences in response to these viruses may also be due to interference in the pathway by the respective NS1 proteins. The influenza NS1 protein is known to modulate the interferon-mediated antiviral response (Garcia-Sastre, 2011; Garcia-Sastre et al., 1998; Zielecki et al., 2010) and the RIG-I pathway is a known target of NS1 through interaction with TRIM25 (Gack et al., 2009) or binding vRNA (Rehwinkel et al., 2010).
While many ISGs were induced more downstream of RIG-I in BC500-infected DF-1 cells, a notable exception was the expression of Th1 cytokines, where IFNG (5.7-fold higher), IL6 and IL1β, were augmented more in the VN1203 infected DF-1 cells. Upregulation of proinflammatory cytokines was also observed in primary human macrophages following infection with A/Vietnam/3212/2004 (H5N1) compared to a seasonal isolate A/HongKong 58/1998 (H1N1) (Lee et al., 2009). However, the profile of upregulated cytokine genes in avian fibroblast cells does not closely mirror human macrophages. TNF is missing from the avian genome, and IL18 and other cytokines (CCL8, CXCL10) are not on the microarray, while others (TNFSF10 and CCL5) are expressed more in BC500 infected cells than VN1203. High expression of proinflammatory cytokines was also seen in chicken lung and spleen tissues at 36 hpi with VN1203 (Karpala et al., 2011a). The induction of proinflammatory cytokines is thought to contribute to the pathology of H5N1 infections in humans and chickens.
Our experiment does not address which genes are upregulated by influenza infection in untransfected chicken cells. However, others have used gene expression profiling by microarray to examine the early transcriptional responses to influenza A viruses in chickens (Reemers et al., 2009; Reemers et al., 2010; Sarmento et al., 2008b) (Sarmento et al., 2008a). An examination of the genes expressed in primary chicken embryonic fibroblasts infected with two H5N1 HPAI strains, A/Ck/Hong Kong/220/97 and A/Egret/Hong Kong/757.2/02 at 4 hpi showed that of 191 genes that demonstrated a twofold induction, 10 were associated with the immune response (Sarmento et al., 2008a). None of these genes appear to be associated with the innate response, and the genes induced in RIG-I transfected chicken cells are notably absent. However, the early 4 hours post-infection timepoint, compared to our later 15 hpi timepoint, is likely to have been too early to detect ISGs. Indeed, in chicken lung tissues at 16 hpi with H9N2, MX, ISG12-2, OASL are upregulated (Reemers et al., 2010) albeit less than in RIG-I transfected epithelial cells. This upregulation is likely due to MDA5 and TLR recognition of influenza. Our experiment also cannot rule out that some genes are upregulated due to the higher expression of TLR3 and MDA5 in chicken DF-1 cells expressing RIG-I. MyD88 was downregulated in all cells relative to uninfected, untransfected control DF-1 cells, therefore it is unlikely that TLR7 is contributing to gene induction observed in our microarrays.
The genes identified by comparison of influenza-infected DF-1 cells expressing RIG-I or empty vector are a subset of the genes identified in the RIG-I knockout mouse compared to wildtype. Of the RIG-I bioset genes, or genes downstream of RIG-I (Loo et al., 2008), some were not observed in our microarrays. Some, like RIG-I itself, appear to be missing from the chicken genome (IRF3, ISG15 and other IFITs) while others were not included in the microarray (RSAD2 and OAS). Among the genes downstream of RIG-I and TNF pathways in human macrophages that are upregulated more than 10-fold by infection with H5N1 (IFIT, OASL, PTGS2) (Lee et al., 2009) these are also upregulated in DF-1 cells with RIG-I. Other genes (PMAIP, GBP4, CCL8, CXCL10) were not on the chicken microarray. Finally, increased expression of IFITM5 was unexpected since it does not inhibit influenza (Huang et al., 2011) and expression is restricted to developing bone in mammals (Moffatt et al., 2008). IFITM1, which has antiviral function, is not on the microarray.
