Abstract
The striking excess of affected males in autism spectrum disorders (ASD) suggests that genes located on chromosome X contribute to the etiology of these disorders. To identify new X-linked genes associated with ASD, we analyzed the entire chromosome X exome by next-generation sequencing in 12 unrelated families with two affected males. Thirty-six possibly deleterious variants in 33 candidate genes were found, including PHF8 and HUWE1, previously implicated in intellectual disability (ID). A nonsense mutation in TMLHE, which encodes the ɛ-N-trimethyllysine hydroxylase catalyzing the first step of carnitine biosynthesis, was identified in two brothers with autism and ID. By screening the TMLHE coding sequence in 501 male patients with ASD, we identified two additional missense substitutions not found in controls and not reported in databases. Functional analyses confirmed that the mutations were associated with a loss-of-function and led to an increase in trimethyllysine, the precursor of carnitine biosynthesis, in the plasma of patients. This study supports the hypothesis that rare variants on the X chromosome are involved in the etiology of ASD and contribute to the sex-ratio disequilibrium.
Keywords: Autism spectrum disorders, carnitine, chromosome X, male excess, next-generation sequencing, TMLHE
Introduction
Autism spectrum disorders (ASD) constitute a common but heterogeneous group of neurodevelopmental disorders characterized by impairment of social interactions and communication, stereotyped behaviors and restricted interests. Although they are probably the most heritable of psychiatric conditions, with a concordance rate of ∼80–90% in monozygotic twins versus ∼10–20% in dizygotic twins, few genes have been reliably associated with ASD.1, 2, 3 Recent studies have highlighted the vast heterogeneity and complexity of the genetics of these disorders. All mutations or copy number variants (CNVs) associated so far with ASD have been rare, with minor allele frequencies <1%. A few de novo or inherited CNVs, some of which are recurrent, such as duplications of the 15q11-q13 or 7q11.23 and deletions of 16p11.2 regions, were shown to confer a highly penetrant risk of autism.4, 5, 6, 7 More recently, de novo mutations in various highly interconnected genes were shown to contribute to ASD, suggesting that abnormalities in different genes could converge to alter common pathways.8, 9, 10 Abnormalities in at least two pathways were repeatedly related to ASD: the first includes mutations in TSC1/TSC2, NF1 or PTEN in the mTOR (mammalian target of rapamycin) pathway; the second is illustrated by mutations in NLGN3–4, SHANK1–3 and NRXN1, all of which encode synaptic proteins.11, 12, 13
A striking feature of ASD is the excess of affected males, with a sex-ratio disequilibrium of 4:1 that reaches 10:1 in patients with normal cognitive abilities.14 This suggests that genes located on sex chromosomes contribute to the etiology of ASD or that the penetrance of autistic traits depends on sex determinants such as hormones. In favor of the first hypothesis, mutations in NLGN4X and NLGN3 on chromosome X have been identified in a few families with ASD.15 Additionally, the analysis of all or selected genes located on the X chromosome successfully identified new candidate genes for intellectual disability (ID),16 ASD and schizophrenia.17 Interestingly, the risk of recurrence of ASD is significantly increased in families with two affected sibs, reaching 32% and more when both affected subjects are males.18 This suggests that highly penetrant forms of ASD with autosomal recessive or X-linked inheritance have been overlooked.
To test the hypothesis that yet undiscovered X-linked genes are associated with highly penetrant forms of ASD, we selected 12 unrelated families with at least two affected males compatible with X-linked inheritance and analyzed all of the coding regions on the X chromosome.
Material and methods
Patients
The entire exome of chromosome X was sequenced in 12 families with two affected males with ASD or ID compatible with X-linked inheritance, recruited from the ‘Centre de Référence Déficiences Intellectuelles de causes rares' (Pitié-Salpêtrière Hospital; Supplementary Figure S1). Index cases were evaluated by specialized geneticists and pediatric neurologists and/or child psychiatrists. Patients were assessed with the Autism Diagnostic Interview-Revised. Nine index cases had autism with ID and three had Asperger syndrome or high-functioning autism based on DSM IV-TR (Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision) criteria. Clinical features of the index cases and the affected relatives are detailed in Supplementary Clinical Table. Normal results were previously obtained by karyotyping, searches for fragile-X syndrome, microarray analysis (CytoSNP-12, Illumina, San Diego, CA, USA) and sequencing of NLGN3–4X and SHANK3 when appropriate, as well as metabolic screening (including at least creatine and guanidinoacetate analysis).
