Abstract
One of the most mysterious aspects of insect clock mechanisms is that some insects, including Hymenoptera and Tribolium, only express a vertebrate-type cryptochrome (cry2). It is unknown whether or not cry2 underwent adaptive evolution in these insects. In the present study, we cloned and sequenced the full-length cry2 from a fig pollinator species, Ceratosolen solmsi (Hymenoptera: Chalcidoidea: Agaonidae), and examined the molecular evolution and daily expression of this gene. Our results suggest that cry2 underwent positive selection in the branch leading to hymenopteran insects. The function of CRY2 might have been fixed since undergoing natural selection in the ancestor of Hymenoptera. Male pollinators showed stronger rhythmicity in the host figs, which reflect an adaptation to their life cycles.
Keywords: fig pollinator, cryptochrome, adaptation, positive selection
1. Introduction
Cryptochrome (CRY) is a photolyase-like flavoprotein that shows no DNA-repair activity [1]. Animal CRY proteins are phylogenetically divided into two clusters: one contains Drosophila-type CRY (CRY1) and the other includes all the vertebrate CRY (CRY2) [2,3]. Drosophila species only possess cry1, mosquitoes and butterflies have both genes, and hymenopteran insects and Tribolium have lost cry1 [2–4]. CRY1 is a blue-light sensor that plays a major role in photic entrainment in Drosophila [5–7]. In contrast, insect CRY2 is an important transcriptional repressor in the circadian clock, which shows no light sensitivity in culture [3,8]. Recently, both CRYs were reported to have light-dependent magnetosensitivity [9–11]. However, we have no knowledge of whether or not the CRYs have different evolutionary patterns in the diverse insect taxa. As the protein encoded by cry1 serves as an important light sensor in the circadian systems of insects, we hypothesize that natural selection might have driven the evolution of cry2 to compensate for loss of cry1 in hymenopteran insects. To test this hypothesis, we amplified and sequenced the full-length cry2 of a fig pollinator species, Ceratosolen solmsi, whose life cycle is strictly synchronized to its host fig tree Ficus hispida, and characterized the daily expression of this gene.
2. Material and methods
Fig fruits of F. hispida, were sampled from Danzhou (19°30′29′ N, 109°29′6′ E), Hainan province, China in October 2011. Both female and male pollinators were collected.
Full-length cry2 of C. solmsi was amplified by RT-PCR and RACE PCR from cDNA samples and sequenced. Additional insect cry2 gene sequences were acquired from NCBI (www.ncbi.nlm.nih.gov), as listed in electronic supplementary material, table S1.
To study sequence evolution of cry2 in holometabolic insects, we reconstructed a gene tree for cry2, and based on which we performed tests for positive selection using maximum-likelihood estimation by CODEML in PAML v. 4.5 [12]. Two types of selection models, Site models and Branch-site models [13–16] were used to test for episodic evolution of cry2 along branches a–g (figure 1a). Since seven separate likelihood ratio tests (LRTs) were performed in this analysis, Bonferroni adjustment was used for multiple testing correction. The results are shown in electronic supplementary material, tables S2 and S3. Detailed methods were given in the electronic supplementary material.
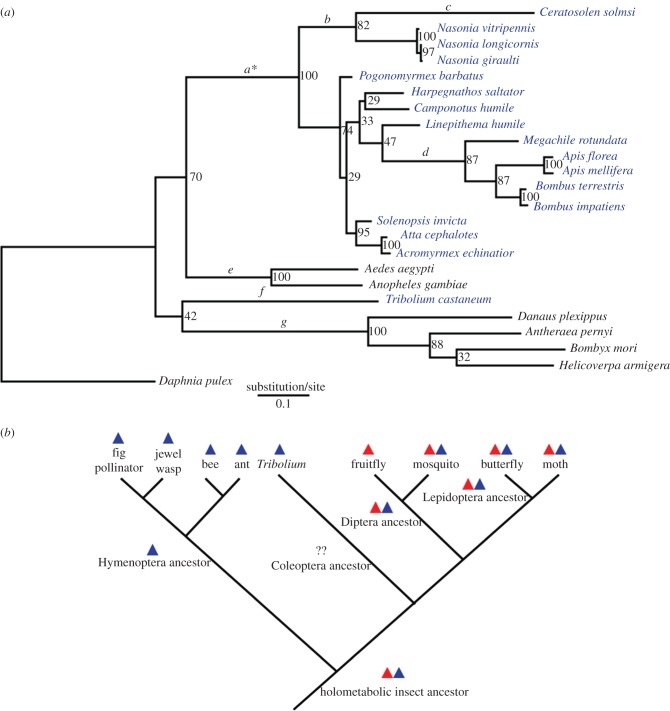
Figure 1.