It is not clear how the loss of RIG-I in chickens contributes to the inability of chickens to defend against highly pathogenic influenza strains. Chickens certainly have an interferon response to influenza, and induce an inflammatory response upon infection with VN1203 (Karpala et al., 2011a). Chickens also have a strong innate immune response to other RNA viruses such as Newcastle Disease Virus (NDV) upregulating many innate immune genes including IFIT5, MX1, ISG12-2 and CCL19 in spleen (Rue et al., 2011). Furthermore, influenza-infected chicken trachea and tracheal organ cultures express numerous innate immune genes (Reemers et al., 2009). Also contributing to the overall response, infection with highly pathogenic influenza consistently induced high levels of Type I interferon in chicken splenocytes, likely through the TLR7 pathway (Moulin et al., 2011). Chicken MDA5 also contributes to influenza detection in chicken epithelial cells (Liniger et al., 2012), but knockdown does not affect the titre of influenza strain A/Wyoming/3/03 (H3N2) (Karpala et al., 2011b), thus it is not completely clear what contribution MDA5 makes to host defense in the chicken. Lack of protection by RIG-I in chicken epithelial cells is undoubtedly detrimental in an acute influenza infection, given that these cells are on the frontline of defense against influenza viruses.
The possibility of augmenting influenza defense in chickens through transgenesis is compelling. Lyall and colleagues successfully used such an approach by creating a transgenic chicken expressing a short-hairpin RNA that inhibits the influenza polymerase. While the chickens still succumbed to infection, importantly, transmission of influenza to uninfected chickens was reduced (Lyall et al., 2011). Because duck RIG-I can induce many known antiviral mediators in chicken cells, a transgenic chicken expressing duck RIG-I could theoretically translate to increased protection to influenza in vivo. Chickens expressing duck RIG-I in tracheal epithelial cells might be expected to control influenza virus replication more efficiently. There are certainly concerns to creating a transgenic chicken with RIG-I, such as creating a chicken that can replicate HPAI strains without symptoms. However, a superior innate immune response may limit viral replication and thus reduce the emergence of highly pathogenic strains.
Here we compare global gene expression by microarray for chicken DF-1 cells infected with low or highly pathogenic strains of influenza in the presence or absence of duck RIG-I. Overall, the microarray results demonstrate that signalling through duck RIG-I can induce important antiviral genes.
Research Highlights.
-
>
44K Agilent microarray analyses of chicken DF-1 cells with duck RIG-I or empty vector
-
>
transcriptome of H5N2 infected cells with duck RIG-I or empty vector
-
>
transcriptome of H5N1 infected cells with duck RIG-I or empty vector
-
>
identification of genes augmented in expression by duck RIG-I
-
>
quantitative real-time PCR confirms upregulation of key innate immune genes
Acknowledgements
The authors would like to thank the Hartwell Center for Bioinformatics and Biotechnology (Granger Ridout) for microarray hybridization and assistance with analysis. This work was supported by the Canadian Institute for Health Research (CIHR grant MOP93561 to KEM); contract HHSN266200700005C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (RGW); and American Lebanese Syrian Associated Charities (RGW). MRWB was supported by a Natural Sciences and Engineering Research Council of Canada Postgraduate Doctoral (PGS-D) Scholarship and a Canadian Poultry Research Council Postgraduate Scholarship Supplement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barber MRW, Aldridge JR, Jr, Webster RG, Magor KE. Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci U S A. 2010;107:5913–5918. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi D, Schultz U, Staeheli P. The interferon-induced Mx protein of chickens lacks antiviral activity. Journal of Interferon & Cytokine Research. 1995;15:47–53. doi: 10.1089/jir.1995.15.47. [DOI] [PubMed] [Google Scholar]
