Abstract
Dispersal by passive oceanic rafting is considered important for the assembly of biotic communities on islands. However, not much is known about levels of population genetic connectivity maintained by rafting over transoceanic distances. We assess the evolutionary impact of kelp-rafting by estimating population genetic differentiation in three kelp-associated invertebrate species across a system of islands isolated by oceanic gaps for over 5 million years, using mtDNA and AFLP markers. The species occur throughout New Zealand's subantarctic islands, but lack pelagic stages and any opportunity for anthropogenic transportation, and hence must rely on passive rafting for long-distance dispersal. They all have been directly observed to survive transoceanic kelp-rafting journeys in this region. Our analyses indicate that regular gene flow occurs among populations of all three species between all of the islands, especially those on either side of the subtropical front oceanographic boundary. Notwithstanding its perceived sporadic nature, long-distance kelp-rafting appears to enable significant gene flow among island populations separated by hundreds of kilometres of open ocean.
Keywords: AFLP, gene flow, island, migration, rafting, mtDNA
1. Introduction
Passive transportation of biota on floating substrates, or rafting dispersal, has had a major role in shaping the species composition of island ecosystems [1,2]. While the most dramatic examples of long-distance rafting are sporadic oceanic dispersals of terrestrial vertebrates, such as lizards and monkeys [3,4], we hypothesize that, for some species that lack both autonomous and anthropogenic dispersal capability, rafting may be a more regular dispersal mechanism, sufficient to drive on-going genetic connectivity between remote island populations.
In cool latitudes of the Southern Hemisphere, diverse marine invertebrates use holdfasts of buoyant bull-kelp, Durvillaea antarctica, for shelter and grazing. Bull-kelp individuals may detach from their rocky substrate during heavy seas and subsequently drift, carrying their epifaunal invertebrate communities for vast distances [5]. Our previous studies have shown that kelp-rafting may facilitate the founding of new invertebrate populations on oceanic islands thousands of kilometres away [6], and that rafting of adults can enhance gene flow over short coastal distances [7]. However, no study has yet assessed the extent to which transoceanic gene flow can be driven solely by kelp-rafting. Here we assess genetic structuring of three kelp-associated intertidal invertebrate species across the New Zealand (NZ) subantarctic region (figure 1). Specifically, we test whether rafting can be the primary driver of gene flow between islands over approximately 500 km oceanic scales, and also whether such rafting events are constrained by a major oceanographic barrier, the subtropical front (STF).
Figure 1.
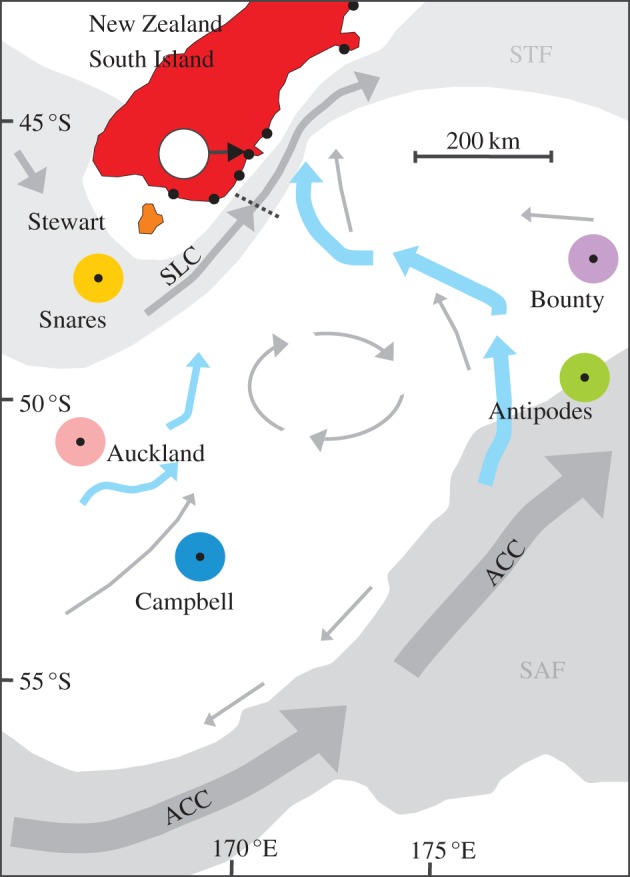
Sampling locations of three intertidal invertebrate species that rely on kelp-rafting for long-distance dispersal (black dots). Detailed sampling information is given in the electronic supplementary material, table S1. STF, subtropical front; SAF, subantarctic front; ACC, antarctic circumpolar current, SLC, southland current; grey arrows, modern current systems; blue arrows, deflection of the ACC towards New Zealand east coast during the LGM. Dashed line shows northern limit of the SLC at LGM. Oceanographic features were drawn following [8]. White circle shows the collection locality of beach-cast, rafted individuals reported in [5].