For TMLHE screening, a cohort of 161 patients (134 patients with autism and ID and 27 patients with Asperger syndrome) recruited at the Pitié-Salpêtrière Hospital (Centre de référence Déficiences Intellectuelles de causes rares or Centre référent diagnostic autisme, Paris, France) and 340 patients from the PARIS (Paris Autism Research International sib pair) study (including 194 patients with autism and ID and 59 patients with Asperger syndrome) were included, for a total of 501 unrelated male patients with ASD. In addition, 765 healthy male controls from North Africa (n=320), Europe (n=350) and Lebanon (n=95) were included to test the new variants.
The analysis of microrearrangements in TMLHE included 178 additional patients with ASD from the PARIS studies previously included in Autism Genome Project (AGP, http://www.autismgenome.org/)7 and 896 healthy male individuals. The control groups included 371 European male subjects from La Pitié-Salpêtrière hospital, 142 from other European laboratories and 383 control individuals from the Study on Addiction Genetics and Environment (n=371) and HapMap CEPH Utah (n=12) series.7 Raw intensities and genotypes were obtained from NHGRI-dbGaP (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1). The sub-set of control data set used in the specific CNV analyses in this paper is composed of control samples that passed all quality control filters (Log R ratios s.d.=0.27; B allele frequency s.d.=0.13; Call Rate >0.99). Informed written consent was obtained from each individual or his/her parents before blood sampling. All experiments were performed in accordance with French guidelines and rules.
Next-generation sequencing
Next-generation sequencing was performed at Integragen SA (Evry, France). Regions of the X chromosome corresponding to coding and 3′–5′ untranslated region (UTR) sequences were captured from genomic DNA using a custom Agilent SureSelect Target Enrichment methodology (Agilent, Santa Clara, CA, USA) with the biotinylated oligonucleotide probe library, followed by paired-end 75 b massively parallel sequencing on Illumina GAIIx (Illumina). For detailed explanations of the process, see Gnirke et al.19 Sequence capture, enrichment and elution were performed precisely according to the manufacturer's instructions and protocols. Briefly, 3 μg of each genomic DNA was fragmented by sonication to yield fragments of 150–200 bp and then purified. Paired-end adapter oligonucleotides from Illumina were ligated on repaired A-tailed fragments, then purified and enriched by six PCR cycles. Then 500 ng of these purified libraries were hybridized to the SureSelect oligo probe capture library for 24 h. After hybridization, washing and elution, the eluted fraction was PCR-amplified for 10–12 cycles, purified and quantified by quantitative PCR to obtain sufficient DNA template for downstream applications. Each eluted-enriched DNA sample was sequenced on an Illumina GAIIx as paired-end 75 b reads. Image analysis and base calling was performed using Real Time Analysis Pipeline version 1.9 (Illumina) with default parameters.
