Abstract
Biochemical testing of hexosaminidase A (HexA) enzyme activity has been available for decades and has the ability to detect almost all Tay-Sachs disease (TSD) carriers, irrespective of ethnic background. This is increasingly important, as the gene pool of those who identify as Ashkenazi Jewish is diversifying. Here we describe the analysis of a cohort of 4,325 individuals arising from large carrier screening programs and tested by the serum and/or platelet HexA enzyme assays and by targeted DNA mutation analysis. Our results continue to support the platelet assay as a highly effective method for TSD carrier screening, with a low inconclusive rate and the ability to detect possible disease-causing mutation carriers that would have been missed by targeted DNA mutation analysis. Sequence analysis performed on one such platelet assay carrier, who had one non-Ashkenazi Jewish parent, identified the amino acid change Thr259Ala (A775G). Based on crystallographic modeling, this change is predicted to be deleterious, as threonine 259 is positioned proximal to the HexA alpha subunit active site and helps to stabilize key residues therein. Accordingly, if individuals are screened for TSD in broad-based programs by targeted molecular testing alone, they must be made aware that there is a more sensitive and inexpensive test available that can identify additional carriers. Alternatively, the enzyme assays can be offered as a first tier test, especially when screening individuals of mixed or non-Jewish ancestry.
Introduction
Tay-Sachs disease (TSD) (MIM ID #272800) carrier screening in the Ashkenazi Jewish (AJ) population, first initiated in the 1970s, has reduced the birthrate of infants with TSD in the AJ community worldwide by 90% (Kaback 2000). The disease is caused by the lack of beta-hexosaminidase A (HexA) enzymatic activity resulting in neurodegeneration and lethality, usually in childhood (Kaback 2006). Since three mutations in the HEXA gene account for 92–98% of AJ carriers (Triggs-Raine et al. 1990), molecular screening is highly sensitive in the more homogenous segments of the AJ population (Bach et al. 2001; Fernandes et al. 1992b). This is in contrast to non-Jewish populations, wherein over 100 TSD-causing mutations have been identified (Kaback 2006). Biochemical testing of HexA enzyme activity has been available for decades and has the ability to detect almost all carriers (Kaback 2006), irrespective of ethnic background. This is increasingly important, as the gene pool of those who identify as Ashkenazi Jewish is diversifying, and more individuals will be tested who have some non-Ashkenazi or non-Jewish heritage (Kotler-Berkowitz et al. 2003).
In a previous report (Schneider et al. 2009), we described initial findings from TSD carrier screening performed on self-identified Ashkenazi Jewish individuals who participated in population-based screening programs. In that report, we suggested that the platelet HexA enzyme assay had the ability to identify Tay-Sachs disease carriers who would have been missed if they had been screened by targeted mutation DNA analysis only. Here we present the results of an expanded cohort and also describe one such carrier with a mutation that was identified by HEXA sequencing. This mutation is predicted to be deleterious based on mutation modeling using the published HexA crystal structure as a template.
Materials and Methods
The cohort comprised 4,325 self-identified Ashkenazi Jewish individuals who participated in screening programs, as described in (Schneider et al. 2009), during the March 2006-December 2010 time period. All clinical samples were tested in the Human Genetics Laboratory at the Jacobi Medical Center by the serum and/or platelet HexA enzyme assays as well as by targeted DNA mutation analysis. Serum enzyme assay was done using heat inactivation methodology while platelet assay was performed using charge separation with DEAE-cellulose columns; both assays employ the artificial substrate 4-methylumbelliferyl-β-D-N-acetylglucosaminide to measure enzyme activity and have been described previously (Nakagawa et al. 1977, 1978; O'Brien et al. 1970). Our established reference ranges for the serum assay are noncarrier >56% HexA, carrier <47% HexA, and inconclusive 48–55% HexA. Our established reference ranges for the platelet assay are noncarrier >57% HexA, carrier <48% HexA, and inconclusive 49–56% HexA.
Genomic DNA was tested for seven common mutations (five disease-causing and two pseudodeficiency mutations) in the HEXA gene using the Tag-It™ Ashkenazi Jewish Panel (Luminex Molecular Diagnostics, Toronto, Canada). The clinical data were retrospectively retrieved, anonymized, and analyzed. Confidence intervals were calculated using the Wilson score method without continuity correction (Newcombe 1998).
