Abstract
Milked venom from cone snails represent a novel biological resource with a proven track record for drug discovery. To strengthen this correlation, we undertook a chromatographic and mass spectrometric study of individual milked venoms from Conus purpurascens. Milked venoms demonstrate extensive peptide differentiation amongst individual specimens and during captivity. Individual snails were found to lack a consistent set of described conopeptides, but instead demonstrated the ability to change venom expression, composition and post-translational modification incorporation; all variations contribute to an increase in chemical diversity and prey targeting strategies. Quantitative amino acid analysis revealed that milked venom peptides are expressed at ranges up to 3.51–121.01 μM within single milked venom samples. This provides for a 6.37–20,965 fold-excess of toxin to induce apparent IC50 for individual conopeptides identified in this study. Comparative molecular mass analysis of duct venom, milked venom and radula tooth extracts from single C. purpurascens specimens demonstrated a level of peptide continuity. Numerous highly abundant and unique conopeptides remain to be characterized. This study strengthens the notion that approaches in conopeptide drug lead discovery programs will potentially benefit from a greater understanding of the toxinological nature of the milked venoms of Conus.
Keywords: Cone Snail, toxins, Conus purpurascens, envenomation, conotoxins, conopeptides, mass spectrometry, milked venom, peptide quantification, amino acid analysis, radula harpoon
1. Introduction
Cone snails represent a highly valuable natural resource for drug-lead discovery programs. With an estimated >75,000 individual bioactive peptides within the genus of some +500 species (Olivera, 2002), the venom peptides from these carnivorous marine predators have provided numerous highly selective ligands that target a myriad of ion channels (as reviewed in Terlau and Olivera, 2004). Several of these targets have direct implications in human healthcare. As a result, cone snail peptides, commonly referred to as conotoxins or conopeptides, are attracting much attention from the medical research and pharmaceutical communities.
One peptide in particular, ω-conotoxin MVIIA, also cited as SNX-111 or Ziconotide has received US Food and Drug Administration approval as Prialt™ (Primary Alternative to morphine), a potent neuropathic pain analgesic, specifically targeting N-type voltage-gated calcium channels (Miljanich, 2004). Other conopeptides have now transitioned into clinical trials (see Han et al., 2008), providing a clear indication attesting to their pharmaceutical and therapeutic worth.
Unfortunately, such developments are often restricted by difficulties in accessing native cone snail venoms. This problem is compounded by concerns regarding depletion of the limited marine resource via bioprospecting (see Chivian et al., 2003 and Duda et al., 2004). To offset these concerns, combined efforts encompassing genomic and proteomic approaches have both increased the diversity of conopeptide sequences and minimized animal usage (Galyer et al. 2005, Livett et al., 2006, Jakubowski et al., 2006). Sustainable research efforts have also been illustrated by utilization of pooled milked venoms as an alternative source of novel lead-compounds (see Shon et al., 1995; Hopkins et al., 1995). Direct evidence of their success is established by the milked venom-drug lead correlation, as illustrated with the expression of ω-conotoxin MVIIA (Prialt™) in individual specimens of Conus magus (see Bingham 1998, Bingham et al., 2010). Such observations necessitate the expansion of our limited biological understanding of the milked venoms of Conus. This in turn will maximize the drug-lead potential of milked venoms and provide a platform to ensure future biosustainable research supply.
Conus purpurascens (the purple cone; Fig. 1) represents a unique research candidate within the cone snail genus. Being an Eastern Pacific piscivore (fishing eating species), its mammalian bioactive peptides have been well described – from their isolation and characterization (Shon et al., 1995; 1997; 1998a; 1998b; Jacobsen et al., 1998; 1999; Teichert et al., 2007 and Gowd et al, 2008), pharmacological specificity (Shon et al., 1997; Jacobsen et al., 2000; Dowell et al., 2003; Teichert et al., 2007; López-Vera et al., 2007; Gowd et al., 2008), to 3-Dimensional structural analysis (Savarin et al., 1998; Mitchell et al., 1998; Van Wagoner et al., 2003; Chi et al., 2005). What differentiates these studies from previous conopeptide research is the source of material; the majority of these conopeptides have been isolated directly from the pooled milked venom (see Table 1) and not from the crude dissected venom duct gland. C. purpurascens presently represents the species with the best-characterized milked venom of the genus. Using this established knowledge base in turn provides: (i) an avenue to further expand our general understanding of milked venom biochemistry, and; (ii) a solid foundation to examine alternative and novel approaches in conopeptide research.
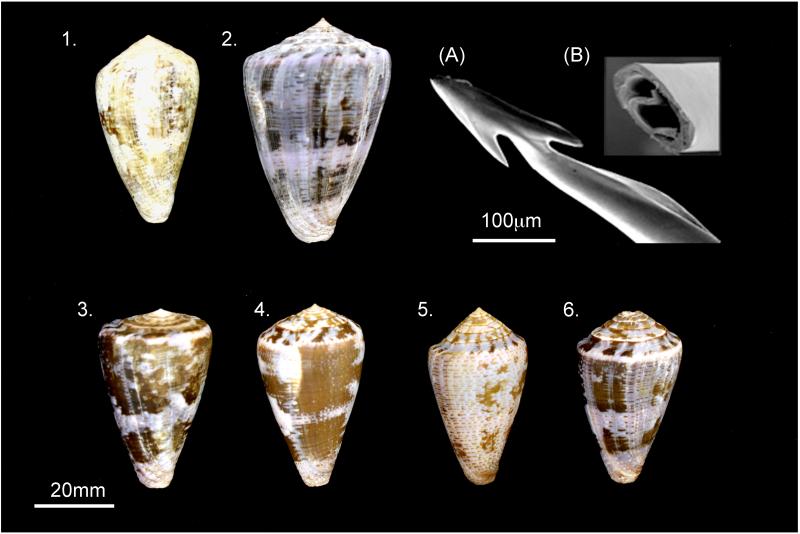
Fig. 1. Representative specimens of Conus purpurascens.
Captive specimens 1–2 (representatives used in this study) and locality specimens 3–6. 1. Panama, long-term captive specimen - notice chalky appearance, loss of shell color and gloss; 2. Panama, long-term captive specimen - notice growth ridges and bands, shell was originally 23.9 mm in diameter and grew to 37.9 mm - at the broadest point; 3. Horseshoe Bay, Costa Rica, non-captive specimen; 4. Cocos Is. Costa Rica, non-captive specimen; 5. Gobernadora Is. Panama, non-captive specimen; 6. Jaco, Panama, non-captive specimen. (A) Scan Electron Microscopy of C. purpurascens radula harpoon showing apex, 1st and 2nd barbs – each radula is unique to each species of Conus; Insert (B) shows cross section of harpoon providing access to lumen and peptide venom remnants within.
Table 1.
Conus purpurascens conopeptides: Potency, pharmacological targeting and source location
| Conopeptide | Sequence | IC50 | Target | Source | Ref. | |
|---|---|---|---|---|---|---|
| Neuromuscular | αA-PIVA | GCCGSYONAACHOCSCKDROSYCGQ* | 2.3 nM | adult mouse muscle nAChR |
MV | Hopkins et al. 1995 |
| α-PIA | RDPCCSNPVCTVHNPQIC* | 0.95 nM | rat nAChR subtype rα6/α3β2β3 |
cDNA1 | Dowell et al. 2003 | |
| α-PIB | ZSOGCCWNPACVKNRC* | 36 nM | adult mouse muscle nAChR |
MV | Lopez-Vera et al. 2007 | |
| μ-PIIIA | ZRLCCGFOKSCRSRQCKOHRCC* | 44 nM | mammalian Nav | cDNA1 | Shon et al. 1998a | |
| κ-PVIIA | CRIONQKCFQHLDDCCSRKCNRFNKCV | 57 ± 4 nM | K+ shaker channel |
MV | Shon et al. 1998 | |
| ψ-PIIIE | HOOCCLYGKCRRYOGCSSASCCQR | 127 nM 14 μM |
torpedo nAChR adult mouse muscle nAChR |
MV | Shon et al. 1997 | |
| ψ-PIIIF | GOOCCLYGSCROFOGCYNALCCRK* | 19 μM ~ 1 mM |
torpedo nAChR adult mouse muscle nAChR |
MV | Wagoner et al. 2003 | |
|
|
||||||
| Excitotoxic | δ-PVIA | EACYAOGTFCGIKOGLCCSEFCLPGVCFG* | Excitatory activity2 |
voltage-sensitive Nav | MV | Shon et al. 1995 |
| κA-PIVE | DCCGVKLEMCHPCLCDNSCKNYGK* | Excitatory activity2 |
- | MV | Teichert et al. 2007 | |
| κA-PIVF | DCCGVKLEMCHPCLCDNSCKKSGK* | Excitatory activity2 |
- | MV | Teichert et al. 2007 | |
|
|
||||||
| Unclassified |
d-Trp Contryphan-P |
GCOWDPWC* | - | - | DV | Jacobsen et al. 1998 |
|
d-Leu Contryphan-P |
GCVLLPWC | - | - | MV | Jacobsen et al. 1998 | |
| PVA | GCCPKQMRCCTL | - | - | MV | Jacobsen et al. 1998 | |
| P1.9 | CRWLQHSCLQ | - | - | cDNA1 | - | |
| Conantokin-P | GEγγHSKYQγCLRγlRVNKVQQγC | 0.3-2.3nM | NMDA Receptor NR2A/NR2B |
cDNA1 | Gowd et al., 2008 | |
*, C-terminal amidated; W, d-Tryptophan; L, d-Leucine; Z, pyroglutamic acid; O, 4-trans-Hydroxyproline; γ, γ-carboxylglutamic acid; MV, Milked Venom; DV, Duct Venom;
cDNA was prepared by reverse transcription of RNA isolated from the C. purpurascens venom duct
IC50 on target channel is unknown.
