Abstract
Major depressive disorder (MDD) is a common psychiatric illness characterized by low mood and loss of interest in pleasurable activities. Despite years of effort, recent genome-wide association studies (GWAS) have identified few susceptibility variants or genes that are robustly associated with MDD. Standard single-SNP (single nucleotide polymorphism)-based GWAS analysis typically has limited power to deal with the extensive heterogeneity and substantial polygenic contribution of individually weak genetic effects underlying the pathogenesis of MDD. Here, we report an alternative, gene-set-based association analysis of MDD in an effort to identify groups of biologically related genetic variants that are involved in the same molecular function or cellular processes and exhibit a significant level of aggregated association with MDD. In particular, we used a text-mining-based data analysis to prioritize candidate gene sets implicated in MDD and conducted a multi-locus association analysis to look for enriched signals of nominally associated MDD susceptibility loci within each of the gene sets. Our primary analysis is based on the meta-analysis of three large MDD GWAS data sets (total N=4346 cases and 4430 controls). After correction for multiple testing, we found that genes involved in glutamatergic synaptic neurotransmission were significantly associated with MDD (set-based association P=6.9 × 10−4). This result is consistent with previous studies that support a role of the glutamatergic system in synaptic plasticity and MDD and support the potential utility of targeting glutamatergic neurotransmission in the treatment of MDD.
Keywords: Genome-wide association study, glutamatergic synaptic neurotransmission, major depressive disorder, pathway analysis
Introduction
Major depressive disorder (MDD) is a common, disabling and frequently recurrent mood disorder that affects up to one in six individuals worldwide during their lifetime.1 A number of antidepressant treatments exist, but due to modest rates of remission and substantial rates of recurrence, there is a pressing demand for new interventions with better efficacy. Identification of specific genetic risk factors could help to elucidate the neurobiological basis of MDD, which would facilitate the development of novel treatment and possibly even prevention strategies.2
To date, efforts to identify specific susceptibility genes in MDD have had limited success. Linkage studies have suggested several regions in the genome that may harbor susceptibility genes for MDD but generally without replication.3 Of note, two recent linkage studies4, 5 have reported genome-wide significant linkage to chromosome 3p26-3p25, with a peak signal near the metabotropic glutamate receptor 7 gene GRM7. Still, due to the small sample sizes and the discrepancy in the phenotype classification in the two studies, larger family-based samples will be needed to confirm this finding.6 Genome-wide association studies (GWAS) of MDD have likewise reported largely inconclusive results.2, 7, 8, 9, 10, 11, 12, 13, 14 The pathogenesis of MDD is likely to be characterized by extensive etiological heterogeneity,11, 15 complex interaction of genetic and non-genetic factors,16 as well as substantial polygenic contribution of individually weak effects,12, 17 all of which are difficult to capture with single-SNP (single nucleotide polymorphism)-based association tests typically used in current GWAS analyses.
Here we utilize a multi-locus, gene-set-based association analysis—also known as pathway analysis18 or functional gene group analysis19—that uses previous biological knowledge of molecular function and cellular processes to facilitate the genetic dissection of MDD. A number of statistical strategies and methods exist for conducting gene-set-based analysis,18, 20, 21 but the basic goal of the proposed methods is to look for sets of biologically related genetic variants that, in aggregate, show enriched association with a target phenotype beyond what would be expected by chance. Typically, well-established canonical pathways or functional annotations of genes and proteins are used to define these sets of biologically related genetic variants. Of note, recent applications of gene-set-based GWAS analysis have provided new biological insights into the genetic basis of several major psychiatric disorders, including schizophrenia,21, 22 bipolar disorder20 and autism.23, 24
Gene-set-based association analysis has three major advantages that complement standard single-locus-based GWAS analysis. First, gene-set-based analysis has improved statistical power to detect subtle and collective effects of multiple genetic variants over single-locus-based tests under various polygenic disease models.25 This is particularly noteworthy given that recent GWAS studies have indicated substantial polygenic contribution of thousands of common genetic variants to major psychiatric disorders, including MDD.17, 26 We presume that such polygenic risk variants underlying MDD would involve dysfunction of specific molecular networks or cellular pathways, as appears to be the case in other complex disorders.21, 23 Secondly, gene-set-based analysis is robust to the extensive genetic heterogeneity of complex disorders.20 Different combinations of genetic variants may alter the function or the activity levels of a given pathway or a molecular mechanism, making it difficult to replicate associations with specific genes or variants across multiple studies. Dysfunction at the level of biological pathways or molecular functions might be detected more consistently regardless of heterogeneity in individual susceptibility variants or risk genes. Finally, gene-set-based analysis allows direct insight into disease mechanism and biology by translating a list of statistically ascertained disease-associated genomic regions into sets of cellular functions or molecular pathways that may underlie disease pathogenesis.
In this work, we applied an integrative gene-set-based GWAS analysis on MDD that utilizes statistical text-mining analysis27 to prioritize the list of pathways and functional gene sets to be tested for aggregated association with MDD. We focused on biological mechanisms indexed by previous MDD candidate genes selected from biological studies of the pathogenesis of MDD. Although these genes have not individually demonstrated genome-wide association with MDD at a single variant/gene level,28 they have been implicated in the etiology of MDD in a variety of biological and pharmacological studies. As such, they may index biological pathways that are relevant to genetic risk. Application of the statistical text-mining tool enabled us to elucidate the key biological relationships among the MDD candidate genes, which were then used to compile a list of relevant Gene Ontology29 (GO) gene sets. We then tested the enrichment of these gene sets in a meta-analysis of our MDD GWAS data sets. Such prioritized gene-set-based analysis thus avoids the pitfall of reduced statistical power entailed by examining the full range (tens of thousands) of GO gene sets.20, 30
The primary goals of this analysis, thus, were: (i) to prioritize biological pathways or molecular functions (that is, gene sets) that might be involved in the pathogenesis of MDD using text-mining analysis; and (ii) to test enriched association of the compiled gene sets using a meta-analysis of three large MDD GWAS studies (4346 cases and 4430 controls in total).
Materials and methods
Study samples
NESDA/NTR. The NESDA/NTR sample consisted of 1738 MDD cases, mainly from the Netherlands Study of Depression and Anxiety, and 1802 controls mainly from the Netherlands Twin Registry.11 Cases met criteria for a lifetime diagnosis of DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, fourth edition) MDD, assessed with the Composite International Diagnostic Interview (CIDI), and did not have a primary diagnosis of schizophrenia or schizoaffective disorder, obsessive compulsive disorder, bipolar disorder or severe substance use dependence. Controls had no lifetime diagnosis of MDD or anxiety disorder and were also assessed to carry a low genetic liability for neuroticism, anxiety and depressive symptoms based on a factor score derived from longitudinal phenotyping measures. Genotype data from the Perlegen platform were obtained from dbGaP (http://www.ncbi.nlm.nih.gov/gap), which consisted of 435,291 SNPs. All subjects were of self-reported western European ancestry.
