Abstract
Ploidy is predicted to influence adaptation directly, yet whether single mutations behave the same in different ploidy backgrounds has not been well studied. It has often been assumed theoretically that aside from dominance, selective parameters do not differ between cells of varying ploidy. Using the budding yeast Saccharomyces cerevisiae, I compared the effect size of 20 adaptive mutations in haploids and homozygous diploids and found, surprisingly, that the same mutations often had a much larger effect in haploids than homozygous diploids. This empirical result demonstrates that it cannot be assumed that mutations will have the same effect in haploids and homozygous diploids.
Keywords: effect size, fitness, growth rate, dose–response, IC50
1. Introduction
The dynamics of evolution are expected to vary between populations that differ in ploidy [1]. In haploid individuals, composed of a single set of chromosomes, all novel adaptive mutations are immediately ‘seen’ by evolution, and selection is very efficient. In diploids (composed of two sets) or polyploids (multiple sets), mutations generally arise in a single copy that can be partially or completely masked by wild-type alleles. The efficacy of selection depends on the dominance properties of the mutation in question [2]. Selection may act quickly in the case of a fully dominant allele, or slowly if the mutation is recessive and must appear in the same genome in multiple copies before its effects are exposed. This may reduce the rate of adaptation in diploid populations [3] and may prevent the establishment of recessive or partially recessive adaptive mutations [4].
A factor that has been left out of this (and similar) discussions is whether all else is equal between individuals of different ploidy. Although direct empirical tests are relatively sparse, the existing data suggest that ploidy background may well affect the properties of mutations. The effect sizes of both deleterious [5] and adaptive [6] mutations have been found to be larger in haploids compared with homozygous diploids in yeast, although this is not always the case [7]. The study design of those experiments makes it difficult to interpret differences in effect size of single mutations, however, as the studies compared lines that contained many mutations [6,7] or assayed fitness in a manner that included the spore germination phase in haploids but not diploids [5]. Here, I sought to directly assess whether single mutations that confer tolerance to a fungicide, nystatin, have the same effect size in homozygous diploids as in haploids.
2. Material and methods
(a). Mutation acquisition
Twenty unique mutations that confer tolerance to 4 µM nystatin were acquired in Saccharomyces cerevisiae haploid lines of genotype BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) [8] (see the electronic supplementary material). Data deposited in the Dryad Repository: see http://dx.doi.org/10.5061/dryad.h51jg. All mutations are in one of four genes that act late in the ergosterol biosynthesis pathway (one mutation in ERG7; one in ERG5; seven unique mutations in ERG6 and 11 unique mutations in ERG3). The majority of mutations are non-synonymous substitution or premature stop codons, but the mutation set also includes three deletions and a duplication. We previously showed that the type of mutation within a gene had no detectable effect and thus present results without differentiating between mutation type. Details on the mutations can be found in table 1 of Gerstein et al. [8].
(b). Creation of homozygous diploids
Homozygous MATa/MATa diploids were created through a plasmid selection regime to avoid the potentially confounding effect of a heterozygous mating (MAT) locus [9]. Two colonies for each mutation line were transformed using standard protocols [10], one with a plasmid containing LEU2 and one with a plasmid containing URA3 and MATα. Colonies containing each plasmid were mated, and diploids were selected on –leu/–ura plates. To test for an effect of the MAT locus genotype, BMN1, BMN9, BMN32 and BMN35 (one line for each gene hit in the screen) were backcrossed to BY4739 (MATα leu2Δ0 lys2Δ0 ura3Δ0). The resulting diploids were sporulated, and MATα nystatin-tolerant spores were mated to the original MATa haploid mutation lines. MATa/MATα homozygous diploids were then selected on –his/–lys plates.
(c). Nystatin dose–response assay
The effect of ploidy was examined, using a dose–response assay to measure three parameters: the tolerance of each line to nystatin measured as the half-maximal inhibitory concentration (IC50), the rate of decline in 72 h culture density at IC50 (slope, m50) and the asymptotic level of 72 h density reached at low levels of nystatin (a). The experiment was repeated twice. For each, haploid (MATa) and diploid (MATa/MATa) mutation lines were streaked from freezer stock onto yeast extract, peptone and dextrose (YPD) plates and grown for 72 h. Culture was inoculated into 10 ml YPD and grown for 24 h. The optical density was standardized across lines, and 10 µl was inoculated into 1 ml YPD plus one of 10 levels of nystatin in deep well boxes, maintained shaking at 30°C. Four replicates were measured for each line in each experiment. After 72 h, wells were mixed and the optical density of 200 µl culture was measured on a BioTek plate reader. Data from both experiments were combined for analysis.
