Abstract
Nanoscale assemblies are a unique class of materials, which can be synthesized from inorganic, polymeric or biological building blocks. The multitude of applications of this class of materials ranges from solar and electrical to uses in food, cosmetics and medicine. In this review, we initially highlight characteristic features of polymeric nanoscale assemblies as well as those built from biological units (lipids, nucleic acids and proteins). We give special consideration to protein nanoassemblies found in nature such as ferritin protein cages, bacterial microcompartments and vaults found in eukaryotic cells and designed protein nanoassemblies, such as peptide nanofibres and peptide nanotubes. Next, we focus on biomedical applications of these nanoscale assemblies, such as cell targeting, drug delivery, bioimaging and vaccine development. In the vaccine development section, we report in more detail the use of virus-like particles and self-assembling polypeptide nanoparticles as new vaccine delivery platforms.
Keywords: nanoparticle, nanoscale assemblies, virus-like particles, peptide nanoparticles, drug delivery, vaccines
1. Introduction
Nanotechnology encompasses the understanding and control of matter at dimensions between 1 and 100 nm (http://www.nano.gov). At those dimensions, materials may acquire unusual physical, chemical and biological properties, and functions that are remarkably different from those observed at the macro scale. For example, at the macro scale, one of carbon's allotropes graphite is characterized by softness, whereas at the nanoscale, carbon nanotubes show unique strength of attraction [1]. Fullerene is another example of a nanomaterial composed of carbon. Other examples of nanomaterials are quantum dots (QDs), liposomes, dendrimers, paramagnetic nanoparticles and virus-like particles (VLPs).
The future implications of nanotechnology are outstanding as it can offer more solutions to technological problems than conventional systems. Nanotechnology is a multi-disciplinary field, which can create materials and devices that can be applied to aerospace, medicine, information technology, agriculture and energy production. The aim of this review is twofold: first, to describe new methods of producing nanoscale assemblies and their characteristic features; and second, to report novel developments in nanoscale assemblies that can be applied to medical treatments involving diagnosis, prevention and treatment of disease.
Initially, polymer nanosystems will be reviewed in the light of their potential biomedical applications. Following that, nanomaterials composed of biomolecules, such as nucleic acids and proteins (nanoscale bioassemblies), and their potential medical applications will be described. A special focus is given to nanobioassemblies as these are biocompatible and thus avoid toxicity issues, which may be present in polymer nanosystems. Another important point when linking nanotechnology to biology is that those nanoparticles have the same size as biological entities such as ribosomes and viruses [2].
2. Polymer-based nanoscale assemblies
Polymer nanocapsules have various useful applications. They can act as drug transporters, constricted reaction vessels, shielding casings for enzymes or cells, gene delivery systems, protective shells for heterogeneous catalysis, dye dispersants or as mediums for the displacement of contaminated waste. Here we focus on the biomedical applications of polymer nanocapsules only. For example, a modified oil-in-water emulsion technique was used to fabricate poly(u-pentadecalactone-co-p-dioxanone) (poly(PDL-co-DO)) copolyester nanoparticles that were encapsulated with an anticancer drug, doxorubicin (Dox), or an oligonucleotide, siRNA [3]. The poly(PDL-co-DO) copolyesters were being investigated as new materials for biomedical applications. Both the Dox and the siRNA encapsulated nanoparticles showed a biphasic release profile over many weeks. It was also found that the physical properties and biodegradation rate could be adjusted over a broad range by varying the copolymer composition. The authors concluded that poly(PDL-co-DO) copolymers which are enzymatically synthesized are promising biomaterials.
Poly(lactic-co-glycolic acid) (PLGA) is a biomaterial that was approved by the US Food and Drug Administration (FDA) in 1969 and, since then, has been in continuous, safe clinical use. It has also been approved by the European Medicine Agency in several drug delivery systems in humans. For a very comprehensive review of PLGA-based nanoparticles and their biomedical applications, see [4]. PLGA nanoparticles have been shown to be an adequate vehicle for the delivery of siRNA [5]. Recently, there has been much attention given to siRNA for the development of a new class of therapeutic agents. However, a major hurdle for its clinical utility has been inefficient in vivo delivery. By creating a multi-functional PLGA nanoparticle encapsulated with siRNA, Zhou et al. [5] showed that target genes could be knocked down and tumour growth could be controlled in vivo. Notably, eight separately controlled functions were incorporated in the PLGA nanoparticles. Just as in vivo delivery of siRNA is challenging, so is any nucleic acid delivery. Poly(glycidyl methacrylate) is an interesting polymer because its pendant epoxide groups can be opened with different functional groups to fabricate poly(glycerol methacrylate) (PGOHMA) derivatives [6]. Gao and co-workers reported that some aminated PGOHMAs readily complexed with an antisense oligonucleotide and high transfection efficacy was obtained. Some other water-insoluble PGOHMAs could form pH-sensitive nanoassemblies. PGOHMAs represent thus another interesting alternative to oppositely charged delivery vehicles, such as cationic polymers and lipids traditionally used in nucleic acid delivery.
In order to create polymer nanoparticles, several approaches are being investigated, including template synthesis, self-assembly, emulsion polymerization and core removal of dendrimers [7]. Foster et al. [8] used the collapse of single polymer chains by UV irradiation to create polymer nanoparticles. In an effort inspired by biology, hydrogen bonding was the driving force responsible for the reversible folding into a nanoparticle. The nanoparticles were created from poly(norbornene) diblock copolymers in which the minor block has either a urea or urethane pendant group with an ureidopyrimidinone moiety. These polymer nanoparticles could be used for drug delivery.
Kim et al. [9] departed from the traditional approaches of synthesizing polymer nanocapsules, such as the use of an emulsifier or template, by directly creating a polymer nanocapsule using traditional chemical reactions. The building blocks of these polymer nanocapsules are cucurbit[6]uril (CB), which are rigid, disc-shaped host molecules with a cavity and multiple polymerizable groups at the periphery. When CB molecules reacted with dithiols, the CB units were brought together to form the shell of a nanosphere. These polymer nanocapsules had a highly stable structure with diameters ranging from approximately 60 to 600 nm. Their hollow interiors lend themselves to a wide range of applications, such as imaging and drug delivery. Recently, Sakai et al. [10] prepared polymeric nanocapsules made of mixtures of poly(ε-caprolactone) (PCL) and PLGA using an electrocapillary emulsification method. The nanometre level was obtained as the proportion of PLGA added to the mixture increased. The duration of glucose release from the PCL/PLGA nanocapsules was longer compared with PCL capsules.
As discussed by Kuykendall & Zimmerman [11], with all the aforementioned polymer nanosystems, it must be pointed out that their usefulness in a clinical setting of drug delivery depends on demonstrating that they are non-toxic, non-immunogenic and possess an appropriate biodistribution profile. Chitosan, a cationic polysaccharide, is biodegradable, innately biocompatible and non-toxic to living tissues [12]. Chitosan nanoparticles have been prepared by ionic gelation [13]. Another challenge is finding a suitable drug delivery vehicle that does not by itself enhance inflammation. Sy et al. [14] attempted to solve this problem by an emulsion/solvent-evaporation procedure to fabricate large poly(cyclohexane-1,4-diylacetone dimethylene ketal) (PCADK) microspheres encapsulated with a hydrophobic drug to treat cardiac dysfunction. Even though the obtained spheres are not at the nanoscale, the new biomaterial PCADK used in this study is of great distinction because its degradation products are neutral. This in turn prevents a grave foreign-body response by the immune system. For myocardial infarction, this is of utmost importance, as cardiac dysfunction is characterized by an inflammatory response.
Another issue to consider regarding drug delivery using polymer nanoparticles is the amount of time the polymer nanosystem circulates in the blood. Normally, the polymer nanospheres are modified with polyethylene glycol (PEG) to increase their circulation time in the bloodstream. Geng et al. [15] show that filamentous polymeric micelles known as filomicelles (which are between 22 and 60 nm in diameter, and 2–8 μm in length) stay in the blood longer than PEG vesicles and 10 times longer than spheres of similar chemistry. This line of research was inspired by filamentous viruses that infect animals, and it showed the importance of size and shape when designing a nanocarrier.
3. Nanoscale bioassemblies and their characteristic features
In contrast to polymer nanoparticles, nanoscale bioassemblies are nanomaterials created from biological building blocks, which can be used for biomedical applications, such as drug delivery, gene therapy, vaccination and bioimaging. Liposomes, DNA, proteins and VLPs have been extensively investigated as materials in bionanotechnology. VLPs consist of the virus capsid devoid of genetic material. For nucleic acids, proteins and viruses, the driving forces for self-assembly are multiple non-covalent interactions, which lead to the formation of exceptionally organized nanostructures with a variety of sizes and shapes.
Lee & Wang [16] reported the distinctive features of bionanoparticles making them attractive biomaterials compared with synthetic nanoparticles as follows:
— well-organized architectures with a broad selection of sizes at the nanometre scale;
— monodispersed particles with uniform size and shape;
— three-dimensional structures resolved at atomic or near-atomic levels;
— economic large-scale production in gram or even kilogram quantities;
— availability of genomic sequence, through which the composition and surface properties can be controlled through recombinant technology (applicable only in the case of proteins and VLPs); and
— in particular, both genetic and chemical modification techniques can be used to mould the scaffolds, thereby allowing theoretically unlimited alterations of the nanomaterials with submolecular precision.
3.1. Liposomes
Liposomes are lipid bilayer vesicles (figure 1e). They consist of single or multiple concentric lipid bilayers with an internal aqueous core. In the case of a single lipid bilayer, liposomes are classified as unilamellar vesicles (50–250 nm); when there are multiple bilayers, they are classified as multi-lamellar vesicles (500–5000 nm). Liposomes were first discovered by Bangham [21] and were initially used as models for plasma membranes. However, because of their versatility, liposomes started being used in biomedical applications, such as drug delivery and imaging. Their internal aqueous compartments also make them suitable as nanoscale reaction vessels [22]. The versatility of liposomes is derived from their hydrophobic outer shell (lipid bilayer) and internal hydrophilic compartments. Drugs or macromolecules that are hydrophobic can be embedded in the lipid bilayer, while those that are hydrophilic can be encapsulated in the central aqueous cavity. As a third way to engineer liposomes drug molecules can be covalently coupled or physically adsorbed to the liposome surface. When chemical conjugation occurs the mean vesicle diameter can be altered. Besides their versatility, liposomes offer the extra advantages of biocompatibility, biodegradability, low toxicity and ability for surface and size modification.
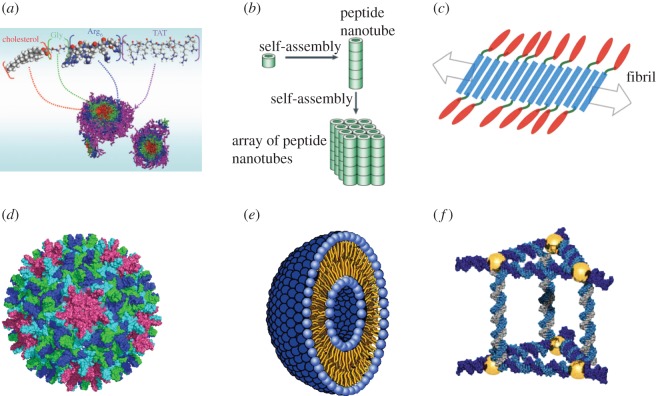
Figure 1.