QIMR. The Queensland Institute of Medical Research (QIMR) sample consisted of 1450 cases and 1703 controls from the Australian Twin Registry. The sample represents the QIMR cases and controls included in the analyses of the Psychiatric GWAS Consortium for MDD13 and overlaps considerably with the QIMR samples of the MDD2000+ study.12 MDD cases and controls were identified through psychiatric questionnaires administered to adult twins and their families recruited through the Australian Twin Registry. All cases met DSM-IV lifetime criteria for MDD based on the shortened CIDI or the Semi-Structured Assessment for the Genetics of Alcoholism-OZ interview instrument. The interviews were administered by trained telephone interviewers, closely supervised by a clinical psychologist. In cases where more than one family member met the criteria for being included in the control group, the individual with lowest neuroticism score was selected. Samples were genotyped as part of multiple QIMR projects31 on Illumina platforms (San Diego, CA, USA; 317K, CNV370-Quadv3 and Human610-Quad). A set of 270,799 genotyped SNPs common across the three Illumina platforms that survived quality control (QC) was carried forward to the imputation phase.
STAR*D. The Sequenced Treatment Alternatives to Relieve Depression (STAR*D) was a multisite, randomized clinical trial of outpatients with MDD that enrolled 4041 participants.10 Blood samples were collected from 1953 patients and 1204 psychiatrically screened controls from the National Institute of Mental Health (NIMH)-Genetics Repository.10 Cases met DSM-IV criteria for MDD and had a Hamilton Depression Rating Scale score of at least 14. Controls were screened for a range of psychiatric disorders based on DSM IV criteria and were excluded if they reported a previous diagnosis of schizophrenia, psychosis or bipolar disorder, or met criteria for a history of major depressive episodes. We included 1500 cases who were self-identified Caucasians. After quality-control procedures, genotyping data included 284,889 SNPs for 1223 MDD patients and 970 controls common across three Affymetrix (Santa Clara, CA, USA) genotyping platforms (Affymetrix Genome-Wide Human SNP 6.0, Affymetrix Genome-Wide Human SNP 5.0 and Affymetrix GeneChip Human Mapping 500K Array).
Data QC and imputation
QC procedures for the individual data sets are described in the original GWAS studies.10, 11, 12 In brief, subjects were excluded if they exhibit missing call rates >0.05, inconsistency in a genotype-based gender with a self-reported one, incompatible levels of autosomal heterozygosity (|F inbreeding coefficient estimates| >0.05), high identity by descent estimates with other subjects (PI_HAT >0.25) or are population outliers based on five nearest neighboring clustering on information by similarity (|Z-score| >3σ). All study subjects were of self-reported non-Hispanic European ancestry. SNPs were excluded if they exhibit missing rates >0.05, minor allele frequencies (MAF) <0.01 and the Hardy–Weinberg equilibrium deviation P<1 × 10−6 in controls. SNPs were further excluded if they show significantly different missingness between cases and controls (P<1 × 10−6) or non-random missingness based on genotypes (P<1 × 10−10). All QC steps were conducted using PLINK.32 To remove potential biases from discrepancies of SNP coverage across the three samples, we imputed non-genotyped SNPs using BEAGLE33 version 3.11 and Hapmap34 II CEU data (release 23, forward strand), except for the QIMR data set, which was imputed using MACH.35 Further details on the imputation procedure for the QIMR data set are given here.31 After QC, STAR*D, QIMR and NESDA/NTR include 1223/970, 1450/1703, and 1673/1757 cases/controls, respectively. Imputation was conducted within each sample. After filtering out SNPs with imputation quality scores <0.8, 1,930,980 SNPs remained across the three data sets. Our data analysis was based on 1,386,571 autosomal SNPs that exist in all of the three data sets.
Primary association analysis
PLINK32 was used to conduct single-SNP-based logistic regression of imputed allele dosage within individual samples (that is, NESDA/NTR, QIMR and STAR*D). To control for potential population sub-stratification, 10 multi-dimensional scaling analysis components were calculated using PLINK. The first four multi-dimensional scaling components showed inflated association with genetic data and thus were included as covariates in regression analysis. Meta-analysis was conducted using a fixed effect model on the dosage analysis results from the three data sets using PLINK.
Candidate gene set compilation
To compile a list of gene sets that are potentially most relevant to MDD pathogenesis, we analyzed a list of well-known MDD candidate genes using the text-mining analysis method GRAIL (Gene Relationships Among Implicated Loci).27 The detailed gene-set-compilation procedure is as follows: First, we assembled the list of 188 MDD candidate genes examined in five previous GWAS studies of MDD.2, 7, 10, 12, 28 The authors compiled their MDD candidate genes through literature search either because the genes have been implicated in susceptibility to MDD due to their biological function and/or association with MDD in previous candidate gene association studies. The list was fed to the GRAIL analysis to identify functional keywords that index the common biological relationships among the MDD candidate genes. The GRAIL analysis parameters were set as follows: genome assembly: release 22/hg18; HapMap population: CEU; functional data-source: PubMed text (December 2006); gene-size correction: on; gene list: all human genes within database. Basically, GRAIL quantified the functional relationships between the input candidate genes using a text-based similarity metric and extracted a list of functional keywords that occur significantly more often than would be expected by chance in the text literature describing the functionally related candidate genes. We used each of the identified keywords as a search term to retrieve relevant candidate gene sets from the GO resource (http://www.geneontology.org).29
Interval-based gene-set-enrichment analysis
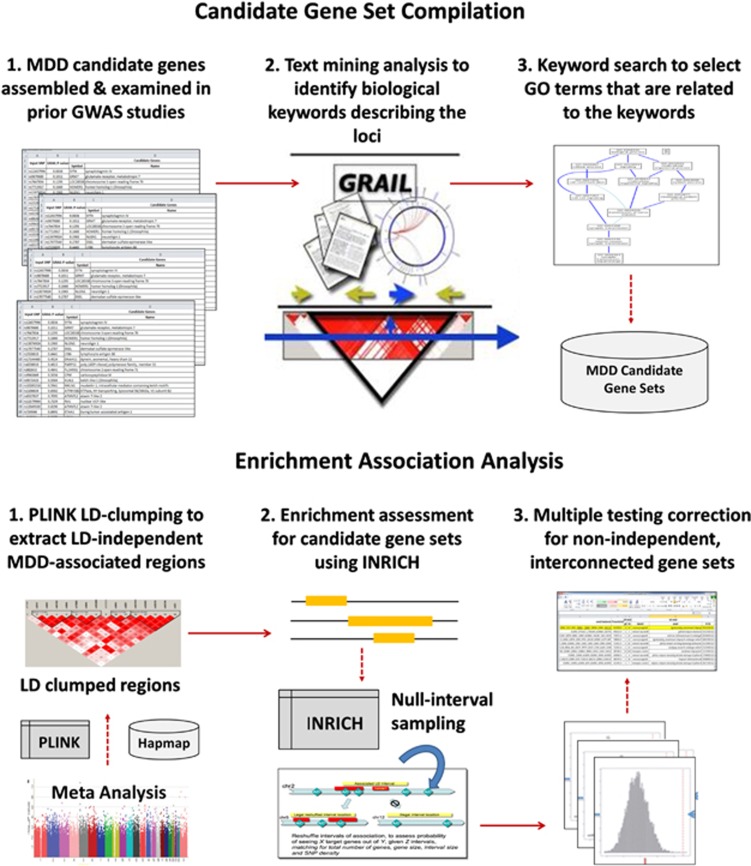
To quantify the statistical significance of aggregated MDD association signals for candidate gene sets, we used a GWAS-specific set-based association analysis tool, INRICH36 (http://atgu.mgh.harvard.edu/inrich). The INRICH analysis procedure comprises three major steps: (i) linkage disequilibrium (LD)-based interval data generation to identify unique regions of association; (ii) empirical enrichment calculation using an interval-based permutation strategy; and (iii) second-step permutation for multiple testing correction at the gene-set level. First, PLINK LD-based clumping was used to generate a list of LD-independent-associated genomic regions from the meta-analysis results (clump-p1=0.005; clump-p2=0.05; clump-r2=0.5; clump-kb=250). Next, the enrichment statistics of each target gene set was calculated as a P-value that indicates the probability of observing a given number of overlaps between the MDD-associated genomic intervals and the reference genes mapped to the examined gene set under the null hypothesis of no-disease association. We defined gene regions as 20 kb up/downstream of the RefSeq transcription starting/ending sites for 17,529 genes on autosomal chromosomes based on the Human Genome Browser build hg18 (Note that recent expression quantitative trait loci studies have shown that 95% of common genetic variation affecting transcript expression resides within 20 kb of the transcription start and end sites of genes37). Finally, resampling-based second-step permutation was conducted to adjust the pathway-level empirical P-values for testing multiple candidate gene sets. Figure 1 illustrates our data analysis procedure.
