Abstract
Dengue virus currently causes 50-100 million infections annually. Comprehensive knowledge about the evolution of Dengue in response to selection pressure is currently unavailable, but would greatly enhance vaccine design efforts. In the current study, we sequenced 187 new dengue virus serotype 3(DENV-3) genotype III whole genomes isolated from Asia and the Americas. We analyzed them together with previously-sequenced isolates to gain a more detailed understanding of the evolutionary adaptations existing in this prevalent American serotype. In order to analyze the phylogenetic dynamics of DENV-3 during outbreak periods; we incorporated datasets of 48 and 11 sequences spanning two major outbreaks in Venezuela during 2001 and 2007-2008 respectively. Our phylogenetic analysis of newly sequenced viruses shows that subsets of genomes cluster primarily by geographic location, and secondarily by time of virus isolation. DENV-3 genotype III sequences from Asia are significantly divergent from those from the Americas due to their geographical separation and subsequent speciation. We measured amino acid variation for the E protein by calculating the Shannon entropy at each position between Asian and American genomes. We found a cluster of 7 amino acid substitutions having high variability within E protein domain III, which has previously been implicated in serotype-specific neutralization escape mutants. No novel mutations were found in the E protein of sequences isolated during either Venezuelan outbreak. Shannon entropy analysis of the NS5 polymerase mature protein revealed that a G374E mutation, in a region that contributes to interferon resistance in other flaviviruses by interfering with JAK-STAT signaling was present in both the Asian and American sequences from the 2007-2008 Venezuelan outbreak, but was absent in the sequences from the 2001 Venezuelan outbreak. In addition to E, several NS5 amino acid changes were unique to the 2007-2008 epidemic in Venezuela and may give additional insight into the adaptive response of DENV-3 at the population level.
Keywords: Dengue 3 virus, phylogeny, E protein, NS5 protein, Venezuela, whole genome sequence
Introduction
Dengue virus serotypes 1 through 4 (DENV-1-4) are arthropod-borne viruses (arboviruses) of the Flavivirus genus and the Flaviviridae family. Dengue is the most prevalent arthropod-borne viral disease found in tropical and sub-tropical regions of the world. DENV-1-4 are known to cause dengue fever ranging in severity, from asymptomatic, to mild or life-threatening disease. Dengue illness is a serious health burden with between 50 to 100 million cases estimated to occur annually and is considered an urban zoonosis of epidemic and hyperendemic proportions (WHO, 2009). Currently, there is no approved vaccine or therapeutic treatment for this disease. The difficulties in producing a viable vaccine are compounded by the alternate circulation of the four DENV serotypes over long periods of time. Life-long immunity to a specific serotype does not protect against infection from the remaining serotypes; but rather increases the pathological risks including antibody-dependent enhancement (Halstead et al., 2010; Miller, 2010; Thomas et al., 2006). The prophylactic vaccine approach may therefore decrease the number of human cases and disease burden of DENV, but this approach will require a considerable amount of time and effort.
Consequently, it is imperative that the evolution of the viral genome be monitored and studied at the population level, as DENV variants that appear in nature could impact vaccine effectiveness. In particular, the re-emergence of DENV-3 represents a major clade replacement that has become one of the most important epidemiological events related to dengue in the Americas.
DENV-3 (genotype IV) was first detected in the Americas in 1963 and last isolated in 1977 (Pinheiro and Corber, 1997). After 17 years of apparent absence of DENV-3 in the region, the DENV-3 genotype III was isolated in 1994 during outbreaks in Nicaragua and Panama. This new taxon in the Americas spread rapidly throughout the Central American and Caribbean countries between 1995 and 2001, including Puerto Rico in 1998, and caused dengue fever (DF) outbreaks with sporadic dengue hemorrhagic fever/dengue shock syndrome (DHF/ DSS) (Gubler, 2005; Pinheiro and Corber, 1997; Rigau-Perez et al., 2002; Wilson and Chen, 2002). As the prevalence of DENV-3 increased, the virus continued to spread throughout South America. In 2000, the virus simultaneously appeared in Brazil, Ecuador, Venezuela, and Peru (Fajardo et al., 2009; Kochel et al., 2008; Nogueira et al., 2001; Uzcategui et al., 2003), and then spread to Paraguay, Argentina, and Bolivia between 2002 and 2007 (Aquino et al., 2008; Aquino et al., 2006; Barrero and Mistchenko, 2008; de Mora et al., 2009).
Two different introductions of DENV-3 in the Americas in the last 10 years have been previously hypothesized (Aquino et al., 2009; Aquino et al., 2006; Barcelos Figueiredo et al., 2008; Vilela et al., 2010). Although all four serotypes of dengue cocirculated throughout Central- and South America, DENV-3 became the dominant serotype causing extensive DHF/ DSS epidemics in the region in Colombia and Venezuela and was the cause of two consecutive epidemics in Venezuela during 2001 and 2007-2008 (Mendez and Bernal, 2002; Ocazionez et al., 2006; Ospina, 2004).
The aim of this work is to provide a temporal and spatial distribution analysis of DENV-3 using whole genome sequences from two consecutive DENV-3 outbreaks in Venezuela together with a phylogenetic analysis that incorporates representative American and recent Asian sequences. The DENV-3 viruses we isolated, sequenced, and analyzed from South America were categorized as genotype III by homology to predefined taxonomic designations (Lanciotti et al., 1994). Sequence variation analyses were performed on the E and NS5 regions, which are responsible for viral entry and genome replication respectively. These two mature peptides were specifically chosen to highlight the contrast existing between sequence variation occurring within a protein known to have a relatively high substitution rate (E), and a protein with a relatively low substitution rate (NS5). The rationale for focusing on the substitutions observed within these two proteins between the two DENV-3 genotype 3 (DENV-3 III) Venezuelan outbreaks (2001 and 2007-2008) was to identify residues that were capable of undergoing change while maintaining viral replication. Our results show evidence of amino acid substitutions that are linked to the different Venezuelan epidemics of DENV-3 and may help to elucidate how the virus responds to immunological, epidemiological, and other effects. Such residues could subsequently be characterized more fully and exploited in antiviral drug or vaccine research.