The three parameters were fit by maximizing the likelihood of observing the data [8]. It was determined whether haploids and homozygous diploids significantly differed by fitting a likelihood model to the combined data, allowing each ploidy to have its own parameter values. The fit of this ‘full’ model was compared with a constrained model with a single value for the parameter of interest (other parameters were allowed to vary), using a likelihood ratio test. If the drop in log-likelihood between the full and constrained model was greater than χ1,0.052/2 = 1.92, then the hypothesis that the parameter was the same was rejected. An overall effect of ploidy and gene was determined using the maximum likelihood parameter estimates from the full model as data in a two-way ANOVA. Out of concern that the MATa/MATa diploid is an unnatural diploid state, a second independent set of tolerance assays were conducted to compare MATa/MATa and MATa/MATα homozygous diploids with MATa and MATα haploid lines using the same methods as mentioned earlier. Results are presented for the MATa versus MATa/MATa comparison, except where noted.
(d). Growth rate assay
The growth rates of all lines were measured at three nystatin concentrations (2, 4 and 8 µM). For each level of nystatin, each line was cultured in four non-adjacent wells containing 150 µl in the Bioscreen C Microbiological Workstation (Thermo Labsystems). Bioscreen plates were grown at 30°C for 48 h, with constant shaking; optical density readings were taken automatically every 30 min. The maximal growth rate for each well was determined as the spline with the highest slope using an analysis program written by Richard FitzJohn in R [11]. The entire assay was conducted at each level of nystatin on two separate occasions. Significance of the difference in growth rates between haploid and diploid lines was examined for each mutation separately by t-tests that use the Welch approximation for degrees of freedom. The overall effect of ploidy and gene was determined by a two-way ANOVA, as mentioned earlier.
3. Results and discussion
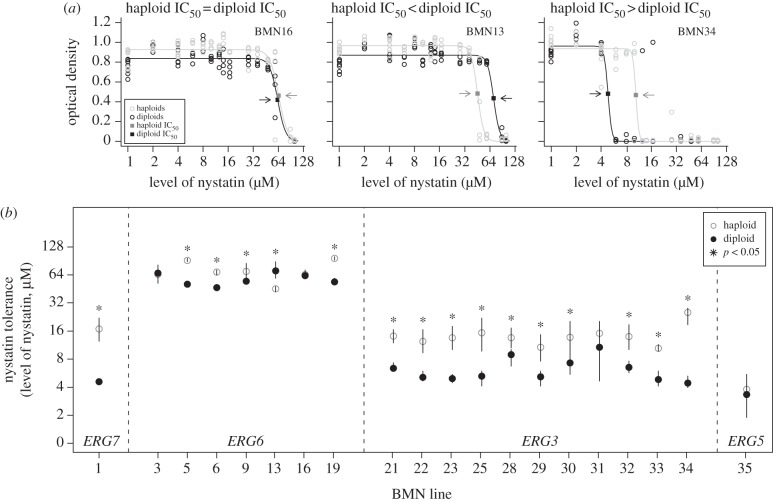
The effect size of nystatin adaptive mutations depends strongly on ploidy level. Mutations in a haploid background were more tolerant to nystatin than the same mutations in a homozygous diploid background (figure 1). Tolerance (IC50) was significantly affected by gene (F3 = 221.8, p < 0.0001) and ploidy (F1 = 62.1, p < 0.0001), and there was a significant interaction between the two (F3,32 = 7.19, p = 0.0008). Haploid lines reached a higher asymptote than diploids (see the electronic supplementary material, figure S1; ploidy: F1 = 24.4, p < 0.0001; gene: F3 = 15.4, p < 0.0001; interaction: F3,32 = 1.74, p = 0.19), and the slope at IC50 was also significant for these factors (see the electronic supplementary material, figure S1; ploidy: F1 = 4.5, p = 0.04; gene: F1 = 3.9, p = 0.018; interaction: F3,32 = 0.84, p = 0.48). A second set of tolerance experiments (see §2) indicated that the genotype at the MAT locus did not significantly affect tolerance (MAT genotype: F1 = 0.6, p = 0.46, gene: F3 = 487, p < 0.0001, ploidy: F1 = 31, p = 0.0024).
Figure 1.
Nystatin adaptive mutations generally yield higher tolerance in haploids than homozygous diploids. (a) The dose–response relationship was measured for each adaptive mutation in haploids and homozygous diploids to determine IC50 (indicated by arrows). (b) Significant differences for tolerance were found between ploidy levels and among lines that carried mutations in different genes. Line-specific statistical results are presented in the electronic supplementary material, table S2.