HexA sequencing was performed by the Ambry Genetics Tay-Sachs Plus Test (Ambry Genetics, Aliso Viejo, CA) (all 14 exons and intron/exon boundaries in sense and antisense directions). Models were generated using PyMOL (Schrodinger 2010) employing the 2.8 Å crystallographic structure of human HexA (Lemieux et al. 2006) as a template.
Results
Platelet enzyme assays were performed on a total of 2,596 of the 4,325 individuals screened. For the most part, serum enzyme assays were performed first, with a reflex to the platelet assay for carriers and inconclusive samples, although, in some cases, (including but not limited to pregnant women) only the platelet assay was performed (780 samples did not have serum testing). The inconclusive rate for the platelet assay continued to be low (1.35% (35/2,596); 95% CI 0.1–1.9%), compared to a 29% (95% CI 28–31%) inconclusive rate for all serum tests performed (1,034 inconclusive samples of 3,545 samples tested by the serum assay).
Targeted DNA analysis for the seven mutations on all 4,325 samples showed that there were 188 carriers, with mutation distribution frequencies similar to published results (Monaghan et al. 2008; Scott et al. 2010) (Table 1). Of the 175 carriers of disease-causing mutations, 172 were platelet carriers, while three were platelet inconclusive. Of the 13 pseudodeficiency mutations carriers, 50% were designated as carriers by the platelet assay and the other 50% as inconclusive. Finally, no platelet (or serum) noncarriers (0/4,150; 95% CI 0–0.1%) were shown to be carriers by targeted DNA analysis.
Table 1.
Carrier and mutation frequency for 4,325 screenees over a 4.7-year period and correlation with platelet enzyme assay results
| Mutation | Number of times observed | % of total DNA carriers | % that were platelet carriers | % that were platelet inconclusives |
|---|---|---|---|---|
| 1278+TATC | 137 | 72.9 | 98.5 | 1.5 (n = 2) |
| IVS12+1G>C | 27 | 14.4 | 96.3 | 3.7 (n = 1) |
| G269S | 10 | 5.3 | 100 | 0 |
| del 7.6 kb | 0 | 0 | ||
| IVS9+1G>A | 1 | 0.5 | 100 | 0 |
| R247W | 12 | 6.4 | 50 | 50 (n = 6) |
| R249W | 1 | 0.5 | 100 | 0 |
| Total carriers by targeted DNA analysis | 188 | 95.2 | 4.8 (n = 9) | |
| Enzyme positive and targeted DNA negative | 17 | 100 | 0 |
Importantly, there were individuals (n = 17) who were shown to be carriers by the platelet assay but did not carry any of the common DNA mutations (Table 2) (note that these individuals were never serum noncarriers). The providers associated with the screening programs offered sequencing to these individuals, but only one individual (patient #4 in Table 2) (with HexA levels of 42% in the platelet assay; carriers defined as <48%) agreed to have the follow-up HEXA gene sequencing. This sequence analysis showed the heterozygous presence of an A > G base pair substitution in HEXA exon 7 at position 775 of the HexA coding region (GenBank accession number NM_000520). This base pair change results in a Thr to Ala change at amino acid position 259 of the HexA alpha subunit (GenBank accession number NP_000511). Enzyme testing performed on the parents of this carrier showed that the mother also was a carrier, with % HexA activity levels highly comparable to those of the proband (platelet HexA levels of 46%). An oligonucleotide ligation assay on the Prism Genetic Analyzer was developed by the laboratory to interrogate position 775, and this confirmed the same nucleotide change in the mother but not the father (data not shown). This A775G (Thr259Ala) mutation also was uncovered recently after HEXA sequencing of a non-AJ carrier (Park et al. 2010), suggesting that it may not be a rare familial mutation.
Table 2.