Results
The E coding region is translated into one of the main surface protein of the virus and is known to have higher relative variability when compared to other regions of the polyprotein. The NS5 coding region gives rise to the viral RNA-dependent RNA polymerase and methyltransferase, and is known to be a highly conserved region of the genome. The resulting amino acid substitutions that consistently differed either between the two outbreaks in Venezuela or between the New- and Old World sequences are reported in Tables 1 and 2.
Table 1.
Amino Acid Residues in E Protein (domains I, II, and III) with high Shannon Entropy Scores.
| Position | Consensus | Entropy |
|---|---|---|
| 81 | I | 0.455 |
| 123 | E | 0.089 |
| 124 | P | 0.161 |
| 132 | H | 0.451 |
| 154 | D | 0.398 |
| 160 | V | 0.384 |
| 169 | V | 0.398 |
| 222 | A | 0.123 |
| 301 | L | 0.398 |
| 320 | I | 0.108 |
| 329 | A | 0.593 |
| 360 | E | 0.185 |
| 380 | V | 0.123 |
| 383 | K | 0.442 |
| 384 | A | 0.161 |
Table 2.
Amino Acid Residues in NS5 Protein with high Shannon Entropy Scores.
| Position | Consensus | Entropy |
|---|---|---|
| 50 | T | 0.458 |
| 188 | T | 0.361 |
| 190 | I | 0.259 |
| 229 | S | 0.443 |
| 288 | S | 0.443 |
| 365 | P | 0.485 |
| 371 | K | 0.443 |
| 374 | E | 0.544 |
| 389 | R | 0.443 |
| 422 | R | 0.443 |
| 429 | E | 0.493 |
| 585 | K | 0.493 |
| 619 | V | 0.443 |
| 631 | V | 0.443 |
| 637 | H | 0.259 |
| 639 | L | 0.443 |
| 656 | R | 0.458 |
| 749 | K | 0.458 |
| 835 | D | 0.443 |
| 864 | L | 0.53 |
| 876 | D | 0.443 |
Using the whole genome information, a phylogenetic tree consisting of nucleotide DENV-3 sequence data from 187 recently sequenced DENV-3 genotype III isolates was constructed (Figure 1, Supplemental Table 1). The phylogenetic tree shows a high amount of divergence between the Asian and the American DENV-3 III sequences, as shown by the amount of separation and distance between the two main branches, or clades, of the tree. In the main upper branch containing sequences from the Western hemisphere, we found DENV-3 III to be prevalent not only in Venezuela but in the rest of the Western hemisphere and discovered that subsets of these isolates formed primary clades according to the geographical location where each virus was obtained. Sub-lineages, or secondary branches of these primary clades, also existed and were found to correlate well with the time of virus isolation. Within the branches representing American sequences, we found several instances in which a virus from one country was closely related to one or more viruses isolated from a second country and likely represent specific instances of international transmission. According to this phylogenetic reconstruction the BR_R01_02 sequence, obtained from Brazil in 2002, appears to have been introduced independently from Asia. Since there is no other sequence closely related to this isolate in Brazil, it is possible that this Asian introduction was transmitted only transiently. The second introduction of DENV-3 III into the New World seems to have occurred by isolates either directly related to, or highly similar to those found in Sri Lanka and Mozambique in the 1980’s and 1990’s. Based on the tree topology, it is likely that the 1994 Nicaraguan strain, BID V2420, spread rapidly throughout other countries in Central- and South America and became the ancestor of the other New World DENV-3 III viruses, which were isolated in the 1990’s and 2000’s and included in this study. The majority of the Venezuelan and Colombian sequences, which lie in the bottom half of the upper branch of the tree, together with several Brazilian sequences appear to be closely related to DENV-3 III viruses isolated from Puerto Rico in 1999, 2000, 2002, and 2003 (sequences BID V1455, BID V2104, BID V1477, and BID V1490 respectively), indicating a possible origin of the Venezuelan and Colombian viruses.
Figure 1. Whole Genome Bayesian Phylogenetic Tree.
A Bayesian phylogenetic tree reconstruction of DENV-3 whole genomes isolated from various times and places. Sequence names are color-coded to represent the time of isolation for each virus. The large topological separations correlate with the geographical location where each virus was isolated (indicated by black brackets and text). The smaller topological separations within any geographical clade appear to be dependent on the temporal point of viral isolation. Branch lengths are proportional to distance (the number of nucleotide substitutions per site), and the distance scale for the number of changes is provided at the bottom of the figure.
Using the time of isolation metadata associated with only the New World whole genome sequences, we used BEAST to calculate the mutation rate for these viruses. This analysis revealed the mean rate of nucleotide mutation in this subset of viruses to be 1.698×10−5 or 2.881×10−5 nucleotide substitutions per site per year when using a relaxed or strict molecular clock respectively.
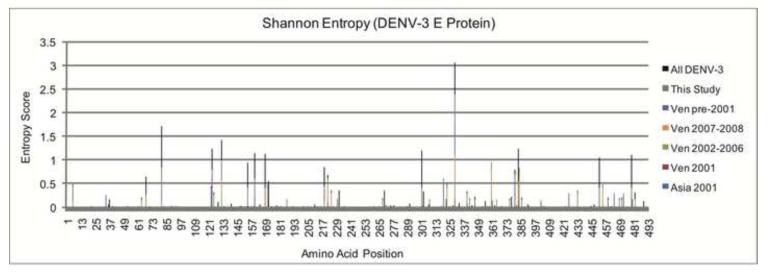
In order to observe how the genotypic changes affect the amino acid sequence in these mature proteins we calculated the Shannon entropy for the amino acid sequences of the DENV-3 mature E protein from these, and other representative sequences (Supplemental Table 2). Shannon entropy determines the amount of uncertainty (i.e. variability), and takes the residue frequency and the number of possible residues into account while performing the calculations for each column of a multiple sequence alignment (Cover and Thomas, 1991). We found that while the changes appeared to be scattered throughout the entire protein, the highest scoring (i.e. most variable) amino acid substitutions within the E protein were located at residue 329 (Table 1). This position is a Valine or Alanine in the isolates from Venezuela and Colombia included in this study, as well as those from Perú and Ecuador (de Mora et al, 2009). A region of high-scoring positions was also found between residues 380 and 385 of the E protein (Figure 2).