Bioinformatics analysis
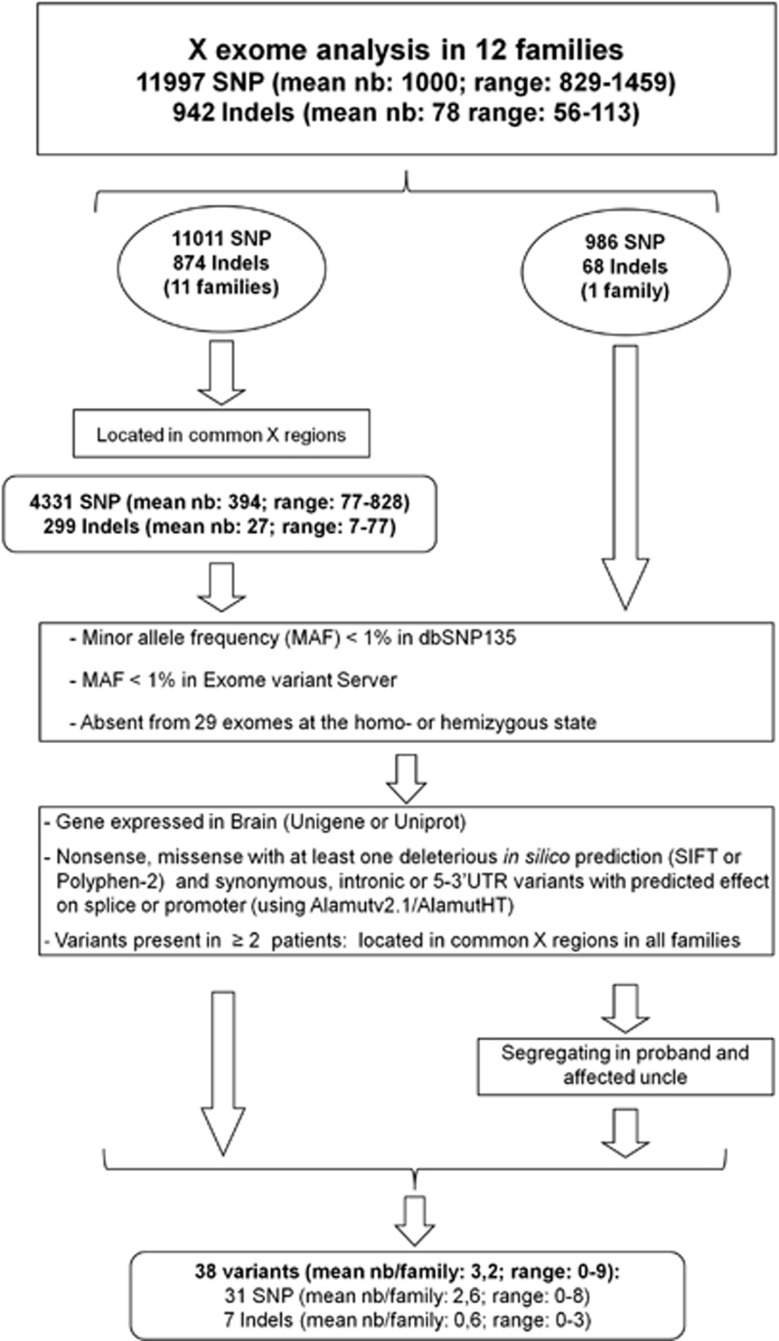
Sequencing data was analyzed according to the Illumina pipeline (CASAVA1.7) and aligned with the Human reference genome (Hg19) using the ELANDv2 algorithm. Variant annotation (RefSeq gene annotation) identification of known polymorphisms (referenced in dbSNP or 1000 Genome) and analysis of the position and consequences of the variants (for example, exonic, intronic, silent and nonsense), was determined with an in-house pipeline from the positions included in the bait coordinates. The frequencies (in the homozygous or heterozygous state) were determined from all exomes sequenced at Integragen and from exome results provided by HapMap. Results per sample were obtained in tabulated text files, and coverage/depth statistical analyses were performed for each bait. The 13,464 single nucleotide polymorphisms (SNPs) and 1532 indels in the 12 male index cases included 1467 SNPs (10.9%) and 590 indels (38.5%) predicted to be at the heterozygous state. Eleven of these variants were tested by Sanger sequencing and proved to be false-positives. Further analysis focused, therefore, on variants predicted to be hemizygous. A total of 171 hemizygous variants were tested and confirmed by Sanger sequencing. The strategy used for selecting potentially pathogenic variants is detailed in Figure 1. They: (i) were located in chromosome X regions common to the two affected sibs (4331 SNPs and 299 indels in 11 families); (ii) had with a minor allele frequency <1% in dbSNP135 (http://www.ncbi.nlm.nih.gov/projects/SNP/), Exome variant server (http://evs.gs.washington.edu/EVS/) and 29 other exomes; (iii) were found in genes expressed in brain according to the Unigene (http://www.ncbi.nlm.nih.gov/unigene) or Uniprot (http://www.uniprot.org) databases; and (iv) were predicted to have an impact on the gene or the protein (nonsense variants, missense variants, predicted at least once in silico to be deleterious, and synonymous, intronic or 5–3′UTR variants with possible effects on splice sites or promoters using Alamutv2.1/AlamutHT). For variants present in at least two index cases, only those segregating in all the affected members of all families were retained. Mutation interpretation and amino-acid conservation in orthologs and paralogs were assessed using the Alamutv2.1/AlamutHT softwares (Interactive Biosoftware, Rouen, France). Prediction of pathogenicity was assessed using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (scale-invariant feature transform) (http://sift.bii.a-star.edu.sg/), Mutpred (http://mutpred.mutdb.org/) and SNPs&GO (http://snps-and-go.biocomp.unibo.it/snps-and-go/). Frequencies were compared with the Fisher's exact test.