Demographics of platelet assay-positive/targeted-DNA-panel-negative individuals
| Patient # | Gender | Serum result | Platelet result | Family history | Ancestry |
|---|---|---|---|---|---|
| 1 | F | Inconca | Carrier | No | Israel/Romania/Russia/Spain |
| 2 | M | Inconc | Carrier | Adopted | Adopted |
| 3 | F | Inconc | Carrier | No | Yugoslavia/Hungary/Serbia/Lithuania |
| 4 | M | Carrier | Carrier | No | Russia (AJb); Ireland/England/Nova Scotia (non-AJ) |
| 5 | F | Not done | Carrier | No | Austria |
| 6 | F | Not done | Carrier | Mother is a carrier | USA |
| 7 | F | Inconc | Carrier | No | Poland |
| 8 | F | Not done | Carrier | No | Russia |
| 9 | F | Inconc | Carrier | No | Hungary/Russia |
| 10 | M | Inconc | Carrier | No | Poland/Hungary |
| 11 | F | Carrier | Carrier | Father and brother are carriers | Russia/Germany |
| 12 | M | Inconc | Carrier | No | England/Poland/Greece |
| 13 | F | Inconc | Carrier | No | England/Russia |
| 14 | F | Carrier | Carrier | Yes, but does not specify | Russia/Poland/Lithuania |
| 15 | M | Inconc | Carrier | No | Russia |
| 16 | F | Carrier | Carrier | Mother is a carrier | USA/Russia/Poland |
| 17 | F | Carrier | Carrier | No | Russia/Romania |
aInconc=inconclusive
bAJ=Ashkenazi Jewish
We analyzed further the Thr259Ala change to gain insight as to whether it could represent a deleterious mutation. This residue is located in the catalytic core domain of the HexA alpha subunit (beta-hexosaminidase subunit alpha; EC = 3.2.1.52), a highly conserved domain that belongs to glycoside hydrolase family 20 (interpro IPR015883, residues 167–488 of human HexA alpha subunit) (Henrissat 1991). HexA orthologues from chicken to man bear a threonine at this position, while the comparable residue in the paralogous HexB proteins is also a threonine or the structurally similar amino acid serine. This suggests that maintaining a polar/hydrophilic residue at this position of HexA and HexB may have important functional implications for these enzymes.
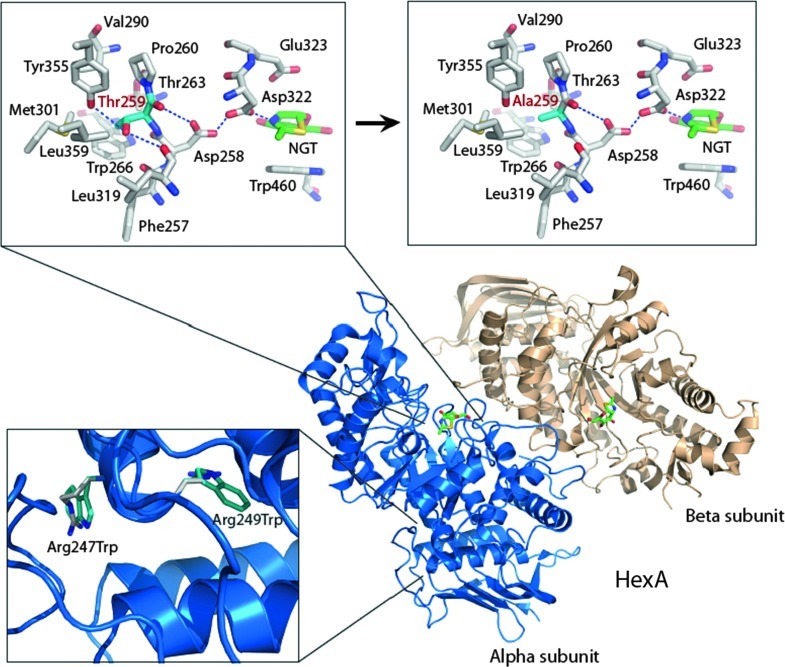
Notably, the crystallographic structure of human HexA (Lemieux et al. 2006) reveals that Thr259 is proximal to the alpha subunit active site, where it appears to help stabilize the position of Asp258, a residue that when mutated to His causes infantile Tay-Sachs (Fernandes et al. 1992a) (Fig. 1, upper left; Thr259 is shown with cyan-colored carbon atoms). Asp258 shares a proton with Asp322 via a short (2.35 Å) hydrogen bonding interaction in the HexA structure, which keeps the carboxylate of Asp322 rigidly positioned within the active site and negative. Asp322 is crucial to the catalytic mechanism of the HexA alpha subunit, as the negatively charged carboxylate on the side chain of this highly conserved residue stabilizes the positive charge that develops on the oxazolinium ion intermediate that occurs during the reaction (Lemieux et al. 2006).