Figure 2. Histogram of the Shannon Entropy Calculated for the DENV-3 E Protein.
A stacked column graph showing the Shannon entropy scores calculated for each amino acid position in the DENV-3 E protein. Subsets of sequences were divided based on time and/or place of isolation as follows: Asian sequences from Thailand in 2001 (Asia 2001), Venezuelan sequences isolated before 2001 (Ven pre-2001), the first Venezuelan DENV-3 outbreak in 2001 (Ven 2001), 2002-2006 (Ven 2002-2006), the second Venezuelan DENV-3 outbreak in 2007-2008 (Ven 2007-2008), all new Venezuelan and Asian sequences characterized in the present study (This Study), and all DENV-3 sequences combined (All DENV-3). The height of each color within each peak represents the Shannon entropy for that sequence set alone at the specified position in the E protein. The height of each peak represents the amount of amino acid variability for each individual position in all sequence sets for the E mature protein. Any column that is conserved in the multiple sequence alignment will not have an entropy score. No consistent amino acid substitutions were found to exist between sequences from the two outbreaks.
When these positions were mapped onto the DENV-3 E crystal structure, along with the color-coded Shannon entropy values, we saw that while there were some substitutions in all three domains, there was a definitive cluster of residues with high Shannon entropy values (i.e. high variability) specifically in domain III (Supplementary Figure 1). This domain in the E protein is essential for entry into the host cell and is responsible for serotype-specific neutralization escape mutations that prevent host antibodies from effectively neutralizing the virus (Modis et al., 2005). These variable amino acid positions were then compared to those obtained from a previous study that included 361 E protein sequences (Amarilla et al., 2009). We found that most of the positions identified in our analysis overlapped with those that were previously reported, including the Alanine and Valine substitutions present at position 329, and have been shown to exhibit polymorphism (de Mora et al., 2009). A comparison between the substitutions existing in the E protein sequences from New- and Old World isolates revealed several residues with mutual entropy at the same position and others with entropies that differed between the same position in sequences taken from geographically-distinct areas. Interestingly, although substitutions occurred in the sequences included in this study, there were no amino acid substitutions within the E protein that completely differed between the 2001 and 2007-2008 Venezuelan outbreaks. Similarly, there were also no novel E protein amino acid substitutions when compared against all other DENV-3 whole genome sequences presently available. This result suggests that diversity in the DENV-3 E protein is only tolerable in a limited number of amino acid positions that do not negatively affect the function of the E protein.
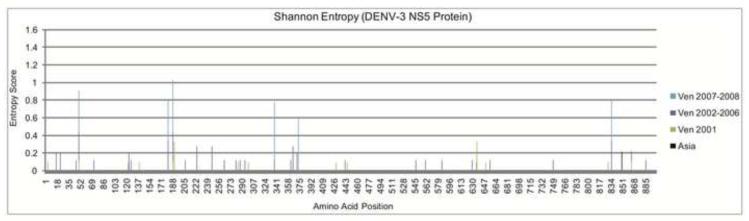
Since the enzymatic functions of NS5 include viral methyltransferase and viral RNA-dependent RNA polymerase (RdRP), we next calculated the Shannon entropy to determine the amount of variability present within this mature protein. Examination of the entropy scores from sequences spanning both Venezuelan outbreaks show low variability existing throughout the NS5 protein as a whole with relatively high peaks, classified as having scores greater than 0.47, scattered throughout the mature peptide (Figure 3). The majority of hits are located in the N-terminal third of the protein, which has been characterized as having methyltransferase activity. The remaining hits are located within the C-terminal two-thirds of the protein, which is known to contain the RdRP (Mazzon et al., 2009). In contrast, when sequences from both hemispheres were combined, only 21 positions had entropy scores higher than 0.25. Position 374 of the NS5 mature protein was the only high-scoring peak found to differ specifically between the two Venezuelan outbreaks. For the 2001 Venezuelan outbreak, Glycine was the only amino acid residue found at position 374. In contrast, Glutamic acid was found to occupy this position at an 8:3 ratio over Glycine in the 2007-2008 outbreak sequences. Interestingly, Glutamic acid, not Glycine, was the residue found at position 374 in all of the Asian sequences isolated from Thailand in 2001. No sequences from the 2010 Puerto Rico outbreak were present in our dataset. However, one strain from 2006 (DENV-3/US/BID-V2126) possessed the same G374E mutation as the 2007 Venezuelan sequences, with phylogenetic tree topology showing this sequence to be an ancestor to those from Venezuela. The other five Puerto Rican sequences included in our dataset, which contained the more common Glycine residue, were isolated between 1999 and 2005 and were not directly related to the 2007 outbreak sequences. The G374E mutation is located in a region that has been shown in Langat virus, another Flavivirus, to interfere with host JAK-STAT signaling and resulted in increased resistance to the host interferon response (Park et al., 2007). The region surrounding residue 374 was also shown to inhibit the interferon-alpha (IFN-α) response in host cells infected with DENV serotype 2 (Mazzon et al., 2009). Our entropy results show that most of the amino acid substitutions present in the NS5 protein of the sequences being investigated in this study were not shared between viruses isolated from Asian and those from the Western hemisphere. Although additional amino acid substitutions were identified throughout the NS5 protein in this analysis, the phenotypic importance of these substitutions is still unknown and further investigation is needed to ascertain whether any phenotypic changes specifically result from these substitutions. The region surrounding position 374 in the NS5 protein was examined for 51 representative sequences from the four different DENV serotypes. For serotypes 1 and 4, no intra-typic variation was observed in this region. In serotype 2, the homologous position was found to be either an Arginine or a Lysine with no apparent distinction between continents, time periods, or other criteria. This control is helpful in validating our observations for uniqueness of the dengue 3 mutations and further confirms previous reports that although the four DENV serotypes belong to the same species, each is distinct and must be studied separately.