The marine snail Cantharidus roseus, the chiton Onithochiton neglectus and the amphipod Parawaldeckia karaka are regular and abundant, facultative grazer-inhabitants of holdfasts in bull-kelp beds surrounding NZ and its subantarctic islands. The chiton and the amphipod brood their eggs until fully developed juveniles emerge [9,10]; the snail attaches its fertilized eggs on a surface where they develop into crawl-away larvae [11]. As these taxa lack pelagic dispersal capacity, their autonomous dispersal ability is limited to crawling on hard surfaces, covering only the scale of metres in the intertidal zone and clearly insufficient for between-island migration in this region. Apart from Stewart Island and New Zealand South Island (NZSI), none of the study islands, which all formed more than 5 million years ago and are isolated by mesopelagic waters, have ever been connected [12]. Additionally, as the species do not foul ships, and as there is very limited ship traffic between the remote islands [12], rafting is their only plausible long-distance dispersal mechanism. Moreover, all three species’ ability to survive extensive kelp-rafting journeys is confirmed by our data from beach-cast bull-kelp holdfasts that had rafted from the subantarctic to mainland NZ with live epifauna [5] (the discovery location and mtDNA haplotypes of these rafted individuals are colour-coded white in figures).
2. Material and methods
We sampled invertebrates at low tide from inner surfaces of bull-kelp holdfasts that had been prised off rock, or from rock surfaces adjacent to the bull-kelp beds. Animals from each island (figure 1) were selected from five to 18 different holdfasts, and usually from multiple localities (see the electronic supplementary material, table S1), to capture the local genetic diversity. The samples from NZSI originated from multiple locations along the east coast (figure 1).
We used mtDNA sequencing and genome-wide AFLP fingerprinting to estimate levels of genetic differentiation between islands. DNA was extracted from ethanol-preserved animals and a fragment of the mitochondrial COI gene was amplified using universal primers following the protocols reported in [6]. Sequencing was done by Massey Genome Service (New Zealand) and Macrogen Ltd. (Korea) using the forward primer. AFLP-fingerprinting and phenotype calling were carried out as in [9], with the required dataset-specific modifications to the semi-automated phenotype calling [13] (see the electronic supplementary material, table S2). Phenotype-calling thresholds were set individually for each species, and resulted in 94–164 polymorphic loci per species, with an overall replicability rate of 93.6 per cent. The main sample sets consisted of 81–90 animals per species distributed across all islands, 82 per cent of them successfully genotyped with both marker types (see the electronic supplementary material, table S1). In addition, the amphipod mtDNA data included 68 additional sequences from older NZSI and Campbell Island samples that were not available for AFLP fingerprinting. The AFLP phenotype data were deposited in the Dryad Repository (http://dx.doi.org/10.5061/dryad.p5k2r) and the COI sequences in GenBank (accessions JX121210–JX121228, JX123091–JX123127). Estimates of global and among-island fixation indices (ΦPT, an analogue of FST for dominant data) and AFLP-distances between individuals, and their PCA-plots, were generated in GenAlEx [14]. The island-pairwise ΦPT estimates were converted into estimates of effective number of migrants per generation (Nm) using equation Nm = (1 − ΦPT)/4ΦPT [15]. Maximum-parsimony (MP) networks of mtDNA haplotypes were constructed using Network [16].