To date, 15 peptides have been isolated, sequenced, synthesized and pharmacologically classified from C. purpurascens (see Table 1). The majority of these peptides have been found to selectively target sub-types of the acetylcholine receptor (α-, αA- and ψ-conotoxins; López-Vera et al., 2007, Teichert et al., 2007 and Van Wagoner et al., 2003); isoforms of the voltage-gated sodium channel (μ- and δ-conotoxins; Shon et al., 1998a and 1995); block potassium channels (κ-conotoxin; Savarin et al., 1998) and antagonize the N-methyl-d-aspartate receptor (conantokins; Gowd et al., 2008). These peptides either inhibit neuromuscular transmission, causing flaccid paralysis, or increase excitability at the target ion channel (see Terlau et al., 1996). Interestingly enough, ω-conotoxins have yet to be isolated from C. purpurascens venom, further differentiating this species from most other piscivorous species studied to date; this provides the first indication to pharmacological diversification within the envenomation and prey immobilization strategies within Conus.
Toxinological characterization or quantitative biochemical analysis of individual Conus milked venoms is absent from current literature. Questions relating to venom production (Newcomb et al., 1995; Tayo et al., 2010), duct venom variability (Vianna Braga et al., 2005) and synergistic pharmacological strategies within Conus (Terlau et al., 1996) have arisen. Those regarding the presence of peptides in the venom organ/apparatus (see Biggs et al., 2008) and their correlation within the envenomation process are further compounded by comments surrounding the extent of venom peptide differentiation amongst individual specimens as well as the effects of captivity on this expression (Jakubowski et al., 2005; Dutertre et al., 2010).
In this work, we lay a foundation for addressing some of these questions by providing the first comprehensive quantitative analysis of milked venom peptides within C. purpurascens. Their assignment represents a novel development in the understanding of the envenomation strategies within Conus. This work provides the proof-of-concept working with a well-established and studied Conus species. We commence with the examination of the expressive nature of milked venom and its differentiation in individual long-term captive animals via Reverse-Phase Liquid Chromatographic/Ultraviolet (RP-HPLC/UV) profiling. We then undertake a comparative molecular mass analysis, via Matrix Assisted Laser Desorption/Ionization Time of flight Mass Spectrometry (MALDI-TOF-MS), to identify milked venom peptide constituents, specifically their presence within the lumen of individual radula harpoons, and their expression within whole crude dissected duct venom extracts. We conclude with determining the precise concentration of individual peptides delivered in a single envenomation. Comparison of expressed milked venom concentrations against previously reported IC50 (half maximal inhibitory concentration) values indicates that high potency candidates can be found in low abundance peaks. Combined, these findings provide for a unique alternative route for identifying and then correlating compounds/molecular masses of potential pharmacological interest within Conus. These findings add to our basic toxinological understanding of the milked venoms of Conus.
2. Materials and methods
2.1 Materials
Chromatographic solvents used in this work were HPLC grade or higher – these being subjected to high vacuum filtration to aid degassing prior to use. All buffers and solvents were filtered through 0.22 μm Nylon filters (Millipore). Chemical reagents, including matrices, ion-pairing agents were supplied from manufacturers, as indicated – these being stored and used as per manufacturer’s recommendations.
2.2 Cone snail housing, feeding, milking and venom extraction
Nine specimens of C. purpurascens were collected near the Smithsonian Tropical Research Institute, Panama. Specimens were transported to the USA, acclimated and housed in a temperature controlled environment in a single 30-gallon artificial saltwater tank fitted with a Fuval™ 402 biological filtration system. Specimens were fed weekly, using Carassius auratus auratus (goldfish; weight 2-5 grams) and consequentially milked of venom using a modified method previously described in Hopkins et al. (1995) and Bingham et al. (2005). Individual milked venom volumes were collected, measured, then lyophilized. Specimens were later sexed upon dissection for duct venom and radula harpoons. Milked venoms, together with radula harpoons were stored at −20°C until required.
Secrertory venom ducts were dried by Speed-Vac, then homogenized into a fine powder and weighed. Peptide extraction was achieved with 95% Solvent A (0.1% v/v TFA/aq.) and 5% Solvent B (90/10 v/v MeCN/0.08% v/v TFA/aq.), with samples typically representing 1 mg mL−1. Samples were vortexted and then sonicated for 10 min. Extracts were then centrifuged at 12,000g for 10 min. The resulting supernatant was removed, dried via Speed-Vac, weighed and then stored at −20°C until required. All extracts were resuspended in the above solvent at 1 mg mL−1, sonicationated (5 mins.) and re-centitrifuged (at 12,000g for 10 min.) prior to chromatographic separation and MS analysis.
2.3 RP-HPLC/UV - Peptide purification
Milked venom peptides and duct venom extracts were separated and purified using C18 Narrow-bore RP-HPLC/UV (Vydac; 5 mm, 300 Å, 2.1 × 250 mm) and later quantified using a capillary bore RP-HPLC/UV (Phenomenex; 5 mm, 300 Å, 1.0 × 250 mm). A Waters 2695 Alliance HPLC System interfaced with a 996 Waters Photo Diode Array (PDA) Detector was used for automated sample analysis and detection. Data was acquired and analyzed using Waters Millennium32 (v3.2) software. Samples were eluted using a standard linear 1% min.−1 gradient of Acetonitrile (HPLC grade, Fisher Scientific; MeCN; 90/10 MeCN /0.08% v/v aq. TFA; Solvent B) against 0.1% v/v aqueous Trifluoroacetic acid (Spectrophotometric grade, Sigma-Aldrich; aq. TFA; Solvent A) at a flow rate of 250μL min.−1 (narrow-bore) or 100μL min.−1 (capillary-bore) for a period of 65 min., as shown in Figs. 2 and 3. RP-HPLC column was pre-equilibrated with 5% solvent B, prior to sample injection. Elutant profiles were extracted at 214 nm. Samples for later amino acid quantification were fractionated manually, and subjected to repeated RP-HPLC/UV purification when necessary.
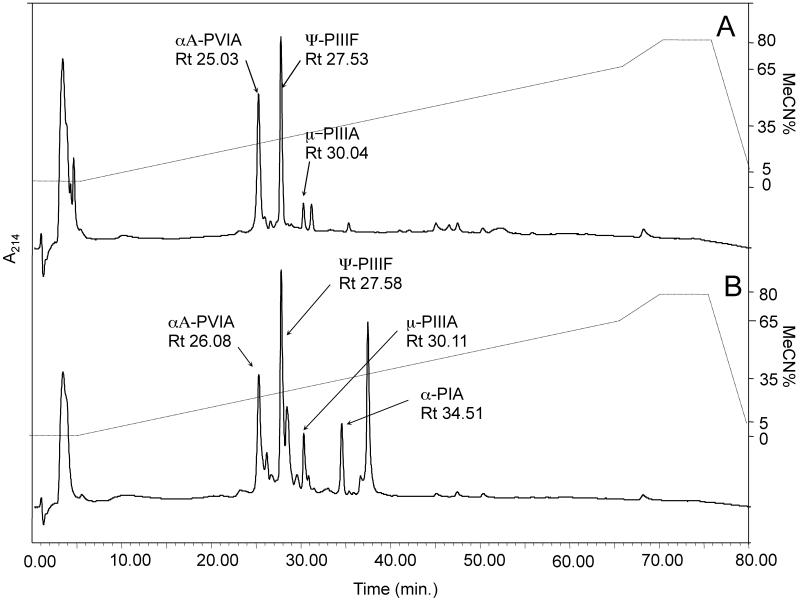
Fig. 2. Representative RP-HPLC/UV profile of time differentiated milked venoms obtained from a single Conus purpurascens specimen (Panama, specimen No. 2 – Fig. 1).