Figure 1.
Multi-locus association analysis procedure for major depressive disorder (MDD). Our analysis consisted of two major steps: (1) candidate gene-set compilation and (2) enrichment association analysis. The gene-set-compilation step aims to prioritize groups of biologically relevant genes that are most relevant to MDD pathogenesis. First, a list of widely studied MDD candidate genes was assembled from previous GWAS (genome-wide association studies) studies. The text-mining tool GRAIL (Gene Relationships Among Implicated Loci) was used to identify biological relationships between the selected genes. We used the GRAIL biological keywords to select relevant gene sets from the Gene Ontology database. The second enrichment analysis step aims to identify gene sets that show aggregated association with major depression than would be expected by chance. In order to control for different gene sizes, SNP density and linkage disequilibrium (LD) across genomic region and LD-independent MDD association regions were generated using PLINK. Enrichment of the candidate gene sets was examined using GWAS-specific multi-locus association analysis tool, INRICH, based on the LD-independent regions. The second-step permutation strategy in INRICH was conducted for correcting the testing of multiple gene sets that are not independent of each other.
Results
Primary association analysis
No SNP demonstrated genome-wide significant association with MDD in a single-SNP-based meta-analysis of NESDA/NTR, QIMR and STAR*D. Supplementary Figures S1 and S2 show the Manhattan and quantile-quantile plots for the meta-analysis of these data sets, respectively. The genomic control inflation factor λ (that is, the ratio of the observed median χ2 to that expected by chance) was 1.046, while λ1000 was 1.011 (that is, λ rescaled to a sample size of 1000 cases and 1000 controls38), indicating negligible inflation of association test statistics due to potential confounding factors.
Candidate gene-set compilation
Supplementary Table S3.1 summarizes the list of 188 MDD candidate genes assembled from five previous MDD studies.2, 7, 10, 12, 28 Using these genes as input, GRAIL quantified their functional relationships using a text-based similarity measure (Supplementary Tables S3.2 and S3.3), and identified 11 keywords that are commonly associated with the MDD candidate genes in the literature: ‘serotonin', ‘dopamine', ‘NMDA (N-methyl-𝒟-aspartate)', ‘glutamate', ‘neuron', ‘GABA', ‘adrenergic', ‘agonist', ‘cGMP', ‘synaptic' and ‘phosphodiesterase'. Table 1 lists these 11 keywords and their associated MDD candidate genes. Using each of these keywords as a search term, we identified 178 GO terms (that is, gene sets) with at least three human reference genes in the domains of biological process, molecular function and cellular complex (Supplementary Table S3.4).
Table 1. List of text-mining-based biological keywords representing MDD candidate genes.
| Serotonin (72 genes ) |
|---|
| GRIA3, GPR50, MAOA, MAOB, HTR6, OPRD1, LEPR, CREB1, HTR2B, PER2, POMC, TACR1, ADRA2B, SLC6A1, GSK3B, CCK, ITPR1, NFKB1, ADRA2C, CLOCK, DRD5, NR3C1, SLC6A3, HTR4, GRIA1, ADRA1B, GABRA6, DRD1, HTR1A, OPRM1, DTNBP1, TNF, HTR1B, CNR1, LEP, NOS3, HTR5A, IL6, NPY, CRHR2, TAC1, ADRA1A, GRIN1, NTRK2, ADRA2A, ADRB1, HTR7, CYP2C9, DRD2, HTR3B, HTR3A, TPH1, TH, BDNF, DRD4, NOS1, AVPR1A, GNB3, TPH2, HTR2A, ESR2, CHRNA7, SLC6A2, GRIN2A, CRHR1, NGFR, ACE, PER1, GRIN2D, GNAS, COMT, ADORA2A |
| Dopamine (92 genes ) |
| HTR2C, GRIA3, GABRA3, MAOA, MAOB, AVPR1B, OPRD1, GRIK3, IL1B, GAD1, CREB1, HTR2B, PER2, POMC, TACR1, ADRA2B, SLC6A1, GSK3B, AGTR1, CCK, ITPR1, CCKAR, ADRA2C, CLOCK, DRD5, PAM, NR3C1, SLC6A3, HTR4, GRIA1, ADRA1B, GABRA6, DRD1, HTR1A, CRHBP, ESR1, OPRM1, GABBR1, TNF, HTR1B, CNR1, LEP, CHRM2, NOS3, HTR5A, IL6, NPY, CRHR2, ABCB1, TAC1, ADRA1A, FGFR1, OPRK1, PENK, GRIN1, NTRK2, ADRA2A, ADRB1, HTR7, GRIA4, CD3E, GRIK4, TPH1, TH, BDNF, CCKBR, DRD4, NOS1, CACNA1C, LRP1, TPH2, KCNC2, HTR2A, AKT1, ESR2, CHRNA7, NTRK3, ADCY9, SLC6A2, GRIN2A, SLC6A4, CRHR1, NGFR, ACE, GRIN2C, GNAL, GRIK5, GRIN2D, GNAS, COMT, ADORA2A, SYN3 |
| N-methyl-𝒟-aspartate (NMDA) (57 genes ) |