Figure 1.
Strategy used for the selection of rare and possibly deleterious variants. Data from NGS and single nucleotide polymorphism (SNP) arrays were combined to conserve only variants located in X regions shared by the affected sibs (families 1–11). Further filters included a minor allele frequency (MAF) <1%, expression of the corresponding genes in brain and in silico predictions compatible with an effect of the variant on the gene or the protein (nonsense variants, missense variants with at least one prediction in silico by SIFT (scale-invariant feature transform) or Polyphen-2 that it is deleterious and synonymous, intronic or 5–3′UTR variants with possible effects on splice sites or promoters using Alamutv2.1/AlamutHT). For variants present in at least two index cases, only those that segregated in all affected members of all families were conserved. For one family (family 12), microarray data were unavailable for the affected uncle; segregation of variants found in the index case was performed at a later time.
High-density SNP arrays
Index cases and affected relatives were screened using Illumina cytoSNP-12 arrays to search for CNVs and identify regions on the X chromosome shared by the affected sibs. Illumina microarray experiments were automated and performed at the P3S platform (Pitié-Salpêtrière Hospital), according to the manufacturer's specifications (Illumina, San Diego, CA, USA). Image acquisition was performed using a BeadArray Reader (Illumina). Image data analysis and automated genotype calling was performed using GenomeStudiov2011.1 (Illumina). Genomic positions were based on the UCSC and Ensembl Genome Browsers. Genotypes on chromosome X were analyzed in affected relatives of each family to identify shared X regions (with the exception of family 12 for which SNP microarray data were unavailable for the affected uncle). Shared regions were defined as identical genotypes spanning at least 2 Mb.
For the analysis of TMLHE microrearrangements, control individuals (n=525) and ASD cases (n=356) were genotyped using Illumina Human 1M-single BeadChip arrays. Samples were processed using the manufacturer's recommended protocol, and BeadChips were scanned on the Illumina BeadArray Reader using default settings. Analysis and intra-chip normalization were performed using Illumina's BeadStudio software v.3.3.7, with a GenCall cutoff of 0.1. The quality-control criteria were selected: the Array with call rate >95% standard deviation for log R ratio values in the autosomes <0.35; and standard deviation of the B Allele frequency values (that is, allelic ratios within the 0.25–0.75 ranges) >0.13. For the samples that passed the above SNP and intensity quality-control filters, we used the QuantiSNP20 and visualizator SnipPeep CNV calling algorithms. The required data for CNV analysis, that is, within-sample normalized fluorescence (that is, X and Y normalized values), between-sample normalized fluorescence (that is, Log R ratios and B allele frequency values) and genotypes for each sample, were exported directly from Illumina's Beadstudio software. We excluded CNVs when they failed stringent quality control criteria: <5 consecutive probes covering 1 kb of sequence were merged using outside probe boundaries (that is, union of the CNVs) and low confidence score log Bayes factor <15.
Sanger sequencing
Specific primer pairs were designed to amplify 182 variants detected by next-generation sequencing. In addition, eight primer pairs were designed to amplify the coding exons and adjacent intron–exon boundaries of the TMLHE gene. Primer sequences are provided in supplemental data. Forward and reverse sequence reactions were performed with the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA) using the same primers. G50-purified sequence products were run on an ABI 3730 automated sequencer (Applied Biosystems) and data were analyzed with Seqscape v2.6 software (Applied Biosystems). The mutation nomenclature is based on the TMLHE cDNA reference sequence (NM_018196.3).