Self-assembly strategies and architectures. (a) Formation of core–shell-structured nanoparticles created by linking a hydrophobic block of cholesterol to a hydrophilic block of cell-penetrating peptide TAT and six arginine residues. A linker of three glycines is used to separate the two moieties. Adapted from [17]. (b) Diagram illustrating how a cyclic peptide molecule self-assembles into peptide nanotubes through hydrogen bonding interactions followed by further self-assembly into arrays of nanotubes. Reprinted with permission from [18]. (c) Scheme showing functionalized peptide nanofibres. The blue domain self-assembles into fibrils exposing the epitope (red) by means of a linker (green). Reprinted with permission from [19]. (d) Virus-like particle of the human hepatitis B viral capsid (PDB code 1QGT). The four monomers of the asymmetric unit of the T4-icosahedron are shown in magenta, blue, green and cyan. The view is down the fivefold symmetry axis. (e) Schematic of a liposome showing the double layer of phospholipid molecules that assemble into a hollow sphere. The polar head groups at the inner and outer surface of the hollow sphere are shown in blue. The lipid chains (yellow) are directed towards the inside of the double layer. (f) Schematic showing the self-assembly of DNA molecules into the shape of a triangular prism that encapsulates six gold particles (yellow). Reprinted with permission from [20].
Liposomal technology has been clinically approved for cancer treatment [23]. Doxil is an example of liposomes loaded with the drug Dox (http://www.doxil.com/assets/DOXIL_PI_Booklet.pdf). Doxil is a stealth liposome because its surface was covalently modified with PEG to improve its circulation time. Besides short blood circulation half-lives, other shortcomings of conventional liposomes are steric stability and poor control of drug release over a prolonged period of time. Researchers from different areas, such as materials science, pharmaceuticals and cell biology, have had to collaborate in order to overcome the limitations of conventional liposome technology. One way of achieving local drug retention and sustained release over a prolonged period of time is the triggered release of bioactive molecules by environmental or external stimuli, such as pH, magnetism, temperature and electromagnetic radiation at different radiofrequencies. For instance, Thermodox is a thermally sensitive liposome loaded with Dox being studied for primary liver cancer and recurrent chest wall breast cancer (http://celsion.com/docs/technology_thermodox).
Because of their clinical success and physico-chemical flexibility, liposomes have generated interest in hybrid constructs. These constructs could obtain multiple functionalities, such as therapeutics and diagnostics. This would allow for personalization of treatment. Indeed, several liposome-hybrid systems are in preclinical development for theranostics applications. As an example, TOPO-capped QDs were incorporated in lipid bilayers, and the internal cavity was loaded with Dox with an efficiency of at least 97 per cent to create a theranostic vector [24].
3.2. Nucleic acid-based nanoassemblies
The particular size of the DNA molecule is one of its most attractive characteristics for use in nanotechnology. DNA has a diameter of approximately 2 nm, a short structural repeat of around 3.4–3.6 nm and a persistence length (a measure of stiffness) of about 50 nm. In order to create DNA-based nanostructures, the specific bonding between DNA base pairs has been explored. Shih et al. [25] reported that a 1.7 kb single-stranded DNA folded into an octahedral structure by a simple denaturation–renaturation procedure. Another method for creating specific DNA structures is DNA origami. DNA origami (named after the Japanese art of paper folding; figure 1f) is an incredibly versatile and popular method for creating DNA nanostructures. This method was pioneered in 2006 by Paul Rothemund from the California Institute of Technology, and it basically consists of taking a long single-stranded DNA and mixing it with short, complementary ‘staple’ strands [26]. Afterwards, the mixture is heated then cooled, allowing the single-stranded DNA to bind with the staples creating different shapes, such as five-pointed stars and smiley faces. A computer program is used to predict the sequences of the short ‘staple’ strands.
In 2009, Douglas et al. [27] expanded the DNA origami method by using it to create three-dimensional shapes, which turned DNA into an accessible and attractive nanomaterial for the ‘bottom-up’ fabrication of synthetic devices. Andersen et al. [28] designed and synthesized a DNA box with a programmable lid that has the potential to both ‘sense and act’. In 2012, Douglas et al. [29] advanced the DNA origami technology one step further. Douglas and colleagues created a DNA nanorobot with cell-targeting capabilities, which was capable of delivering molecular payloads such as antibody fragments and gold nanoparticles. These results have important implications in cancer treatment. For a more in-depth discussion of the DNA origami technology, see the study of Torring et al. [30].
The creation of DNA nanoparticles with other materials has been used in gene therapy as an alternative to viral vectors, which are limited due to safety concerns. The following advantages can be attributed to these non-viral delivery systems: repeated administration with the ability to evade interception by the immune system, targeting potential, long-term storage stability and easy mass-production. A folate–chitosan–DNA nanoparticle has been characterized, and it has been shown to combine the biocompatibility of chitosan with increased transfection efficiency by using folic acid as a ligand for targeting cell membranes [31]. While the current DNA delivery systems are safe and versatile, they do not have the transfection yield of viruses. It is likely that the integration of the smart mechanisms of infection by viruses into forthcoming DNA delivery systems will allow these systems to function similar to viral DNA delivery.
3.3. Virus-like particles
VLPs are composed of viral capsid proteins in their authentic conformation devoid of their genetic material (figure 1d). Because VLPs lack the DNA or RNA genome of the virus, issues relating to reversion to virulence or recombination with the wild-type or related viruses are avoided [32]. Viral particles possess the following exciting features: nanometre-sized, robust protein shells with defined geometry and astonishing uniformity making them well suited for nanoscale production; knowledge of the atomic structures of many viruses which enables researchers to mutate amino acids in the viral capsid for bioconjugation; and site-directed mutagenesis can be easily used to create cysteines and lysines in surface-accessible regions of the viral capsid, allowing for chemical conjugation of molecules to those amino acids in different regions of the viral capsid.
Because of the good accessibility of lysine and cysteine residues on VLPs, bioconjugation has been achieved using commercially available homo- or hetero-bifunctional linkers [33–36]. For example, three foreign proteins were chemically conjugated to the surface of cowpea mosaic virus (CPMV) using appropriate bifunctional cross-linkers [34]. It must be pointed out, however, that there are different chemistries being used for bioconjugation. Recently, Patel & Swartz [37] reported the use of a cell-free protein synthesis platform and a one-step, direct conjugation scheme to synthesize VLPs with ligands, such as proteins and nucleic acids, attached to their surface. Direct conjugation was done using azide-alkyne click chemistry. This synthesis platform allows the production of custom-designed VLP bioconjugates for biomedical applications such as vaccines, diagnostics and therapeutics, along with its use in other nanotechnological applications.
Another interesting application of VLPs is the presentation of fullerenes (C60 or buckyballs) on their surface. Recently, there has been much interest in the use of fullerene in biomedicine but the fact that it is insoluble in water limits its application. VLPs could have a dual purpose: first to act as hydrophilic ‘chaperones’ for C60 to improve fullerene's aqueous biocompatibility, and second, to serve as a template for the methodical organization of several C60 units in conjunction with other functional molecules. The hybrid C60–VLPs show promise in photoactivated tumour therapy [38].
VLPs have also been used to generate anti-tumour responses. McKee et al. [39] showed that by conjugating a glycolipid to a rabbit haemorrhagic disease VLP, potent anti-tumour responses were obtained from a single intravenous vaccination. Avogadri et al. [40] investigated the effect of VLP technology on melanoma, one of the most dangerous forms of skin cancer. For this immunotherapy study, they selected alphavirus-based virus-like replicon particles (VRP) expressing different antigens of melanoma differentiation antigen tyrosinase. They identified tyrosinase-related protein 2 (TRP-2) as the most potent antigen. VRP-TRP-2 showed potent therapeutic effect by eliciting TRP-2 specific antibodies and cellular immunity. It is interesting to note that the first two preventive cancer vaccines are VLP based (anti-hepatitis B virus (HBV), to prevent HBV-associated hepatocellular carcinoma [41], and anti-human papillomavirus (HPV), to prevent HPV-associated cervical carcinoma [42]).
While VLPs have therapeutic potential, it can be challenging to produce them when the functional virus coat is from a virus with a lipid envelope and is used to carry an artificial cargo. This is the case with alphaviruses, which are enveloped viruses. Dragnea and co-workers [43] were able to self-assemble an alphavirus capsid around a functionalized gold nanoparticle. While the membrane layer was absent, the self-assembly of the alphavirus capsid protein shell was a first step towards the creation of VLPs from enveloped viruses. Another challenge for VLP production is that the engineering ‘rules’ for protein self-assembly into virus capsids are still not always clear. Virus capsid folding, such as that of the hepatitis B core (HBc), is not always well understood [44]. Janssens et al. investigated improvements in the HBc particle assembly for vaccination research and found that ‘in silico predictions do not ensure assembly into particles’.
3.4. Ferritin protein cages
Ferritins are globular protein complexes that are vital to iron homeostasis and that are present throughout the microbial, plant and animal kingdoms. The ferritin protein family can self-assemble into protein cages of two types. Maxi-ferritins have 24 nearly identical protein subunits that self-assemble into a spherical cage with an octahedral symmetry [45]. According to the crystal structure solved by Lawson et al. [46], ferritin has an outer diameter of 12 nm and an inner cavity diameter of 8 nm. The iron cluster is located within the protein layer and has a dimension of 1–2 nm, which positions it between the inner and the outer shell surface at a distance of about 1 nm from each of these surfaces. The other types of nano-cage are the mini-ferritins that have tetrahedral symmetry, hollow assemblies composed of 12 monomers.
Since ferritins belong to a class of mineralization proteins, their hollow spheres have been used as size-constrained reaction containers for the synthesis of bioinorganic nanomaterials with controlled dimensions. For example, amorphous iron sulphide minerals were synthesized in situ using horse spleen ferritin [47]. Generally, the mineralization of ferritin using high-oxidation-state metal ions has been unattainable. Recently, however, Klem et al. [48] reported the photoinduced mineralization of ferritin using high-oxidation-state metal ions as precursors. They turned to the natural sequestration mechanism observed in some marine siderophore systems to photochemically synthesize stable europium, titanium and iron oxyhydroxide nanoparticles in the ferritin protein cage. A different approach to synthesize inorganic nanoparticles is to genetically engineer metal-binding peptides into the ferritin's inner cavity, creating chimeric cages. This was the approach used by Kramer et al. [49] to synthesize silver nanocrystals.
Human ferritin cages can also be used in diagnostics as magnetic resonance imaging agents for in vivo detection of vascular macrophages [50]. This is possible because ferritin accumulates in human plaque macrophages and iron oxide (magnetite) nanoparticles can be encapsulated in its cavity. Uchida et al. [51] incorporated multiple functionalities in ferritin by genetically engineering cell-specific targeting peptide on its exterior surface and synthesizing magnetite nanoparticles within the interior cavity. This multi-functional ferritin protein cage shows potential as a novel diagnostic imaging agent.
3.5. Protein-based organelles in bacteria and eukaryotic cells
Most metabolic pathways that occur in eukaryotic cells occur in membrane-enclosed organelles, such as mitochondria and lysozomes. However, approximately 20 per cent of all bacteria and all eukaryotic cells also contain protein bound organelles [52]. Two types of these natural proteinaceous complexes are bacterial microcompartments (BMCs) and vaults, present in eukaryotic cells.