| GRIA3, GABRA3, OPRD1, GRIK3, IL1B, GAD1, CREB1, TACR1, SLC6A1, GSK3B, ITPR1, GRM7, NFKB1, GRIA2, DRD5, NR3C1, SLC6A3, GRIA1, DRD1, CRHBP, GRIK2, ESR1, OPRM1, GABBR1, HTR1B, CNR1, LEP, NOS3, IL6, NPY, CRHR2, TAC1, PENK, GRIN3A, NTRK2, GRIA4, DRD2, HTR3A, GRIK4, BDNF, CCKBR, DRD4, NOS1, GRIN2B, CACNA1C, LRP1, DAOA, HTR2A, AKT1, CHRNA7, GRIN2A, CCL2, NGFR, GRIK5, GRIN2D, GRIK1, ADORA2A |
| Glutamate (70 genes ) |
| GABRA3, PLA2G2A, OPRD1, GRIK3, IL1B, GAD1, CREB1, PER2, TACR1, ADRA2B, CD47, SLC6A1, GSK3B, ITPR1, GRIA2, CCKAR, ADRA2C, NR3C1, SLC6A3, GRIA1, GABRA6, DRD1, HTR1A, GRIK2, ESR1, OPRM1, DTNBP1, GABBR1, TNF, HTR1B, CNR1, CHRM2, HTR5A, NPY, ABCB1, TAC1, ADRA1A, PENK, GRIN3A, GRIN1, NTRK2, HTR7, GRIA4, DRD2, HTR3A, GRIK4, TH, BDNF, CNTF, DRD4, NOS1, P2RX7, P2RX4, GRIN2B, CACNA1C, AVPR1A, DAOA, HTR2A, AKT1, CHRNA7, SLC6A2, GRIN2A, NGFR, GRIN2C, PER1, GRIK5, GRIN2D, GNAS, GRIK1, ADORA2A |
| Neuron (118 genes ) |
| HTR2C, GRIA3, GABRA3, MAOA, MAOB, HTR6, PLA2G2A, AVPR1B, OPRD1, GRIK3, LEPR, PER3, IL1B, GAD1, CREB1, HTR2B, PER2, POMC, TACR1, ADRA2B, CD47, SLC6A1, DRD3, GSK3B, CCK, ITPR1, GRM7, GRIA2, FGFR3, CCKAR, ADRA2C, CLOCK, DRD5, PAM, SLC6A3, HTR4, GRIA1, ADRA1B, GABRA6, DRD1, FGFR4, HTR1A, CRHBP, GRIK2, OLIG3, ESR1, OPRM1, DTNBP1, PDE10A, GABBR1, TNF, HTR1B, CNR1, LEP, CHRM2, HTR5A, IL6, NPY, CRHR2, TAC1, ADRA1A, FGFR1, OPRK1, PENK, GRIN3A, GRIN1, NTRK2, ADRA2A, FGFR2, HTR7, GRIA4, DRD2, HTR3B, HTR3A, GRIK4, TPH1, BDNF, CNTF, DRD4, PDE2A, NOS1, P2RX7, P2RX4, GRIN2B, CACNA1C, CCND2, LRP1, AVPR1A, TPH2, KCNC2, M6PR, HTR2A, AKT1, ESR2, GABRA5, CHRNA7, NTRK3, ADCY9, SLC6A2, GRIN2A, SLC6A4, STAT3, CRHR1, GRIN2C, PER1, GNAL, GRIK5, GSK3A, APOE, GRIN2D, GNAS, GRIK1, OLIG2, OLIG1, PDE9A, COMT, ADORA2A, SYN3 |
| GABA (62 genes ) |
| GRIA3, GABRA3, OPRD1, GRIK3, IL1B, GAD1, POMC, TACR1, DRD3, CCK, GRM7, GRIA2, ADRA2C, DRD5, SLC6A3, HTR4, GRIA1, ADRA1B, GABRA6, DRD1, HTR1A, CRHBP, GRIK2, ESR1, OPRM1, HTR1B, CNR1, LEP, CHRM2, HTR5A, IL6, NPY, CRHR2, GRIN1, NTRK2, DRD2, HTR3A, TH, BDNF, CNTF, CCKBR, DRD4, NOS1, P2RX7, GRIN2B, AVPR1A, KCNC2, HTR2A, AKT1, GABRA5, CHRNA7, SLC6A2, GRIN2A, SLC6A4, GRIN2C, PER1, GNAL, GRIK5, GRIN2D, GRIK1, OLIG2, ADORA2A |
| Adrenergic (69 genes ) |
| HTR2C, MAOA, MAOB, OPRD1, LEPR, IL1B, GAD1, CREB1, TACR1, ADRA2B, DRD3, GSK3B, AGTR1, ITPR1, GRM7, PDE5A, ADRA2C, DRD5, NR3C1, HTR4, GRIA1, DRD1, HTR1A, ESR1, OPRM1, GABBR1, HTR1B, CNR1, LEP, CHRM2, NOS3, IL6, NPY, CRHR2, TAC1, ADRA1A, OPRK1, GRIN1, NTRK2, ADRA2A, HTR7, CYP2C9, DRD2, HTR3A, TH, BDNF, CCKBR, DRD4, PDE2A, NOS1, P2RX4, CACNA1C, AVPR1A, GNB3, HTR2A, AKT1, ESR2, CHRNA7, NTRK3, ADCY9, MMP2, SLC6A2, CRHR1, ACE, PER1, APOE, GNAS, COMT, ADORA2A |
| Agonist (102 genes ) |
| HTR2C, GRIA3, GPR50, GABRA3, MAOA, MAOB, AR, PLA2G4A, HTR6, PLA2G2A, AVPR1B, OPRD1, GRIK3, LEPR, IL1B, GAD1, CREB1, HTR2B, POMC, TACR1, ADRA2B, CD47, DRD3, GSK3B, AGTR1, CCK, ITPR1, GRM7, NFKB1, GRIA2, CCKAR, ADRA2C, DRD5, NR3C1, HTR4, GRIA1, ADRA1B, GABRA6, HTR1A, GRIK2, ESR1, OPRM1, GABBR1, TNF, HTR1B, CNR1, LEP, CHRM2, NOS3, HTR5A, IL6, NPY, CRHR2, TAC1, ADRA1A, OPRK1, PENK, GRIN3A, GRIN1, NTRK2, ADRA2A, HTR7, GRIA4, DRD2, HTR3B, HTR3A, CD3E, TH, BDNF, CCKBR, DRD4, PDE2A, P2RX7, P2RX4, GRIN2B, CACNA1C, AVPR1A, GNB3, HTR2A, AKT1, PRKCH, ESR2, GABRA5, CHRNA7, ADCY9, MMP2, SLC6A2, GRIN2A, SLC6A4, CCL2, STAT3, CRHR1, NGFR, GRIN2C, PER1, GNAL, GRIK5, APOE, GRIN2D, GNAS, GRIK1, ADORA2A |
| cGMP (29 genes ) |
| LEPR, IL1B, PDE11A, PDE1A, CREB1, HTR2B, POMC, ADRA2B, ITPR1, NFKB1, HTR1A, PDE10A, NOS3, ADRB1, PDE6C, CNTF, NOS1, GNB3, HTR2A, AKT1, CHRNA7, SLC6A4, CRHR1, ACE, GRIN2C, GSK3A, APOE, GNAS, PDE9A |
| Synaptic (78 genes ) |
| HTR2C, GRIA3, GABRA3, PLA2G4A, DISC1, GRIK3, IL1B, GAD1, CREB1, PER2, TACR1, CD47, SLC6A1, CCK, ITPR1, GRM7, GRIA2, ADRA2C, DRD5, SLC6A3, HTR4, GABRA6, DRD1, HTR1A, CRHBP, GRIK2, OPRM1, DTNBP1, PDE10A, GABBR1, HTR1B, CNR1, CHRM2, NOS3, NPY, CRHR2, TAC1, GRIN3A, GRIN1, NTRK2, ADRB1, HTR7, PDE6C, GRIA4, DRD2, HTR3A, GRIK4, TPH1, TH, BDNF, DRD4, PDE2A, NOS1, P2RX7, P2RX4, GRIN2B, CACNA1C, LRP1, KCNC2, HTR2A, AKT1, ESR2, GABRA5, CHRNA7, NTRK3, SLC6A2, GRIN2A, SLC6A4, CRHR1, NGFR, GRIN2C, GRIK5, APOE, GRIN2D, GNAS, OLIG1, ADORA2A, SYN3 |
| Phosphodiesterase (22 genes ) |
| DISC1, LEPR, PDE11A, PDE1A, CREB1, ESR1, LEP, NOS3, HTR7, PDE6C, CNTF, PDE2A, NOS1, CCND2, GNB3, AKT1, ADCY9, MMP2, PER1, GNAS, PDE9A, ADORA2A |
Abbreviation: MDD, major depressive disorder.