Fig. 1.
Crystal structure of wild-type HexA (PDB 2GK1) and model of the Thr259Ala mutant. The wild-type active site (boxed upper left) and model of the T259A-containing active site (boxed upper right) are shown with the carbon atoms of T259 and the predicted A259 residues colored in cyan. The catalytic acid/base residue of the HexA alpha subunit (Glu323) is positioned above the mechanism-based inhibitor NAG-thiazoline (NGT; green carbon atoms). Bottom left box shows positions of the two pseudodeficiency mutations. Images were generated by PyMOL (Schrodinger 2010)
Based on the alpha subunit structure, we predict that mutation to an alanine at position 259 (Fig. 1, upper right; Ala259 is shown with cyan-colored carbon atoms) causes the loss of stabilizing hydrogen bonds that occur between the side chain OH group of Thr259 and the backbone carbonyl group of Phe257 and side chain OH group of Tyr355 (Fig. 1, upper boxes). There would also be additional loss of favorable van der Waals interactions between the side chain methyl group of Thr259 and surrounding hydrophobic residues (Fig. 1, upper boxes). Loss of these interactions is predicted to destabilize the structure in this area, causing Asp322 not to remain appropriately positioned during catalysis and thereby reducing the rate of substrate turnover. The change may also, or alternatively, cause a more serious misfolded state within this region of the protein. Accordingly, we believe that the A775G (Thr259Ala) change in the proband and mother is the likely basis for reduced HexA activity as uncovered by the platelet assay. Of note, there are benign mutations (Arg247Trp and Arg249Trp; also called pseudodeficient mutations) that have been described that lack activity against the synthetic substrates used in biochemical screening approaches but not against the natural GM2 ganglioside substrate (Triggs-Raine et al. 1992). Although Thr259 is located close to Arg247 and Arg249 in the HexA primary sequence, the tertiary structure places Thr259 much closer to the active site than Arg247 and Arg249. The arginine residues are instead located on the periphery of the catalytic domain, near an additional domain that comprises the remainder of the alpha subunit (Fig. 1, bottom left box, interdomain region of the alpha subunit).
Discussion
Retrospective analysis of our cohort of 4,325 individuals who participated in screening programs shows a TSD carrier frequency of 1/23 (95% CI 1/20 to 1/26) if we only include the 188 known DNA mutation carriers or 1/21 (95% CI 1/18 to 1/24) if we also include the set of carriers who were enzyme positive/DNA negative. These carrier rates are similar to, albeit slightly higher than, previously published rates (Broide et al. 1993; Eng et al. 1997; Schneider et al. 2009; Scott et al. 2010). This could be explained by a self-selection bias in our screened population, since 38% (77/205) of the carriers had listed that they had known TSD carriers or affected individuals in their families. Importantly, up to 8% (95% CI 5–13%) of carriers (17 platelet assay carriers/205 carriers total) of possible disease-causing mutations would have been missed if TSD testing was performed by targeted DNA mutation analysis alone. Also, given that three carriers of disease-causing mutations had inconclusive results in the platelet assay, the percentage of missed carriers could become even higher if the 26 platelet inconclusive/targeted-DNA-negative samples also were tested further.
Accordingly, if individuals are screened for TSD in broad-based programs by molecular testing alone, they must be made aware that there is a more sensitive and inexpensive test available that can identify additional carriers. This is in line with a recent position statement by the National Tay-Sachs and Allied Disease Association (NTSAD 2009) and reflects the importance of having an accurate and sensitive method to screen individuals who may have mixed ancestry. Ashkenazi Jewish individuals participating in population-based screening programs appear to be becoming more genetically diverse, in keeping with demographic studies that reflect exogamy, higher intermarriage rates, adoption, egg/sperm donation, and individuals uncertain about their ancestral origins (Kotler-Berkowitz et al. 2003). Indeed, the one enzyme positive-DNA negative sample that we were able to follow up by sequence analysis has one non-AJ parent. As mentioned, the Thr259Ala mutation that we identified in this family was reported in a study of non-AJ TSD carriers and late-onset patients (Park et al. 2010). Although in that report the authors speculated that this Thr259Ala could be a benign polymorphism, we suggest that it may instead be disease causing. While the precise nature of this mutation remains to be demonstrated formally, employing data such as that derived from crystallographic structures can prove to be very valuable for contemplating the possible clinical significance of novel changes observed by gene sequencing (see also (Richards et al. 2008)).