Profiles A and B represent a time difference of approximately 8 months in captivity. Peaks labeled correspond to the individual conopeptides observed and their retentions (Rt), as listed in Table 2. Profiles demonstrate a level of chromatographic simplicity. Note: ψ-conotoxin PIIIF (Fig. 2B) is represented in two separate, closely eluting forms. Molecular mass analysis assigns these two peptides a PTM variants differing by a single trans-4-hydroxyproline modification.
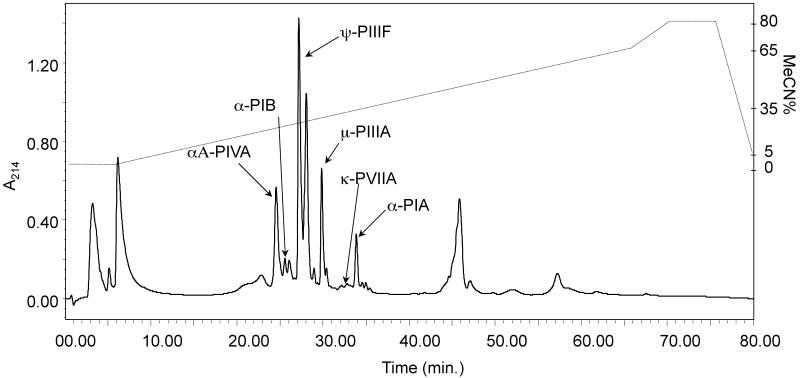
Fig. 3. Maximal milked venom complexity observed a single captive specimen Conus purpurascens (Panama, specimen No. 1 – Fig. 1).
RP-HPLC/UV showing the dominant peptide peaks being expressed in separate captive specimen of C. purpurascens milked venom, MV #41. Six C. purpurascens conopeptides were observed by retention and MS analysis (see Table 4). This specific profile is used to illustrate quantitative amino acid analysis, as in reference to Table 2.
2.4 Conopeptide reduction
Speed-Vac dried RP-HPLC/UV purified peptides were resuspended 100 mM Tris(2-carboxyethyl)phosphine (TCEP; Pierce Chemicals) in 25 mM NH4OAc, pH 4.5. To aid reduction, samples were heated at 60°C for 15 min. Upon reduction, samples were either re-purified using capillary-bore RP-HPLC/UV (as described above) or desalted by using Millipore Zip-Tip™, as described by the manufacturer, prior to mass spectrometric analysis.
2.5 Conopeptide quantification
Amino acid analyses of RP-HPLC/UV purified peptides were performed at the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University. Samples were hydrolyzed in vacuo with 6N HCl/0.2% v/v phenol for 16 hours at 115°C. Analysis was undertaken using a Hitachi L-8900 PH amino acid analyzer (ion-exchange separation, post-column derivatization with ninhydrin for detection at 570 nm and 440 nm). Data was collected using EZChrom Elite for Hitachi software and tabulated in a Microsoft Excel spreadsheet.
2.6 Mass Spectrometry conopeptide analysis
2.6.1 Electrospray Ionization Mass Spectrometry (ESI-MS)
Speed-Vac dried RP-HPLC/UV purified peptides were resuspended in 0.1% v/v aqueous Formic acid (LC/MS grade, Sigma-Aldrich). Samples were delivered to the ionization source of an API 3000 Mass Spectrometer (Thorn hill, Ontario, Canada) via a Redone 8125 Injector (20μL external loop; Rheodyne, Cotati, CA, USA) and infused with carrier solvent (50% MeCN/0.1% v/v aq. Formic acid; 50μL min.−1) as provided by an ABI 140B Dual Syringe Pump. Full-scan single MS experiments were typically obtained by scanning quadrupole-3 (Q-3) from m/z 400-2200 in 2-3 s with a scan step size of 0.1-0.5 Da. Data was acquired using Analyst Software (v.1.4.1) (Applied Biosystems/MDS Sciex, Thornhill, Ontario, Canada). ESI-MS system was calibrated manually in positive mode with PPG 3000/Mass Standards Kit (AB/MDS-Sciex), to achieve <5-ppm mass accuracy, as per manufacturer’s protocol.
Collision Induced Dissociation (CID) was performed by directly infusing the TCEP reduced RP-HPLC/UV purified peptide into the ESI-MS at 5μL min.−1, using the same Formic acid/MeCN carrier solvent as mentioned above. Bombardment was confined to quadrupole-2 (Q-2) with a collision cell gas thickness of 3×1014 atoms/cm2 and a collision energy (Q-0 to Q-2 rod offset voltage) typically set at ~20-40 eV. The resulting CID (daughter ion) spectra were obtained by scanning quadrupole-3 (Q-3) from m/z 50-3000 in 5 s with a step size of 0.1 Da. MS/MS data analysis was assisted with the use of Mac BioSpec v1.01 (PE Sciex, Thornhill, Ontario, Canada).
2.6.2 Matrix Assisted Laser Desorption/Ionization Time of Flight/(Time of Flight) Mass Spectrometry [MALDI-TOF/(TOF)-MS]
RP-HPLC/UV purified peptides fractions in 0.1% v/v aq. TFA were mixed 1:1 with matrix solution (2, 5-dihydroxybenzoic acid [DHB] in 1:1 0.1% v/v aq. TFA: MeCN; Ultra pure, Sigma-Aldrich) and spotted onto a thin layer of dried DHB matrix saturated in methanol on the target plate. The spots were dried under a stream of N2 gas. Parent ions were identified on the Ultraflex III MALDI-TOF/(TOF)-MS (Bruker Daltonics), controlled by the Compass 1.2 SR1 software package (Bruker Daltonics), in positive reflector mode. Peptide II Calibration Mix (Bruker Daltonics) was used for external calibration, with a <5-ppm mass accuracy. Analysis of the spectra was done using flexAnalysis v3.0 (Bruker Daltonics).
2.7 Mass Spectrometry radula harpoon analysis – MALDI-TOF-MS
C. purpurascens radula harpoons were collected from either (i) spent radula harpoons during the milking process, or (ii) from dissected radula sacs. Radula harpoons were prepared for MALDI-TOF-MS analysis in the following manner: (a) externally dH2O washed air dried radula harpoons were repeatedly cut into sections in an eppendorf and extracted with 100μL extracting solution (1:1 MeCN:0.1% v/v aq. TFA), sonicated for 20 min., centrifuged at 12,000 g. The resulting solvent was Millipore Zip-Tip™ (containing C18 reversed-phase media) desalted and the elute mixed 1:1 with MALDI-MS matrix solvent (40 mg mL−1 DHB in 1:2 MeCN:0.1% v/v aq. TFA) directly on the MALDI-MS target plate and allowed to air dry prior to analysis, or; (b) externally dH2O washed-air dried whole radula harpoons were cut in halves, taped onto the target plate via removable double-coated Scotch Tape® and then flooded with matrix solvent by dropping 1μL at the luminal position of the mounted radula harpoon. Matrix solvent was allowed to dry with the assistance of a stream of N2 prior to analysis.
2.8 Scanning Electron-Microscopy of radula harpoons
Imaging by Scanning Electron-Microscopy (SEM) was performed with a Hitachi S-4800 Field Emission Scanning Electron Microscope with Oxford INCA X-Act EDS System. Dissected radula harpoons were prepared for imaging by dip washing in 100% methanol, followed by sub sequential washes in dH20. Dry samples were mounted on aluminum stubs with conductive silver paint. The samples were then sputter-coated with 10-12 nm gold/palladium with a Cressington 203 sputter coater. Imaging was performed at a beam accelerating voltage of 5 kV. Images were obtained at magnifications ranging from 10-100x; this was dependent on the feature to be tracked.