Gene-set enrichment analysis
PLINK LD-clumping of the MDD meta-analysis results (4346 cases and 4430 controls in total) yielded 1477 genomic regions with independent association signals; 539 intervals among them overlap with annotated genic regions according to the NCBI Entrez RefSeq Gene database (hg18, Homo Sapiens). After gene-set-based association analysis and multiple testing correction, only one of the 178 target gene sets, the glutamatergic synaptic transmission set, remained significant (GO:0035249; empirical enrichment P=6.90 × 10–4; corrected enrichment P=2.89 × 10–2). This glutamatergic synaptic transmission set comprised of 16 genes, of which 6 genes include LD-independent MDD-associated genomic regions at a P-value<5 × 10–3. Varying the regulatory genic regions from 0kb, 20kb, 50kb to 100kb of up/downstream of transcription starting/ending sites resulted in similar enrichment results (data not shown). Table 2 summarizes the set-based association analysis results for the top 10 gene sets (see Supplementary Table S4.1 for the top 30 gene sets).
Table 2. Gene-set-based association analysis results of the top 10 gene sets for major depressive disorder.
| Gene set | Gene number | Enrichment P-value | Annotated genes | ||
|---|---|---|---|---|---|
|
GO term |
Name |
All |
Sig |
|
|
| GO:0035249 | Synaptic transmission, glutamatergic | 16 | 6 | 6.90e-04 | NAPB, GRM8, CACNA1A, GRID2, PLAT, UNC13A, UNC13B, GRIN2D, PARK2, ADRB2, SLC1A4, P2RX1, CDK5, GRIN1, CNIH2, SHC3 |
| GO:0021953 | Central nervous system neuron differentiation | 7 | 2 | 3.66e-02 | MET, NR4A2, TULP3, MNX1, LMX1A, SMO, BCL11B, |
| GO:0021522 | Spinal cord motor neuron differentiation | 6 | 2 | 5.77e-02 | GIGYF2, CACNA1A, NKX2–2, SOX4, MDGA2, ABT1, |
| GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity | 10 | 3 | 7.70e-02 | PDE1B, PDE4B, PDE8A, PDE4A, PDE4C, PDE4D, PDE3B, PDE7A, PDE3A, PDE7B |
| GO:0035254 | Glutamate receptor binding | 5 | 2 | 1.16e-01 | CAMK2A, ATP2B2, GRIN2A, RASGRF1, C16orf70 |
| GO:0051968 | Positive regulation of synaptic transmission, glutamatergic | 12 | 3 | 3.01e-01 | ADORA2A, DRD1, OXTR, CCKBR, PTGS2, GRIA4, NRXN1, NLGN1, NLGN2, NTRK2, TNR, SHANK3 |
| GO:0005234 | Extracellular-glutamate-gated ion channel activity | 18 | 4 | 3.26e-01 | GRIA2, GRIK3, GRIA1, GRIA3, GRID1, GRIK1, GRID2, GRIN2D, GRIA4, STX1B, GRIN1, GRIN2A, GRIK2, GRIN2B, GRIN2C, GRIK4, GRIK5, GRIN3A |
| GO:0043525 | Positive regulation of neuron apoptosis | 33 | 2 | 3.27e-01 | AGRN, ATM, BAX, BCL2L11, NCF2, NR3C1, CASP2, CASP6, CASP7, MAP3K11, RHOC, HRK, MAP2K7, MUSK, TNF, TP53, JUN, PTPRF, NQO1, NF1, FASLG, ASCL1, ITGA1, UBE2M, TGFB2, SRPK2, CDK5, RAPSN, PMAIP1, IL18, CDK5R1, EPHA7, PCSK9 |
| GO:0030672 | Synaptic vesicle membrane | 41 | 4 | 3.47e-01 | AMPH, BCAN, C16orf70, DMXL2, DOC2A, DTNBP1, GAD2, ICA1, OTOF, RPH3A, SCAMP5, SEMA4C, SLC17A6, SLC17A7, SLC17A8, SLC30A3, SLC32A1, SNAPIN, STX1A, SV2A, SV2B, SV2C, SVOP, SYN1, SYN3, SYNGR1, SYNPR, SYT1, SYT10, SYT11, SYT12, SYT2, SYT3, SYT4, SYT5, SYT6, SYT7, SYT9, VAMP1, VAMP2, ZNRF1 |
| GO:0004972 | N-methyl-𝒟-aspartate selective glutamate receptor activity | 6 | 2 | 3.81e-01 | GRIN1, GRIN2A, GRIN2B, GRIN2C, GRIN3A, GRIN2D |
Abbreviations: AMP, adenosine monophosphate; MDD, major depressive disorder.For each of the examined gene sets, Gene Ontology (GO) identifier, name, the number of all annotated genes, the number of genes that overlap with MDD-associated genomic regions, set-based association significance and the list of member genes are listed. Note that the genes intersecting with MDD-associated genomic regions (P<1 × 10−3) are in bold.
GO term: Gene Ontology identifier.
Name: Gene Ontology name.
Gene number: (i) All: the total number of genes annotated by the corresponding GO gene sets; and (ii) Sig: the number of genes that overlap with LD-independent MDD-associated genomic regions at a MDD meta-analysis P-value <5 × 10−3.
Enrichment P-value: statistical significance of aggregated set-based association for gene sets, calculated by INRICH.
Annotated genes: list of genes associated with each gene set.