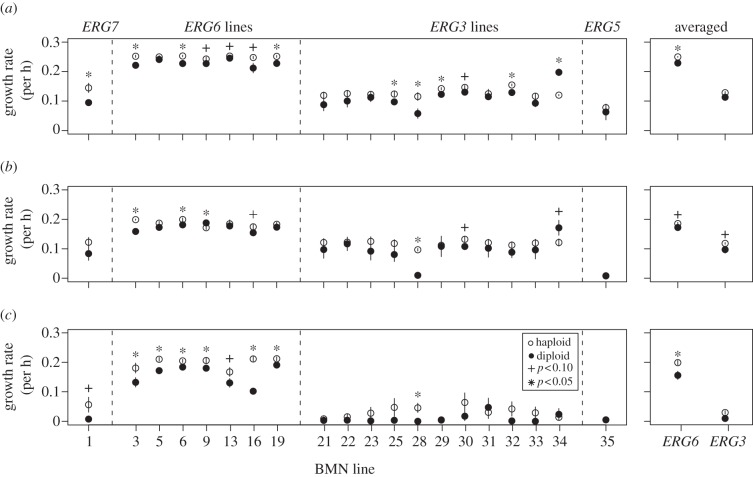
Maximal growth rate was also significantly affected by ploidy background and gene (figure 2). This was not the result of an inherent growth advantage to haploids, as no significant difference was found between wild-type haploids and diploids in 2 µM nystatin (t15 = 0.52, p = 0.61, haploid mean: 0.04 ± 0.01, diploid mean: 0.03 ± 0.01). Wild-type cells could not be tested at 4 µM or higher levels of nystatin, as by design they were unable to grow without acquiring a mutation. Haploids grew significantly faster than diploids across mutant lines in 2 µM and 4 µM nystatin (2 µM—ploidy: F1 = 7.8, p = 0.009; gene: F1 = 101.3, p < 0.0001; interaction: F3,32 = 0.41, p = 0.75; 4 µM—ploidy: F1 = 6.4, p = 0.017; gene: F1 = 48.6, p < 0.0001; interaction: F3,32 = 0.33, p = 0.80). Similar results were obtained when the data were analysed with a linear mixed-effect model that controlled for batch effects (see the electronic supplementary material, table S1). At 8 µM nystatin, only lines that carried mutations in ERG6 (the most tolerant lines; figure 1) were able to grow consistently, yet haploids again grew significantly faster (t6 = 4, p = 0.01). Lines that carried mutations in ERG3 and ERG7 grew stochastically in 8 µM nystatin, i.e. many lines did not grow, whereas other lines showed rapid growth starting at different time points. This is reminiscent of the growth pattern used to isolate mutation lines in our initial experiments [8] (see the electronic supplementary material, figure S2), and we interpret this growth as the appearance of new mutations. Of the 96 haploid and 96 diploid replicates assayed in 8 µM nystatin, 46 haploid and only five diploid replicates showed growth; this difference in the rate of mutation acquisition among lines is significant (χ12 = 42.72, p < 0.0001). A ploidy-specific difference in mutation acquisition illustrates one of the fundamental differences between lines of different ploidy. When novel mutations arise in diploid form, they are often partially masked by the wild-type alleles and thus less able to confer a fitness advantage unless present in more than one copy. I thus predict that the diploid lines which showed growth contain either rare dominant mutations or a recessive mutation in homozygous form.
Figure 2.
Nystatin adaptive mutations tend to have a higher growth rate in haploids than homozygous diploids in (a) YPD + 2 µM nystatin, (b) YPD + 4 µM nystatin, and (c) YPD + 8 µM nystatin. This pattern is also true when averaged across lines that carried different mutations in the same gene (right-hand column, ERG6 and ERG3 mutation lines). Line-specific statistical results are presented in the electronic supplementary material, table S3.
Combined, these results demonstrate that the adaptive nystatin alleles examined here confer a greater tolerance on haploids than homozygous diploids. McBride et al. [12] also found that diploid S. cerevisiae were more affected than haploids by toxins in their environment. These complementary results may indicate that physical- or expression-level differences exist between ploidy levels of otherwise isogenic yeast that alter the response of cells to either environmental or genetic perturbations. It has long been posited that the fitness of haploid : diploid cells should depend on cell geometry rather than ploidy per se [13]. It may be that resistance to nystatin, which acts by binding to ergosterol in the cell membrane, is directly correlated to total surface area. Thus, diploid cells, which are larger than haploid cells, could be more affected by the same concentration of stressor in their environment.