3. Results
3.1 Comments on snail husbandry
A number of observations were made during long-term C. purpurascens captivity: (i) Snails were able to consume fish equal to their own shell size and had ability to feed weekly; (ii) If snails were not fed regularly or did not discharge their pre-loaded radula harpoons from the proboscis (<2 times/month), they would singularly lose the ability to penetrate the receptacle membrane, resulting in loss of milked venom – recovery occurred naturally with ‘spent’ radula harpoon expulsion and then re-arming; (iii) Specimens seem to be sturdy and resilient to captive conditions, as illustrated by the ability to produce viable veliger upon mating – a situation that occurred twice during 48 months of captivity. No specimen mortalities were recorded during these experiments, until specimens were terminated for duct venom extracts.
3.2 RP-HPLC/UV milked venom profiling
Milked venoms from C. purpurascens (n = 100) were measured to have volume ranges of 6–480μL (Mean ± Std Dev.; 56.21μL ± 105.21μL). Chromatographically milked venom differentiation was observed both in the expression of individual conopeptides and in their peak area integration, the latter reflecting differences in concentrations – given that the peaks of interest demonstrate peptide homogeneity by LC/MS, this being in contrast to the crude duct venom extracts (not shown). The maximal difference in milked venom peptide expression within a single ~8 month time differentiated captive individual is illustrated in Fig. 2. This represented our most differentiated milked venom profile observed in this study. It was common to see milked venom transition to a more complex constituent containing profile – this including the appearance and disappearance of major profile contributing peaks.
Expressional conopeptide differentiation in some individual specimens was seen as a gradual process demonstrating minor changes on a week-to-week basis (see supplemental Fig. S1), this continued throughout the 48 months of captivity (see below). Oviposting females demonstrated the absence of peptides within their milked venoms, although the motions of venom capture were successful (i.e. recorded milked venom volume, radula harpoon discharge and prey engulfment). These profiles later recovered to re-expressing conopeptides some weeks later. There were a number of other unexplained instants in which dramatic RP-HPLC/UV profile changes were noted – in most revolved around the single event expression of newly abundant peptide peaks. We also observed ‘blank’ milkings (~7% of all milkings) – in which ejaculate was collected with no apparent peptide profile being resolved chromatographically. These isolated events demonstrated no correlation to animal sex, feeding success, or season. They occurred between normal milkings that exhibited typical elution patterns. Their occurrence may represent a dynamic flux of expressional or secretional events that occur within the secretory venom duct itself.
The majority of captive milked venom peaks were observed to chromatographically elute between 20-40% MeCN. The presence of low to moderately abundant hydrophobic peaks (eluting >50% MeCN) were observed, these did not equate to the hydrophobic δ-conotoxins (see below). Near baseline resolution was apparent for the majority of individual milked venom constituents (see Fig. 2A), this could be further extended with TCEP reduction. Individual chromatographically resolvable peaks of interest, as examined in this study, demonstrated a high level of peptide homogeneity, with minimal co-eluents being observed. Peak homogeneity was assessed by: (i) UV spectrum extraction via PDA; (ii) ESI-MS infusion of RP-HPLC/UV fractions; (iii) Liquid Chromatography Interfaced Mass Spectrometry profiling (LC/MS); (iv) TCEP reduction of whole milked venoms and isolated fractions followed by RP-HPLC/UV analysis, and; (v) MALDI-TOF-MS of RP-HPLC/UV venom fractions (not shown).
The identified conopeptides previously isolated from C. purpurascens, their presence being assigned by ESI-MS/MALDI-TOF-MS molecular mass analysis and quantitative amino acid analysis – as reported in this study, demonstrated an RP-HPLC/UV elution pattern of αA-conotoxin PIVA, α-conotoxin PIB, ψ-conotoxin PIIIF, μ-conotoxin PIIIA, κ-conotoxin PVIIA and lastly α-conotoxin PIA (see Table 2, Fig. 3). Individual Post-translational (PTM) variant forms of parent conopeptide sequences were observed but limited. From this single population of C. purpurascens, as throughout this study, the hydrophobic δ-conotoxin PVIA (‘lock-jaw peptide’; Table 1) was absent, as determined by RP-HPLC/UV and by molecular mass analysis (see below). Its absence also extends to the reported PTM variants – which represent differential hydroxylated proline forms (see Shon et al., 1995).
Table 2.
Identification and assigned concentrations of Conus purpurascens conopeptides observed within a single milked venom
| Conopeptide | Retention Time (min.) |
Conc. (pMoles)a |
[MV peptide] (μM)b |
IC50 (nM)c |
[MV peptide] /IC50d |
|---|---|---|---|---|---|
| αA-PIVA | 24.61 | 359 | 48.22 | 2.3 | 20965.21 |
| α-PIB | 26.26 | 214 | 28.74 | 36 | 798.33 |
| ψ-PIIIF | 27.46 | 901 | 121.01 | 19000 | 6.37 |
| μ-PIIIA | 30.06 | 450 | 60.44 | 44 | 1373.63 |
| κ-PVIIA | 32.77 | 36 | 4.84 | 57 | 84.91 |
| α-PIA | 33.86 | 26.1 | 3.51 | 0.95 | 3694.73 |
As observed by initial RP-HPLC/UV integration with peptide standards – without taking into consideration dilution factor (50%), amount loaded (100/110μL) and total volume of original milked venom (16.38μL – specific sample, see Fig. 3).
Corrected peptide concentration as observed in total milked venom sample.
As reported in literature, see Table 1.
Calculated from milked venom peptide concentrations present(b) and reported IC50.
3.3 Molecular mass comparison of crude duct extract, milked venom and radula harpoon lumen
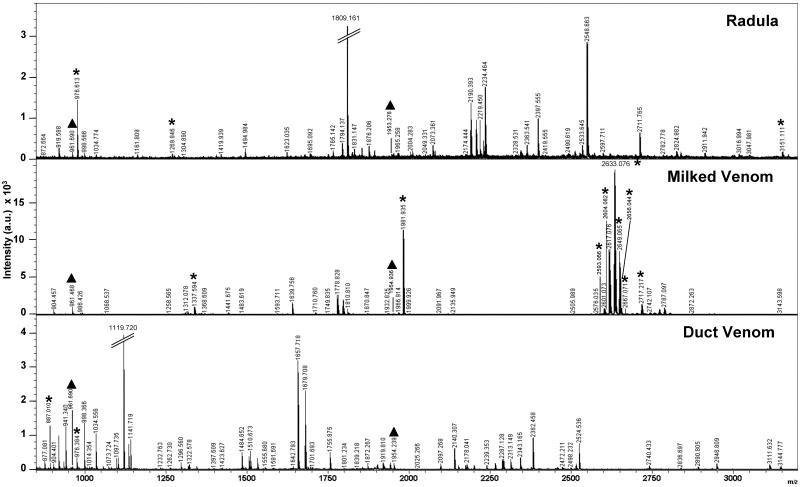
Zip-Tip™ processed venom source extracts from duct venom, milked venom and radula harpoon lumen, obtained from single C. purpurascens specimens, in part contained common m/z as observed by MALDI-TOF-MS (see Fig 4). Table 3 summarizes the molecular masses observed for single C. purpurascens representative that typifies this level of molecular mass continuity in these extracts. The observed molecular mass continuity was restricted to 27 m/z in total, and encompassed either 2 or all 3 of the venom source extracts. These do not represent the only m/z observed within these venom source extracts, with numerous relative abundant m/z being unassigned (Fig. 4).
Fig. 4. MALDI-TOF mass spectra of radula harpoon (top), captive milked venom (middle), and crude duct venom (bottom) obtained from a single specimen of Conus purpurascens.
Molecular Masses (m/z) common to all three are designated by a triangle (σ). An asterisk (M) indicates peaks that correspond to previously published conopeptides derived from C. purpurascens. 2,5-dihydroxybenzoic acid (DHB) was used as the matrix. One advantage of using DHB is that there is a low occurrence of matrix peaks and matrix adducts. To further ensure that common masses observed are not matrix peaks, mass spectra were compared to ‘matrix only’ mass spectra. No matrix peaks were detected.
Table 3.