Discussion
In recent years, genome-wide association analysis has transformed our understanding of the genetic etiology of various common and complex human disorders, but progress has been less rapid for studies of MDD. Along with several practical issues (for example, insufficient sample size, substantial phenotypic heterogeneity or large environmental factors), we presume that common genetic variants confer individually very small effects on risk of MDD, and therefore, standard single-locus-based association analysis is likely to have limited power to dissect the pathogenesis of MDD. Using a text-mining-based statistical analysis and the GO resource, we compiled a collection of 178 GO sets that represent potential biomolecular mechanisms underlying MDD pathogenesis. Multi-locus enrichment analysis on these gene sets uniquely implicated genes involved in glutamatergic synaptic neurotransmission (enrichment P=6.9 × 10–4).
A pathophysiological role of the glutamate system in MDD has been suggested consistently in pre-clinical and clinical studies. The glutamatergic system is a critical mediator of stress responses through regulating the hypothalamic–pituitary–adrenal axis function.39, 40, 41 In animal models of depression, alterations in glutamatergic neurotransmission proteins induce depressive-like behaviors.42, 43 Abnormal activity of the glutamatergic system has been attributed to impairments in synaptic and neural plasticity often observed in patients with severe or recurrent mood disorders.44 Post-mortem MDD studies have reported dysregulation of glutamate levels and glutamate signaling genes, as well.45, 46, 47 Perhaps the most convincing evidence comes from studies suggesting rapid antidepressant effects of glutamatergic interventions, including riluzole48 and, more recently, intravenous ketamine.49, 50, 51
Our analysis supports these previous findings by demonstrating the statistical enrichment of glutamatergic synaptic genes in a meta-analysis of over 8700 MDD cases/controls. The glutamatergic synaptic transmission gene set (GO:0035249) includes 16 genes (Table 3), of which six genes, SLC1A4, CACNA1A, GRM8, PARK2, UNC13A and SHC3, showed nominal association with MDD in our meta-analysis (P<5e-03). Two genes GRM8 and SHC3 have previously been implicated in depression-related phenotypes. The metabotropic glutamate receptor GRM8 inhibits presynaptic glutamate release.52 Nominal association was reported for GRM8 with trait depression in a recent GWAS meta-analysis of European and US samples.53 Multiple studies reported that GRM8 knock-out mice exhibit anxiety-related phenotypes.54, 55, 56 SHC3 is a signaling adaptor involved in the signal transduction pathways in neurons. It has been implicated in modulating hippocampal synaptic plasticity underlying learning and memory57 and associated with nicotine dependence.58
Table 3. MDD association results of 16 genes involved in glutamatergic synaptic transmission GO gene set.
| Symbol |
Minimum association P-value |
Gene name |
Genomic locus (20 kb) |
MDD-associated Region |
|||
|---|---|---|---|---|---|---|---|
| GAIN | STAR*D | QIMR | META | ||||
| SLC1A4 | 0.06729 | 0.03046 | 0.05934 | 0.00089 | Solute carrier family 1 (glutamate/neutral amino acid | chr2:65050097.65124503 | chr2:65098869.65233861 |
| GRID2 | 0.00035 | 0.02806 | 0.00760 | 0.01973 | Glutamate receptor, ionotropic, delta 2 | chr4:93424572.94932672 | none |
| ADRB2 | 0.03455 | 0.19090 | 0.04661 | 0.2975 | Adrenergic, beta-2, receptor, surface | chr5:148166348.148208381 | none |
| PARK2 | 0.00074 | 0.00176 | 0.00389 | 0.00317 | Parkinson disease (autosomal recessive, juvenile) 2, parkin | chr6:161668579.163088824 | chr6:162001468.162009689 |
| GRM8 | 0.00460 | 0.01781 | 0.00379 | 0.00464 | Glutamate receptor, metabotropic 8 | chr7:125845887.126699664 | chr7:126471344.126629287 |
| CDK5 | 0.09839 | 0.47840 | 0.07756 | 0.03576 | Cyclin-dependent kinase 5 | chr7:150361831.150405929 | none |
| PLAT | 0.05833 | 0.09617 | 0.12330 | 0.27460 | Plasminogen activator, tissue | chr8:42131392.42204351 | none |
| GRIN1 | 0.37960 | 0.89950 | 0.14820 | 0.66710 | Glutamate receptor, ionotropic, N-methyl-𝒟-aspartate 1 | chr9:139133429.139203029 | none |
| UNC13B | 0.04341 | 0.05830 | 0.19580 | 0.18260 | Unc-13 homolog B (Caenorhabditis elegans) | chr9:35131988.35415332 | none |
| SHC3 | 0.05547 | 0.02948 | 0.01609 | 0.00427 | SHC (Src homology 2 domain containing) transforming | chr9:90797888.91003502 | chr9:90802459.90826962 |
| CNIH2 | 0.18710 | 0.11400 | 0.07725 | 0.20100 | Cornichon homolog 2 (Drosophila) | chr11:65782271.65828259 | none |
| P2RX1 | 0.01168 | 0.68050 | 0.08987 | 0.05608 | Purinergic receptor P2X, ligand-gated ion channel, 1 | chr17:3726633.3786709 | none |
| CACNA1A | 0.01359 | 0.03251 | 0.00223 | 0.00059 | Calcium channel, voltage-dependent, P/Q type, alpha 1A | chr19:13158256.13498274 | chr19:13304770.13360045 |
| UNC13A | 0.08735 | 0.01120 | 0.10920 | 0.00409 | Unc-13 homolog A (C. elegans) | chr19:17553136.17680401 | chr19:17589502.17595848 |
| GRIN2D | 0.32020 | 0.12270 | 0.19300 | 0.17740 | Glutamate receptor, ionotropic, N-methyl-𝒟-aspartate 2D | chr19:53569943.53660000 | none |
| NAPB | 0.23100 | 0.20230 | 0.20040 | 0.51580 | N-ethylmaleimide-sensitive factor attachment protein, beta | chr20:23283164.23370081 | none |
Abbreviations: GO, Gene Ontology; LD, linkage disequilibrium; MDD, major depressive disorder; SNP, single nucleotide polymorphism.
Symbol: HUGO gene symbol.
Minimum association P-value: most significant association P-value of the SNPs mapped to each genic region (including 20 kb up/downstream region).
Gene name: gene name.
Genomic locus: chromosomal location of gene, including 20 kb upstream and downstream regions.
MDD-associated region: LD-independent (r2<0.5) genomic regions that are associated with MDD at a nominal P-value threshold of 5 × 10−3 in the meta-analysis.
Some limitations should be noted when interpreting the current findings. First, although our analysis indicates statistically enriched association of glutamatergic synaptic genes, we were unable to determine how these genes confer risk of MDD. Functional analysis of glutamatergic genes in MDD is necessary to decipher the underlying disease biology. Secondly, our analysis resorts to predefined GO sets that are indexed by biological keywords representing previous MDD candidate genes. This type of targeted subset analysis increases statistical power to detect true biological gene sets if true sets are included in the examined list. However, other disease gene sets not related to previous MDD candidate genes could have been missed in our analysis. Another caveat is that by using established GO sets, previously unknown relationships among genes could not be examined. Lastly, the present work defines regulatory SNP regions to 20kb up/downstream of transcription starting/ending sites. Most common genetic variations affecting gene expression are known to reside close to transcription starting and ending sites, but the optimal boundaries for defining regulatory regions likely vary by genes. Nevertheless, we observed similar results for a range of gene region windows (0–100 kb up/downstream of coding regions). As regulatory regions are further refined, additional findings may emerge. For example, incorporating known expression quantitative trait loci or methylation quantitative trait loci data is a promising way to assign non-genic, functional SNPs to gene loci.