- Cormican P, Lloyd AT, Downing T, Connell SJ, Bradley D, O'Farrelly C. The avian Toll-Like receptor pathway--subtle differences amidst general conformity. Developmental & Comparative Immunology. 2009;33:967–973. doi: 10.1016/j.dci.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt OG, Sirma H, Pandolfi PP, Haller O. Mx1 GTPase accumulates in distinct nuclear domains and inhibits influenza A virus in cells that lack promyelocytic leukaemia protein nuclear bodies. J. Gen. Virol. 2004;85:2315–2326. doi: 10.1099/vir.0.79795-0. [DOI] [PubMed] [Google Scholar]
- Ewald SJ, Kapczynski DR, Livant EJ, Suarez DL, Ralph J, McLeod S, Miller C. Association of Mx1 Asn631 variant alleles with reductions in morbidity, early mortality, viral shedding, and cytokine responses in chickens infected with a highly pathogenic avian influenza virus. Immunogenetics. 2011;63:363–375. doi: 10.1007/s00251-010-0509-1. [DOI] [PubMed] [Google Scholar]
- Fleming-Canepa X, Brusnyk C, Aldridge JR, Ross KL, Moon D, Wang D, Xia J, Barber MR, Webster RG, Magor KE. Expression of duck CCL19 and CCL21 and CCR7 receptor in lymphoid and influenza-infected tissues. Mol. Immunol. 2011;48:1950–1957. doi: 10.1016/j.molimm.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nature Reviews. Immunology. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host & Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A. Induction and evasion of type I interferon responses by influenza viruses. Virus Res. 2011;162:12–18. doi: 10.1016/j.virusres.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Grant CE, Vasa MZ, Deeley RG. cIRF-3, a new member of the interferon regulatory factor (IRF) family that is rapidly and transiently induced by dsRNA. Nucleic Acids Res. 1995;23:2137–2146. doi: 10.1093/nar/23.12.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L, Longobardi LE, Boltz D, Kuhn JH, Elledge SJ, Bavari S, Denison MR, Choe H, Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Path. 2011;7 doi: 10.1371/journal.ppat.1001258. e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das S, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher E, Neumann G, Kawaoka Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, Wakita T, Esumi M. 2',5'-Oligoadenylate synthetase-like gene highly induced by hepatitis C virus infection in human liver is inhibitory to viral replication in vitro. Biochemical & Biophysical Research Communications. 2010;392:397–402. doi: 10.1016/j.bbrc.2010.01.034. [DOI] [PubMed] [Google Scholar]
- Karpala AJ, Bingham J, Schat KA, Chen LM, Donis RO, Lowenthal JW, Bean AG. Highly pathogenic (H5N1) avian influenza induces an inflammatory T helper type 1 cytokine response in the chicken. Journal of Interferon & Cytokine Research. 2011a;31:393–400. doi: 10.1089/jir.2010.0069. [DOI] [PubMed] [Google Scholar]
- Karpala AJ, Stewart C, McKay J, Lowenthal JW, Bean AG. Characterization of chicken Mda5 activity: regulation of IFN-beta in the absence of RIG-I functionality. Journal of Immunology. 2011b;186:5397–5405. doi: 10.4049/jimmunol.1003712. [DOI] [PubMed] [Google Scholar]
- Khaperskyy DA, Hatchette TF, McCormick C. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J. 26:1629–1639. doi: 10.1096/fj.11-196915. [DOI] [PubMed] [Google Scholar]
- Ko JH, Asano A, Kon Y, Watanabe T, Agui T. Characterization of the chicken PKR: polymorphism of the gene and antiviral activity against vesicular stomatitis virus. Jap. J. Vet. Res. 2004a;51:123–133. [PubMed] [Google Scholar]