Figure 3. Histogram of the Shannon Entropy in the DENV-3 NS5 Protein by Time of Isolation.
A stacked column graph showing the Shannon entropy scores calculated for each amino acid position in the DENV-3 NS5 protein. Subsets of sequences were divided based on time and/or place of isolation and include: the Asian sequences isolated from Thailand in 2001 (Asia), and various sequences isolated from Venezuela during the first DENV-3 outbreak in 2001 (Ven 2001), 2002-2006 (Ven 2002-2006), the second DENV-3 outbreak in 2007-2008 (Ven 2007-2008), and all Venezuelan DENV-3 sequences combined (Venezuela All). The NS5 G374E mutation occurred between the 2001 and 2007-2008 outbreaks and has been associated with viral resistance to the host interferon response. The height of each color within each peak represents the Shannon entropy for that sequence set alone at the specified position in the NS5 protein. The height of each peak represents the amount of amino acid variability for each individual position in all sequence sets for the NS5 mature protein. Any column that is conserved in the multiple sequence alignment will not have an entropy score.
Upon seeing the high degree of variability that exists in E and NS5 proteins, we used the Synonymous Nonsynonymous Analysis Program (SNAP) to calculate the ratio of nonsynonymous-to-synonymous amino acid substitutions within these coding regions. The results show all codons, consisting of the coding regions of strains isolated from Venezuela, the Western hemisphere, and Asia have a cumulative average behavior per codon score of less than 1.0 (data not shown), which indicates that positive selection pressure is not only lacking, but that neutral and/or purifying selection pressures are present in the DENV-3 III sequences included in this study.
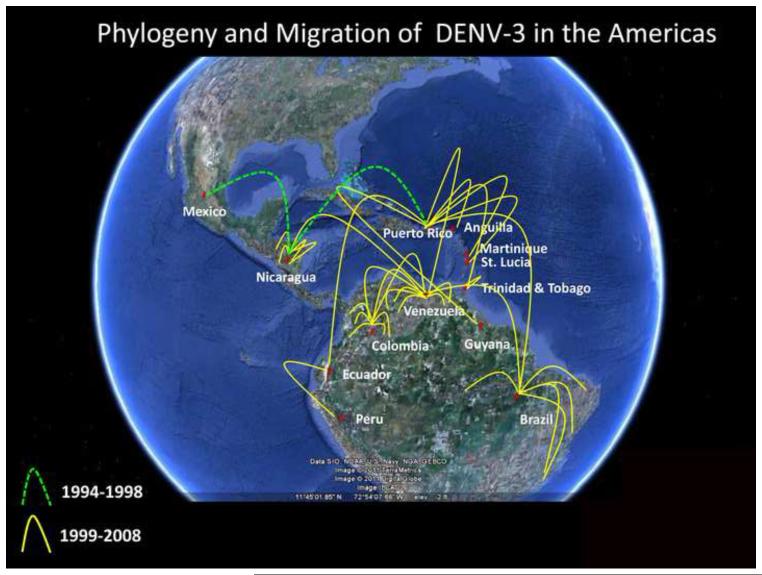
The number of whole genome sequences that were included in the phylogenetic reconstruction also allowed us to estimate the evolutionary history and the migration patterns that were associated with the introduction of DENV-3 III into the Americas (Figure 4). The Puerto Rican isolates lie basal to the rest of all other DENV-3 viruses isolated after 1998 in the Americas. Before 1998, the ancestral DENV-3 corresponded to the Nicaraguan isolate.
Figure 4.
Historical dissemination of DENV-3 in the Americas. The genomic associations portrayed in the phylogenetic analysis were plotted over this satellite image of the American continents. Each line connects an immediate ancestor with its succeeding viral isolate. Red pins indicate the countries that originated DENV-3 isolates. The green dashed lines represent transmission in and prior to 1998. Yellow lines represent transmission in and after 1999. According to the temporal and phylogenetic data, DENV-3 was re-introduced in Nicaragua in 1994. There is no evidence showing transmission of the virus until its re-appearance in 1998, where it was isolated in Nicaragua and Puerto Rico. Phylogenetic relationships show that the Nicaraguan strain is ancestral to the Puerto Rican strain but the latter experienced sustained transmission in the island. The Puerto Rican isolates lie basal to the rest of all other DENV-3 viruses isolated after 1998 in the Americas, suggesting that Puerto Rico was the springboard that launched the re-emergence of DENV-3 in the Americas.
Discussion
In this study we generated de novo full length virus genomic information and used in silico methods, including phylogenetic tree reconstruction and Shannon entropy, to examine the sequence variation existing between DENV-3 III viruses isolated during the past decade on two continents and spanning the course of two virus outbreaks in Venezuela. We found that the evolutionary phylogenetic relatedness for subsets of these viruses is primarily dependent on the geographical location of isolation, and that individual sub-lineages of each geographical region exist, which correlate with the time of isolation.
We found that a high amount of variability exists within domain III of the DENV-3 E protein, lower amounts of variability are present in the NS5 protein, and that specific amino acid positions in each of these two proteins may contribute to viral pathogenesis in the context of consecutive outbreaks.
In addition, the geo-temporal separation of our sequences in a phylogenetic tree agrees with previous studies, which showed that the geographical and temporal point of isolation are common factors in separating related sequences from the same species (Auguste et al., 2008; Bragstad et al., 2008). The phylogeny demonstrated a profound preponderance of DENV-3 III, and lack of other DENV-3 genotypes, both in Venezuela and throughout the rest of the Americas, which cannot be solely accounted for by sampling bias; indicating high viral fitness for this genotype. Higher fitness allows a given strain within a species to not only replicate or propagate more efficiently than another strain, but to persist within a subpopulation of the species in a given environment through time (da Silva et al., 2010; Domingo, 2010; Perez et al., 2009).