(a) Phylogenetic reconstruction of cry2 in holometabolic insects. The ML tree is constructed from full-length coding sequences of cry2. Bootstrap values are shown at the nodes. The branches tested for positive selection are labelled as a–g. Asterisk (*) indicates that positively selected sites are detected. The species that lack cry1 are highlighted in blue. (b) Phylogeny of the holometabolic insects, character mapping of possession of cryptochrome genes and proposed ancestral state. Triangles indicate cry1 (red) and cry2 (blue).
RT-qPCR was used to analyse daily transcript levels of cry2 in female and male wasps under different light conditions (females in natural light-treated figs, females in dark-treated figs, females exposed to natural light, males in natural light-treated figs, and males in dark-treated figs).
3. Results
(a). Episodic evolution of cry2
We obtained the 1704 bp full-length cry2 sequence (Genbank accession no. JX409893) from C. solmsi. Combined with published data from other holometabolic insect species, we reconstructed the gene tree of cry2 (figure 1a). The sequences of cryptochrome genes have been mapped onto the species phylogeny of holometabolic insects based on previous studies [17]. We also inferred the distribution of both genes in the ancestor of the four main holometabolic insect orders: Hymenoptera, Coleoptera, Lepidoptera and Diptera (figure 1b). Results of the evolutionary analysis of site models indicated that both cry1 and cry2 experienced strong purifying selection (cry2: ω = 0.01536 for all taxa; cry1: ω = 0.02130 for mosquitoes and Lepidoptera), although there is evidence that cry2 (cry2: ω = 0.01624 for mosquitoes and Lepidoptera) is even more constrained than cry1 (p = 0.015; electronic supplementary material, table S2). Branch-site models identified a group of amino acid sites that underwent positive selection along the branches leading to Hymenoptera (17 sites for branch a) (see the electronic supplementary material, table S2, S3). However, we found no evidence of positive selection along other branches we tested.
(b). Daily expression of cry2 in Ceratosolen solmsi
Daily levels of cry2 mRNA in both female and male C. solmsi varied as a function of the time × light interaction (table 1). Cosinor analyses [18] indicated that the cry2 was rhythmically expressed in emerged females exposed to natural light (emerged female light; one way ANOVA: p < 0.001; Cosinor: p = 0.016) and males in both natural light-treated (fig-male light; one way ANOVA: p = 0.001; Cosinor: p < 0.001) and dark-treated (fig-male dark; one way ANOVA: p < 0.001; Cosinor: p = 0.004) figs (figure 2).
Table 1.
Statistical values of cry2 expression in C. solmsi. d.f., degree of freedom; F, value of F-test; p-value, probability.
| source | d.f. | F | p-value | % of total variation |
|---|---|---|---|---|
| female: | ||||
| light | 2 | 190.9 | <0.0001 | 44.73 |
| time | 7 | 9.772 | <0.0001 | 15.31 |
| light × time | 14 | 9.524 | <0.0001 | 29.85 |
| male: | ||||
| light | 1 | 412.4 | <0.0001 | 49.58 |
| time | 7 | 5.837 | =0.0003 | 13.30 |
| light × time | 7 | 12.08 | <0.0001 | 27.52 |
Figure 2.
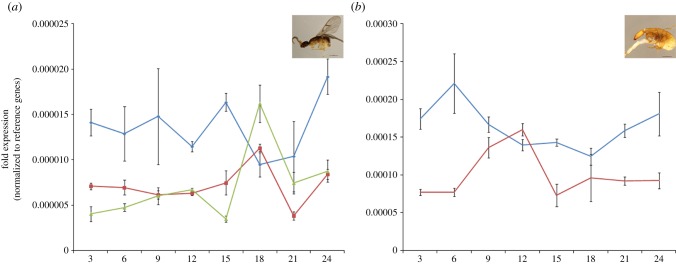
(a) Daily expression of cry2 in female and (b) male C. solmsi. Fold expression of cry2 (mRNA abundance of opsin genes relative to reference genes) at each time point are represented by bars. LF (blue), fig-female light; DF (red), fig-female dark; F (green), emerged females light; LM (blue), fig-male light; DM (red), fig-male dark. Asterisk (*) indicates significant rhythmic expression. Morphological dimorphism between female and male of C. solmsi is present.