That some mutations have different effects in haploid and homozygous diploid backgrounds is an important result, as theoretical studies often assume that all else is equal when modelling the effects of ploidy, including models about ploidy evolution [14,15], rates of adaptation [16] and host–parasite interactions [17]. A better understanding about when (and why) mutational parameters differ between individuals of varying ploidy may help explain why one ploidy-level predominates in a group of organisms, a longstanding question in evolutionary biology. Here, I have provided one empirical example where the same adaptive mutations yield a greater fitness increase in haploid individuals compared with homozygous diploids.
Acknowledgments
I thank the Pellman laboratory for generous donation of plasmids; J. Ono for assistance with strain construction; D. Lo, A. Kuzmin, W. Li and T. Hinder for laboratory assistance. I particularly thank S. Otto for her encouragement, discussions and comments on the manuscript. Funding was provided by the National Science and Engineering Research Council of Canada, a Killam Trusts Predoctoral Fellowship, and a Faculty of Science graduate fellowship from the University of British Columbia.
References
- 1.Kondrashov A. S., Crow J. F. 1991. Haploidy or diploidy—which is better? Nature 351, 314–315 10.1038/351314a0 (doi:10.1038/351314a0) [DOI] [PubMed] [Google Scholar]
- 2.Orr H. A., Otto S. P. 1994. Does diploidy increase the rate of adaptation? Genetics 136, 1475–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerstein A. C., Otto S. P. 2011. Cryptic fitness advantage: diploids invade haploid populations despite lacking any apparent advantage as measured by standard fitness assays. PLoS ONE 6, e26599. 10.1371/journal.pone.0026599 (doi:10.1371/journal.pone.0026599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson J. B., Sirjusingh C. 2004. Haploidy, diploidy and evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics 168, 1915–1923 10.1534/genetics.104.033266 (doi:10.1534/genetics.104.033266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szafraniec K., Wloch D. M., Sliwa P., Borts R. H., Korona R. 2003. Small fitness effects and weak genetic interactions between deleterious mutations in heterozygous loci of the yeast Saccharomyces cerevisiae. Genet. Res. 82, 19–31 10.1017/S001667230300630X (doi:10.1017/S001667230300630X) [DOI] [PubMed] [Google Scholar]
- 6.Zeyl C. 2003. An evolutionary advantage of haploidy in large yeast populations. Science 299, 555–558 10.1126/science.1078417 (doi:10.1126/science.1078417) [DOI] [PubMed] [Google Scholar]
- 7.Korona R. 1999. Unpredictable fitness transitions between haploid and diploid strains of the genetically loaded yeast Saccharomyces cerevisiae. Genetics 151, 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerstein A. C., Lo D. L., Otto S. P. 2012. Parallel genetic changes and non-parallel gene–environment interactions characterize the evolution of drug resistance in yeast. Genetics 192, 241–252 10.1534/genetics.112.142620 (doi:10.1534/genetics.112.142620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birdsell J., Wills C. 1996. Significant competitive advantage conferred by meiosis and syngamy in the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 93, 908–912 10.1073/pnas.93.2.908 (doi:10.1073/pnas.93.2.908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amberg D. C., Burke D. J., Strathern J. N. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Long Island, NY: CSHL Press [Google Scholar]
- 11.R Development Core Team 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org (ISBN: 3-900051-07-0) [Google Scholar]
- 12.Mcbride R., Greig D., Travisano M. 2008. Fungal viral mutualism moderated by ploidy. Evolution 62, 2372–2380 10.1111/j.1558-5646.2008.00443.x (doi:10.1111/j.1558-5646.2008.00443.x) [DOI] [PubMed] [Google Scholar]
- 13.Weiss R. L., Kukora J. R., Adams J. 1975. The relationship between enzyme activity, cell geometry, and fitness in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 72, 794–798 10.1073/pnas.72.3.794 (doi:10.1073/pnas.72.3.794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paquin C., Adams J. 1983. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature 302, 495–500 10.1038/302495a0 (doi:10.1038/302495a0) [DOI] [PubMed] [Google Scholar]
- 15.Otto S. P., Goldstein D. B. 1992. Recombination and the evolution of diploidy. Genetics 131, 745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orr H. A. 2003. The distribution of fitness effects among beneficial mutations. Genetics 163, 1519–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M'Gonigle L. K., Otto S. P. 2011. Ploidy and the evolution of parasitism. Proc. R. Soc. B 278, 2814–2822 10.1098/rspb.2010.2146 (doi:10.1098/rspb.2010.2146) [DOI] [PMC free article] [PubMed] [Google Scholar]