The average rate of mutation of 1.698×10−5 or 2.881×10−5 nucleotide substitutions per site per year is similar to the rate calculated for related viruses that also use an RdRP enzyme for genome replication (Castet et al., 2002). Since the complete DENV-3 genome is approximately 1.07×104 bases in length, this mutation rate would result in the introduction of a minimal number of nucleotide polymorphisms in any individual genome during any given year. Notwithstanding the observed mutation rate, the presence of purifying and/or neutral selection pressure in these sequences is not surprising as it has previously been shown that arthropod-borne viruses (arboviruses) undergo purifying selection pressure due to the fact that their life cycle involves alternating replication between human and insect hosts (Weaver, 2006). Even with a minimal number of changes in the viral RNA genome resulting in nonsynonymous amino acid substitutions within any individual virion, it is important to note that the overall diversity observed between the sequences included in this study is at the population scale. Consequently, the high replication rate and viral titers that result during multiple host infections throughout an epidemic greatly increases the overall genetic variation of the viruses.
Previous studies involving DENV-3 focused specifically on the E protein due to the amount of sequence data that was available for this coding region (Amarilla et al., 2009; Araujo et al., 2009a; Araujo et al., 2009b). In contrast, we used whole viral genomes in order to gain a more comprehensive idea of viral mutation and evolution. While the Shannon entropy calculations of the E protein identified several residues with intermediate to high amounts of diversity scattered throughout the amino acid sequence, a cluster of such high-scoring residues was especially apparent within domain III. Since this domain of the E protein has multiple functions, the Alanine or Valine residues that are present at position 329 in these sequences could either contribute to viral escape from the host immune response or affect viral entry into the cell. While the cluster of amino acids having high Shannon entropy scores were found at the tip of domain III of the E protein, other residues having low- to intermediate levels of Shannon entropy were also observed to be scattered randomly when mapped onto the E protein crystal structure and have yet to be characterized. The positions having elevated Shannon entropy scores for the E protein show no novel epitopes emerging between sequences pertaining to the two Venezuelan outbreaks and those from the rest of the world. This implies that the second Venezuelan outbreak was not likely attributable to a lack of “herd immunity” against a novel viral epitope that had materialized during the interim between the two outbreaks.
The NS5 protein has been shown to be not only the largest, but also the most highly conserved mature protein coded for by the DENV genome (Rawlinson et al., 2006). However, individual residues or regions can still exhibit higher-than-expected numbers of substitutions and represent areas of the protein that, when changed, allow for “fine-tuning” of protein function. The reappearance of Glutamic acid at position 374, which is present in Old World strains but absent in New World sequences between the time of DENV-3 III introduction to the Americas from Asia and shortly before the 2007 Venezuelan outbreak, suggests the presence of a stimulus in Asia that was similarly absent in the New World prior to the second Venezuelan outbreak. This stimulus could include host immunity, amino acid covariance, protein-protein interactions, and other external or internal virus selection pressures.
In addition, a homologous region of the Langat virus (Flavivirus) NS5 protein from amino acids 342 through 734 has previously been implicated in regulating IFN function (Park et al., 2007). In that study, point mutations that inhibited nuclear accumulation of STAT1 protein were screened in an in vitro system after treatment with IFN-beta (Park et al., 2007). Similar in vitro tests could be used to compare representative viruses from the 2001 and 2007 epidemics and elucidate the precise contribution of the G374E mutation to viral fitness and/or pathogenesis.
Scores from nonsynonymous-to-synonymous analysis have previously been used to identify whether positive-, neutral-, or purifying selection pressure is acting on the sequences (Ganeshan et al., 1997; Mindell, 1996; Yang, 1998). Positive selection pressure occurs when environmental factors, such as host immunity or anti-viral drugs, increase the rate of virus sequence variation and therefore the overall fitness of a virus sub-population (Kuntzen et al., 2007; von Hahn et al., 2007). The opposite of positive selection is purifying selection, which conserves a functional motif at the RNA and/or protein level(s) (Koonin; Tang et al., 2009). Neutral selection on the other hand, results from randomly-occurring mutations that have minimal effect on the evolution or fitness of the virus (Gordo et al., 2009; Ngandu et al., 2008). Given that these genome sequences display neutral and/or purifying selection pressure overall, it might be expected that several positions show evidence of positive selection pressure. However, such was not the case in this dataset as no individual position obtained a high enough score to be categorized as undergoing positive selection.
Consequently, after conducting a thorough analysis of DENV-3 genotype III sequences obtained from Venezuela during two subsequent outbreaks as well as from other regions throughout the world, we conclude that a measurable amount of divergence exists between the Old- and New World virus sequences. This is not unusual since dengue virus may rely on a multiplicity of amino acid substitutions found throughout the polyprotein to preserve its life cycle, continue its replication and hostvirus protein interactions and at the same time create immunological diversity. Moreover, when analyzed together using whole genome diversity, the American DENV-3 IIIs shows geographical evolution and possible routes of introduction to South America via the Caribbean.
The evolution of DENV-3 reported in this work depended not only on the extent to which nucleotide polymorphisms were tolerated, but also on the overall fitness of the viruses and their capability of producing viable polymorphism-containing progeny. This study has potential ramifications in the effectiveness of vaccines and anti-viral drugs in particular because of the high amount of diversity existing between Asian and American DENV-3 III sequences. Therefore, following the genetic diversity of the dengue virus will be necessary in the future since the degree of protection and efficacy that results from vaccines would depend on the representative isolate(s) that are used to develop such vaccines.
Materials and Methods
Whole genome DENV-3 sequences were obtained from either Brazil (PV, AM, MN), Puerto Rico (JM-G) or Venezuela (GC, NB) or obtained from the Broad Institute genome project directly (http://www.broadinstitute.org/annotation/viral/Dengue/). Published complete dengue virus sequences were also obtained from the Viral Bioinformatics Resource Center (www.vbrc.org). The metadata associated with these sequences was obtained or verified using either the Broad Institute website (www.broadinstitute.org), or the NIAID Virus Pathogen Database and Analysis Resource (ViPR) website (www.viprbrc.org). The sequences were aligned using the MUSCLE program (Edgar, 2004), and were then codon-aligned manually. Alignments of smaller numbers of sequences were constructed in a similar manner for isolates taken from different geographical locations and/or time points of isolation.