4. Discussion
The crustacean species Daphnia pulex has both cry1 and cry2, suggesting that the gene duplication of cryptochrome occurred before the emergence of insects. Therefore, possession of only one cryptochrome in some holometabolic insects should be attributed to lineage-specific gene loss events. Since all the hymenopteran species (ants, bees and wasps) that we sampled only possess cry2, it is likely that loss of cry1 occurred in the ancestor of Hymenoptera. Tribolium castaneum also lacked cry1, but we could not determine when the gene loss event occurred in the coleopteran lineage. Based on our selective pressure test of cryptochrome in holometabolic insects, cry2 seemed to have undergone stronger purifying selection than cry1 in the same set of species (mosquitos and lepidopteran insects). This was predictable because the function of CRY2 as a transcriptional repressive component was relatively conservative. However, branch-site tests suggested that during the evolutionary history of holometabolic insects, at least some residues of CRY2 experienced positive selection in the ancestor of hymenopteran species, after gene loss of cry1. This evolutionary pattern supports our hypothesis that cry2 was subjected to more positive selection in the absence of the function conferred by CRY1. We did not find evidence of positive selection for branches leading to extant hymenopteran species such as bees, wasps and the fig pollinator, C. Solmsi. It seems that the function of CRY2 has been conserved since undergoing natural selection in the ancestor of Hymenoptera. Rhythmic expression of cry2 has been recorded in several holometabolic species [2,19,20]. In some cases, expression of this gene showed stronger plasticity than cry1 not only across species [19] but also in different tissues within the same organism [20]. This suggests that the expression pattern of cry2 has the potential to reveal species-specific adaptation to the light environment or lifestyle, especially in the insects that lack photosensitive CRY1.
Figs (Ficus: Moraceae) and their pollinators (Family Agaonidae) form one of the best-known examples of obligate mutualism [21]. The wasps develop in the galls formed within the syconia of figs. Upon maturation, males emerge from the galls before females, and they chew holes in the galls containing females so that the latter can disperse to pollinate in other receptive fig fruits [22]. Apparently, males initiate the life cycles in the fig pollinators. Because males seldom leave the fig fruits and the females are responsible for colonizing new hosts [22,23], it is reasonable to anticipate that males will show stronger rhythmicity within the fig fruits than females. Although it remains unclear how CRY2 functions, hymenopteran's cry2 mRNA levels should reflect some rhythmicity as a core clock-element [2,3]. This prediction allows for testing our hypothesis on the adaptation of rhythms of fig pollinators to their host. Consistent with our hypothesis, the results from RT-qPCR suggested that males maintain rhythmic expression of their cry2, yet females maintain the rhythmicity only when they are outside the fig fruits. Such sex-specific expression patterns of cry2 could be the result from the adaptation of pollinators to their host figs during their long-term coevolution [24,25].
Acknowledgements
This project was supported by the National Natural Science Foundation of China (NSFC grant nos 31090253, 31172072), partially by Major Innovation Program of Chinese Academy of Sciences (KSCX2-EW-Z-2), a grant (no. O529YX5105) from the Key Laboratory of the Zoological Systematics and Evolution of the Chinese Academy of Sciences and National Science Fund for Fostering Talents in Basic Research (Special Subjects in Animal Taxonomy, NSFC-J0930004). We thanks Dr Wen Xin and TransGen Biotech for providing most of the reagents used in the study. We thank the anonymous reviewers for their valuable comments and suggestions.
References
- 1.Ozturk N, Song SH, Ozgur S, Selby CP, Morrison L, Partch C, Zhong D, Sancar A. 2007. Structure and function of animal cryptochromes. Cold Spring Harb. Symp. Quant. Biol. 72, 119–131 10.1101/sqb.2007.72.015 (doi:10.1101/sqb.2007.72.015) [DOI] [PubMed] [Google Scholar]
- 2.Rubin EB, Shemesh Y, Cohen M, Elgavish S, Robertson HM, Bloch G. 2006. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 16, 1352–1365 10.1101/gr.5094806 (doi:10.1101/gr.5094806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan Q, Metterville D, Briscoe AD, Reppert SM. 2007. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 24, 948–955 10.1093/molbev/msm011 (doi:10.1093/molbev/msm011) [DOI] [PubMed] [Google Scholar]