Quantitative multiplex PCR and long-range PCR
The presence of a deletion of exon 2 in TMLHE was tested in the 501 patients with ASD and 371 male control individuals by quantitative multiplex PCR. One hundred and seventy-eight patients with ASD were also screened using Illumina Human 1 M-single BeadChip arrays with the same results. Two primer pairs were used in the quantitative multiplex assay: one specific of exon 2 of TMLHE (final concentration: 0.5 μℳ) and one amplifying exon 2 of GPR128 (final concentration: 0.08 μℳ). PCR conditions were as follows: 96 °C 5 min, 20 cycles: 94 °C 30 s, 60 °C−0.5 °C/cycle 30 s, 72 °C 40 s, and 15 cycles: 94 °C 30 s, 50 °C 30 s, 72 °C 40 s, followed by 7 min at 72 °C. PCR products were quantified on a Caliper LabChip system (Caliper Life Sciences, Hopkinton, MA, USA). In addition, the presence of the deletion of exon 2 in TMLHE was confirmed by long-range PCR using the SequalPrep Long PCR kit (Invitrogen, Life Technologies Corporation, Carlsbad, CA, USA) according to the manufacturer's recommendations. The PCR conditions were as follows: 2 min at 94 °C, 10 cycles: 94 °C 10 s, 58 °C 30 s, 68 °C 18 min (1 min kb−1), 25 cycles: of 94 °C 10 s, 58 °C 30 s, 68 °C 18 min (1 min kb−1)+20 s/cycle, followed by 5 min at 72 °C. Frequencies were compared with the Fisher's exact test.
Cell culture and mRNA experiments
Lymphoblasts from the two affected brothers and the mother of family 9 were isolated from peripheral blood cells using standard procedures. Fibroblasts were taken from skin cells of the affected brothers. Lymphoblastic cells and fibroblasts were pretreated, or not, overnight with 10 μg ml−1 emetin, an inhibitor of nonsense-mediated decay.
Total RNA from lymphoblasts and fibroblasts was isolated using the Qiagen RNeasy Mini kit (Invitrogen). cDNAs were synthesized from 1 μg of total RNA using the SuperScript III First-Strand Kit (Invitrogen). The reverse-transcribed TMLHE cDNA was amplified and sequenced using specific primers located in exons 2 (forward) and 4 (reverse). The PCR products were run on 2% agarose gels. TMLHE mRNA was quantified using the Qiagen QuantiTect primer assays for TMLHE (forward and reverse primers located in exons 7 and 8 of TMLHE). PPIA was used as the control gene. Each sample was run in triplicate on a Lightcycler 480 (Roche, Applied Sciences, Penzberg, Germany). Forty-five two-step cycles (15 s at 95 °C and 30 s at 60 °C) were performed and analyzed using Lightcycler 480 software release 1.5.0. Relative abundance was calculated using the formula r=2−ΔΔCt, where ΔΔCt=(Ct Gene tested −Ct control genes) TMLHE – (Ct Gene tested −Ct control genes) PPIA.
Chromatography and mass spectrometry
A mixture of internal standard solution was prepared by dissolving 30 μℳ of carnitine-(N-methyl-d3) (Cambridge Isotopes Lab, Andover, MA, USA) and 15 μℳ of ɛ-N-trimethyl-(13C3)-L-Lysine (Sigma-Aldrich, St Louis, MO, USA) in methanol. In all, 20 μl of internal standard mixture was added to 30 μl of plasma or urine after mixing, 100 μl of methanol was added and mixed with vortex for protein precipitation. The mixture was incubated in ice for 15 min and centrifuged at 15000 g for 10 minutes at +4 °C. The supernatant was transferred into the vials and 5 μl were injected into tandem LCMS/MS system. Calibration curves were performed by serial dilution of stock solution containing 46.8 μℳ of l-carnitine and 20 μℳ of ɛ-N-trimethyllysine (Sigma-Aldrich) in methanol. An Acquity UPLC (ultra performance liquid chromatography) chromatographic system equipped with a BEH C18 RP column (1.7 μ, 50 mm × 2) maintained at 45 °C was coupled to a TQD (tandem quadrupole detector) MS/MS system (Waters, Guyancourt, France) and used as an LCMS/MS (liquid chromatography–mass spectrometry/mass spectrometry) system for trimethyllysine (TML) and carnitine measurement. The mobile phases were: eluent A, ultrapure water; eluent B, acetonitrile. The elution gradient was as follows: flow rate 0.8 ml min−1, 0–1 min, 0% A; 1–1.2, 0–100% A; 1.2–2 min, 100% 2–2.2, 100–0% A; 2.2–5, column equilibration with 100% B. The detector was used in multiple-reaction monitoring to detect the transition of a specific precursor to daughter ions 189.1/84.1 and 162.2/103.1 for N6-trimethyllysine and carnitine, respectively.