Based on our findings, we propose an approach to TSD screening in self-reported AJ ancestry populations that would start with enzyme assays. Normal results would not require further testing. Targeted DNA analysis would be required in the case of abnormal and inconclusive results, and gene sequencing analysis should be considered if none of the frequent mutations are detected. Such an approach, with possible expansion of the targeted mutation panel, may prove beneficial if TSD enzyme screening were to be expanded to non-Jewish and/or non-Ashkenazi Jewish populations.
Acknowledgments
We thank the Jonas Ehrlich Charitable Foundation for their generous support to the Carrier Testing Program at the Jacobi Human Genetics Laboratory, as well as the generous donors who subsidized the screening programs. We thank the National Tay-Sachs and Allied Diseases Association for their grant for HEXA sequence analysis. We thank Raffi Sturm for his early contributions to data analysis.
Take Home Message
The platelet HexA enzyme assay is a highly effective method for Tay-Sachs disease carrier screening, with a low inconclusive rate and the ability to detect putative disease-causing carriers who would have been missed by targeted DNA mutation analysis alone.
Competing Interests
Steven Keiles and Anja Kammesheidt are employees and own stock in Ambry Genetics, and Tina Hambuch was also an employee there at the time of the study.
Authors’ Contributions
Sachiko Nakagawa • Jie Zhan • Wei Sun: directed the laboratory component of this study and generated all the cohort data under the direct supervision of SJG and NSA
Steven Keiles • Tina Hambuch • Anja Kammesheidt: responsible for the HEXA sequence analysis and mutation identification
Brian L. Mark: responsible for mutation modeling and interpretation
Adele Schneider: responsible for recruitment of participants and, in particular, the proband
Sachiko Nakagawa • Jose Carlos Ferreira • Susan Gross • Nicole Schreiber-Agus: interpreted the data and drafted and revised the article
Nicole Schreiber-Agus is the guarantor for the article
Footnotes
Competing interests: None declared
References
- Bach G, Tomczak J, Risch N, Ekstein J. Tay-Sachs screening in the Jewish Ashkenazi population: DNA testing is the preferred procedure. Am J Med Genet. 2001;99(1):70–75. doi: 10.1002/1096-8628(20010215)99:1<70::AID-AJMG1120>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Broide E, Zeigler M, Eckstein J, Bach G. Screening for carriers of Tay-Sachs disease in the ultraorthodox Ashkenazi Jewish community in Israel. Am J Med Genet. 1993;47(2):213–215. doi: 10.1002/ajmg.1320470214. [DOI] [PubMed] [Google Scholar]
- Eng CM, Schechter C, Robinowitz J, Fulop G, Burgert T, Levy B, Zinberg R, et al. Prenatal genetic carrier testing using triple disease screening. JAMA. 1997;278(15):1268–1272. doi: 10.1001/jama.1997.03550150072038. [DOI] [PubMed] [Google Scholar]
- Fernandes M, Kaplan F, Natowicz M, Prence E, Kolodny E, Kaback M, et al. A new Tay-Sachs disease B1 allele in exon 7 in two compound heterozygotes each with a second novel mutation. Hum Mol Genet. 1992;1(9):759–761. doi: 10.1093/hmg/1.9.759. [DOI] [PubMed] [Google Scholar]
- Fernandes MJ, Kaplan F, Clow CL, Hechtman P, Scriver CR. Specificity and sensitivity of hexosaminidase assays and DNA analysis for the detection of Tay-Sachs disease gene carriers among Ashkenazic Jews. Genet Epidemiol. 1992;9(3):169–175. doi: 10.1002/gepi.1370090303. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback MM. Population-based genetic screening for reproductive counseling: the Tay-Sachs disease model. Eur J Pediatr. 2000;159(Suppl 3):S192–S195. doi: 10.1007/PL00014401. [DOI] [PubMed] [Google Scholar]