Individual Shannon entropies were calculated by either the Los Alamos HIV sequence database resource (www.hiv.lanl.gov/content/sequence/ENTROPY/entropy_one.html), or were calculated and mapped onto the E crystal structure (PDB: 1UZG) by the H2PDB tool (http://bio.dfci.harvard.edu/Tools/entropy2pdb.html) and Chimera (Pettersen et al., 2004). The location of the non-zero (i.e. non-conserved) values within the polyprotein were then compared and contrasted between the various alignments to find mutations unique to any subset(s) of sequences.
To optimize the parameters for phylogenetic tree reconstruction, jModeltest was used to identify Symmetrical Model plus gamma (SYM+G) as the best-fit nucleotide substitution model to use for the sequences being analyzed (Posada, 2008). Phylogenetic tree reconstructions were made with MrBayes using the SYM+G model and all other parameters set to default (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). The algorithm was allowed to run for 700,000 generations until the standard deviation between both runs and the Potential Scale Reduction Factor (PSRF) approached 0.01 and 1.0 respectively. Tree samples were taken every 100 generations with the first 25% (1,750) of trees discarded as “burn-in”. Support values for the consensus tree were then calculated from the remaining 5,250 trees. The leaves of the trees were color-coded and labeled according to the temporal and geographical point of isolation.
The mutation rate analysis was performed using BEAST version 1.5.3b (Drummond and Rambaut, 2007). Both strict and uncorrelated exponential relaxed molecular clocks were used to calculate the mutation rate for the DENV-3 genotype III sequences isolated from the Western hemisphere (Drummond et al., 2006). A general time reversible (GTR) substitution model with estimated base frequencies and gamma + invariant sites heterogeneity model was used with four gamma categories. Default priors and auto-optimized operators were set to calculate the mutation rate with a total chain length of one million and sampling every one thousand generations.
The synonymous nonsynonymous analysis program (SNAP) (www.hiv.lanl.gov), which uses a codon-by-codon pairwise comparison analysis using the method described by Nei and Gojobori (Nei and Gojobori, 1986), was run locally to calculate the synonymous/nonsynonymous substitution ratios from the codon-aligned nucleotide sequences (Korber, 2000). Regions having high synonymous mutation values (SNAP score ≤ 1.0) were categorized as undergoing either neutral or purifying selection pressure.
Sequencing was performed by extracting viral RNA from plasma samples or cell supernatants using the QiaAmp Viral RNA extraction kit (Qiagen). We created cDNA with a 20ul reverse transcription reaction containing: 1ul Superscript III Reverse Transcriptase (Invitrogen); random hexamers (1ul of 50ng/ul stock); Specific 3′ reverse primer (see below) (1ul of 10uM stock) and 5ul of template RNA. We used primer 5′AGAACCTGTTGATTCAACAGCAC3′ for specific priming of the RT reaction of samples of Dengue 3. We used 20 ul of viral cDNA, diluted to 800 ul in water, as template for 96 specific PCR reactions (supplementary table 3). Into each 10 ul PCR reaction we put: 3ul of template; 0.03 ul of pfuUltra II polymerase (5U/ul) (Stratagene); 100 mM dNTPs (Applied Biosystems) and 4 ul of a mixture of forward and reverse primers (0.5 uM stock). Our primers were synthesized with M13 sequence tags (forward primers with 5′GTAAAACGACGGCCAGT3′ and reverse primers with 5′CAGGAAACAGCTATGACC3′) so that PCR amplicons could be sequenced using universal M13 forward and reverse primers. Our PCR reactions produced 96 overlapping amplicons, each 500-900 nucleotides in length, which were subsequently sequenced bidirectionally using the Big Dye chemistry on ABI3730×l DNA sequencers (Applied Biosystems).
Supplementary Material
Supplementary Table 1: Shows the sequence names, GenBank accession numbers, and other metadata for the newly sequenced isolates from the Western hemisphere included in the phylogenetic analysis presented in this study.
Supplementary Table 2: Shows the sequence names and GenBank accession numbers for all DENV-3 sequences included in the Shannon Entropy calculations included in this study.
Supplementary Table 3: Sequencing primers
Supplementary Figure 1: Crystal Structure Showing Shannon Entropy: The Shannon entropy scores from an amino acid multiple sequence alignment containing the Venezuelan and Asian sequences obtained for the current study were calculated. These entropy values are mapped onto a stick model of the crystallized DENV-3 E protein (PDB: 1UZG) with the amino acid positions having measureable Shannon entropy scores color-coded according to their entropy score (red > yellow > green > blue > purple). The Shannon entropy scores are only shown on one of the monomers that make up the E homodimer. There is a definitive cluster of residues located in domain III of the E protein that have high Shannon entropy scores, including amino acid 329. No unique consistent amino acid substitutions existed between the 2001 and 2007-2008 Venezuelan DENV-3 outbreaks.
Acknowledgments
We thank our colleagues Dr. Aranvinda de Silva, Eva Harris, Duane Gubler and Richard Jarman for viral sequence information. Our thanks to Dr. Angel Balmaseda from the Nicaraguan Ministry of Health of Nicaragua and to the Banco de Sangre de Caracas, for clinical sample collection.