Molecular masses shared between radula, milked venom, and duct venom extracts derived from a single specimen of Conus purpurascens
| Obs. m/z | ||
|---|---|---|
| Radula Extract | Milked Venom | Duct Venom |
| – | 904.5 | 904.4 |
| 919.6 | – | 919.4 |
| – | 925.4 | 925.3 |
| – | 940.4 | 941.3 |
| 961.7a | 961.5 | 961.4 |
| 976.6 | – | 976.4 |
| 998.6 | – | 998.4 |
| 1034.8 | – | 1034.6 |
| 1262.9 | – | 1262.7 |
| – | 1344.6 | 1344.6 |
| 1495.0 | – | 1495.7 |
| 1695.1 | – | 1695.7 |
| 1750.1 | 1749.8 | – |
| 1765.1 | 1764.8 | – |
| 1794.1 | 1794.8 | – |
| 1825.2 | 1825.8 | – |
| – | 1872.9 | 1872.3 |
| 1953.3 | 1954.9 | 1954.2 |
| 1965.3 | 1965.9 | – |
| 1968.3 | 1968.9 | – |
| 2004.3 | 2003.9 | – |
| 2174.4 | – | 2174.0 |
| 2343.5 | – | 2343.2 |
| 2382.5 | – | 2382.5 |
| 2490.6 | 2490.0 | – |
| 2836.9 | – | 2836.7 |
| 3330.3 | – | 3330.8 |
Peaks common to all three are in bold; Common peaks have a Δm/z = 0 ± 1 Da.
Examining Fig. 4 revealed that different venom source extracts demonstrated different clusters of molecular masses and possess peptides of differing relative intensities, as seen specifically with the milked venom source extract in the 2600 – 2750 m/z range. In the duct venom source extract the most relative intense m/z was seen as 1119.72 Da, while in the milked venom source extract the dominant m/z was assigned as 2633.07 Da. This specific molecular mass corresponds to the previously reported PTM variant (Hyp deletion) of [Pro13]αA-conotoxin PIVA (see Hopkins et al., 1995). The parent sequence of αA-conotoxin PIVA is also observed, m/z 2649.07 Da (Calc. m/z 2649.94 Da). In this specific illustrated specimen, the dominant ion present in the radula harpoon source extract was observed as m/z 1809.16 Da. Of the all the observed m/z, two masses were shown to be common across all three venom source extracts, m/z 961.7 and 1954.3 Da, as indicated in Fig. 4 and Table 3. Examining common masses between individual venom source extracts we see: (i) radula harpoon – milked venom source extract contains 10 common m/z; (ii) duct venom – milked venom source extract contains 7 common m/z, while; (iii) duct venom – radula harpoon source extract possesses the largest number with 14 common m/z (see Table 3).
Of the previously published/patented C. purpurascens conopeptides, 12 m/z were observed to correlate to known conopeptides as indicated in Fig. 4 and illustrated in Table 4. The majority of these conopeptides, corresponding to 8 molecular masses, were observed in the milked venom source – the original venom source for their isolation (see Table 1).
Table 4.
Molecular masses identified in radula, milked venom, and duct venom samples that correspond to known conopeptides from Conus purpurascens
| Conopeptide | Calc. m/z | Obs. m/z |
||
|---|---|---|---|---|
| Radula | MV | DV | ||
| D-Leu Contryphan-P | 888.4 | 887.01 | ||
| Contryphan-P | 976.34 | 976.61 | 976.38 | |
| P1.9 | 1271.57 | 1269.85 | ||
| PVA | 1336.52 | 1337.59 | ||
| α-PIA | 1980.82 | 1980.94 | ||
| κA-PIVF | 2594.05 | 2593.07 | ||
| μ-PIIIA | 2604.11 | 2604.08 | ||
| αA-PIVA | 2649.94 | 2649.07 | ||
| κA-PIVE | 2656.03 | 2656.04 | ||
| ψ-PIIIF | 2665.06 | 2667.07 | ||
| ψ-PIIIE | 2717.06 | 2717.22 | ||
| Conantokin-P | 3152.31 | 3151.11 | ||
3.4 Example of standardized procedure for quantitative analysis of milked venom peak concentrations
A detailed and standardized procedure for determining milked venom constituent concentrations is illustrated for future reference. αA-conotoxin PIVA (Rt 24.61 min.) from MV #41 (Fig. 3) was quantified via assigning its peak area integration and correlating this to a specific αA-conotoxin PIVA concentration standard curve. The αA-conotoxin PIVA standard was isolated and RP-HPLC/UV purified from pooled milked venom. This peak demonstrated: (i) molecular mass correlation to αA-conotoxin PIVA via ESI-MS (Obs. MH+ 2647.85; Calc. MH+ 2647.93 Da; Obs. Reduced MH+ 2653.79; Calc. Reduced MH+ 2653.93 Da); (ii) partial sequence conformation via collision-induced dissociation (CID) analysis (not shown), and; (iii) the corresponding amino acid composition to αA-conotoxin PIVA.
Quantitative amino acid hydrolysis was undertaken using a 1/3 of this sample. Remaining (100μL, 2/3) non-hydrolyzed material was assigned the calculated concentration – thus becoming the ‘αA-conotoxin PIVA standard’ stock. Five different concentrations of αA-conotoxin PIVA (10, 50, 100, 500 and 1000 pMoles) resolved on a RP-HPLC capillary C18 column and peak areas integrated to graphically establish a standard curve to then assign its unknown concentrations, as observed in various milked venoms. αA-Conotoxin PIVA milked venom profile identification was achieved by RP-HPLC/UV retention time analysis, using the same RP-HPLC capillary C18 column as above.
In our example, (Fig. 3 and Table 2) 359 pMoles of αA-conotoxin PIVA was found to be present upon RP-HPLC/UV profile analysis of sample C. purpurascens MV #41. Sample concentration correction was undertaken to give the final amount of peptide produced in whole milked venom, as injected by the animal. This was achieved by using our established protocol: (i) collected milked venom is measured (16.38μL), (ii) lyophilized and, (iii) resuspended in 100μL of solvent A to make a standardized milked venom working stock. 50μL (50%) of this milked venom working solution was removed and further diluted with 60μL of solvent A, giving Tv =110μL. A total of 100μL of this sample was analyzed by RP-HPLC/UV, thus representing 90.9% of total RP-HPLC sample volume aliquot, or a physical representation of 45.45% of the whole milked venom sample. This is then calculated to represent an equivalent aliquot of 7.44μL whole milked venom (i.e. 45.45% of 16.38μL). Thus 359 pMole of αA-conotoxin PIVA, as determined by standard curve, was equal to 7.44μL equivalents of the original milked venom volume. The total concentration of αA-conotoxin PIVA present in sample MV #41 (Fig. 3) is calculated to be 48.22 pMoles μL−1 = 48.22μM (359 pMoles/7.44μL). This same procedure was followed for RP-HPLC/UV purified native milked venom peptide standards of α-conotoxin PIB, ψ-conotoxin PIIIF, μ-conotoxin PIIIA, α-conotoxin PIA and κ-conotoxin PVIIA; all demonstrated (i) correct molecular mass correlation in native and TCEP reduced state and (ii) expected amino acid composition. These allowed for the expressed concentration to be assigned to individual conopeptides within C. purpurascens (see Table 2).
3.5 Milked venom peptide concentrations and IC50 values
Within a single milked venom from C. purpurascens, conopeptide concentrations are present within μM ranges, i.e. from 3.51 μM (α-conotoxin PIA) to 121.01 μM (ψ-conotoxin PIIIF), as listed in Table 2. This specific milked venom profile exemplifies the greatest coverage of known individual conopeptides quantified in our present studies of C. purpurascens (see Fig. 3).
In this single milked venom sample, the ratios of individual conopeptide concentrations expressed with respect to their experimental IC50 values ranged from 6.37 to 20,965.21 (Table 2). This data potentially provides an indication to the observed pharmacological excess in venom production, as examined with current IC50 data, and strengthens the viability of milked venom as a source of potent bioactive constituents. Within the gathered data, αA-conotoxin PIVA (48.22 μM) demonstrates the highest ratio value, partially attributed to its low IC50 value (2.3 nM), Table 2. This ratio is in contrast to the highly concentrated ψ-conotoxin PIIIF (121.01 μM), having a diminished ratio due to its relatively high IC50 value (19 μM). The conopeptide with the second lowest concentration in the milked venom, as an added example to strengthen this seemingly inverse relationship, α-conotoxin PIA (3.51 μM), exhibited the second highest ratio as a result of its low IC50 value (0.95 nM; Table 2).
4. Discussion
The pharmaceutical potential of cone snail venoms is well recognized (as reviewed by Lewis, 2009). Venom complexity and demonstrated isoform selectivity in ion channel targeting makes these extracts a natural pharmacopoeia for biological testing and drug lead discovery programs. However, this is a limited marine resource, to which few researchers have access. Novel approaches in conopeptide research, including venom procurement, are needed to maximize their full potential. Our approach has been to emphasize the integration of traditional biochemical and advanced mass spectrometric technologies, together focusing on biosustainable resource management – via animal husbandry and cone snail venom milking, in an endeavor to place these rare commodities in the hands of pharmacologists and neuroscientists alike.