Dietary assessment
In order to determine prospective and retrospective dietary intakes, the patient and their parents had an interview with a dietician. A dietary questionnaire listing all the food and beverages consumed by the patient for a period of 3 days was completed by the parents. Nutritional intakes were estimated with the DSMS software. Carnitine intakes were calculated based on the reference table of the Linus Pauling Institute Oregon State University.
Results
Chromosome X exome sequencing
The coding regions of all genes on chromosome X, including 5′–3′ UTRs, were sequenced in 12 index cases using next-generation sequencing (pedigrees are shown in Supplementary Figure S1). A mean number of 1000 SNPs (829–1459) and 78 indels (56–113; Table 1 and Supplementary Table SA and SB) were identified per patient. In parallel, chromosome X regions common to the affected sibs, ranging from 15 to 109 Mb, were identified in 11 families using Illumina cytoSNP-12 arrays (Supplementary Figure S2). The mean numbers of SNPs and indels located in shared X regions were 394 (77–828) and 27 (7–77) per patient, respectively. Further analysis focused on variants that were (i) absent or rare in databases (minor allele frequency <1%), (ii) located in genes expressed in brain and (iii) predicted to be deleterious (Figure 1). Thirty-eight possibly deleterious variants (mean number per family: 3.2 range: 0–9), all confirmed by Sanger sequencing, were detected (Table 2). Analysis of matched control populations excluded two variants that had a frequency ⩾1%. The variants were present in 15/22 asymptomatic male relatives. Altogether, these results identified 36 rare, possibly deleterious variants in 33 different genes in 9 families.
Table 1. Summary of all SNPs and indels detected by exome analysis of chromosome X in the index cases of families 1–12 and those located in chromosome X regions shared by families 1–11.
| Family | Nb SNPs (novel) | Nb indels (novel) | Linked regions (Mb) | SNPs in linked regions |
Indels in linked regions
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Nonsense (novel) | Missense (novel) | Synonymous (novel) | Non-coding (novel) | Total | Exons (novel) | Non-coding (novel) | ||||
| Family 1 | 1233 (54) | 108 (45) | 109 | 828 | 1 (0) | 122 (3) | 137 (3) | 568 (23) | 77 | 5 (1) | 72 (30) |
| Family 2 | 1039 (40) | 82 (37) | 37 | 369 | 1 (0) | 59 (3) | 48 (1) | 261 (8) | 19 | 0 | 19 (6) |
| Family 3 | 871 (16) | 56 (22) | 33 | 251 | 1 (0) | 36 (0) | 40 (1) | 174 (2) | 15 | 0 | 15 (9) |
| Family 4 | 861 (27) | 67 (23) | 57 | 340 | 1 (0) | 72 (4) | 58 (1) | 209 (2) | 21 | 2 (1) | 19 (8) |
| Family 5 | 892 (30) | 81 (34) | 76 | 412 | 1 (0) | 57 (1) | 66 (0) | 288 (15) | 36 | 2 (1) | 34 (14) |
| Family 6 | 989 (33) | 83 (37) | 27 | 319 | 0 | 54 (2) | 46 (2) | 219 (7) | 28 | 3 (2) | 25 (14) |
| Family 7 | 829 (18) | 66 (27) | 36 | 200 | 1 (0) | 37 (1) | 41 (0) | 121 (2) | 14 | 2 (1) | 12 (6) |
| Family 8 | 1459 (63) | 113 (60) | 42 | 614 | 1 (0) | 71 (3) | 101 (1) | 441 (22) | 42 | 4 (3) | 38 (24) |
| Family 9 | 1098 (35) | 82 (33) | 81 | 656 | 1 (1) | 93 (2) | 90 (2) | 472 (21) | 23 | 1 (0) | 22 (7) |
| Family 10 | 835 (31) | 63 (24) | 15 | 77 | 0 | 6 (0) | 10 (1) | 61 (2) | 7 | 0 | 7 (1) |
| Family 11 | 905 (24) | 73 (32) | 53 | 265 | 3 (0) | 38 (1) | 38 (1) | 186 (6) | 17 | 1 (1) | 16 (5) |
| Family 12 | 986 (24) | 68 (19) | Unknown | ||||||||
Abbreviation: SNP, single-nucleotide polymorphism.