- Ko JH, Jin HK, Asano A, Takada A, Ninomiya A, Kida H, Hokiyama H, Ohara M, Tsuzuki M, Nishibori M, Mizutani M, Watanabe T. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res. 2002;12:595–601. doi: 10.1101/gr.210702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Takada A, Mitsuhashi T, Agui T, Watanabe T. Native antiviral specificity of chicken Mx protein depends on amino acid variation at position 631. Animal Genetics. 2004b;35:119–122. doi: 10.1111/j.1365-2052.2004.01096.x. [DOI] [PubMed] [Google Scholar]
- Koerner I, Kochs G, Kalinke U, Weiss S, Staeheli P. Protective role of beta interferon in host defense against influenza A virus. Journal of Virology. 2007;81:2025–2030. doi: 10.1128/JVI.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchipudi SV, Dunham SP, Nelli R, White GA, Coward VJ, Slomka MJ, Brown IH, Chang KC. Rapid death of duck cells infected with influenza: a potential mechanism for host resistance to H5N1. Immunol. Cell Biol. Mar. 2011;22:1–8. doi: 10.1038/icb.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne M, Etchart N, Goubier A, Lira SA, Sirard JC, van Rooijen N, Caux C, Ait-Yahia S, Vicari A, Kaiserlian D, Dubois B. Dendritic cells rapidly recruited into epithelial tissues via CCR6/CCL20 are responsible for CD8+ T cell crosspriming in vivo. Immunity. 2006;24:191–201. doi: 10.1016/j.immuni.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Lee SM, Gardy JL, Cheung CY, Cheung TK, Hui KP, Ip NY, Guan Y, Hancock RE, Peiris JS. Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 influenza viruses in primary human macrophages. PLoS ONE [Electronic Resource] 2009;4:e8072. doi: 10.1371/journal.pone.0008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liniger M, Summerfield A, Zimmer G, McCullough KC, Ruggli N. Chicken cells sense influenza A virus infection through MDA5 and CARDIF signaling involving LGP2. Journal of Virology. 2012;86:705–717. doi: 10.1128/JVI.00742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. Journal of Virology. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall J, Irvine RM, Sherman A, McKinley TJ, Nunez A, Purdie A, Outtrim L, Brown IH, Rolleston-Smith G, Sang H, Tiley L. Suppression of avian influenza transmission in genetically modified chickens. Science. 2011;331:223–226. doi: 10.1126/science.1198020. [DOI] [PubMed] [Google Scholar]
- Marques J, Anwar J, Eskildsen-Larsen S, Rebouillat D, Paludan SR, Sen G, Williams BR, Hartmann R. The p59 oligoadenylate synthetase-like protein possesses antiviral activity that requires the C-terminal ubiquitin-like domain. J. Gen. Virol. 2008;89:2767–2772. doi: 10.1099/vir.0.2008/003558-0. [DOI] [PubMed] [Google Scholar]
- Marsland BJ, Battig P, Bauer M, Ruedl C, Lassing U, Beerli RR, Dietmeier K, Ivanova L, Pfister T, Vogt L, Nakano H, Nembrini C, Saudan P, Kopf M, Bachmann MF. CCL19 and CCL21 induce a potent proinflammatory differentiation program in licensed dendritic cells. Immunity. 2005;22:493–505. doi: 10.1016/j.immuni.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Melchjorsen J, Kristiansen H, Christiansen R, Rintahaka J, Matikainen S, Paludan SR, Hartmann R. Differential regulation of the OASL and OAS1 genes in response to viral infections. Journal of Interferon & Cytokine Research. 2009;29:199–207. doi: 10.1089/jir.2008.0050. [DOI] [PubMed] [Google Scholar]
- Moffatt P, Gaumond MH, Salois P, Sellin K, Bessette MC, Godin E, de Oliveira PT, Atkins GJ, Nanci A, Thomas G. Bril: a novel bone-specific modulator of mineralization. J Bone Miner Res. 2008;23:1497–1508. doi: 10.1359/jbmr.080412. [DOI] [PubMed] [Google Scholar]