BMCs are enzyme-containing proteinaceous compartments which are similar to viruses in size and shape [53]. There are three known types of BMCs: the carboxysome, the propanediol utilizing (Pdu) and the ethanolamine utilizing (Eut). The most studied BMC is the carboxysome, which is present in photosynthetic bacteria [54]. Carboxysomes contain two enzymes responsible for carbon fixation reactions: carbonic anhydrase and RuBisCo. Because of the encapsulation of the two enzymes, CO2 fixation is enhanced [55,56]. When carboxysomes were first visualized by electron microscopy decades ago, they were thought to be viruses. Indeed, there are strong resemblances between carboxysomes and viruses [57]. Carboxysomes have regular, polyhedral structures with a size of 80–120 nm [58]. A few thousand protein subunits form the shell with the basic building block being a hexameric unit. Pdu and Eut are also polyhedral but they are less regular than carboxysomes. Another difference is that Pdu and Eut encapsulate many more enzymes than the two that carboxysome encapsulates. Both Pdu and Eut have aldehydes as toxic intermediates in the metabolic pathways they are responsible for. Thus, it is thought that one of the functions of these BMCs is to limit cytosol exposure of aldehyde. In view of all these unique features, BMCs provide the possibility of being engineered as nano-bioreactors for biosynthesis and biocatalysis [59] and also for molecular delivery of drugs and other biomolecules [60].
Vaults are another example of protein-enclosed compartments, which are found in nearly all eukaryotic cells. While the function of vaults has not been clearly identified, their structure is well characterized [61,62]. A crystal structure at 3.5 Å resolution showed that rat liver vaults are ovoid spheres with overall dimensions of approximately 70 nm in length and 40 nm in width [63]. While vaults are ribonucleoprotein particles containing several copies of RNA and multiple copies of three protein species, it is possible to obtain a recombinant vault nanoparticle from only its most abundant protein (the 97 kDa major vault protein) [64]. Vault nanoparticles have a central cavity that can be used to encapsulate chemically diverse proteins simply by fusing the non-vault protein to a vault-targeting peptide [65]. This strategy allows for encapsulation of biologically active materials within the vault central cavity [65], which is of vital importance for bionanotechnology. Furthermore, studies have revealed that vaults are non-immunogenic [66]. Vaults’ structural features, in vitro stability and non-immunogenicity make them well suited for biomedical applications involving protection and encapsulation of cargo—whether it be a drug or a biomolecule [67]. Even immunogenic proteins can be encapsulated generating vaccines [66].
3.6. Designed protein nanoassemblies
Proteins are biopolymers that have been used as versatile building blocks in the construction of nanosystems. There are several favourable characteristics of proteins for the bottom-up approach of nanostructure synthesis: (i) no external catalysts are needed in construction as all the molecular information necessary for self-assembly is already found in the proteins’ three-dimensional structures, (ii) functionality can easily be increased by de novo design or directed evolution using well-known genetic and chemical procedures, and (iii) proteins show good chemical compatibility with inorganic materials, creating opportunities for applications which go beyond traditional biological applications.
From just knowing a protein's amino acid sequence, it is very challenging to predict its three-dimensional structure and to understand protein folding. Molecular dynamics can aid in our knowledge of protein folding but so far it has been limited to time scales that are inferior to the time in which biological processes occur. Shaw et al. [68,69] have constructed a custom-built supercomputer called Anton that can simulate a biological process up to 1 ms. While this is an advancement of great importance, it remains inaccessible to most research groups. Therefore, researchers have attempted to design protein nanosystems solely based on the knowledge of the structure of the peptide chains used as building blocks in these bottom-up approaches. The following two criteria are needed to successfully engineer nanostructures from naturally occurring building blocks: first, the building block of choice must have the majority of its population in the required conformation; and second, from an energistic point of view the interactions between the building blocks should be favourable enough, so as to achieve an energy gap between the desired fully folded self-assembly and other possible unfolded forms.
MacKay et al. [70] used their knowledge of elastin-like polypeptides (ELP) to create a chimeric polypeptide made up of an ELP segment and a short Cys-rich segment, which can be used to covalently attach a cancer chemotherapeutic. The conjugation of the drugs to the short Cys-rich segment triggers the self-assembly of the nanoparticles. This creates a chimeric nanoparticle with a drug-rich core surrounded by a hydrophilic peptide corona. These nanoparticles can induce nearly complete tumour regression after a single dose. Another way of triggering the self-assembly of peptide nanoparticles is by attaching a hydrophobic block of cholesterol to a hydrophilic block of cell-penetrating peptides, such as six Arg residues [17] (figure 1a). The cationic peptide has antimicrobial properties, and the nanoparticles were able to terminate bacterial growth in infected brains of rabbits.
In nature, self-assembly into well-ordered structures is ubiquitous [71]. While nature produces these well-ordered structures in a reproducible and effortless manner, in the laboratory it is very challenging for scientists to mimic nature and obtain these self-assembled structures. Nonetheless, the concept of self-assembly has been increasingly used when creating nanostructures from peptide building blocks. Molecular self-assembly process is mainly mediated by non-covalent interactions, such as ionic bonds, hydrogen bonds, hydrophobic interactions and van der Waals interactions.
Ryadnov [72] created a self-assembling peptide polynanoreactor from the non-covalent dendrimer-like assembly of short leucine-zipper sequences. The nanoreactor possessed multiple cavities and could synthesize silver nanoparticles. Padilla et al. [73] engineered self-assembling nanomaterials such as protein cages and filaments by combining naturally symmetric protein oligomers into a fusion protein. Haghpanah et al. [74] studied the fusion of two distinct self-assembling domains and found that assembly was dependent on the block orientation and number of domains. Recently, spherical virus-inspired peptide nanoassemblies have been created using a novel C3-symmetric molecular design of peptides [75].
3.6.1. Peptide nanotubes
Peptide nanotubes, or cyclic peptides, are other peptide architectures created by peptide self-assembly (figure 1b). Cyclic peptide subunits made up of alternating even numbers of d- and l-amino acid residues are the building blocks for stable, one-dimensional hollow cyclic peptide nanotubes [76]. The driving force for peptide self-assembly is the generation of an extensive network of intersubunit hydrogen bonds. Gobeaux et al. [77] revealed how counterions can modulate peptide nanotubes’ diameter by examining the structural role of counterions in the self-assembly of lanreotide, a cationic octapeptide. Potential applications of these structures are as novel antibacterial agents [78] and as antiviral agents that target HBV entry at a post-binding step [79]. Another useful application of peptide nanotubes is in drug delivery systems, because they could mimic artificial transmembrane ion channels [80]. Hourani et al. [81] found an easy route to create peptide nanotubes with tunable interiors by adding 3-amino-2-methyl benzoic acid in the d,l-alternating primary sequence of the cyclic peptide. Hourani's results are important in the light of mimicking transmembrane proteins as it leads to enhanced selectivity in molecular recognition, transport and separation processes. For a review on nanotubes and their biomedical applications, see the study of Martin & Kohli [18].
3.6.2. Peptide nanofibres
There are several ways to obtain peptide nanofibres. For example, peptide nanofibres can be obtained from a modified amyloid-β peptide (AAKLVFF) when assembly is done in water [82]. When assembly is done in methanol, nanotubes are formed [82]. The different morphologies arise from changes in the hydrogen-bonding capacity of the solvent, which may modify the propensity for β-sheets to twist. Yet another method of obtaining peptide nanofibres is using peptides with alternating hydrophobic and hydrophilic amino acid residues (self-complementary repeats) that have a tendency to adopt a β-sheet structure [83]. In physiological conditions, these short fibrillizing peptides self-assemble to form β-sheet-rich nanofibres [19]. These self-assembling peptide nanofibres can also be functionalized by extending the sequence with B-cell and T-cell epitopes (figure 1c) [19]. When administered in saline, the functionalized nanofibres raised antibodies in mice to levels similar to the epitope peptide delivered in complete Freund's adjuvant (CFA). The interesting aspect of this study was that the nanofibres alone were not immunogenic even when administered with CFA, and the antibody response was dependent on self-assembly. Recently, the molecular determinants and immunological mechanisms of the previously mentioned functionalized nanofibres [19] were investigated [84]. It was found that protection lasted at least a year and that the potent antibody response was T-cell dependent. Modifying the T-cell epitope and/or mutating the self-assembling domain, which would prevent the formation of fibres, could attenuate immunogenicity.
Apart from β-sheet peptide motifs as building blocks for the assembly of peptide nanofibres [19,82–84], α-helices can also be used to create self-assembling peptide nanofibres. Coiled coils are α-helices typically composed of a repetitive heptad amino acid sequence pattern (abcdefg)n (figure 2a). These α-helices wind around each other in a supercoil. Hydrophobic amino acids typically occupy positions a and d which create the hydrophobic core of the coiled coil. The amino acids located at positions e and g are often charged residues that form interhelical salt bridges. Coiled coils are one of the principal subunit oligomerization motifs in proteins [90].
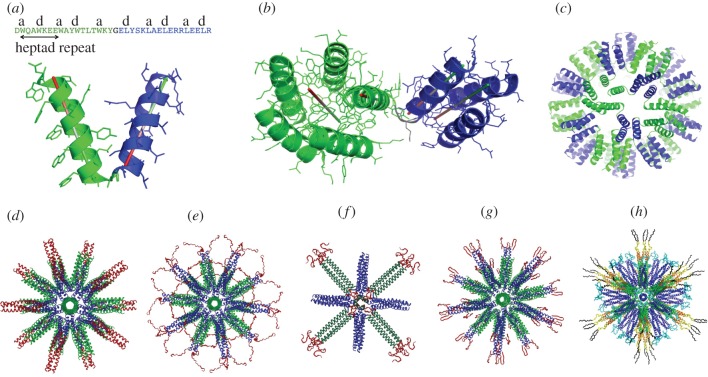
Figure 2.
Coiled coils as building blocks for the self-assembly of SAPNs. (a) Peptide sequence and structural model of the monomeric building block of the SAPN consisting of a pentameric tryptophan-zipper (green) and a trimeric leucine-zipper (blue). (b) Structural model of one pentameric and one trimeric coiled coil of the SAPN. One of the helical axes each, as well as the two coiled-coil symmetry axes are shown. (c) Computer model of the complete SAPN assembled into an icosahedron. (d) Computer model of an SAPN functionalized with a trimeric coiled-coil epitope from SARS (red). The calculated diameter is 23 nm. Adapted from [85]. (e) Computer model of an SAPN functionalized with an unstructured malaria epitope (red). Adapted from [86]. (f) Computer model of an octahedral SAPN functionalized with the tetrameric M2e epitope from avian influenza (red). The view is down the fourfold symmetry axis of the octahedron. Adapted from [87]. (g) Computer model of an SAPN functionalized with the hairpin structure of a poorly antigenic actin determinant (red). Adapted from [88]. (h) Computer model of an SAPN functionalized with the 2F5 and 4E10 epitopes from HIV. Adapted from [89].
When coiled coils are adequately designed they can self-assemble into higher-order and larger supramolecular assemblies, such as fibrils and nanoparticles. The Woolfson group first described the design and creation of peptide nanofibres using α-helical coiled coils by engineering specific charged interactions [91]. Papapostolou et al. [92] designed a dual-peptide system that coassembles in water to form protein fibres. The two peptides are complementary leucine-zipper peptides, of de novo design. The leucine-zipper motif is one type of coiled-coil architecture. The two complementary peptides were designed to assemble into offset dimeric coiled coils with complementary sticky ends to promote longitudinal assembly into fibres.
Using such coiled-coil fibres, the Woolfson group rationally designed and engineered hydrogelating self-assembling fibres (hSAFs), which differ from the previous design by weaker interactions at residues that are exposed on the surface of coiled-coil assemblies [93]. Interestingly, hSAFs support cell growth and differentiation. It must be pointed out, however, that in drug delivery applications hydrogels suffer from the limitation of rapid drug release. Ferstl et al. showed that the addition of an anionic polyelectrolyte to a cationic peptide with hydrophobic side chains formed nanofibres and their binding mode might be useful as a tool to expand the time scale of drug release in hydrogel drug delivery systems [94].