- 4.Zhu H, Yuan Q, Froy O, Casselman A, Reppert SM. 2005. The two CRYs of the butterfly. Curr. Biol. 15, R953–R954 10.1016/j.cub.2005.11.030 (doi:10.1016/j.cub.2005.11.030) [DOI] [PubMed] [Google Scholar]
- 5.Tomioka K, Matsumoto A. 2010. A comparative view of insect circadian clock systems. Cell. Mol. Life Sci. 67, 1397–1406 10.1007/s00018-009-0232-y (doi:10.1007/s00018-009-0232-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fogle KJ, Parson KG, Dahm NA, Holmes TC. 2011. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science 331, 1409–1413 10.1126/science.1199702 (doi:10.1126/science.1199702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins B, Mazzoni EO, Stanewsky R, Blau J. 2006. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr. Biol. 16, 441–449 10.1016/j.cub.2006.01.034 (doi:10.1016/j.cub.2006.01.034) [DOI] [PubMed] [Google Scholar]
- 8.Ikeno T, Katagiri C, Numata H, Goto SG. 2011. Causal involvement of mammalian-type cryptochrome in the circadian cuticle deposition rhythm in the bean bug Riptortus pedestris. Insect Mol. Biol. 20, 409–415 10.1111/j.1365-2583.2011.01075.x (doi:10.1111/j.1365-2583.2011.01075.x) [DOI] [PubMed] [Google Scholar]
- 9.Foley LE, Gegear RJ, Reppert SM. 2011. Human cryptochrome exhibits light-dependent magnetosensitivity. Nat. Commun. 2, 356. 10.1038/ncomms1364 (doi:10.1038/ncomms1364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gegear RJ, Foley LE, Casselman A, Reppert SM. 2010. Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463, 804–807 10.1038/nature08719 (doi:10.1038/nature08719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gegear RJ, Casselman A, Waddell S, Reppert SM. 2008. Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature 454, 1014–1018 10.1038/nature07183 (doi:10.1038/nature07183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 10.1093/molbev/msm088 (doi:10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479 10.1093/molbev/msi237 (doi:10.1093/molbev/msi237) [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, dos Reis M. 2011. Statistical properties of the branch-site test of positive selection. Mol. Biol. Evol. 28, 1217–1228 10.1093/molbev/msq303 (doi:10.1093/molbev/msq303) [DOI] [PubMed] [Google Scholar]
- 15.Yang Z, Wong WS, Nielsen R. 2005. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118 10.1093/molbev/msi097 (doi:10.1093/molbev/msi097) [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917 10.1093/oxfordjournals.molbev.a004148 (doi:10.1093/oxfordjournals.molbev.a004148) [DOI] [PubMed] [Google Scholar]
- 17.Trautwein MD, Wiegmann BM, Beutel R, Kjer KM, Yeates DK. 2012. Advances in insect phylogeny at the dawn of the postgenomic era. Annu. Rev. Entomol. 57, 449–468 10.1146/annurev-ento-120710-100538 (doi:10.1146/annurev-ento-120710-100538) [DOI] [PubMed] [Google Scholar]
- 18.Nelson W, Tong YL, Lee JK, Halberg F. 1979. Methods for cosinor-rhythmometry. Chronobiologia 6, 305–323 [PubMed] [Google Scholar]
- 19.Gentile C, Rivas GB, Meireles-Filho AC, Lima JB, Peixoto AA. 2009. Circadian expression of clock genes in two mosquito disease vectors: cry2 is different. J. Biol. Rhythms 24, 444–451 10.1177/0748730409349169 (doi:10.1177/0748730409349169) [DOI] [PubMed] [Google Scholar]
- 20.Merlin C, Gegear RJ, Reppert SM. 2009. Antennal circadian clocks coordinate sun compass orientation in migratory monarch butterflies. Science 325, 1700–1704 10.1126/science.1176221 (doi:10.1126/science.1176221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anstett MC, Hossaert-McKey M, Kjellberg F. 1997. Figs and fig pollinators: evolutionary conflicts in a coevolved mutualism. Trends Ecol. Evol. 12, 94–99 (doi:10.1016/s0169-5347(96)10064-1) [DOI] [PubMed] [Google Scholar]
- 22.Cook JM, West SA. 2005. Figs and fig wasps. Curr. Biol. 15, R978–R980 10.1016/j.cub.2005.11.057 (doi:10.1016/j.cub.2005.11.057) [DOI] [PubMed] [Google Scholar]
- 23.Weiblen GD. 2002. How to be a fig wasp. Annu. Rev. Entomol. 47, 299–330 10.1146/annurev.ento.47.091201.145213 (doi:10.1146/annurev.ento.47.091201.145213) [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Vaamonde C, Wikstrom N, Kjer KM, Weiblen GD, Rasplus JY, Machado CA, Cook JM. 2009. Molecular dating and biogeography of fig-pollinating wasps. Mol. Phylogenet. Evol. 52, 715–726 10.1016/j.ympev.2009.05.028 (doi:10.1016/j.ympev.2009.05.028) [DOI] [PubMed] [Google Scholar]
- 25.Ronsted N, Weiblen GD, Cook JM, Salamin N, Machado CA, Savolainen V. 2005. 60 million years of co-divergence in the fig–wasp symbiosis. Proc. R. Soc. B 272, 2593–2599 10.1098/rspb.2005.3249 (doi:10.1098/rspb.2005.3249) [DOI] [PMC free article] [PubMed] [Google Scholar]