Linked regions and the total number of SNPs and indels in linked regions are indicated in bold.
Table 2. Summary of rare, possibly deleterious variants located in shared regions on chromosome X.
| Index case | Position (Hg19) | Gene | NM_number | Nucleotide change | Amino-acid change | Mutation type | rs number (dbSNP135) | rsMAF(dbSNP 135/Exome variant server) (%) | Grantham distance | SIFT | PolyPhen2 | Mutpred | SNPs&GO | Controls | Variant found in healthy man in the family | Causative status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family 1 | 3228257 | MXRA5 | NM_015419 | c.7987G >T | p.Gly2663Trp | Missense | rs143264543 | 0/0.02 | 184 | deleterious | damaging | 0.575 | neutral | 0/93 | NA | possible |
| 16860727 | TXLNG | NM_018360 | c.*838A >G | 3'UTR | rs140169598 | 0/- | NA | unknown | ||||||||
| 18910664 | PHKA2 | NM_000292 | c.*939T >C | 3'UTR | rs142176794 | 0/- | NA | unknown | ||||||||
| 19037757 | GPR64 | NM_001079858 | c.670C >T | p.Pro224Ser | Missense | rs140334931 | 0/0.34 | 74 | tolerated | damaging | 0.268 | neutral | NA | possible | ||
| 23801307 | SAT1 | NM_002970 | c.-162G >A | 5'UTR | rs113458958 | 0.8/- | NA | unknown | ||||||||
| 118250487 | KIAA1210 | NM_020721 | c.622G >T | p.Val208Phe | Missense | — | -/0.18 | 50 | deleterious | damaging | 0.174 | unknown | NA | possible | ||
| 124028159 | ODZ1 | NM_001163278 | c.521C >A | p.Ala174Asp | Missense | rs139486546 | 0/0.1 | 126 | deleterious | benign | 0.396 | neutral | 0/159 | NA | possible | |
| 148081861 | AFF2 | NM_002025 | c.*8999G >C | 3'UTR | rs142697456 | 0/- | NA | unknown | ||||||||
| Family 2 | 17750042 | NHS | NM_198270 | c.4351G >C | p.Ala1451Pro | Missense | — | -/- | 27 | tolerated | damaging | 0.345 | neutral | 0/88 | NA | possible |
| 130411844 | IGSF1 | NM_001170961 | c.2321A >G | p.Glu774Gly | Missense | — | -/- | 98 | deleterious | damaging | 0.484 | unknown | 0/96 | NA | possible | |
| 151092546 | MAGEA4 | NM_001011548 | c.410T >A | p.Leu137Gln | Missense | — | -/- | 113 | deleterious | damaging | 0.827 | disease | 0/96 | NA | possible | |
| 153734897 | FAM3A | NM_021806 | c.*245A >G | 3'UTR | rs145895727 | 0/- | NA | unknown | ||||||||
| Family 4 | 3240238 | MXRA5 | NM_015419 | c.3488G >A | p.Arg1163His | Missense | rs139106444 | 0/0.36 | 29 | deleterious | damaging | 0.399 | neutral | 1/130 male | yes (grand-father) | unlikely |
| 49105970 | CCDC22 | NM_014008 | c.1636G >A | p.Asp546Asn | Missense | rs147222955 | 0/0.048 | 23 | tolerated | damaging | 0.625 | neutral | 1/155 male | yes (grand-father) | unlikely | |
| 49142473 | PPP1R3F | NM_033215 | c.1321G >A | p.Glu441Lys | Missense | — | -/- | 56 | deleterious | benign | 0.153 | neutral | 0/384 | yes (grand-father) | possible | |
| 53630356 | HUWE1 | NM_031407 | c.2849T >A | p.Val950Asp | Missense | — | -/- | 152 | deleterious | damaging | 0.702 | neutral | 0/329 male | no (grand-father) | probable | |
| 75649236 | MAGEE1 | NM_020932 | c.913A >T | p.Ser305Cys | Missense | rs142080557 | 0/0.16 | 112 | tolerated | damaging | 0.309 | neutral | yes (grand-father) | unlikely | ||
| Family 5 | 117251283 | KLHL13 | NM_033495 | c.-132040G >A | 5'UTR | — | -/- | 0/71 male | NA | unknown | ||||||