Indeed, nature creates fibres from collagen due to its triple-helix structure, which is clearly distinct from the α-helical structure. Luo & Tong [95] have advanced the field of synthetic collagen-mimetic peptides by creating a novel collagen-mimetic peptide amphiphile that self-assembles into a nanofibre with both structural and biological properties of native collagen. This is of great importance in the field of tissue regeneration, as native collagen can lead to autoimmune reactions and pathogenic side effects.
Peptide nanofibres can also be formed in response to changes in the environment. Ghosh et al. [96] designed smart, self-assembling peptide amphiphiles that changed the morphology from linear/spherical to fibrous upon lowering the pH. Potential applications of such a system are drug delivery and in vivo imaging. Another well-known and attractive fibre fabrication technique is electrospinning. Protein- and peptide-based electrospun nanofibres have recently been reviewed by Khadka & Haynie [97].
3.6.3. Peptide nanoparticles
One strategy for obtaining peptide nanoparticles is to use coiled coils as building blocks. The main features of coiled coils, which render them suitable as building blocks of nanoparticles to be used for biological applications, are their potential for modification using external stimuli, such as temperature or pH, and their highly selective and specific binding characteristics.
Boato et al. [98] designed and produced a spherical particle with a mean diameter of 17–20 nm. The basic building block was a designed self-assembling coiled-coil lipopeptide. The coiled-coil sequence was derived from a fragment of the F1 glycoprotein of respiratory syncytial virus, which has been shown to form a parallel three-helix bundle. The assumption is that self-assembly into parallel helical bundles is driven by the coiled-coil motif. The coiled-coil bundles then self-assemble into nanoparticles by clustering of the lipid chains.
Another powerful tool that can be exploited to build peptide nanoparticles is symmetry. As mentioned in the designed protein nanoassemblies section of this review, Padilla et al. [73] first harnessed the power of symmetry to design self-assembling protein cages. They fused a dimeric protein to a trimeric protein with a nine-residue rigid helical linker forming a tetrahedral protein cage.
In our group, we have combined the coiled-coil structural motif with the power of symmetry to engineer self-assembling peptide/protein nanoparticles (SAPNs) [99]. Our design is inspired by the architecture of virus capsids and capsid-like protein cages. Our SAPNs [99] aim to mimic the structural design of viruses and their icosahedral symmetry. Virus capsids of small viruses are normally composed of one to four capsid proteins that self-assemble into a protein shell, thus protecting the genomic material. Therefore, the design is based on a single polypeptide chain (figure 2a) that self-assembles into a protein nanoparticle. The icosahedron has twofold, threefold and fivefold symmetry axes, and two of them were incorporated in the design by creating a fusion protein. A pentameric coiled coil was genetically joined to a trimeric coiled coil (figure 2b). The linker between them was only one/two glycine residues, which allowed flexibility between the two domains. The alignment of these two oligomerization domains along the corresponding symmetry axes of the icosahedron and application of the symmetry elements create a nanoparticle with icosahedral symmetry (figure 2c). Identical copies of the protein chain self-assemble to produce the nanoparticle. At least 60 protein chains are required to form a nanoparticle where every chain occupies an identical environment.
We first used solid-phase peptide synthesis to obtain the peptide chain composed of two oligomerization domains with different oligomerization states joined by a short linker segment [99]. More homogeneous nanoparticles were obtained when refolding was done using lower peptide concentrations and complete denaturation of the peptide. Biophysical characterization of the peptide nanoparticles showed that nanoparticles with a 16 nm diameter were formed.
We also produced protein nanoparticles from oligomerization domains other than coiled coils, such as the globular foldon domain from fibritin with a trimeric β-sheet conformation [100]. Recombinant expression methods were used to obtain the protein chain, as chemical synthesis for these longer sequences would be too expensive. In this study, eight different proteins were expressed, purified, refolded and analysed by transmission electron microscopy and analytical ultracentrifugation. Nanoparticles of the expected icosahedral symmetry formed only when the foldon domain was extended with an additional trimeric coiled-coil domain as a combined trimerization domain that is linked to the pentameric coiled coil.
4. Biomedical applications
4.1. Cell targeting
Cell targeting is aimed at achieving high uptake of therapeutic and/or diagnostic reagent in a preferential location such as a tumour and high tumour to blood/normal tissue ratio. In this way, it is possible to reduce potential side effects and increase therapeutic/diagnostic efficiency, which are important goals in the treatment of cancer and other diseases. One way of accomplishing targeting is to change the physico-chemical properties (surface topography and charge) of the nanoscale assemblies to facilitate their intracellular delivery [101]. Targeting can also be accomplished using proteins (mainly antibodies and their substructures), peptides, nucleic acids (aptamers), small molecules or vitamins and carbohydrates. By attaching targeting moieties, specificity for cell targeting is obtained by receptor-mediated endocytosis. For example, bacteriophage MS2 VLPs were chemically conjugated to a targeting peptide (SP94) that binds human hepatocellular carcinoma (HCC) [102]. This allowed for the selective delivery of nanoparticles, chemotherapeutic drugs, siRNA cocktails and protein toxins to HCC.
VLPs have a natural affinity to target host cells, and this has been investigated for cell-targeting applications [103,104]. Other nanobioassemblies such as CPMV simply interact with different cell lines and tissues showing low cell specificity. Thus, it becomes necessary to modify them chemically or genetically for targeting purposes. For example, using a copper-catalysed azide-alkyne cyclo-addition reaction, folic acid-PEG conjugates of CPMV particles were synthesized [105]. The folic acid-PEG conjugated CPMVs were efficiently targeted to cell-surface folic acid receptors. Additionally, this study showed that higher density loading of targeting ligands on CPMV might not be necessary for efficient targeting to tumour cells. Recently, the chemical conjugation of human epidermal growth factor (EGF) to simian virus 40 VLPs allowed for cell selective targeting [106]. Simian virus 40 VLPs have gained attention in gene delivery due to their low toxicity and high stability in the blood. The cell selectivity of the simian virus 40 VLPs was evaluated with carcinoma cells that over-express the EGF.
The discovery of targeting ligands, which enhance cellular uptake, is a major task in the cell-targeting field. Recently, a novel cell-uptake selection strategy was developed to isolate specific prostate cancer (Pc) internalizing aptamers [107]. Aptamers are short RNA or DNA oligonucleotides that fold into three-dimensional conformations with high binding affinity and specificity. In simple terms, in this novel cell-uptake selection strategy, an RNA library is incubated with normal cell lines, and the unbound RNA is collected in a counter-selection fashion. Next, in an internalizing-selection step the unbound RNA is incubated with Pc cell lines. The cells are lysed and the internalized RNA is collected, reverse transcribed and amplified by polymerase chain reaction. The cycle is repeated 12 times. Not only were internalizing aptamers selected from this strategy, but proof-of-concept was also demonstrated by conjugating the aptamer to docetaxel-encapsulated nanoparticles. This kind of strategy deserves merit as it allows for the design and engineering of targeting moieties in the absence of information on the target antigens.
4.2. Drug delivery
The hollow cavity of some nanoassemblies is useful for the encapsulation of anticancer chemotherapeutics, immunotherapeutics or nucleic acids. Whether the nanoassembly be rod-shaped or spherical, genetic and chemical modifications are at the core of successful encapsulation for drug delivery applications. For example, vault nanocapsules have hollow cavities that can be used as carrier of the lymphoid chemokine CCL21 [108]. After a single intra-tumoural administration in mice, these CCL21-carrying vaults inhibit lung cancer growth [108]. It must be pointed out that there are many specific issues to consider when dealing with drug delivery, such as route of administration, drug transport in cells and tissues, clearance from the body, drug resistance, concentration distribution and total accumulation of drug in target tissues, toxicity and antigenicity and lastly dose, dose rate and time schedule of administration. It is especially challenging to deliver drugs to tumour cells because of the following factors: heterogeneous blood supply and vascular permeability, inadequate interstitial penetration, intracellular transport barriers, limited transport of hydrophilic drugs across cell membranes, and in relevant cases transport across nuclear envelopes. Taking into consideration all these factors, we will highlight some of the most recent findings of nanoscale assemblies being used in drug delivery.
pH-responsive polymeric micelles are a type of smart nanocarrier system. The change in pH allows for controlled drug release [109]. However, the inherent toxicity of the polymers used to make the micelles has been an issue when considering their use in clinical settings. Recently, hyperbranched double hydrophilic block copolymer micelles of poly(ethylene oxide) and polyglycerol have been designed and synthesized [109]. The study showed that not only were the micelles pH-responsive but that the polymers were biocompatible when cytotoxicity was evaluated with HeLa cells.
Another type of intracellular stimulus being used for drug delivery is the high redox potential difference between the reducing intracellular space and the oxidizing extracellular space. Recently, redox-responsive polymeric nanoparticles were prepared using a new disulphide bond-containing redox-sensitive polymer, poly(ethylene glycol)-b-poly(lactic acid) [110]. Paclitaxel was encapsulated in the polymeric nanoparticle using an optimized oil-in-water emulsion/solvent-evaporation method and scanning electron microscopy showed that the nanoparticles were rice shaped. The reducing environment of the cellular cytoplasm provided a triggered and continuous release of the drug in tumour cells resulting in pronounced cytotoxicity. The polymeric carrier itself had low cytotoxicity. It remains to be seen, with both pH- and redox-responsive polymeric nanoparticles, how the introduction of targeting molecules would affect assembly, in vitro drug release, and finally in vivo assays.
Photoswitchable nanoparticles are another type of smart nanocarrier system; they harness the power of physics, chemistry and biology [111]. Hybrid spiropyran/lipid-PEG nanoparticles change size from 150 to 40 nm when illuminated with UV light. The volume change is reversible and allows for spatio-temporal control of drug release and improved tissue penetration, which is advantageous in many diseases, including cancer. Multi-stage nanoparticles are another type of nanoparticle that changes size [112]. Originally, these nanoparticles are 100 nm in diameter but upon encounter with proteases that are highly expressed in the tumour microenvironment they break up into smaller 10 nm nanoparticles. The smaller size allows for greater diffusion throughout the tumour and thus deeper penetration into tumour tissue. Examples of FDA-approved nanoparticle-based therapeutics are Doxil (≈100 nm PEGylated liposomal form of Dox) and Abraxane (≈130 nm albumin-bound paclitaxel nanoparticle). They readily accumulate into solid tumours by the enhanced permeability and retention effect. In addition, they have shown improved pharmacokinetics and reduced adverse effects, but only modest survival benefits to patients. This may be due to poor tissue penetration [113].
4.3. Bioimaging
Nanoscale assemblies, and particularly nanobioassemblies, serve as excellent scaffolds for the loading of fluorescent dyes or other probes for in vitro or in vivo imaging due to their high surface area-to-volume ratio. High local concentrations of dyes and/or probes are obtained, as they can be loaded onto the surface of the nanoassembly or inside its hollow interior. Furthermore, fluorescence quenching can to some degree be avoided when the dyes are loaded in a highly ordered and structured manner. In the case of polymeric nanoassemblies, chemical modification is needed to conjugate the dye or probe. However, in the case of nanobioassemblies, chemical or genetic modification can be used for bioconjugation of fluorescent dyes or other probes. Another advantage of nanobioassemblies such as VLPs for bioimaging is their biological compatibility.