MLN is supported by a FAPESP grant and AM is supported by INCT-CNpq, Brazil. BEP and EJL are supported by NIH/NIAID Contract No. HHSN266200400036C. This project was funded in part by the National Institutes of Health, contract HHSN266200400001C (B.W.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amarilla AA, de Almeida FT, Jorge DM, Alfonso HL, de Castro-Jorge LA, Nogueira NA, Figueiredo LT, Aquino VH. Genetic diversity of the E protein of dengue type 3 virus. Virology journal. 2009;6:113. doi: 10.1186/1743-422X-6-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino JD, Tang WF, Ishii R, Ono T, Eshita Y, Aono H, Makino Y. Molecular epidemiology of dengue virus serotypes 2 and 3 in Paraguay during 2001-2006: the association of viral clade introductions with shifting serotype dominance. Virus research. 2008;137:266–270. doi: 10.1016/j.virusres.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Aquino VH, Amarilla AA, Alfonso HL, Batista WC, Figueiredo LT. New genotype of dengue type 3 virus circulating in Brazil and Colombia showed a close relationship to old Asian viruses. PloS one. 2009;4:e7299. doi: 10.1371/journal.pone.0007299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino VH, Anatriello E, Goncalves PF, EV DAS, Vasconcelos PF, Vieira DS, Batista WC, Bobadilla ML, Vazquez C, Moran M, Figueiredo LT. Molecular epidemiology of dengue type 3 virus in Brazil and Paraguay, 2002-2004. The American journal of tropical medicine and hygiene. 2006;75:710–715. [PubMed] [Google Scholar]
- Araujo JM, Bello G, Schatzmayr HG, Santos FB, Nogueira RM. Dengue virus type 3 in Brazil: a phylogenetic perspective. Memorias do Instituto Oswaldo Cruz. 2009a;104:526–529. doi: 10.1590/s0074-02762009000300021. [DOI] [PubMed] [Google Scholar]
- Araujo JM, Nogueira RM, Schatzmayr HG, Zanotto PM, Bello G. Phylogeography and evolutionary history of dengue virus type 3. Infect Genet Evol. 2009b;9:716–725. doi: 10.1016/j.meegid.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Auguste AJ, Pybus OG, Carrington CV. Evolution and dispersal of St. Louis encephalitis virus in the Americas. Infect Genet Evol. 2008 doi: 10.1016/j.meegid.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Barcelos Figueiredo L, Batista Cecilio A, Portela Ferreira G, Paiva Drumond B, Germano de Oliveira J, Bonjardim CA, Peregrino Ferreira PC, Kroon EG. Dengue virus 3 genotype 1 associated with dengue fever and dengue hemorrhagic fever, Brazil. Emerging infectious diseases. 2008;14:314–316. doi: 10.3201/eid1402.070278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero PR, Mistchenko AS. Genetic analysis of dengue virus type 3 isolated in Buenos Aires, Argentina. Virus research. 2008;135:83–88. doi: 10.1016/j.virusres.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Bragstad K, Nielsen LP, Fomsgaard A. The evolution of human influenza A viruses from 1999 to 2006: a complete genome study. Virology journal. 2008;5:40. doi: 10.1186/1743-422X-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castet V, Fournier C, Soulier A, Brillet R, Coste J, Larrey D, Dhumeaux D, Maurel P, Pawlotsky JM. Alpha interferon inhibits hepatitis C virus replication in primary human hepatocytes infected in vitro. Journal of virology. 2002;76:8189–8199. doi: 10.1128/JVI.76.16.8189-8199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TM, Thomas JA. Elements of Information Theory. John Wiley & Sons, Inc; New York: 1991. [Google Scholar]
- da Silva J, Coetzer M, Nedellec R, Pastore C, Mosier DE. Fitness Epistasis and Constraints on Adaptation in a Human Immunodeficiency Virus Type 1 Protein Region. Genetics. 2010 doi: 10.1534/genetics.109.112458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mora D, Andrea LD, Alvarez M, Regato M, Fajardo A, Recarey R, Colina R, Khan B, Cristina J. Evidence of diversification of dengue virus type 3 genotype III in the South American region. Arch Virol. 2009;154:699–707. doi: 10.1007/s00705-009-0343-7. [DOI] [PubMed] [Google Scholar]
- Domingo E. Mechanisms of viral emergence. Veterinary research. 2010;41:38. doi: 10.1051/vetres/2010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC evolutionary biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo A, Recarey R, de Mora D, L DA, Alvarez M, Regato M, Colina R, Khan B, Cristina J. Modeling gene sequence changes over time in type 3 dengue viruses from Ecuador. Virus research. 2009;141:105–109. doi: 10.1016/j.virusres.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Ganeshan S, Dickover RE, Korber BT, Bryson YJ, Wolinsky SM. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. Journal of virology. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo I, Gomes MG, Reis DG, Campos PR. Genetic diversity in the SIR model of pathogen evolution. PloS one. 2009;4:e4876. doi: 10.1371/journal.pone.0004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler D. The emergence of epidemic dengue fever and dengue hemorrhagic fever in the Americas: a case of failed public health policy. Rev Panam Salud Publica. 2005;17:221–224. doi: 10.1590/s1020-49892005000400001. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Mahalingam S, Marovich MA, Ubol S, Mosser DM. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: disease regulation by immune complexes. Lancet Infect Dis. 2010;10:712–722. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kochel T, Aguilar P, Felices V, Comach G, Cruz C, Alava A, Vargas J, Olson J, Blair P. Molecular epidemiology of dengue virus type 3 in Northern South America: 2000--2005. Infect Genet Evol. 2008;8:682–688. doi: 10.1016/j.meegid.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Taming of the shrewd: novel eukaryotic genes from RNA viruses. BMC biology. 8:2. doi: 10.1186/1741-7007-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B. HIV Signature and Sequence Variation Analysis. In: Rodrigo AG, Learn GH, editors. Computational Analysis of HIV Molecular Sequences. Kluwer Academic Publishers; Dordrecht, Netherlands: 2000. pp. 55–72. [Google Scholar]
- Kuntzen T, Timm J, Berical A, Lewis-Ximenez LL, Jones A, Nolan B, Schulze zur Wiesch J, Li B, Schneidewind A, Kim AY, Chung RT, Lauer GM, Allen TM. Viral sequence evolution in acute hepatitis C virus infection. Journal of virology. 2007;81:11658–11668. doi: 10.1128/JVI.00995-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Lewis JG, Gubler DJ, Trent DW. Molecular evolution and epidemiology of dengue-3 viruses. The Journal of general virology. 1994;75(Pt 1):65–75. doi: 10.1099/0022-1317-75-1-65. [DOI] [PubMed] [Google Scholar]
- Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. The Journal of infectious diseases. 2009;200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- Mendez J, Bernal M. Serotipos de dengue aislados por semana epidemiológica dentro de la vigilancia de enfermedades febriles. Laboratorio de Virología, Instituto Nacional de Salud; Bogota, Colombia: 2002. [Google Scholar]
- Miller N. Recent progress in dengue vaccine research and development. Curr Opin Mol Ther. 2010;12:31–38. [PubMed] [Google Scholar]
- Mindell DP. Positive selection and rates of evolution in immunodeficiency viruses from humans and chimpanzees. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3284–3288. doi: 10.1073/pnas.93.8.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. Journal of virology. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular biology and evolution. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Ngandu NK, Scheffler K, Moore P, Woodman Z, Martin D, Seoighe C. Extensive purifying selection acting on synonymous sites in HIV-1 Group M sequences. Virology journal. 2008;5:160. doi: 10.1186/1743-422X-5-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira RM, Miagostovich MP, de Filippis AM, Pereira MA, Schatzmayr HG. Dengue virus type 3 in Rio de Janeiro, Brazil. Memorias do Instituto Oswaldo Cruz. 2001;96:925–926. doi: 10.1590/s0074-02762001000700007. [DOI] [PubMed] [Google Scholar]
- Ocazionez RE, Cortes FM, Villar LA, Gomez SY. Temporal distribution of dengue virus serotypes in Colombian endemic area and dengue incidence. Reintroduction of dengue-3 associated to mild febrile illness and primary infection. Memorias do Instituto Oswaldo Cruz. 2006;101:725–731. doi: 10.1590/s0074-02762006000700004. [DOI] [PubMed] [Google Scholar]
- Ospina MC. Vigilancia epidemiológica del dengue en Antioquia., 1er. Simposio Nacional de Virología; Iatreia, Medellín: 2004. [Google Scholar]
- Park GS, Morris KL, Hallett RG, Bloom ME, Best SM. Identification of residues critical for the interferon antagonist function of Langat virus NS5 reveals a role for the RNA-dependent RNA polymerase domain. Journal of virology. 2007;81:6936–6946. doi: 10.1128/JVI.02830-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DR, Sorrell E, Angel M, Ye J, Hickman D, Pena L, Ramirez-Nieto G, Kimble B, Araya Y. Fitness of pandemic H1N1 and seasonal influenza A viruses during co-infection: evidence of competitive advantage of pandemic H1N1 influenza versus seasonal influenza. PLoS currents. 2009:RRN1011. doi: 10.1371/currents.RRN1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pinheiro FP, Corber SJ. Global situation of dengue and dengue hemorrhagic fever, and its emergence in the Americas. World Health Stat Q. 1997;50:161–169. [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular biology and evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Rawlinson SM, Pryor MJ, Wright PJ, Jans DA. Dengue virus RNA polymerase NS5: a potential therapeutic target? Current drug targets. 2006;7:1623–1638. doi: 10.2174/138945006779025383. [DOI] [PubMed] [Google Scholar]
- Rigau-Perez JG, Ayala-Lopez A, Garcia-Rivera EJ, Hudson SM, Vorndam V, Reiter P, Cano MP, Clark GG. The reappearance of dengue-3 and a subsequent dengue-4 and dengue-1 epidemic in Puerto Rico in 1998. The American journal of tropical medicine and hygiene. 2002;67:355–362. doi: 10.4269/ajtmh.2002.67.355. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Tang WF, Ogawa M, Eshita Y, Aono H, Makino Y. Molecular evolution of Japanese encephalitis virus isolates from swine in Oita, Japan during 1980-2009. Infect Genet Evol. 2009 doi: 10.1016/j.meegid.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Thomas S, Redfern JB, Lidbury BA, Mahalingam S. Antibody-dependent enhancement and vaccine development. Expert Rev Vaccines. 2006;5:409–412. doi: 10.1586/14760584.5.4.409. [DOI] [PubMed] [Google Scholar]
- Uzcategui NY, Comach G, Camacho D, Salcedo M, Cabello de Quintana M, Jimenez M, Sierra G, Cuello de Uzcategui R, James WS, Turner S, Holmes EC, Gould EA. Molecular epidemiology of dengue virus type 3 in Venezuela. The Journal of general virology. 2003;84:1569–1575. doi: 10.1099/vir.0.18807-0. [DOI] [PubMed] [Google Scholar]
- Vilela AP, Figueiredo LB, dos Santos JR, Eiras AE, Bonjardim CA, Ferreira PC, Kroon EG. Dengue virus 3 genotype I in Aedes aegypti mosquitoes and eggs, Brazil, 2005-2006. Emerging infectious diseases. 2010;16:989–992. doi: 10.3201/eid1606.091000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hahn T, Yoon JC, Alter H, Rice CM, Rehermann B, Balfe P, McKeating JA. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology. 2007;132:667–678. doi: 10.1053/j.gastro.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Weaver SC. Evolutionary influences in arboviral disease. Current topics in microbiology and immunology. 2006;299:285–314. doi: 10.1007/3-540-26397-7_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Dengue and dengue haemorrhagic fever. 2009
- Wilson ME, Chen LH. Dengue in the Americas. Dengue Bulletin. 2002;26:44–61. [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular biology and evolution. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Shows the sequence names, GenBank accession numbers, and other metadata for the newly sequenced isolates from the Western hemisphere included in the phylogenetic analysis presented in this study.
Supplementary Table 2: Shows the sequence names and GenBank accession numbers for all DENV-3 sequences included in the Shannon Entropy calculations included in this study.
Supplementary Table 3: Sequencing primers
Supplementary Figure 1: Crystal Structure Showing Shannon Entropy: The Shannon entropy scores from an amino acid multiple sequence alignment containing the Venezuelan and Asian sequences obtained for the current study were calculated. These entropy values are mapped onto a stick model of the crystallized DENV-3 E protein (PDB: 1UZG) with the amino acid positions having measureable Shannon entropy scores color-coded according to their entropy score (red > yellow > green > blue > purple). The Shannon entropy scores are only shown on one of the monomers that make up the E homodimer. There is a definitive cluster of residues located in domain III of the E protein that have high Shannon entropy scores, including amino acid 329. No unique consistent amino acid substitutions existed between the 2001 and 2007-2008 Venezuelan DENV-3 outbreaks.