- Moulin HR, Liniger M, Python S, Guzylack-Piriou L, Ocana-Macci M, Ruggli N, Summerfield A. High interferon type 1 responses in the lung, plasma, and spleen during highly pathogenic H5N1 infection of chicken. Vet. Res. 2011;42:6–12. doi: 10.1186/1297-9716-42-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K, Jogi M, Yoo J-S, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, Matsumiya T, Namiki H, Yoneyama M, Fujita T. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PloS one. 2012;7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantin-Jackwood MJ, Swayne DE. Pathogenesis and pathobiology of avian influenza virus infection in birds. Revue Scientifique et Technique. 2009;28:113–136. [PubMed] [Google Scholar]
- Pasick J, Handel K, Robinson J, Copps J, Ridd D, Hills K, Kehler H, Cottam-Birt C, Neufeld J, Berhane Y, Czub S. Intersegmental recombination between the haemagglutinin and matrix genes was responsible for the emergence of a highly pathogenic H7N3 avian influenza virus in British Columbia. J. Gen. Virol. 2005;86:727–731. doi: 10.1099/vir.0.80478-0. [DOI] [PubMed] [Google Scholar]
- Pavlovic J, Arzet HA, Hefti HP, Frese M, Rost D, Ernst B, Kolb E, Staeheli P, Haller O. Enhanced virus resistance of transgenic mice expressing the human MxA protein. Journal of Virology. 1995;69:4506–4510. doi: 10.1128/jvi.69.7.4506-4510.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic J, Zurcher T, Haller O, Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. Journal of Virology. 1990;64:3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, Rulicke T, Weber F, Colinge J, Muller M, Superti-Furga G. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nat. Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- Pietila TE, Veckman V, Lehtonen A, Lin R, Hiscott J, Julkunen I. Multiple NF-kappaB and IFN regulatory factor family transcription factors regulate CCL19 gene expression in human monocyte-derived dendritic cells. Journal of Immunology. 2007;178:253–261. doi: 10.4049/jimmunol.178.1.253. [DOI] [PubMed] [Google Scholar]
- Reemers SS, Groot Koerkamp MJ, Holstege FC, van Eden W, Vervelde L. Cellular host transcriptional responses to influenza A virus in chicken tracheal organ cultures differ from responses in in vivo infected trachea. Veterinary Immunology & Immunopathology. 2009;132:91–100. doi: 10.1016/j.vetimm.2009.04.021. [DOI] [PubMed] [Google Scholar]
- Reemers SS, van Leenen D, Koerkamp MJ, van Haarlem D, van de Haar P, van Eden W, Vervelde L. Early host responses to avian influenza A virus are prolonged and enhanced at transcriptional level depending on maturation of the immune system. Mol. Immunol. 2010;47:1675–1685. doi: 10.1016/j.molimm.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Rosebeck S, Leaman DW. Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis. 2008;13:562–572. doi: 10.1007/s10495-008-0190-0. [DOI] [PubMed] [Google Scholar]
- Rue CA, Susta L, Cornax I, Brown CC, Kapczynski DR, Suarez DL, King DJ, Miller PJ, Afonso CL. Virulent Newcastle disease virus elicits a strong innate immune response in chickens. J. Gen. Virol. 2011;92:931–939. doi: 10.1099/vir.0.025486-0. [DOI] [PubMed] [Google Scholar]
- Russell CA, Fonville JM, Brown AE, Burke DF, Smith DL, James SL, Herfst S, van Boheemen S, Linster M, Schrauwen EJ, Katzelnick L, Mosterin A, Kuiken T, Maher E, Neumann G, Osterhaus AD, Kawaoka Y, Fouchier RA, Smith DJ. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science. 2012;336:1541–1547. doi: 10.1126/science.1222526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmento L, Afonso CL, Estevez C, Wasilenko J, Pantin-Jackwood M. Differential host gene expression in cells infected with highly pathogenic H5N1 avian influenza viruses. Veterinary Immunology & Immunopathology. 2008a;125:291–302. doi: 10.1016/j.vetimm.2008.05.021. [DOI] [PubMed] [Google Scholar]
- Sarmento L, Pantin-Jackwood M, Kapczynski DR, Swayne DE, Afonso CL. Immediate early responses of avian tracheal epithelial cells to infection with highly pathogenic avian influenza virus. Developments in Biologicals. 2008b;132:175–183. doi: 10.1159/000317158. [DOI] [PubMed] [Google Scholar]