CPMV, which is a small plant virus with an inert nature, is an example of a nanobioassembly used for bioimaging [114]. CPMV was fluorescently labelled with a variety of commercially available fluorescent dyes using N-hydroxysuccinimide ester chemistry. The fluorescent CPMV was injected into adult mice and chick embryos. The fluorescently conjugated viral nanoparticles allowed the visualization of the vasculature and blood flow to a depth of up to 500 μm and also enabled the long-term vascular mapping of tumours. The rod-like virus M13 bacteriophage also shows promise in cancer cell imaging when modified with cell-targeting motifs, such as folic acid and fluorescent dyes [115].
It is not only fluorescent dyes that are used for imaging. QDs and green fluorescent protein (GFP) have been used extensively for in vitro and in vivo imaging as alternatives to labelling. For example, fluorescent chimeric VLPs of canine parvovirus were expressed in insect cells [116]. To create the fluorescent chimeric VLPs of canine parvovirus, enhanced GFP was genetically engineered onto the N-terminus of the viral protein VP2. The attachment of GFP did not disturb self-assembly or susceptibility to infection, as was demonstrated with mammalian cells. Thus, the chimeric VLPs show great potential as a visualization tool to understand mechanisms related to canine parvovirus infection. GFP has also been used to create chimeric HIV VLPs that allow protein to be tracked during assembly and transmission using live-cell imaging [117]. Another alternative to fluorescently labelling nanoscale assemblies are QDs, which are colloidal nanocrystals with unique optical properties [118]. For more in-depth information on the applications of QDs as multi-modal contrast agents in bioimaging, see the study of Michalet et al. [119].
4.4. Vaccine development
For all the different classes of nanoscale assemblies we described in §2 (polymers) and in §3 (liposomes, nucleic acid-based, etc.), there are numerous reports in the literature of how they have been used in new vaccine technologies [120–122]. By way of illustration, in the case of vault nanoparticles, encapsulation of the major outer membrane protein of Chlamydia muridarum in the vaults' cavity and subsequent intranasal immunization in mice induced cell-mediated immune responses at mucosal surfaces [66]. Additionally, recombinant vault nanoparticles encapsulated with the model antigen ovalbumin (OVA) generated greater cellular immunity than OVA packaged in liposomes [123]. A complete literature survey on nanoscale assemblies and carriers, such as keyhole limpet haemocyanin, cholera toxin B subunit and flagellin, used for vaccine applications would be beyond the scope of this review. Here we focus solely on VLPs and SAPN.
4.4.1. Virus-like particles
Considering vaccine applications, VLPs are attractive because they are considered safe. Unlike attenuated or inactivated live virus vaccines, there is no risk of the development of disease in vaccinated individuals because they lack the genomic material needed for the replication and hence the spread of the virus. However, when used as vaccines VLPs also have limitations. For example, there are issues with their mass-production. Furthermore, there is a restriction on the size of antigen that can be attached which is counterproductive to vaccine efficacy. Despite these limitations, there are a number of prophylactic VLP-based vaccines currently being commercialized worldwide: Merck and Co., Inc.'s Recombivax HB (HBV) and Gardasil (HPV) and GlaxoSmithKline's Engerix (HBV) and Cervarix (HPV).
VLP vaccine technology has also been employed to develop vaccines against chronic diseases such as hypertension, Alzheimer's disease and rheumatoid arthritis or vaccines against drug addiction [124]. The VLP vaccines for chronic diseases or drug addiction differ from conventional vaccines as they have an immunotherapeutic nature and use. These VLP vaccines are synthesized by covalently conjugating self-antigens to VLPs to induce autoantibody responses. For example, the only hypertension vaccine tested in clinical trials so far has been an angiotensin II vaccine [125] and was based on VLP technology. A peptide derived from angiotensin II was synthesized by solid-phase chemistry and was chemically conjugated to the RNA bacteriophage Qβ VLP capsid. The angiotensin II vaccine was administered to spontaneously hypertensive rats and resulted in decreased blood pressure. One advantage of such vaccine treatment in humans would be better patient compliance, as daily dosing would not be required.
4.4.2. Self-assembling peptide/protein nanoparticles
Our SAPNs (figure 2d–h) have great potential as repetitive antigen display systems for the development of vaccines. For one thing, antigens from either the surface or from within the pathogen can be genetically engineered into the peptide sequence of the nanoparticle. Secondly, the architectural similarity to virus capsids allows the peptide nanoparticles to elicit a strong immune response while obtaining the purity and high specificity of peptide-based vaccines. This in turn provides two safety features as the inherent risks of live attenuated vaccines are avoided and the need for toxic adjuvants present in peptide-based vaccines is eliminated. Compared with VLPs on the other hand, the SAPNs are ideally suited to present conformation-specific oligomeric epitopes. For example, a fragment of the surface protein of severe acute respiratory syndrome coronavirus (SARS-CoV) (figure 2d) was repetitively displayed on the SAPN in its native trimeric conformation [85]. The SARS epitope selected was a coiled coil too, and hence the epitope was incorporated into the SAPN in such a way that it remained in coiled-coil register with the trimeric coiled coil. Biophysical characterization showed satisfactory self-assembly and that there were about 110 peptide chains per nanoparticle. The SARS-functionalized SAPNs were injected into mice and qualitative enzyme-linked immunosorbent assay showed that conformation-specific antibodies were elicited. Not only was the conformation of the epitope properly maintained but the antibodies elicited could also inhibit infection of cells in vitro. More recently also, a similar study on HIV (figure 2h) has been carried out [89].
Another example of such SAPNs being used as repetitive antigen display systems for the development of a vaccine is the peptide nanoparticles functionalized with a fragment of the circumsporozoite protein of rodent malaria-causing agent Plasmodium berghei (figure 2e) [86]. Kaba and colleagues administered the functionalized peptide nanoparticles in saline to mice that had genetically very different immune backgrounds. The immunizations with these malaria functionalized nanoparticles produced T-cell dependent, high-avidity antibodies. These antibodies were able to protect mice for up to 15 months against a second challenge after an initial challenge had successfully been cleared. Additionally, the SAPN platform was employed for avian influenza (AI) (figure 2f) immunization [87]. The SAPNs were functionalized with the immunogenic epitope from the external domain of matrix protein 2 (M2e). In one set of SAPN constructs M2e was monomeric, and in another set the M2e viral surface was tetrameric. Chickens were used as the test animals and SAPNs with tetrameric M2es showed better protection than their monomeric counterparts. These results show the superior performance of the SAPN platform to display oligomeric epitopes in their native conformation and suggest that the SAPN technology can be a powerful platform for AI vaccination.
A study has also been done to improve the biophysical properties of SAPNs [126]. SAPNs are purified under denaturing conditions, and refolding occurs upon gradual removal of urea. Aggregation can occur if the refolding conditions are not favourable. Therefore, we investigated the optimal refolding conditions (ionic strength, pH and glycerol concentration) to create an adequate buffer formulation that allows for improvement of the immunological properties of the SAPNs. The superior biophysical properties of the SAPNs confirmed by the results of this study would allow for better antigen display, size uniformity and long-term stability.
The SAPNs’ feature of a repetitive antigen display system can be used not only for vaccination but also to elicit an immune response against poor antigens [88]. Schroeder and co-workers wanted to obtain specific actin antibodies in order to study the form of nuclear actin. However, owing to its high degree of conservation in higher animals and humans, actin is a poor immunogen in mice. By genetically engineering actin sequences from the highly conserved ‘hydrophobic loop’ onto the peptide nanoparticles and then immunizing mice, a number of monoclonal antibodies were generated which bound to the hydrophobic loop in vitro and in situ. The immunogenicity of this actin loop was thus improved by its presentation in the nanoparticle (figure 2g) in an ordered, repetitive array.
5. Conclusions and perspectives
This review has summarized characteristic features of nanoscale assemblies. Special focus was given to nanobioassemblies and their biomedical applications. We highlighted how nanobioassemblies can be used in diverse biomedical applications by either using their natural features and affinities or broadening their capabilities by chemical and/or genetic modifications. Given these considerations, it is evident that the study of nanoscale assemblies is at the interface of biology, chemistry and physics and relies on the expertise and collaboration of scientists trained in a range of diverse disciplines from organic synthesis to protein engineering and beyond.
Whether the building block for a nanoscale assembly be a polymer or a biomolecule, the wealth of sizes and shapes that can be created are impressive. This is important not only for scientific curiosity and proof of principles but also for the possible biomedical applications of the nanoscale assemblies. There is a clear tendency towards the development of ‘smart’, multi-functional nanoparticles for biomedical applications. Thus, the nanoparticle should have targeting and sensing capabilities, diagnostic and imaging power, drug-loading abilities—all while maintaining non-toxicity and integrity until the right moment. The DNA nanorobot with cell-targeting abilities [29] and the octa-functional PLGA nanoparticles [5] seem to point in this direction. Finally, in the area of vaccine development whether it be for traditional diseases, chronic diseases or antidrug, nanoscale assemblies have proved to be very useful.
Acknowledgements
This work was supported by the NIH awards 1P01GM096971 and 1DP1DA033524.