We believe our findings provide a unique perspective into the basic toxinology of the cone snail milked venoms. As we have illustrated in this study, there are a number of important observations that have significant implications in the transition potential of venom peptides to drugs. The use of this information may impact how we view and use cone snail milked venoms successfully as a primary source of novel lead compounds.
4.1 Milked venoms demonstrate expressional variability
Venom peptide differentiation within C. purpurascens has been previously alluded to with the expression of the highly hydrophobic δ-conotoxin PVIA (see Shon et al., 1995). However, the nature and extent of peptide variability within the time differentiated milked venom, as a whole, was basically unrealized. Our evidence indicates that hydrophobic δ-conotoxins are not the only conopeptides that demonstrate a high level of expressional regulation in the milked venom, and that peptide differentiation is compounded by captive conditions. Milked venom differentiation increases the number of bioactive peptides present; as seen in Figs. 2 and 3 individual specimens may express different peptide profiles. Pooling vast quantities of venoms may risk masking and/or diluting minor or unique expressional constituents. Here, simplicity maybe the best approach, which may require treating individual snails or individual populations of snails as unique venom producers.
4.2 Post-translational modifications increase the chemical diversity of the milked venoms
In our study of C. purpurascens, we observe only a single proline variant of αA-conotoxin PIVA, corresponding to the [Pro13] derivative. This peptide was observed as a major milked venom constituent (see Fig. 4), which may illustrate a conservation of PTM expressional resources under extended captive conditions, as suggested by Hopkins et al. (1995). Whether the production of this PTM variant represents a highly selective or random event remains to be determined. However, the illustrated differential PTM behavior adds an additional dimension to the chemical diversity achievable within a single milking. Furthermore, we believe that its expressional dominance, as seen by MALDI-TOF-MS (Fig. 4), is not without toxinological purpose, as the occurrence of differential hydroxylation has been observed in other Conus venoms, as illustrated by Franco et al. (2006) and Rivera-Ortiz et al. (2011).
PTM conopeptide variants potentially offer a combinatorial-like approach to maximize phyla-selective pharmacological efficacy via increase functional group complexity, without the need to change the genetic sequence of the parent toxin sequence. PTM variants can possess differential pharmacological selectivity and targeting (Hopkins, et al, 1995; Chi et al., 2003; Nevin et al., 2007). Within the milked venoms of C. purpurascens differential hydroxylation of proline moieties has the potential to affect 7 different classes of conopeptides (Table 1). Yet to date only 2 pharmacological classes, the δ- and αA-conotoxins, have been affected by this natural occurrence (Shon et al. 1995; Hopkins et al., 1995). Unlike previous reports, our observations of PTM variants within C. purpurascens are limited. This occurrence potentially reflects the small and localized population of snails examined in this study. This may infer that the ability to undertake such modifications relates to unique and possibly localized environmental pressures, similar observations have been observed in the milked venom of C. ermineus (see Rivera-Ortiz et al. 2011).
4.3 A number of different pharmacological strategies are represented within the milked venoms derived from C. purpurascens
Not all published C. purpurascens conopeptides are observed in a single milked venom profile-initially or over its entire time in captivity. Similar observations of differential peptide expression are observed in C. striatus (Jakubowski et al., 2005), C. consors (Dutertre et al., 2010) and C. ermineus (Rivera-Ortiz et al., 2011). To ensure predatory success, C. purpurascens must possess a minimal pharmacological strategy. In this study, we see this with the combination of αA-conotoxin PIVA, ψ-conotoxoin PIIIF and μ-conotoxin PIIIA (see Fig. 2A and Table 1). These toxins target a combination of nAChR isoforms (Shon et al., 1997; Van Wagoner et al. 2003) and NaV ion channels (Shon et al., 1998a). αA-Conotoxin PIVA and ψ-conotoxoin PIIIF provide for a potent and independent targeting of the acetylcholine mediated synaptic transmission system (Hopkins et al. 1995; Shon et al., 1997). This is then compounded with Nav targeting by μ-conotoxin PIIIA, ensuring inhibition of any post-synaptic action potential propagation. This illustrates the most basic neuromuscular ‘motor cabal’ as defined by Terlau and Olivera (2004). Long-term captive milked venoms show expression of additional peptides in also varying concentration levels, as phenomena illustrated by the co-expression of α-conotoxin-PIA (Fig. 2B) and α-conotoxin-PIB (Fig. S1). Their expression synergistically complements the differential AChR targeting strategy illustrated above.
The expression of excitotoxic peptides κA-conotoxins PIVE and PIVF (m/z 2656.04 and 2593.07 Da, respectively), as seen with MALDI-TOF-MS, Fig. 4 and Table 4, conforms to the proposed synergistic ‘lighting strike cabal’ or excitotoxic shock inducement as detailed by Terlau and Olivera (2004). While the expression of excitotoxic κA-conotoxins provides an effective route for rapid prey immobilization (Terlau, et al. 1996), members of the aforementioned neuromuscular ‘motor cabal’ dominate via concentration (see Table 5). However, throughout this study the absence of δ-conotoxin PVIA, a member of the excitotoxic peptide family, and its PTM variants, is noted and we find that this is not a result of peptide handling or storage.
Table 5.
Conopeptide concentration assignment within time captive milked venoms of Conus purpurascens
| αA-PIVA | α-PIA | α-PIB | μ-PIIIA | κ-PVIIA | ψ-PIIIF | ||
|---|---|---|---|---|---|---|---|
| IC50 (nM) | 2.3 | 0.95 | 36 | 44 | 57 | 19000 | |
| Concentration (nM) |
|||||||
| Milking No. | Tv (μL)1 | ||||||
| 14 | 6348 | 2146 | 3242 | 12,006 | 795 | 17,243 | 46.8 |
| 34 | 33,550 | 26,400 | - | 21,175 | - | 57,200 | 8 |
| 36 | 178,200 | - | - | 35,200 | - | 134,200 | 5 |
| 40 | 138,600 | - | 39,600 | 323,400 | 13,200 | 305,800 | 3 |
| 46 | 215,600 | 360,800 | 105,600 | 308,000 | 66,000 | 567,600 | 3 |
Total milked venom volume.
Combined, these observations indicate that individual C. purpurascens specimens can use different pharmacological strategies to ensure successful prey capture and immobilization. What represents the quintessential pharmacological envenomation strategy for C. purpurascens remains to be determined. This is complicated by many uncharacterized milked venom peptides, including dominant peptide peaks, which may contribute to the completion of this pharmacological strategy.
4.4 Milked venoms represent a highly refined source of bioactive constituents
Crude duct venom extracts contain cellular debris that can complicate any aspect of pharmacological assessment – whether using a competitive binding assay or electrophysiological evaluation. Milked venoms offer a simple and direct route to those constituents of biological intent and resulting pharmacological interest. The observed chromatographic resolution provides a major incentive for its use (see Fig. 2 and 3), as well as the peptide concentrations achievable from a single milking – namely milked venom peptide production being within the μM range for this specific species (see Table 5). However, the presence of seawater within the expelled venom may effect pharmacological assessment, an issue corrected by chromatographic desalting. Alternatively, venom sample dilution may minimize any potential changes in solution osmolarity during the venom-assay application process (Huang and Bingham, unpublished results). Thus the direct use of the milked venoms then provides for a biosustainable resource of bioactive peptides that are highly amenable to pharmacological testing such as seen with whole cell patch clamping, and later to chromatographic isolation and MS characterization.
In this study, we illustrate the extent of molecular mass continuity between the whole duct venom extract and milked venom, and the detected differences in conopeptides that are expressed (Fig. 4). We equate these differences in part to the previously observed constituent changes seen with traversing the secretory duct (see Tayo et al., 2010). The combination of these duct sections masks the true complexity of the duct venom extract. These occurrences may be attributed to incomplete and/or differential PTM processing, which would result in diluting the level of peptide continuity observed in the milked venom. However, we would expect that closer comparative inspection of a small duct section that terminates into the pharynx of the cone snail would demonstrate a stronger mass correlation to the milked venom upon dissection, this being due to its anatomical proximity – to be illustrated and discussed elsewhere.