3. Results
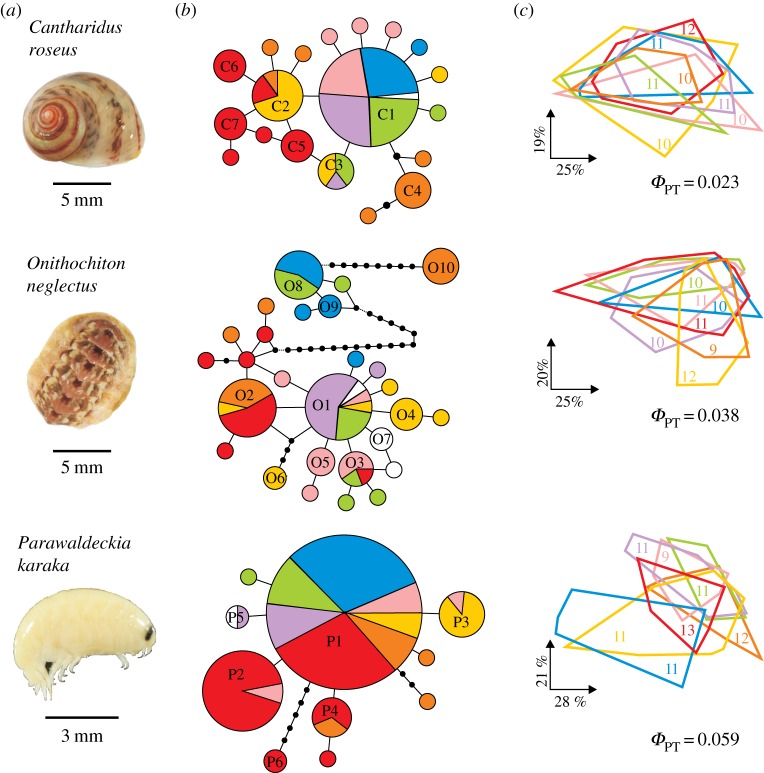
Numerous mtDNA haplotypes were shared between multiple islands in all three species, and in one of the species (P. karaka), a single haplotype was found on all seven islands. Furthermore, several of the island pairs (nine of 21) shared haplotypes for all three species (figure 2 and table 1; electronic supplementary material, table S3). For genome-wide (AFLP) data, all three species showed a low overall level of differentiation between the island populations, as estimated by ΦPT (figure 2). The majority of island pairs (52%) did not exhibit statistically significant differentiation in AFLP markers, while the pairs that did yielded low ΦPT in the range 0.038–0.147 (see the electronic supplementary material, table S4). Notably, samples of all three species from the geographically remote Auckland and Antipodes Islands (740 km apart) were not significantly different from each other for AFLP data, and these samples also shared several mtDNA haplotypes.
Figure 2.
Phylogeographic and population genetic patterns in three kelp-associated invertebrates, pictured in (a), among seven islands. (b) MP networks from 587 to 600 base pair fragment of the COI gene for each species. The area of each circle/sector (see scale) directly reflects the number of individuals detected carrying that haplotype. Black dots denote hypothetical undetected/extinct haplotypes; a connecting line indicates one mutational step. For O. neglectus, dashed lines show where median-joining algorithm joins the MP-networks of the three clades. (c) Principal coordinate (PC) plots of invertebrates based on their pairwise AFLP-fingerprint distances. Each polygon encompasses the smallest possible PC-space that contains all individuals sampled on that island (sample size shown); the degree to which the polygons overlap roughly illustrates the populations' genetic similarity. The proportion of variation explained by PCs and the global among-island fixation index ΦPT are shown for each species. Colour-coding corresponds to figure 1.
Table 1.
Mitochondrial haplotypes shared across ocean gaps in three kelp-associated invertebrates among seven islands. Island pairs separated by the subtropical front are shown in italics. Haplotype names follow figure 2.
| Island | Antipodes | Auckland | Bounty | Campbell | NZ south | Snares |
|---|---|---|---|---|---|---|
| Auckland | C1/O1/O3/P1 | |||||
| Bounty | C1/C3/O1/P1 | C1/O1/P1 | ||||
| Campbell | C1/O8/P1 | C1/P1 | C1/P1 | |||
| NZ South | O3/P1 | O3/P1/P2 | P1 | P1 | ||
| Snares | C3/O1/P1 | O1/P2/P3 | C3/O1/P1 | P1 | C2/O2/P1 | |
| Stewart | P1 | P1 | P1 | P1 | C2/O2/P1/P4 | C2/O2/P2 |
In assessing the role of an oceanographic barrier in constraining rafting events, we note that cases of mtDNA-haplotype sharing (summed over all species) were nearly twice as frequent among island pairs not separated by the STF (3.11 shared haplotypes on average) as among those that were (1.75). Specifically, NZSI, Stewart and Snares Islands samples shared haplotypes for all species, as did the Auckland, Antipodes and Bounty Island samples, whereas haplotype sharing between these groups—across the STF—was rarer (table 1). In contrast, the average estimates of genome-wide differentiation calculated over all species were equally low for island pairs separated by the STF and those not (ΦPT 0.041 and 0.042, respectively). Still, statistical significance of ΦPT-estimates was slightly more frequent among island pairs separated by the STF (50% versus 33% of ΦPT-estimates, electronic supplementary material, table S4).