References
- 1.Gomez-Gualdron DA, Burgos JC, Yu J, Balbuena PB. 2011. Carbon nanotubes: engineering biomedical applications. Progr. Mol. Biol. Transl. Sci. 104, 175–245 10.1016/B978-0-12-416020-0.00005-X (doi:10.1016/B978-0-12-416020-0.00005-X) [DOI] [PubMed] [Google Scholar]
- 2.Raman S. 2008. Design and analysis of peptide based nanoparticles. PhD thesis, University of Basel, Switzerland: 10.5451/unibas-004642021 (doi:10.5451/unibas-004642021) [DOI] [Google Scholar]
- 3.Liu J, Jiang Z, Zhang S, Liu C, Gross RA, Kyriakides TR, Saltzman WM. 2011. Biodegradation, biocompatibility, and drug delivery in poly(omega-pentadecalactone-co-p-dioxanone) copolyesters. Biomaterials 32, 6646–6654 10.1016/j.biomaterials.2011.05.046 (doi:10.1016/j.biomaterials.2011.05.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Preat V. 2012. PLGA-based nanoparticles: an overview of biomedical applications. J. Control. Rel. 161, 505–522 10.1016/j.jconrel.2012.01.043 (doi:10.1016/j.jconrel.2012.01.043) [DOI] [PubMed] [Google Scholar]
- 5.Zhou J, Patel TR, Fu M, Bertram JP, Saltzman WM. 2012. Octa-functional PLGA nanoparticles for targeted and efficient siRNA delivery to tumors. Biomaterials 33, 583–591 10.1016/j.biomaterials.2011.09.061 (doi:10.1016/j.biomaterials.2011.09.061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao H, Elsabahy M, Giger EV, Li D, Prud'homme RE, Leroux JC. 2010. Aminated linear and star-shape poly(glycerol methacrylate)s: synthesis and self-assembling properties. Biomacromolecules 11, 889–895 10.1021/bm901241k (doi:10.1021/bm901241k) [DOI] [PubMed] [Google Scholar]
- 7.Meier W. 2000. Polymer nanocapsules. Chem. Soc. Rev. 29, 295–303 10.1039/a809106d (doi:10.1039/a809106d) [DOI] [Google Scholar]
- 8.Foster EJ, Berda EB, Meijer EW. 2009. Metastable supramolecular polymer nanoparticles via intramolecular collapse of single polymer chains. J. Am. Chem. Soc. 131, 6964–6966 10.1021/ja901687d (doi:10.1021/ja901687d) [DOI] [PubMed] [Google Scholar]
- 9.Kim D, et al. 2007. Direct synthesis of polymer nanocapsules with a noncovalently tailorable surface. Angew. Chem. Int. Ed. Engl. 46, 3471–3474 10.1002/anie.200604526 (doi:10.1002/anie.200604526) [DOI] [PubMed] [Google Scholar]
- 10.Sakai H, Sekita A, Tanaka K, Sakai K, Kondo T, Abe M. 2011. Preparation and properties of nanosized biodegradable polymer capsules. J. Oleo Sci. 60, 569–573 10.5650/jos.60.569 (doi:10.5650/jos.60.569) [DOI] [PubMed] [Google Scholar]
- 11.Kuykendall DW, Zimmerman SC. 2007. Nanoparticles: a very versatile nanocapsule. Nat. Nanotechnol. 2, 201–202 10.1038/nnano.2007.90 (doi:10.1038/nnano.2007.90) [DOI] [PubMed] [Google Scholar]
- 12.Dash M, Chiellini F, Ottenbrite RM, Chiellini E. 2011. Chitosan—a versatile semi-synthetic polymer in biomedical applications. Progr. Polym. Sci. 36, 981–1014 10.1016/j.progpolymsci.2011.02.001 (doi:10.1016/j.progpolymsci.2011.02.001) [DOI] [Google Scholar]
- 13.Xu Y, Du Y. 2003. Effect of molecular structure of chitosan on protein delivery properties of chitosan nanoparticles. Int. J. Pharm. 250, 215–226 10.1016/S0378-5173(02)00548-3 (doi:10.1016/S0378-5173(02)00548-3) [DOI] [PubMed] [Google Scholar]
- 14.Sy JC, Seshadri G, Yang SC, Brown M, Oh T, Dikalov S, Murthy N, Davis ME. 2008. Sustained release of a p38 inhibitor from non-inflammatory microspheres inhibits cardiac dysfunction. Nat. Mater. 7, 863–868 10.1038/nmat2299 (doi:10.1038/nmat2299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. 2007. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2, 249–255 10.1038/nnano.2007.70 (doi:10.1038/nnano.2007.70) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee LA, Wang Q. 2006. Adaptations of nanoscale viruses and other protein cages for medical applications. Nanomed. Nanotechnol. Biol. Med. 2, 137–149 10.1016/j.nano.2006.07.009 (doi:10.1016/j.nano.2006.07.009) [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Xu K, Wang H, Tan PK, Fan W, Venkatraman SS, Li L, Yang YY. 2009. Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nat. Nanotechnol. 4, 457–463 10.1038/nnano.2009.153 (doi:10.1038/nnano.2009.153) [DOI] [PubMed] [Google Scholar]
- 18.Martin CR, Kohli P. 2003. The emerging field of nanotube biotechnology. Nat. Rev. Drug Disc. 2, 29–37 10.1038/nrd988 (doi:10.1038/nrd988) [DOI] [PubMed] [Google Scholar]
- 19.Rudra JS, Tian YF, Jung JP, Collier JH. 2010. A self-assembling peptide acting as an immune adjuvant. Proc. Natl Acad. Sci. USA 107, 622–627 10.1073/pnas.0912124107 (doi:10.1073/pnas.0912124107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, McLaughlin CK, Aldaye FA, Hamblin GD, Rys AZ, Rouiller I, Sleiman HF. 2009. Metal–nucleic acid cages. Nat. Chem. 1, 390–396 10.1038/nchem.290 (doi:10.1038/nchem.290) [DOI] [PubMed] [Google Scholar]
- 21.Bangham AD. 1995. Surrogate cells or Trojan horses. The discovery of liposomes. Bioessays 17, 1081–1088 10.1002/bies.950171213 (doi:10.1002/bies.950171213) [DOI] [PubMed] [Google Scholar]
- 22.Sun B, Lim DS, Kuo JS, Kuyper CL, Chiu DT. 2004. Fast initiation of chemical reactions with laser-induced breakdown of a nanoscale partition. Langmuir 20, 9437–9440 10.1021/la048444m (doi:10.1021/la048444m) [DOI] [PubMed] [Google Scholar]
- 23.Verma S, Dent S, Chow BJ, Rayson D, Safra T. 2008. Metastatic breast cancer: the role of pegylated liposomal doxorubicin after conventional anthracyclines. Cancer Treat. Rev. 34, 391–406 10.1016/j.ctrv.2008.01.008 (doi:10.1016/j.ctrv.2008.01.008) [DOI] [PubMed] [Google Scholar]
- 24.Tian B, Al-Jamal WT, Al-Jamal KT, Kostarelos K. 2011. Doxorubicin-loaded lipid-quantum dot hybrids: surface topography and release properties. Int. J. Pharm. 416, 443–447 10.1016/j.ijpharm.2011.01.057 (doi:10.1016/j.ijpharm.2011.01.057) [DOI] [PubMed] [Google Scholar]
- 25.Shih WM, Quispe JD, Joyce GF. 2004. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 427, 618–621 10.1038/nature02307 (doi:10.1038/nature02307) [DOI] [PubMed] [Google Scholar]
- 26.Rothemund PW. 2006. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 10.1038/nature04586 (doi:10.1038/nature04586) [DOI] [PubMed] [Google Scholar]
- 27.Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. 2009. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 10.1038/nature08016 (doi:10.1038/nature08016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen ES, et al. 2009. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 10.1038/nature07971 (doi:10.1038/nature07971) [DOI] [PubMed] [Google Scholar]
- 29.Douglas SM, Bachelet I, Church GM. 2012. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 10.1126/science.1214081 (doi:10.1126/science.1214081) [DOI] [PubMed] [Google Scholar]
- 30.Torring T, Voigt NV, Nangreave J, Yan H, Gothelf KV. 2011. DNA origami: a quantum leap for self-assembly of complex structures. Chem. Soc. Rev. 40, 5636–5646 10.1039/c1cs15057j (doi:10.1039/c1cs15057j) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansouri S, Cuie Y, Winnik F, Shi Q, Lavigne P, Benderdour M, Beaumont E, Fernandes JC. 2006. Characterization of folate-chitosan-DNA nanoparticles for gene therapy. Biomaterials 27, 2060–2065 10.1016/j.biomaterials.2005.09.020 (doi:10.1016/j.biomaterials.2005.09.020) [DOI] [PubMed] [Google Scholar]
- 32.Noad R, Roy P. 2003. Virus-like particles as immunogens. Trends Microbiol. 11, 438–444 10.1016/S0966-842X(03)00208-7 (doi:10.1016/S0966-842X(03)00208-7) [DOI] [PubMed] [Google Scholar]
- 33.Brown WL, et al. 2002. RNA bacteriophage capsid-mediated drug delivery and epitope presentation. Intervirology 45, 371–380 10.1159/000067930 (doi:10.1159/000067930) [DOI] [PubMed] [Google Scholar]
- 34.Chatterji A, Ochoa W, Shamieh L, Salakian SP, Wong SM, Clinton G, Ghosh P, Lin T, Johnson JE. 2004. Chemical conjugation of heterologous proteins on the surface of cowpea mosaic virus. Bioconjug. Chem. 15, 807–813 10.1021/bc0402888 (doi:10.1021/bc0402888) [DOI] [PubMed] [Google Scholar]
- 35.Wang Q, Kaltgrad E, Lin T, Johnson JE, Finn MG. 2002. Natural supramolecular building blocks. Wild-type cowpea mosaic virus. Chem. Biol. 9, 805–811 10.1016/S1074-5521(02)00165-5 (doi:10.1016/S1074-5521(02)00165-5) [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, Lin T, Johnson JE, Finn MG. 2002. Natural supramolecular building blocks. Cysteine-added mutants of cowpea mosaic virus. Chem. Biol. 9, 813–819 10.1016/S1074-5521(02)00166-7 (doi:10.1016/S1074-5521(02)00166-7) [DOI] [PubMed] [Google Scholar]
- 37.Patel KG, Swartz JR. 2011. Surface functionalization of virus-like particles by direct conjugation using azide-alkyne click chemistry. Bioconjug. Chem. 22, 376–387 10.1021/bc100367u (doi:10.1021/bc100367u) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinmetz NF, Hong V, Spoerke ED, Lu P, Breitenkamp K, Finn MG, Manchester M. 2009. Buckyballs meet viral nanoparticles: candidates for biomedicine. J. Am. Chem. Soc. 131, 17 093–17 095 10.1021/ja902293w (doi:10.1021/ja902293w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKee SJ, Young VL, Clow F, Hayman CM, Baird MA, Hermans IF, Young SL, Ward VK. 2012. Virus-like particles and α-galactosylceramide form a self-adjuvanting composite particle that elicits anti-tumor responses. J. Control. Rel. 159, 338–345 10.1016/j.jconrel.2012.02.015 (doi:10.1016/j.jconrel.2012.02.015) [DOI] [PubMed] [Google Scholar]
- 40.Avogadri F, Merghoub T, Maughan MF, Hirschhorn-Cymerman D, Morris J, Ritter E, Olmsted R, Houghton AN, Wolchok JD. 2010. Alphavirus replicon particles expressing TRP-2 provide potent therapeutic effect on melanoma through activation of humoral and cellular immunity. PLoS ONE 5, e12670. 10.1371/journal.pone.0012670 (doi:10.1371/journal.pone.0012670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buonaguro L, Petrizzo A, Tornesello ML, Buonaguro FM. 2011. Translating tumor antigens into cancer vaccines. Clin. Vaccine Immunol. 18, 23–34 10.1128/CVI.00286-10 (doi:10.1128/CVI.00286-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.No JH, Kim MK, Jeon YT, Kim YB, Song YS. 2011. Human papillomavirus vaccine: widening the scope for cancer prevention. Mol. Carcinog. 50, 244–253 10.1002/mc.20657 (doi:10.1002/mc.20657) [DOI] [PubMed] [Google Scholar]
- 43.Goicochea NL, De M, Rotello VM, Mukhopadhyay S, Dragnea B. 2007. Core-like particles of an enveloped animal virus can self-assemble efficiently on artificial templates. Nano Lett. 7, 2281–2290 10.1021/nl070860e (doi:10.1021/nl070860e) [DOI] [PubMed] [Google Scholar]