- Kaback MM (2006) Hexosaminidase A deficiency. In: Pagon RA, Bird TD, Dolan CR, Stephens K (eds) GeneReviews [Internet]. 2010/03/20 ed. University of Washington, Seattle, WA
- Kotler-Berkowitz L, Cohen SM, Ament J, Klaff V, Mott F, Peckerman-Neuman D. National Jewish Population Survey 2000–2001. New York: United Jewish Communities; 2003. [Google Scholar]
- Lemieux MJ, Mark BL, Cherney MM, Withers SG, Mahuran DJ, James MN. Crystallographic structure of human beta-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of GM2 ganglioside hydrolysis. J Mol Biol. 2006;359(4):913–929. doi: 10.1016/j.jmb.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan KG, Feldman GL, Palomaki GE, Spector EB. Technical standards and guidelines for reproductive screening in the Ashkenazi Jewish population. Genet Med. 2008;10(1):57–72. doi: 10.1097/GIM.0b013e31815f6eac. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Kumin S, Nitowsky HM. Human hexosaminidase isozymes: chromatographic separation as an aid to heterozygote identification. Clin Chim Acta. 1977;75(2):181–191. doi: 10.1016/0009-8981(77)90189-9. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Kumin S, Fox D, Nitowsky HM. Human hexosaminidase isozymes. III. Distribution and activity of isozymes in peripheral blood leukocytes and platelets. J Lab Clin Med. 1978;91(6):922–928. [PubMed] [Google Scholar]
- Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17(8):857–872. doi: 10.1002/(SICI)1097-0258(19980430)17:8<857::AID-SIM777>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- NTSAD (2009) NTSAD Position Statement – “Standards for Tay-Sachs Carrier Screening”
- O'Brien JS, Okada S, Chen A, Fillerup DL. Tay-sachs disease. Detection of heterozygotes and homozygotes by serum hexosaminidase assay. N Engl J Med. 1970;283(1):15–20. doi: 10.1056/NEJM197007022830104. [DOI] [PubMed] [Google Scholar]
- Park NJ, Morgan C, Sharma R, Li Y, Lobo RM, Redman JB, et al. Improving accuracy of Tay Sachs carrier screening of the non-Jewish population: analysis of 34 carriers and six late-onset patients with HEXA enzyme and DNA sequence analysis. Pediatr Res. 2010;67(2):217–220. doi: 10.1203/PDR.0b013e3181c6e318. [DOI] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10(4):294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- Schneider A, Nakagawa S, Keep R, Dorsainville D, Charrow J, Aleck K, et al. Population-based Tay-Sachs screening among Ashkenazi Jewish young adults in the 21st century: Hexosaminidase A enzyme assay is essential for accurate testing. Am J Med Genet A. 2009;149A(11):2444–2447. doi: 10.1002/ajmg.a.33085. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC (2010) The PyMOL Molecular Graphics System, Version 1.3r1
- Scott SA, Edelmann L, Liu L, Luo M, Desnick RJ, Kornreich R. Experience with carrier screening and prenatal diagnosis for 16 Ashkenazi Jewish genetic diseases. Hum Mutat. 2010;31(11):1240–1250. doi: 10.1002/humu.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triggs-Raine BL, Feigenbaum AS, Natowicz M, Skomorowski MA, Schuster SM, Clarke JT, et al. Screening for carriers of Tay-Sachs disease among Ashkenazi Jews. A comparison of DNA-based and enzyme-based tests. N Engl J Med. 1990;323(1):6–12. doi: 10.1056/NEJM199007053230102. [DOI] [PubMed] [Google Scholar]
- Triggs-Raine BL, Mules EH, Kaback MM, Lim-Steele JS, Dowling CE, Akerman BR, et al. A pseudodeficiency allele common in non-Jewish Tay-Sachs carriers: implications for carrier screening. Am J Hum Genet. 1992;51(4):793–801. [PMC free article] [PubMed] [Google Scholar]