4.5 Dominant RP-HPLC/UV milked venom profile peaks do not necessarily correlate to the most pharmacologically potent constituents
Attention is drawn to those conopeptides of low expressional abundance in the individual milked venoms, as illustrated with α-conotoxin PIA (Fig. 2B; Table 2). This conopeptide has the lowest IC50 of any of the C. purpurascens peptides reported. We observed the opposite with the highly abundant RP-HPLC/UV peaks, as illustrated with ψ-conotoxin PIIIF, which is reported to have an IC50 of 19μM or ~1mM, depending on which nAChR isoform is being evaluated (Table 1). Concentrating on the abundant peaks within the duct and milked venom extracts has been a common trend in conopeptide research.
The relative abundance of uncharacterized low intensity molecular masses within the milked venom indicates that C. purpurascens still has much to offer investigators. These observations indicate that each individual peptide needs equal pharmacological consideration. This process is potentially complicated with specific target isoform or phyla specificity. This is illustrated with ψ-conotoxin PIIIE having IC50 of 127 nM for Torpedo AChR subtype, compared to an IC50 of 14 μM for adult mouse muscle nAChR (see Table 1; Shon et al. 1997). Many such examples of this pharmacological phenomenon exist in Conus. Combining this with low peptide abundance may provide a reliance on synthetic production for their analysis.
4.6 Mass spectrometric surveying demonstrates peptide abundance in radula harpoon extracts
A novel idea being introduced in this study is the correlation of molecular masses that are present in the radula harpoon extracts with those found in duct and milk venoms (Fig. 4 and Table 3). The majority of observed radula harpoon masses are likely a result of radula sac specific expression of conopeptides. Evidence for radula sac specific expression of conopeptide-like mRNA was previously characterized (see Biggs et al. 2008). While the molecular mass continuity observed potentially represents an accumulated ‘back-wash’ or leakage into the radula sac itself, as milked venom expulsion is undertaken with great velocity and pressure (Schulz et al., 2004).
Defining these molecular mass correlations may have a number of interesting research implications. As we observe there are unidentified molecular masses present in the radula harpoon (see Fig. 4 and Table 3), which represents a new highly refined source of conopeptides of potential biological interest. As not all cone snails can be milked successfully, this established correlation between radula harpoon extracts and milked venom may assist in streamlining efforts to identifying lead-compound candidates. These assigned molecular masses could be isolated from crude duct venoms, if no milked venom source were available, or correlated to sequence information derived from cDNA-derived libraries. The relative abundance of these same radula harpoon molecular masses could allow for direct sequence analysis by MALDI-TOF/TOF-MS. The MS analysis of the radula harpoon represents another dimension to conopeptide discovery.
4.7 The number of bioactive peptides within Conus may exceed present estimations
Use of mass spectrometric technology, providing molecular mass correlation within Conus, opens an alternative perspective into the venom complexity and the adopted chemical strategies to enhance pharmacological diversity. All these factors indicate the extent of biodiversity observable within Conus – an area that remains poorly addressed toxinologically. We believe that our initial mass spectrometric observations within the milked and duct venoms, as well as radula harpoon extracts (see Fig. 4), together with the combined ability to differentially express venom constituents (see Fig. 2), illustrates that the present 75,000-100,000 individual conopeptide constituents may represent a conservative estimate to the true abundance of biologically active peptides within these gastropods. This same statement is echoed by Davis et al. (2009), Biass et al. (2009) and Rivera-Ortiz et al. (2011), noting that these authors also utilize a mass spectrometric approach in their investigations. Along similar lines, additional analysis of other cone snail species, as we have illustrated here, will add to the debate regarding the actual physical size of the native conopeptide library.
5.0 Concluding remarks and outlook
Enhancement of natural resource management, increased knowledge about venom production and secretion, provides for a focused approach in maximizing the venom potential of Conus. Understanding these natural trends enhances our ability to find novel lead compounds of potential therapeutic use – based on their biochemical and predatory use. Without investigative investments into the basic toxinological understanding of Conus, we are potentially missing important biological aspects of their venom, as it is these facets that differentiate cone snails from other venom producers.
Supplementary Material
Fig. S1 Sequential minor differences in the milked venom of Conus purpurascens RP-HPLC/UV profile. Post 2 week milked venom RP-HPLC profiles which continue directly form Fig. 3B.
Acknowledgements
We are indebted to Dr. Tom Duda for the collection of live C. purpurascens specimens. The authors would like to acknowledge the financial support of the American Heart Association (Scientist Development Award 0530204N to J-P.B.), University of Hawaii Sea Grant College Program (J-P.B.) and USDA TSTAR (# 2009-34135-20067) & HATCH (HAW00595-R) (J-P.B) & NIH (2G12RR003061-26) (UH JABSOM Proteomics Core Facility).
Abbreviations
- aq.
aqueous
- aq. TFA
Aqueous Trifluroacetic acid
- Calc. MH+
Calculated Monoisotopic molecular mass
- CID
Collision induced dissociation
- Da
Daltons
- DHB
2, 5-dihydroxybenzoic acid
- ESI-MS
Electrospray Ionization Mass Spectrometry
- Hyp/O
4-trans-hydroxylproline
- IC50
the half maximal inhibitory concentration
- LC/MS
Liquid Chromatography interfaced Mass Spectrometry
- MALDI-TOF/(TOF)-MS
Matrix Assisted Laser Desorption/Ionization Time of Flight/(Time of Flight) Mass Spectrometry
- MeCN
Acetonitrile
- m/z
mass to charge ratio
- Nav
Voltage-gates sodium channel
- nAChR
nicotinic acetylcholine receptor
- Obs. MH+
Observed Monoisotopic molecular mass
- PDA
Photo Diode Array
- PTM
posttranslational modification
- Q-3
Quadrupole-3
- RP-HPLC/UV
Reverse Phase High Performance Liquid Chromatography interface Ultraviolet detection
- Rt
retention time
- SEM
Scanning Electron-Microscopy
- Std Dev.
Standard Derivation
- TCEP
Tris(2-carboxyethyl)phosphine
- Tv
total volume
Footnotes
Conflict of interest: Authors state that there is no conflict of interest.
References
- Biass D, Dutertre S, et al. Comparative proteomic study of the venom of the piscivorous cone snail Conus consors. J Proteomics. 2009;72(2):210–218. doi: 10.1016/j.jprot.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Biggs JS, Olivera BM, et al. Alpha-conopeptides specifically expressed in the salivary gland of Conus pulicarius. Toxicon. 2008;52(1):101–105. doi: 10.1016/j.toxicon.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham JP. PhD Dissertation. University of Queensland; Australia: 1998. Novel Toxins From the Genus Conus – From Taxonomy to toxins. [Google Scholar]
- Bingham JP, Broxton NM, Livett BG, Down JG, Jones A, Moczydlowski EG. Optimizing the connectivity in disulfide-rich peptides: alpha-conotoxin SII as a case study. Anal Biochem. 2005 Mar 1;338(1):48–61. doi: 10.1016/j.ab.2004.10.001. 2005. [DOI] [PubMed] [Google Scholar]
- Bingham JP, Mitsunaga E, et al. Drugs from slugs--past, present and future perspectives of omega-conotoxin research. Chem Biol Interact. 2010;183(1):1–18. doi: 10.1016/j.cbi.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Chi SW, Lee SH, et al. Solution structure of alpha-conotoxin PIA, a novel antagonist of alpha6 subunit containing nicotinic acetylcholine receptors. Biochem Biophys Res Commun. 2005;338(4):1990–1997. doi: 10.1016/j.bbrc.2005.10.176. [DOI] [PubMed] [Google Scholar]
- Chi SW, Park KH, et al. Solution conformation of alphaA-conotoxin EIVA, a potent neuromuscular nicotinic acetylcholine receptor antagonist from Conus ermineus. J Biol Chem. 2003;278(43):42208–42213. doi: 10.1074/jbc.M303342200. [DOI] [PubMed] [Google Scholar]