- 44.Janssens ME, Geysen D, Broos K, De Goeyse I, Robbens J, Van Petegem F, Timmermans JP, Guisez Y. 2010. Folding properties of the hepatitis B core as a carrier protein for vaccination research. Amino Acids 38, 1617–1626 10.1007/s00726-009-0365-1 (doi:10.1007/s00726-009-0365-1) [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Orner BP. 2011. Self-assembly in the ferritin nano-cage protein superfamily. Int. J. Mol. Sci. 12, 5406–5421 10.3390/ijms12085406 (doi:10.3390/ijms12085406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawson DM, et al. 1991. Solving the structure of human H ferritin by genetically engineering intermolecular crystal contacts. Nature 349, 541–544 10.1038/349541a0 (doi:10.1038/349541a0) [DOI] [PubMed] [Google Scholar]
- 47.Douglas T, Dickson DP, Betteridge S, Charnock J, Garner CD, Mann S. 1995. Synthesis and structure of an iron(III) sulfide-ferritin bioinorganic nanocomposite. Science 269, 54–57 10.1126/science.269.5220.54 (doi:10.1126/science.269.5220.54) [DOI] [PubMed] [Google Scholar]
- 48.Klem MT, Mosolf J, Young M, Douglas T. 2008. Photochemical mineralization of europium, titanium, and iron oxyhydroxide nanoparticles in the ferritin protein cage. Inorg. Chem. 47, 2237–2239 10.1021/ic701740q (doi:10.1021/ic701740q) [DOI] [PubMed] [Google Scholar]
- 49.Kramer RM, Li C, Carter DC, Stone MO, Naik RR. 2004. Engineered protein cages for nanomaterial synthesis. J. Am. Chem. Soc. 126, 13 282–13 286 10.1021/ja046735b (doi:10.1021/ja046735b) [DOI] [PubMed] [Google Scholar]
- 50.Terashima M, Uchida M, Kosuge H, Tsao PS, Young MJ, Conolly SM, Douglas T, McConnell MV. 2011. Human ferritin cages for imaging vascular macrophages. Biomaterials 32, 1430–1437 10.1016/j.biomaterials.2010.09.029 (doi:10.1016/j.biomaterials.2010.09.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchida M, et al. 2006. Targeting of cancer cells with ferrimagnetic ferritin cage nanoparticles. J. Am. Chem. Soc. 128, 16 626–16 633 10.1021/ja0655690 (doi:10.1021/ja0655690) [DOI] [PubMed] [Google Scholar]
- 52.Beeby M, Bobik TA, Yeates TO. 2009. Exploiting genomic patterns to discover new supramolecular protein assemblies. Protein Sci. 18, 69–79 10.1002/pro.1 (doi:10.1002/pro.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsai SJ, Yeates TO. 2011. Bacterial microcompartments: insights into the structure, mechanism, and engineering applications. Progr. Mol. Biol. Transl. Sci. 103, 1–20 10.1016/B978-0-12-415906-8.00008-X (doi:10.1016/B978-0-12-415906-8.00008-X) [DOI] [PubMed] [Google Scholar]
- 54.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. 2008. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6, 681–691 10.1038/nrmicro1913 (doi:10.1038/nrmicro1913) [DOI] [PubMed] [Google Scholar]
- 55.Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. 2001. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl. Environ. Microbiol. 67, 5351–5361 10.1128/AEM.67.12.5351-5361.2001 (doi:10.1128/AEM.67.12.5351-5361.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reinhold L, Zviman M, Kaplan A. 1989. A quantitative model for inorganic carbon fluxes and photosynthesis in cyanobacteria. Plant Physiol. Biochem. 27, 945–954 [Google Scholar]
- 57.Drews G, Niklowitz W. 1956. [Cytology of Cyanophycea. II. Centroplasm and granular inclusions of Phormidium uncinatum]. Archiv Mikrobiol. 24, 147–162 10.1007/BF00408629 (doi:10.1007/BF00408629) [DOI] [PubMed] [Google Scholar]
- 58.Shively JM, Ball F, Brown DH, Saunders RE. 1973. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182, 584–586 10.1126/science.182.4112.584 (doi:10.1126/science.182.4112.584) [DOI] [PubMed] [Google Scholar]
- 59.Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. 2012. Engineered protein nano-compartments for targeted enzyme localization. PLoS ONE 7, e33342. 10.1371/journal.pone.0033342 (doi:10.1371/journal.pone.0033342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renggli K, Baumann P, Langowska K, Onaca O, Bruns N, Meier W. 2011. Selective and responsive nanoreactors. Adv. Funct. Mater. 21, 1241–1259 10.1002/adfm.201001563 (doi:10.1002/adfm.201001563) [DOI] [Google Scholar]
- 61.Kedersha NL, Rome LH. 1986. Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J. Cell Biol. 103, 699–709 10.1083/jcb.103.3.699 (doi:10.1083/jcb.103.3.699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suprenant KA. 2002. Vault ribonucleoprotein particles: sarcophagi, gondolas, or safety deposit boxes? Biochemistry 41, 14 447–14 454 10.1021/bi026747e (doi:10.1021/bi026747e) [DOI] [PubMed] [Google Scholar]
- 63.Kong LB, Siva AC, Rome LH, Stewart PL. 1999. Structure of the vault, a ubiquitous cellular component. Structure 7, 371–379 10.1016/S0969-2126(99)80050-1 (doi:10.1016/S0969-2126(99)80050-1) [DOI] [PubMed] [Google Scholar]
- 64.Stephen AG, Raval-Fernandes S, Huynh T, Torres M, Kickhoefer VA, Rome LH. 2001. Assembly of vault-like particles in insect cells expressing only the major vault protein. J. Biol. Chem. 276, 23 217–23 220 10.1074/jbc.C100226200 (doi:10.1074/jbc.C100226200) [DOI] [PubMed] [Google Scholar]
- 65.Kickhoefer VA, et al. 2005. Engineering of vault nanocapsules with enzymatic and fluorescent properties. Proc. Natl Acad. Sci. USA 102, 4348–4352 10.1073/pnas.0500929102 (doi:10.1073/pnas.0500929102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Champion CI, et al. 2009. A vault nanoparticle vaccine induces protective mucosal immunity. PLoS ONE 4, e5409. 10.1371/journal.pone.0005409 (doi:10.1371/journal.pone.0005409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han M, Kickhoefer VA, Nemerow GR, Rome LH. 2011. Targeted vault nanoparticles engineered with an endosomolytic peptide deliver biomolecules to the cytoplasm. ACS Nano 5, 6128–6137 10.1021/nn2014613 (doi:10.1021/nn2014613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shaw DE, et al. 2010. Atomic-level characterization of the structural dynamics of proteins. Science 330, 341–346 10.1126/science.1187409 (doi:10.1126/science.1187409) [DOI] [PubMed] [Google Scholar]
- 69.Service RF. 2010. Computational biology. Custom-built supercomputer brings protein folding into view. Science 330, 308–309 10.1126/science.330.6002.308-a (doi:10.1126/science.330.6002.308-a) [DOI] [PubMed] [Google Scholar]
- 70.MacKay JA, Chen M, McDaniel JR, Liu W, Simnick AJ, Chilkoti A. 2009. Self-assembling chimeric polypeptide-doxorubicin conjugate nanoparticles that abolish tumours after a single injection. Nat. Mater. 8, 993–999 10.1038/nmat2569 (doi:10.1038/nmat2569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang S. 2003. Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol. 21, 1171–1178 10.1038/nbt874 (doi:10.1038/nbt874) [DOI] [PubMed] [Google Scholar]
- 72.Ryadnov MG. 2007. A self-assembling peptide polynanoreactor. Angew. Chem. Int. Ed. Engl. 46, 969–972 10.1002/anie.200604014 (doi:10.1002/anie.200604014) [DOI] [PubMed] [Google Scholar]
- 73.Padilla JE, Colovos C, Yeates TO. 2001. Nanohedra: using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl Acad. Sci. USA 98, 2217–2221 10.1073/pnas.041614998 (doi:10.1073/pnas.041614998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haghpanah JS, et al. 2009. Artificial protein block copolymers blocks comprising two distinct self-assembling domains. ChemBioChem 10, 2733–2735 10.1002/cbic.200900539 (doi:10.1002/cbic.200900539) [DOI] [PubMed] [Google Scholar]
- 75.Matsuura K. 2012. Construction of spherical virus-inspired peptide nanoassemblies. Polym. J. 44, 469–474 10.1038/pj.2012.16 (doi:10.1038/pj.2012.16) [DOI] [Google Scholar]
- 76.Hartgerink JD, Granja JR, Milligan RA, Ghadiri MR. 1996. Self-assembling peptide nanotubes. J. Am. Chem. Soc. 118, 43–50 10.1021/ja953070s (doi:10.1021/ja953070s) [DOI] [Google Scholar]
- 77.Gobeaux F, et al. 2012. Structural role of counterions adsorbed on self-assembled peptide nanotubes. J. Am. Chem. Soc. 134, 723–733 10.1021/ja210299g (doi:10.1021/ja210299g) [DOI] [PubMed] [Google Scholar]
- 78.Fernandez-Lopez S, et al. 2001. Antibacterial agents based on the cyclic d,l-α-peptide architecture. Nature 412, 452–455 10.1038/35086601 (doi:10.1038/35086601) [DOI] [PubMed] [Google Scholar]
- 79.Montero A, Gastaminza P, Law M, Cheng G, Chisari FV, Ghadiri MR. 2011. Self-assembling peptide nanotubes with antiviral activity against hepatitis C virus. Chem. Biol. 18, 1453–1462 10.1016/j.chembiol.2011.08.017 (doi:10.1016/j.chembiol.2011.08.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghadiri MR, Granja JR, Buehler LK. 1994. Artificial transmembrane ion channels from self-assembling peptide nanotubes. Nature 369, 301–304 10.1038/369301a0 (doi:10.1038/369301a0) [DOI] [PubMed] [Google Scholar]
- 81.Hourani R, Zhang C, van der Weegen R, Ruiz L, Li C, Keten S, Helms BA, Xu T. 2011. Processable cyclic peptide nanotubes with tunable interiors. J. Am. Chem. Soc. 133, 15 296–15 299 10.1021/ja2063082 (doi:10.1021/ja2063082) [DOI] [PubMed] [Google Scholar]
- 82.Madine J, Davies HA, Shaw C, Hamley IW, Middleton DA. 2012. Fibrils and nanotubes assembled from a modified amyloid-β peptide fragment differ in the packing of the same β-sheet building blocks. Chem. Commun. 48, 2976–2978 10.1039/c2cc17118j (doi:10.1039/c2cc17118j) [DOI] [PubMed] [Google Scholar]
- 83.Zhang S, Holmes T, Lockshin C, Rich A. 1993. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc. Natl Acad. Sci. USA 90, 3334–3338 10.1073/pnas.90.8.3334 (doi:10.1073/pnas.90.8.3334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rudra JS, Sun T, Bird KC, Daniels MD, Gasiorowski JZ, Chong AS, Collier JH. 2012. Modulating adaptive immune responses to peptide self-assemblies. ACS Nano 6, 1557–1564 10.1021/nn204530r (doi:10.1021/nn204530r) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pimentel TA, Yan Z, Jeffers SA, Holmes KV, Hodges RS, Burkhard P. 2009. Peptide nanoparticles as novel immunogens: design and analysis of a prototypic severe acute respiratory syndrome vaccine. Chem. Biol. Drug Des. 73, 53–61 10.1111/j.1747-0285.2008.00746.x (doi:10.1111/j.1747-0285.2008.00746.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaba SA, Brando C, Guo Q, Mittelholzer C, Raman S, Tropel D, Aebi U, Burkhard P, Lanar DE. 2009. A nonadjuvanted polypeptide nanoparticle vaccine confers long-lasting protection against rodent malaria. J. Immunol. 183, 7268–7277 10.4049/jimmunol.0901957 (doi:10.4049/jimmunol.0901957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Babapoor S, Neef T, Mittelholzer C, Girshick T, Garmendia A, Shang H, Khan MI, Burkhard P. 2011. A novel vaccine using nanoparticle platform to present immunogenic M2e against avian influenza infection. Influenza Res. Treat. 2011, 1–12 10.1155/2011/126794 (doi:10.1155/2011/126794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schroeder U, et al. 2009. Peptide nanoparticles serve as a powerful platform for the immunogenic display of poorly antigenic actin determinants. J. Mol. Biol. 386, 1368–1381 10.1016/j.jmb.2008.11.023 (doi:10.1016/j.jmb.2008.11.023) [DOI] [PubMed] [Google Scholar]