In contrast to the low mtDNA nucleotide diversity levels in the snail and amphipod, the chiton data revealed three deep mtDNA clades with largely overlapping geographical distributions (figure 2). However, no significant differentiation in AFLP-data was detected between any of these mtDNA clades (between-clade ΦPT = 0.004, non-significant), and they were completely overlapping in a PCA-plot, indicating that the O. neglectus samples represent a single species.
4. Discussion
The widespread mtDNA haplotype sharing and low levels of among-island AFLP differentiation point to regular, on-going oceanic gene flow across the NZ subantarctic in all three invertebrate species. As discussed above, rafting is the only plausible between-island dispersal mechanism for these taxa. Indeed, bull-kelp rafts are abundant in the Southern Ocean [17], at a frequency that should easily provide the single effective migrant per generation required to prevent neutral genetic divergence among islands [18]. This abundance also avoids any need to invoke unique dispersal events (e.g. a single major storm mixing island populations after their long-term isolation), which, moreover, would leave very different mtDNA patterns from the ones observed. The genetic evidence presented here confirms that successful rafting events are frequent enough to maintain high genetic connectivity of kelp epifauna across large oceanic gaps. Under Wright's island model of migration and assuming equilibrium conditions [15], the ΦPT estimates would suggest that from three to eight effective migrants per generation have been needed between the Antipodes and Bounty Islands' populations of the studied species to maintain the observed level of genetic similarity (see the electronic supplementary material, table S4). In comparison, an intertidal limpet species with a pelagic larval phase long enough to allow dispersal between these islands (but without special association of adults with bull-kelp or other rafting vectors) shows genetic differentiation between them (ΦPT = 0.330 [19]) that would correspond to just one effective migrant per generation. However, we emphasize that these migration rate estimates should not be taken literally, as the exact migration model among the islands and the populations' genetic equilibrium status are unknown.
While our study suggests that gene flow effects of rafting can be constrained to some extent by oceanographic barriers (evidenced by the distinct mtDNA haplotype composition of C. roseus populations either side of the SFC), it nevertheless shows that gene flow between the islands, and even across SFC, does occur over ecological time scales. In the absence of gene flow, the different island populations would in time evolve diagnostically divergent mtDNA clades. While biological connectivity across the subantarctic is strongly influenced by the antarctic circumpolar current (ACC) today, it is likely that wind- and current-mediated connectivity were even higher during the last glacial maximum (LGM, [8]; figure 1). We conclude that rafting—in addition to permitting colonization events [2,6]—is an important contemporary process for connecting isolated marine ecosystems and may suppress allopatric speciation rates (cf. [18]) in island biota prone to rafting, relative to other intrinsically non-mobile island biota.
Acknowledgements
We thank Ceridwen Fraser, Rebecca Cumming, Henk Haazen, Ross Watters, Tony Whiting, Elaine Leung, Jennifer Lawn, Kirsten Donald and Department of Conservation for assistance with sample collection, Ken Miller for help with photography, three anonymous reviewers for comments on the manuscript and the Allan Wilson Centre for Molecular Ecology and Evolution and the University of Otago for resources.