- Chivian E, Roberts CM, et al. The threat to cone snails. Science. 2003;302(5644):391. doi: 10.1126/science.302.5644.391b. [DOI] [PubMed] [Google Scholar]
- Davis J, Jones A, et al. Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides. 2009;30(7):1222–1227. doi: 10.1016/j.peptides.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Dowell C, Olivera BM, et al. Alpha-conotoxin PIA is selective for alpha6 subunit-containing nicotinic acetylcholine receptors. J Neurosci. 2003;23(24):8445–8452. doi: 10.1523/JNEUROSCI.23-24-08445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Jr., Bingham JP, et al. How much at risk are cone snails? Science. 2004;303(5660):955–957. doi: 10.1126/science.303.5660.955. author reply 955-957. [DOI] [PubMed] [Google Scholar]
- Dutertre S, Biass D, et al. Dramatic intraspecimen variations within the injected venom of Conus consors: an unsuspected contribution to venom diversity. Toxicon. 2010;55(8):1453–1462. doi: 10.1016/j.toxicon.2010.02.025. [DOI] [PubMed] [Google Scholar]
- Franco A, Pisarewicz K, et al. Hyperhydroxylation: a new strategy for neuronal targeting by venomous marine molluscs. Prog Mol Subcell Biol. 2006;43:83–103. doi: 10.1007/978-3-540-30880-5_4. [DOI] [PubMed] [Google Scholar]
- Gayler K, Sandall D, et al. Molecular prospecting for drugs from the sea. Isolating therapeutic peptides and proteins from cone snail venom. IEEE Eng Med Biol Mag. 2005;24(2):79–84. doi: 10.1109/memb.2005.1411352. [DOI] [PubMed] [Google Scholar]
- Gowd KH, Twede V, et al. Conantokin-P, an unusual conantokin with a long disulfide loop. Toxicon. 2008;52(2):203–213. doi: 10.1016/j.toxicon.2008.04.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TS, Teichert RW, et al. Conus venoms - a rich source of peptide-based therapeutics. Curr Pharm Des. 2008;14(24):2462–2479. doi: 10.2174/138161208785777469. [DOI] [PubMed] [Google Scholar]
- Hopkins C, Grilley M, et al. A new family of Conus peptides targeted to the nicotinic acetylcholine receptor. J Biol Chem. 1995;270(38):22361–22367. doi: 10.1074/jbc.270.38.22361. [DOI] [PubMed] [Google Scholar]
- Jacobsen R, Jimenez EC, et al. The contryphans, a D-tryptophan-containing family of Conus peptides: interconversion between conformers. J Pept Res. 1998;51(3):173–179. doi: 10.1111/j.1399-3011.1998.tb01213.x. [DOI] [PubMed] [Google Scholar]
- Jacobsen RB, Jimenez EC, et al. A novel D-leucine-containing Conus peptide: diverse conformational dynamics in the contryphan family. J Pept Res. 1999;54(2):93–99. doi: 10.1034/j.1399-3011.1999.00093.x. [DOI] [PubMed] [Google Scholar]
- Jacobsen RB, Koch ED, et al. Single amino acid substitutions in kappa-conotoxin PVIIA disrupt interaction with the shaker K+ channel. J Biol Chem. 2000;275(32):24639–24644. doi: 10.1074/jbc.C900990199. [DOI] [PubMed] [Google Scholar]
- Jakubowski JA, Kelley WP, et al. Screening for post-translational modifications in conotoxins using liquid chromatography/mass spectrometry: an important component of conotoxin discovery. Toxicon. 2006;47(6):688–699. doi: 10.1016/j.toxicon.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Jakubowski JA, Kelley WP, et al. Intraspecific variation of venom injected by fish-hunting Conus snails. J Exp Biol. 2005;208(Pt 15):2873–2883. doi: 10.1242/jeb.01713. [DOI] [PubMed] [Google Scholar]
- Lewis RJ. Conotoxin venom peptide therapeutics. Adv Exp Med Biol. 2009;655:44–48. doi: 10.1007/978-1-4419-1132-2_5. [DOI] [PubMed] [Google Scholar]
- Livett BG, Sandall DW, et al. Therapeutic applications of conotoxins that target the neuronal nicotinic acetylcholine receptor. Toxicon. 2006;48(7):810–829. doi: 10.1016/j.toxicon.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Lopez-Vera E, Jacobsen RB, et al. A novel alpha conotoxin (alpha-PIB) isolated from C. purpurascens is selective for skeletal muscle nicotinic acetylcholine receptors. Toxicon. 2007;49(8):1193–1199. doi: 10.1016/j.toxicon.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Miljanich GP. Ziconotide: neuronal calcium channel blocker for treating severe chronic pain. Curr Med Chem. 2004;11(23):3029–3040. doi: 10.2174/0929867043363884. [DOI] [PubMed] [Google Scholar]
- Mitchell SS, Shon KJ, et al. Three-dimensional solution structure of conotoxin psi-PIIIE, an acetylcholine gated ion channel antagonist. Biochemistry. 1998;37(5):1215–1220. doi: 10.1021/bi972186t. [DOI] [PubMed] [Google Scholar]
- Nevin ST, Clark RJ, et al. Are alpha9alpha10 nicotinic acetylcholine receptors a pain target for alpha-conotoxins? Mol Pharmacol. 2007;72(6):1406–1410. doi: 10.1124/mol.107.040568. [DOI] [PubMed] [Google Scholar]
- Newcomb R, Gaur S, et al. Structural and biosynthetic properties of peptides in cone snail venoms. Peptides. 1995;16(6):1007–1017. doi: 10.1016/0196-9781(95)00086-y. [DOI] [PubMed] [Google Scholar]
- Olivera BM. Conus venom peptides: reflections from the biology of clades and species. Annual Reviews of Ecological Systematics. 2002;33:25–47. [Google Scholar]
- Rivera-Ortiz JA, Cano H, et al. Intraspecies variability and conopeptide profiling of the injected venom of Conus ermineus. Peptides. 2011;32(2):306–316. doi: 10.1016/j.peptides.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarin P, Guenneugues M, et al. Three-dimensional structure of kappa-conotoxin PVIIA, a novel potassium channel-blocking toxin from cone snails. Biochemistry. 1998;37(16):5407–5416. doi: 10.1021/bi9730341. [DOI] [PubMed] [Google Scholar]
- Schultz JR, Norton AG, et al. The projectile tooth of a fish-hunting cone snail: Conus catus injects venom into fish prey using a high-speed ballistic mechanism. Biological Bulletin. 2004;207(2):77–79. doi: 10.2307/1543581. [DOI] [PubMed] [Google Scholar]
- Shon KJ, Grilley M, et al. A noncompetitive peptide inhibitor of the nicotinic acetylcholine receptor from Conus purpurascens venom. Biochemistry. 1997;36(31):9581–9587. doi: 10.1021/bi970235w. [DOI] [PubMed] [Google Scholar]
- Shon KJ, Grilley MM, et al. Purification, characterization, synthesis, and cloning of the lockjaw peptide from Conus purpurascens venom. Biochemistry. 1995;34(15):4913–4918. doi: 10.1021/bi00015a002. [DOI] [PubMed] [Google Scholar]
- Shon KJ, Olivera BM, et al. mu-Conotoxin PIIIA, a new peptide for discriminating among tetrodotoxin-sensitive Na channel subtypes. J Neurosci. 1998a;18(12):4473–4481. doi: 10.1523/JNEUROSCI.18-12-04473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shon KJ, Stocker M, et al. kappa-Conotoxin PVIIA is a peptide inhibiting the shaker K+ channel. J Biol Chem. 1998b;273(1):33–38. doi: 10.1074/jbc.273.1.33. [DOI] [PubMed] [Google Scholar]
- Tayo LL, Lu B, et al. Proteomic analysis provides insights on venom processing in Conus textile. J Proteome Res. 2010;9(5):2292–2301. doi: 10.1021/pr901032r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert RW, Jacobsen R, et al. Discovery and characterization of the short kappaA-conotoxins: a novel subfamily of excitatory conotoxins. Toxicon. 2007;49(3):318–328. doi: 10.1016/j.toxicon.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84(1):41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- Terlau H, Shon KJ, et al. Strategy for rapid immobilization of prey by a fish-hunting marine snail. Nature. 1996;381(6578):148–151. doi: 10.1038/381148a0. [DOI] [PubMed] [Google Scholar]
- Van Wagoner RM, Jacobsen RB, et al. Characterization and three-dimensional structure determination of psi-conotoxin Piiif, a novel noncompetitive antagonist of nicotinic acetylcholine receptors. Biochemistry. 2003;42(21):6353–6362. doi: 10.1021/bi0272757. [DOI] [PubMed] [Google Scholar]
- Vianna Braga MC, Konno K, et al. Mass spectrometric and high performance liquid chromatography profiling of the venom of the Brazilian vermivorous mollusk Conus regius: feeding behavior and identification of one novel conotoxin. Toxicon. 2005;45(1):113–122. doi: 10.1016/j.toxicon.2004.09.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Sequential minor differences in the milked venom of Conus purpurascens RP-HPLC/UV profile. Post 2 week milked venom RP-HPLC profiles which continue directly form Fig. 3B.