- 89.Wahome N, Pfeiffer T, Ambiel I, Yang Y, Keppler OT, Bosch V, Burkhard P. 2012. Conformation-specific display of 4E10 and 2F5 epitopes on self-assembling protein nanoparticles as a potential HIV vaccine. Chem. Biol. Drug Des. 80, 349–357 10.1111/j.1747-0285.2012.01423.x (doi:10.1111/j.1747-0285.2012.01423.x) [DOI] [PubMed] [Google Scholar]
- 90.Burkhard P, Stetefeld J, Strelkov SV. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11, 82–88 10.1016/S0962-8924(00)01898-5 (doi:10.1016/S0962-8924(00)01898-5) [DOI] [PubMed] [Google Scholar]
- 91.Pandya MJ, Spooner GM, Sunde M, Thorpe JR, Rodger A, Woolfson DN. 2000. Sticky-end assembly of a designed peptide fiber provides insight into protein fibrillogenesis. Biochemistry 39, 8728–8734 10.1021/bi000246g (doi:10.1021/bi000246g) [DOI] [PubMed] [Google Scholar]
- 92.Papapostolou D, Smith AM, Atkins ED, Oliver SJ, Ryadnov MG, Serpell LC, Woolfson DN. 2007. Engineering nanoscale order into a designed protein fiber. Proc. Natl Acad. Sci. USA 104, 10 853–10 858 10.1073/pnas.0700801104 (doi:10.1073/pnas.0700801104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Banwell EF, et al. 2009. Rational design and application of responsive α-helical peptide hydrogels. Nat. Mater. 8, 596–600 10.1038/nmat2479 (doi:10.1038/nmat2479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ferstl M, Strasser A, Wittmann HJ, Drechsler M, Rischer M, Engel J, Goepferich A. 2011. Nanofibers resulting from cooperative electrostatic and hydrophobic interactions between peptides and polyelectrolytes of opposite charge. Langmuir 27, 14 450–14 459 10.1021/la202252m (doi:10.1021/la202252m) [DOI] [PubMed] [Google Scholar]
- 95.Luo J, Tong YW. 2011. Self-assembly of collagen-mimetic peptide amphiphiles into biofunctional nanofiber. ACS Nano 5, 7739–7747 10.1021/nn202822f (doi:10.1021/nn202822f) [DOI] [PubMed] [Google Scholar]
- 96.Ghosh A, Haverick M, Stump K, Yang X, Tweedle MF, Goldberger JE. 2012. Fine-tuning the pH trigger of self-assembly. J. Am. Chem. Soc. 134, 3647–3650 10.1021/ja211113n (doi:10.1021/ja211113n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khadka DB, Haynie DT. 2012. Protein- and peptide-based electrospun nanofibers in medical biomaterials. Nanomed. Nanotechnol. Biol. Med. 8, 1242–1262 10.1016/j.nano.2012.02.013 (doi:10.1016/j.nano.2012.02.013) [DOI] [PubMed] [Google Scholar]
- 98.Boato F, Thomas RM, Ghasparian A, Freund-Renard A, Moehle K, Robinson JA. 2007. Synthetic virus-like particles from self-assembling coiled-coil lipopeptides and their use in antigen display to the immune system. Angew. Chem. Int. Ed. Engl. 46, 9015–9018 10.1002/anie.200702805 (doi:10.1002/anie.200702805) [DOI] [PubMed] [Google Scholar]
- 99.Raman S, Machaidze G, Lustig A, Aebi U, Burkhard P. 2006. Structure-based design of peptides that self-assemble into regular polyhedral nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2, 95–102 10.1016/j.nano.2006.04.007 (doi:10.1016/j.nano.2006.04.007) [DOI] [PubMed] [Google Scholar]
- 100.Raman S, Machaidze G, Lustig A, Olivieri V, Aebi U, Burkhard P. 2009. Design of peptide nanoparticles using simple protein oligomerization domains. Open Nanomed. J. 2, 15–26 10.2174/1875933500902010015 (doi:10.2174/1875933500902010015) [DOI] [Google Scholar]
- 101.Petros RA, DeSimone JM. 2010. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Disc. 9, 615–627 10.1038/nrd2591 (doi:10.1038/nrd2591) [DOI] [PubMed] [Google Scholar]
- 102.Ashley CE, et al. 2011. Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS Nano 5, 5729–5745 10.1021/nn201397z (doi:10.1021/nn201397z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith AE, Lilie H, Helenius A. 2003. Ganglioside-dependent cell attachment and endocytosis of murine polyomavirus-like particles. FEBS Lett. 555, 199–203 10.1016/S0014-5793(03)01220-1 (doi:10.1016/S0014-5793(03)01220-1) [DOI] [PubMed] [Google Scholar]
- 104.Caruso M, Belloni L, Sthandier O, Amati P, Garcia MI. 2003. α4β1 integrin acts as a cell receptor for murine polyomavirus at the postattachment level. J. Virol. 77, 3913–3921 10.1128/JVI.77.7.3913-3921.2003 (doi:10.1128/JVI.77.7.3913-3921.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Destito G, Yeh R, Rae CS, Finn MG, Manchester M. 2007. Folic acid-mediated targeting of cowpea mosaic virus particles to tumor cells. Chem. Biol. 14, 1152–1162 10.1016/j.chembiol.2007.08.015 (doi:10.1016/j.chembiol.2007.08.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kitai Y, Fukuda H, Enomoto T, Asakawa Y, Suzuki T, Inouye S, Handa H. 2011. Cell selective targeting of a simian virus 40 virus-like particle conjugated to epidermal growth factor. J. Biotechnol. 155, 251–256 10.1016/j.jbiotec.2011.06.030 (doi:10.1016/j.jbiotec.2011.06.030) [DOI] [PubMed] [Google Scholar]
- 107.Xiao Z, et al. 2012. Engineering of targeted nanoparticles for cancer therapy using internalizing aptamers isolated by cell-uptake selection. ACS Nano 6, 696–704 10.1021/nn204165v (doi:10.1021/nn204165v) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kar UK, Srivastava MK, Andersson A, Baratelli F, Huang M, Kickhoefer VA, Dubinett SM, Rome LH, Sharma S. 2011. Novel CCL21-vault nanocapsule intratumoral delivery inhibits lung cancer growth. PLoS ONE 6, e18758. 10.1371/journal.pone.0018758 (doi:10.1371/journal.pone.0018758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee S, Saito K, Lee HR, Lee MJ, Shibasaki Y, Oishi Y, Kim BS. 2012. Hyperbranched double hydrophilic block copolymer micelles of poly(ethylene oxide) and polyglycerol for pH-responsive drug delivery. Biomacromolecules 13, 1190–1196 10.1021/bm300151m (doi:10.1021/bm300151m) [DOI] [PubMed] [Google Scholar]
- 110.Song N, Liu W, Tu Q, Liu R, Zhang Y, Wang J. 2011. Preparation and in vitro properties of redox-responsive polymeric nanoparticles for paclitaxel delivery. Colloids Surf. B 87, 454–463 10.1016/j.colsurfb.2011.06.009 (doi:10.1016/j.colsurfb.2011.06.009) [DOI] [PubMed] [Google Scholar]
- 111.Tong R, Hemmati HD, Langer R, Kohane DS. 2012. Photoswitchable nanoparticles for triggered tissue penetration and drug delivery. J. Am. Chem. Soc. 134, 8848–8855 10.1021/ja211888a (doi:10.1021/ja211888a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wong C, et al. 2011. Multistage nanoparticle delivery system for deep penetration into tumor tissue. Proc. Natl Acad. Sci. USA 108, 2426–2431 10.1073/pnas.1018382108 (doi:10.1073/pnas.1018382108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jain RK, Stylianopoulos T. 2010. Delivering nanomedicine to solid tumors. Nat. Rev. Clin. Oncol. 7, 653–664 10.1038/nrclinonc.2010.139 (doi:10.1038/nrclinonc.2010.139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H. 2006. Viral nanoparticles as tools for intravital vascular imaging. Nat. Med. 12, 354–360 10.1038/nm1368 (doi:10.1038/nm1368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li K, Chen Y, Li S, Nguyen HG, Niu Z, You S, Mello CM, Lu X, Wang Q. 2010. Chemical modification of M13 bacteriophage and its application in cancer cell imaging. Bioconjug. Chem. 21, 1369–1377 10.1021/bc900405q (doi:10.1021/bc900405q) [DOI] [PubMed] [Google Scholar]
- 116.Gilbert L, Toivola J, Lehtomaki E, Donaldson L, Kapyla P, Vuento M, Oker-Blom C. 2004. Assembly of fluorescent chimeric virus-like particles of canine parvovirus in insect cells. Biochem. Biophys. Res. Commun. 313, 878–887 10.1016/j.bbrc.2003.11.176 (doi:10.1016/j.bbrc.2003.11.176) [DOI] [PubMed] [Google Scholar]
- 117.Dale BM, McNerney GP, Hubner W, Huser TR, Chen BK. 2011. Tracking and quantitation of fluorescent HIV during cell-to-cell transmission. Methods 53, 20–26 10.1016/j.ymeth.2010.06.018 (doi:10.1016/j.ymeth.2010.06.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. 2005. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 4, 435–446 10.1038/nmat1390 (doi:10.1038/nmat1390) [DOI] [PubMed] [Google Scholar]
- 119.Michalet X, et al. 2005. Quantum dots for live cells, in vivo imaging, and diagnostics. Science 307, 538–544 10.1126/science.1104274 (doi:10.1126/science.1104274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rieger J, Freichels H, Imberty A, Putaux JL, Delair T, Jerome C, Auzely-Velty R. 2009. Polyester nanoparticles presenting mannose residues: toward the development of new vaccine delivery systems combining biodegradability and targeting properties. Biomacromolecules 10, 651–657 10.1021/bm801492c (doi:10.1021/bm801492c) [DOI] [PubMed] [Google Scholar]
- 121.Watson DS, Endsley AN, Huang L. 2012. Design considerations for liposomal vaccines: influence of formulation parameters on antibody and cell-mediated immune responses to liposome associated antigens. Vaccine 30, 2256–2272 10.1016/j.vaccine.2012.01.070 (doi:10.1016/j.vaccine.2012.01.070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Laddy DJ, Weiner DB. 2006. From plasmids to protection: a review of DNA vaccines against infectious diseases. Int. Rev. Immunol. 25, 99–123 10.1080/08830180600785827 (doi:10.1080/08830180600785827) [DOI] [PubMed] [Google Scholar]
- 123.Kar UK, et al. 2012. Vault nanocapsules as adjuvants favor cell-mediated over antibody-mediated immune responses following immunization of mice. PLoS ONE 7, e38553. 10.1371/journal.pone.0038553 (doi:10.1371/journal.pone.0038553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jennings GT, Bachmann MF. 2009. Immunodrugs: therapeutic VLP-based vaccines for chronic diseases. Annu. Rev. Pharmacol. Toxicol. 49, 303–326 10.1146/annurev-pharmtox-061008-103129 (doi:10.1146/annurev-pharmtox-061008-103129) [DOI] [PubMed] [Google Scholar]
- 125.Ambuhl PM, et al. 2007. A vaccine for hypertension based on virus-like particles: preclinical efficacy and phase I safety and immunogenicity. J. Hypertens. 25, 63–72 10.1097/HJH.0b013e32800ff5d6 (doi:10.1097/HJH.0b013e32800ff5d6) [DOI] [PubMed] [Google Scholar]
- 126.Yang Y, Ringler P, Muller SA, Burkhard P. 2012. Optimizing the refolding conditions of self-assembling polypeptide nanoparticles that serve as repetitive antigen display systems. J. Struct. Biol. 177, 168–176 10.1016/j.jsb.2011.11.011 (doi:10.1016/j.jsb.2011.11.011) [DOI] [PMC free article] [PubMed] [Google Scholar]