References
- 1.Thiel M, Haye P. 2006. The ecology of rafting in the marine environment. III. Biogeographical and evolutionary consequences. Oceanogr. Mar. Biol. Annu. Rev. 44, 323–428 10.1201/9781420006391.ch7 (doi:10.1201/9781420006391.ch7) [DOI] [Google Scholar]
- 2.Gillespie RG, Waters JM, Fraser CI, Nikula R, Baldwin BG, Roderick GK. 2012. Long-distance dispersal—a framework for hypothesis testing. Trends Ecol. Evol. 27, 47–56 10.1016/j.tree.2011.08.009 (doi:10.1016/j.tree.2011.08.009) [DOI] [PubMed] [Google Scholar]
- 3.Censky EJ, Hodge K, Dudley J. 1998. Over-water dispersal of lizards due to hurricanes. Nature 395, 556. 10.1038/26886 (doi:10.1038/26886) [DOI] [Google Scholar]
- 4.Ali JR, Huber M. 2010. Mammalian biodiversity on Madagascar controlled by ocean currents. Nature 463, 653–656 10.1038/nature08706 (doi:10.1038/nature08706) [DOI] [PubMed] [Google Scholar]
- 5.Fraser CI, Nikula R, Waters JM. 2011. Oceanic rafting by a coastal community. Proc. R. Soc. B 278, 649–655 10.1098/rspb.2010.1117 (doi:10.1098/rspb.2010.1117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikula R, Fraser CI, Spencer HG, Waters JM. 2010. Circumpolar dispersal by rafting in two subantarctic kelp-dwelling crustaceans. Mar. Ecol. Prog. Series 405, 221–230 10.3354/meps08523 (doi:10.3354/meps08523) [DOI] [Google Scholar]
- 7.Nikula R, Spencer HG, Waters JM. 2011. Evolutionary consequences of microhabitat: population-genetic structuring in kelp- vs. rock-associated chitons. Mol. Ecol. 20, 4915–4924 10.1111/j.1365-294X.2011.05332.x (doi:10.1111/j.1365-294X.2011.05332.x) [DOI] [PubMed] [Google Scholar]
- 8.Lorrey AM, et al. 2012. Palaeocirculation across New Zealand during the last glacial maximum at ∼21 ka. Q. Sci. Rev. 36, 189–213 10.1016/j.quascirev.2011.09.025 (doi:10.1016/j.quascirev.2011.09.025) [DOI] [Google Scholar]
- 9.Creese RG. 1986. Brooding behaviour and larval development in the New Zealand chiton, Onithochiton neglectus de Rochebrune Mollusca: Polyplacophora. NZ J. Zool. 13, 83–91 10.1080/03014223.1986.10422648 (doi:10.1080/03014223.1986.10422648) [DOI] [Google Scholar]
- 10.Lowry JK, Stoddart HE. 1986. Protandrous hermaphrodites among the lysianassoid Amphipoda. J. Crustacean Biol. 6, 742–748 10.2307/1548388 (doi:10.2307/1548388) [DOI] [Google Scholar]
- 11.Lebour MV. 1963. Notes on the eggs and larvae of some Plymouth prosobranchs. J. Mar. Biol. Assoc. UK 20, 547–566 10.1017/S0025315400058124 (doi:10.1017/S0025315400058124) [DOI] [Google Scholar]
- 12.Department of Conservation 2006. Marine protection for the New Zealand subantarctic islands: a background resource document. Wellington, New Zealand: Department of Conservation [Google Scholar]
- 13.Whitlock R, Hipperson H, Mannarelli M, Butlin RK, Burke T. 2008. An objective, rapid and reproducible method for scoring AFLP peak-height data that minimizes genotyping error. Mol. Ecol. Resour. 8, 725–735 10.1111/j.1755-0998.2007.02073.x (doi:10.1111/j.1755-0998.2007.02073.x) [DOI] [PubMed] [Google Scholar]
- 14.Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295 10.1111/j.1471-8286.2005.01155.x (doi:10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright S. 1951. The genetical structure of populations. Ann. Eugenics 15, 323–354 [DOI] [PubMed] [Google Scholar]
- 16.Polzin T, Daneschmand SV. 2003. On Steiner trees and minimum spanning trees in hypergraphs. Oper. Res. Lett. 31, 12–20 10.1016/S0167-6377(02)00185-2 (doi:10.1016/S0167-6377(02)00185-2) [DOI] [Google Scholar]
- 17.Smith SDA. 2002. Kelp rafts in the Southern Ocean. Global Ecol. Biogeogr. 11, 67–69 10.1046/j.1466-822X.2001.00259.x (doi:10.1046/j.1466-822X.2001.00259.x) [DOI] [Google Scholar]
- 18.Slatkin M. 1985. Gene flow and the geographic structure of natural populations. Science 236, 787–792 10.1126/science.3576198 (doi:10.1126/science.3576198) [DOI] [PubMed] [Google Scholar]
- 19.Reisser CMO, Wood AR, Bell JJ, Gardner JPA. 2011. Connectivity, small islands and large distances: the Cellana strigilis limpet complex in the Southern Ocean. Mol. Ecol. 20, 3399–3413 10.1111/j.1365-294X.2011.05185.x (doi:10.1111/j.1365-294X.2011.05185.x) [DOI] [PubMed] [Google Scholar]