In conclusion, the present work provides novel evidence for a potential etiological role of glutamate-mediated synaptic neurotransmission in MDD. Further characterization of the molecular and cellular mechanisms of this glutamatergic synaptic system in the brain will be imperative to understand the disease biology of MDD. Given increasing evidence that modulators of NMDA receptor function has rapid antidepressant effects, glutamatergic systems also merit further investigation as therapeutic targets.
Acknowledgments
We are grateful to all the investigators, clinicians and study participants of the STAR*D, GAIN, QIMR and NESDA/NTR projects. Statistical analyses were carried out on the Broad Institute Cluster Computer (http://www.broadinstitute.org). STAR*D: The Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study was supported by federal funds from NIMH under contract N01 MH-90003 to the University of Texas Southwestern Medical Center at Dallas (A John Rush, MD, principal investigator). Genotyping of STAR*D samples was funded by NIMH (R01 MH-072802; Steven P Hamilton, MD, PhD, principal investigator). Control subjects were from the NIMH Schizophrenia Genetics Initiative (NIMH-GI) data and biomaterials were gathered by the Molecular Genetics of Schizophrenia II (MGS-2) collaboration. NESDA/NTR: Genome-wide association study results for the NESDA/NTR MDD data set were obtained through the Genetic Association Information Network (GAIN), through dbGaP accession number phs000020.v1.p1 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000020.v2.p1). Major funding was from the Genetic Association Information Network of the Foundation for the US National Institutes of Health, the Netherlands Organization for Scientific Research (NWO: MagW/ZonMW Grants 904–61–090, 985–10–002,904–61–193,480–04–004, 400–05–717, 912–100–20, Spinozapremie 56–464–14192, Geestkracht program [grant 10–000–1002]); the Center for Medical Systems Biology (CSMB, NWO Genomics), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI –NL) and VU University's institutes for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA). QIMR: Statistical analyses were carried out on the QIMR GenEpi Cluster, which is financially supported by contributions from grants from the NHMRC (389892; 496682; 496688; 496739; 613672) and ARC(FT0991022 FT0991360). We thank the twins and their families for their participation. We also thank Dixie Statham, Ann Eldridge, Marlene Grace, Kerrie McAloney (sample collection); Lisa Bowdler, Steven Crooks (DNA processing); David Smyth, Harry Beeby and Daniel Park (IT support). Funding was provided by the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498), the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016, DP0343921), the FP-5 Genom EU twin Project (QLG2-CT-2002–01254) and the US National Institutes of Health (NIH Grants AA07535, AA10248, AA13320, AA13321, AA13326, AA14041, MH66206). A portion of the genotyping on which this study was based (Illumina 370K scans on 4300 individuals) was carried out at the Center for Inherited Disease Research, Baltimore (CIDR), through an access award to our late colleague Dr Richard Todd (Psychiatry, Washington University School of Medicine, St Louis). Statistical analyses were carried out on the Genetic Cluster Computer, which is financially supported by the Netherlands Scientific Organization. JWS is supported by NIH Grants MH-079799 and MH-094614. RHP is supported by NIMH MH086026. EMB is supported from NHMRC Grant 613608.
RHP has served on the scientific advisory board or consulted to Genomind, Proteus Biomedical and RID Ventures and has received royalties from Concordant Rater Systems (now UBC/Medco). The other authors declare no conflicts of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Hasin DS, Goodwin RD, Stinson FS, Grant BF. Epidemiology of major depressive disorder: results from the National Epidemiologic Survey on Alcoholism and Related Conditions. Arch Gen Psychiatry. 2005;62:1097–1106. doi: 10.1001/archpsyc.62.10.1097. [DOI] [PubMed] [Google Scholar]
- Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ, et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychaitry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- Lohoff F. Overview of the genetics of major depressive disorder. Curr Psychiatry Rep. 2010;12:539–546. doi: 10.1007/s11920-010-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen G, Webb BT, Butler AW, van den Oord EJ, Tozzi F, Craddock N, et al. A genome-wide significant linkage for severe depression on chromosome 3: the depression network study. Am J Psychiatry. 2011;168:840–847. doi: 10.1176/appi.ajp.2011.10091342. [DOI] [PubMed] [Google Scholar]
- Pergadia M, Glowinski AL, Wray NR, Agrawal A, Saccone SF, Loukola A, et al. 3p26-3p25 genetic linkage finding for DSM-IV major depression in heavy smoking families. Am J Psychiatry. 2011;168:848–852. doi: 10.1176/appi.ajp.2011.10091319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SA. New lead from genetic studies in depressed siblings: assessing studies of chromosome 3. Am J Psychiatry. 2011;168:783–789. doi: 10.1176/appi.ajp.2011.11060835. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, et al. Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- Rietschel M, Mattheisen M, Frank J, Treutlein J, Degenhardt F, Breuer R, et al. Genome-wide association-, replication-, and neuroimaging study implicates HOMER1 in the etiology of major depression. Biol Psychiatry. 2010;68:578–585. doi: 10.1016/j.biopsych.2010.05.038. [DOI] [PubMed] [Google Scholar]
- Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyn SI, Shi J, Kraft JB, Potash JB, Knowles JA, Weissman MM, et al. Novel loci for major depression identified by genome-wide association study of sequenced treatment alternatives to relieve depression and meta analysis of three studies. Mol Psychaitry. 2011;16:202–215. doi: 10.1038/mp.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychaitry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. A mega-analysis of genome-wide association studies for major depressive disorder Mol Psychaitry 2012. advance online publication, 3 April 2012; doi: 10.1038/mp.2012.21PMID: 22472876. [DOI] [PMC free article] [PubMed]
- Kohli M, Lucae S, Saemann PG, Schmidt MV, Demirkan A, Hek K, et al. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron. 2011;70:252–265. doi: 10.1016/j.neuron.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Nestler EJ. The molecular neurobiology of depression. Nature. 2008;455:894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltser-Dubner T, Galili-Weisstub E, Segman RH. Genetics of unipolar major depressive disorder. Isr J Psychiatry Relat Sci. 2010;47:72–82. [PubMed] [Google Scholar]
- Demirkan A, Penninx BW, Hek K, Wray NR, Amin N, Aulchenko YS, et al. Genetic risk profiles for depression and anxiety in adult and elderly cohorts. Mol Psychiatry. 2011;16:773–783. doi: 10.1038/mp.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genome-wide association studies. Am J Hum Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips E, Cornelisse LN, Toonen RF, Min JL, Hultman CM., the International Schizophrenia Consortium, Holmans PA, O'Donovan MC, Purcell SM, Smit AB, Verhage M, Sullivan PF, et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia Mol Psychiatry 2011[Epub ahead of print] Sep20.2011 [DOI] [PMC free article] [PubMed]
- Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P., Wellcome Trust Case-Control Consortium, Owen MJ, O'Donovan MC, Craddock N. Gene ontology analysis of GWA Study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2010;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips ESCL, Toonen RF, Min JL, Hultman CM., the International Schizophrenia Consortium, Holmans PA, O'Donovan MC, Purcell SM, Smit AB, Verhage M, Sullivan PF, et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia Mol Psychaitry 2011[Epub ahead of print]2011Sep20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dushlaine C, Kenny E, Heron E, Donohoe G, Gill M, Morris D. International Schizophrenia Consortium, Corvin A. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2011;16:286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrè A., DIAGRAM Consortium; MAGIC investigators, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., International Schizophrenia Consortium, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC., International Schizophrenia Consortium, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D, et al. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychaitry. 2011;16:516–532. doi: 10.1038/mp.2010.38. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene ontology: tool for the unification of biology Nat Genet 20002525–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Mingyao Li, Lange EM, Watanabe RM. Prioritized subset analysis: improving power in genome-wide association studies. Hum Hered. 2008;65:129–141. doi: 10.1159/000109730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medland SE, Nyholt DR, Painter JN, McEvoy BP, McRae AF, Zhu G, et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am J Hum Genet. 2009;85:750–755. doi: 10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs Nature 2007449851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Abecasis GR. Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- Lee PH, O'Dushlaine C, Thomas B, Purcell SM.INRICH: interval-based enrichment analysis for genome wide association studies In Press2011 [DOI] [PMC free article] [PubMed]
- Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew SJ, Coplan JD, Schoepp DD, Smith EL, Rosenblum LA, Gorman JM.Glutamate-hypothalamic-pituitary-adrenal axis interactions: implications for mood and anxiety disorders CNS Spectr 20016555-556561–564. [DOI] [PubMed] [Google Scholar]
- Mitsukawa K, Mombereau C, Lötscher E, Uzunov DP, van der Putten H, Flor PJ, et al. Metabotropic glutamate receptor subtype 7 ablation causes dysregulation of the HPA axis and increases hippocampal BDNF protein levels: implications for stress-related psychiatric disorders. Neuropsychopharmacology. 2006;31:1112–1122. doi: 10.1038/sj.npp.1300926. [DOI] [PubMed] [Google Scholar]
- Zelena D, Mergl Z, Makara GB. Glutamate agonists activate the hypothalamic-pituitary-adrenal axis through hypothalamic paraventricular nucleus but not through vasopressinerg neurons. Brain Res. 2005;1031:185–193. doi: 10.1016/j.brainres.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Autry AE, Adachi M, Nosyreva E, Na ES, Los MF, Cheng PF, et al. NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature. 2011;475:91–95. doi: 10.1038/nature10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CX, Wang Y, Gao H, Pan WJ, Xiang Y, Huang M, et al. Cerebral metabolic changes in a depression-like rat model of chronic forced swimming studied by ex vivo high resolution 1H magnetic resonance spectroscopy. Neurochem Res. 2008;33:2342–2349. doi: 10.1007/s11064-008-9739-0. [DOI] [PubMed] [Google Scholar]
- Sanacora G, Zarate CA, Krystal JH, Manji HK. Targetting the glutamatergic system to develop novel, improved therapeutics for mood disorders. Nat Rev Drug Discov. 2008;7:426–437. doi: 10.1038/nrd2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard R, Kerman IA, Thompson RC, Jones EG, Bunney WE, Barchas JD, et al. Altered expression of glutamate signaling, growth factor, and glia genes in the locus coeruleus of patients with major depression. Mol Psychaitry. 2010;16:634–646. doi: 10.1038/mp.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Sawa A, Iyo M. Increased levels of glutamate in brains from patients with mood disorders. Biol Psychiatry. 2007;62:1310–1316. doi: 10.1016/j.biopsych.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Duric V, Banasr M, Stockmeier CA, Simen AA, Newton SS, Overholser JC, et al. Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. Int J Neuropsychopharmacol. 2012;17:1–14. doi: 10.1017/S1461145712000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan BP, Hudson JI, Jensen JE, McCarthy J, Roberts JL, Prescot AP, et al. Rapid enhancement of glutamatergic neurotransmission in bipolar depression following treatment with riluzole. Neuropsychopharmacology. 2010;35:834–846. doi: 10.1038/npp.2009.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Vieira R, Salvadore G, Diazgranados N, Zarate CA. Ketamine and the next generation of antidepressants with a rapid onset of action. Pharmacol Ther. 2009;123:143–150. doi: 10.1016/j.pharmthera.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeng S, Zarate CA. The role of glutamate in mood disorders: results from the ketamine in major depression study and the presumed cellular mechanism underlying its antidepressant effects. Curr Psychiatry Rep. 2007;9:467–474. doi: 10.1007/s11920-007-0063-1. [DOI] [PubMed] [Google Scholar]
- Salvadore G, Cornwell BR, Colon-Rosario V, Coppola R, Grillon C, Zarate CA, et al. Increased anterior cingulate cortical activity in response to fearful faces: a neurophysiological biomarker that predicts rapid antidepressant response to ketamine. Biol Psychiatry. 2009;65:289–295. doi: 10.1016/j.biopsych.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepp DD. Unveiling the functions of presynaptic metabotropic glutamate receptors in the central nervous system. J Pharmacol Exp Ther. 2001;299:12–20. [PubMed] [Google Scholar]
- Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, et al. Genome-wide association scan of trait depression. Biol Psychiatry. 2010;68:811–817. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins MJ, Starr KR, Honey A, Soffin EM, Rourke C, Jones GA, et al. Evaluation of the mGlu8 receptor as a putative therapeutic target in schizophrenia. Brain Res. 2007;1152:215–227. doi: 10.1016/j.brainres.2007.03.028. [DOI] [PubMed] [Google Scholar]
- Linden AM, Johnson BG, Peters SC, Shannon HE, Tian M, Wang Y, et al. Increased anxiety-related behavior in mice deficient for metabotropic glutamate 8 (mGlu8) receptor. Neuropharmacology. 2002;43:251–259. doi: 10.1016/s0028-3908(02)00079-5. [DOI] [PubMed] [Google Scholar]
- Duvoisin RM, Zhang C, Pfankuch TF, O'Connor H, Gayet-Primo J, Quraishi S, et al. Increased measures of anxiety and weight gain in mice lacking the group III metabotropic glutamate receptor mGluR8. Eur J Neurosci. 2005;22:425–436. doi: 10.1111/j.1460-9568.2005.04210.x. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Chen L, Sato M, Sokabe M, Nabeshima T, Pawson T, et al. Hippocampal synaptic modulation by the phosphotyrosine adapter protein ShcC/N-Shc via interaction with the NMDA receptor. J Neurosci. 2005;25:1826–1835. doi: 10.1523/JNEUROSCI.3030-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Sun D, Lou XY, Beuten J, Payne TJ, Ma JZ. Linkage and association studies in African- and Caucasian-American populations demonstrate that SHC3 is a novel susceptibility locus for nicotine dependence. Mol Psychiatry. 2007;12:462–473. doi: 10.1038/sj.mp.4001933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.