- Schaefer-Klein J, Givol I, Barsov EV, Whitcomb JM, VanBrocklin M, Foster DN, Federspiel MJ, Hughes SH. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology. 1998;248:305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- Schulz O, Pichlmair A, Rehwinkel J, Rogers NC, Scheuner D, Kato H, Takeuchi O, Akira S, Kaufman RJ, Reis e Sousa C. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host & Microbe. 2010;7:354–361. doi: 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Bernasconi D, Schultz U, Staeheli P. The chicken Mx promoter contains an ISRE motif and confers interferon inducibility to a reporter gene in chick and monkey cells. Virology. 1994;203:144–148. doi: 10.1006/viro.1994.1464. [DOI] [PubMed] [Google Scholar]
- Schusser B, Reuter A, von der Malsburg A, Penski N, Weigend S, Kaspers B, Staeheli P, Hartle S. Mx is dispensable for interferon-mediated resistance of chicken cells against influenza A virus. J Virol. 2011;85:8307–8315. doi: 10.1128/JVI.00535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm-Ramirez KM, Hulse-Post DJ, Govorkova EA, Humberd J, Seiler P, Puthavathana P, Buranathai C, Nguyen TD, Chaisingh A, Long HT, Naipospos TS, Chen H, Ellis TM, Guan Y, Peiris JS, Webster RG. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. 2005;79:11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez DL, Schultz-Cherry S. Immunology of avian influenza virus: a review. Developmental & Comparative Immunology. 2000;24:269–283. doi: 10.1016/s0145-305x(99)00078-6. [DOI] [PubMed] [Google Scholar]
- Swayne DE. Understanding the complex pathobiology of high pathogenicity avian influenza viruses in birds. Avian Diseases. 2007;51:242–249. doi: 10.1637/7763-110706-REGR.1. [DOI] [PubMed] [Google Scholar]
- Swayne DE, Pantin-Jackwood M. Pathogenicity of avian influenza viruses in poultry. Developments in Biologicals. 2006;124:61–67. [PubMed] [Google Scholar]
- Swayne DE, Suarez DL. Highly pathogenic avian influenza. Revue Scientifique et Technique. 2000;19:463–482. doi: 10.20506/rst.19.2.1230. [DOI] [PubMed] [Google Scholar]
- Tatsumi R, Hamada K, Sekiya S, Wakamatsu M, Namikawa T, Mizutani M, Sokawa Y. 2',5'-oligoadenylate synthetase gene in chicken: gene structure, distribution of alleles and their expression. Biochimica et Biophysica Acta. 2000;1494:263–268. doi: 10.1016/s0167-4781(00)00174-3. [DOI] [PubMed] [Google Scholar]
- Tatsumi R, Sekiya S, Nakanishi R, Mizutani M, Kojima S, Sokawa Y. Function of ubiquitin-like domain of chicken 2'-5'-oligoadenylate synthetase in conformational stability. Journal of Interferon & Cytokine Research. 2003;23:667–676. doi: 10.1089/107999003322558809. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Szretter KJ, Van Hoeven N, Katz JM, Kochs G, Haller O, Garcia-Sastre A, Staeheli P. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. Journal of Virology. 2007;81:10818–10821. doi: 10.1128/JVI.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Watanabe C, Takemae N, Hayashi T, Oka T, Ito T, Saito T. Identification of host genes linked with the survivability of chickens infected with recombinant viruses possessing H5N1 surface antigens from a highly pathogenic avian influenza virus. Journal of Virology. 2012;86:2686–2695. doi: 10.1128/JVI.06374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderven H, Petkau K, Ryan-Jean KE, Aldridge JR, Webster RG, Magor KE. Avian influenza rapidly induces antiviral genes in duck lung and intestine. Mol. Immunol. 2012;51:316–324. doi: 10.1016/j.molimm.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielecki F, Semmler I, Kalthoff D, Voss D, Mauel S, Gruber AD, Beer M, Wolff T. Virulence determinants of avian H5N1 influenza A virus in mammalian and avian hosts: role of the C-terminal ESEV motif in the viral NS1 protein. Journal of Virology. 2010;84:10708–10718. doi: 10.1128/JVI.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]