| 138287146 | FGF13 | NM_004114 | c.-493981G >A | 5'UTR | rs185138825 | 0/- | 3/73 male | NA | unlikely | |||||||
| Family 6 | 152960226 | SLC6A8 | NM_005629 | c.1649C >G | p.Thr550Ser | Missense | — | -/0.01 | 58 | tolerated | damaging | 0.284 | neutral | 0/218 male | yes (grand-father) | possible |
| 153210180 | RENBP | NM_002910 | c.-134_-118delins17 | 5'UTR | — | -/- | yes (grand-father) | unknown | ||||||||
| 153701511 | PLXNA3 | NM_017514 | c.*483 G >A | 3'UTR | rs41311390 | 0.3/- | yes (grand-father) | unknown | ||||||||
| 48432832 | RBM3 | NM_006743 | c.-143C >G | 5'UTR | — | -/- | yes (grand-father) | unknown | ||||||||
| 152989468 | BCAP31 | NM_001139457 | c.-350_-349insCA | 5'UTR ins | — | -/- | yes (grand-father) | unknown | ||||||||
| Family 7 | 120009218 | CT47B1 | NM_001145718 | c.291_293del | p.Glu99del | Inframe del | — | -/- | 3/40 male | NA | unlikely | |||||
| Family 8 | 3227525 | MXRA5 | NM_015419 | c.*232 G >T | 3'UTR | rs143753500 | 0/- | no (2 uncles) | unknown | |||||||
| 8499107 | KAL1 | NM_000216 | c.*1929 A >G | 3'UTR | rs6530183 | 0/- | yes (2 uncles) | unknown | ||||||||
| 53117400 | TSPYL2 | NM_022117 | c.*279 A >C | 3'UTR | rs113822334 | 0.9/- | no (2 uncles) | unknown | ||||||||
| 53966801 | PHF8 | NM_001184896 | c.2904_2906del | p.Ser969del | Inframe del | — | — | 0/189 | no (2 uncles) | probable | ||||||
| 63412635 | FAM123B | NM_152424 | c.532C >T | p.Arg178Cys | Missense | — | −/0.01 | 180 | deleterious | damaging | 0.506 | neutral | no (2 uncles) | unlikely | ||
| 153037579 | PLXNB3 | NM_005393 | c.2739–58_2739–57ins28 | Intronic ins | — | -/- | yes (2 uncles) | unlikely | ||||||||
| 47466513 | SYN1 | NM_006950 | c.435+27 G >C | Intronic | — | −/0.26 | no (2 uncles) | unlikely | ||||||||
| 55027965 | APEX2 | NM_014481 | c.158–5del | Intronic del | — | -/- | yes (2 uncles) | unlikely | ||||||||
| 154284285 | FUNDC2 | NM_023934 | c.*1338C >G | 3'UTR | rs146616394 | 0/- | yes (2 uncles) | unknown | ||||||||
| Family 9 | 153210180 | RENBP | NM_002910 | c.-134_-118delins17 | 5'UTR | — | -/- | yes (grand-father) | unknown | |||||||
| 154754246 | TMLHE | NM_018196 | c.229C >T | p.Arg77X | Nonsense | — | -/- | 0/508 male, 1 htz/184 female | no (grand-father and uncle) | probable | ||||||
| Family 11 | 54956008 | TRO | NM_001039705 | c.2851 G >A | p.Gly951Ser | Missense | — | -/- | 56 | deleterious | unknown | 0.174 | neutral | 0/138 male | yes (heathy brother) | possible |
| Family 12 | 151123143 | GABRE | NM_004961 | c.*30C >T | 3'UTR | — | -/- | NA | unknown |
Abbreviation: MAF, minor allele frequency; SIFT, scale-invariant feature transform; UTR, untranslated region.
This list includes variants that are rare or absent from databases (MAFs <1%), are found in genes expressed in brain and have a possible impact on gene or protein function (nonsense, missense, predicted to be deleterious by at least one bioinformatics algorithm or in an intron, 5'UTR or 3'UTR with a predicted effect on a splice site or promoter). Mutpred: score <0.5: benign; 0.5–0.75: possibly disease-associated; and >0.75: probably disease-associated.Grantham distance: conservative (0–50), moderately conservative (51–100), moderately radical (101–150) or radical (⩾151).
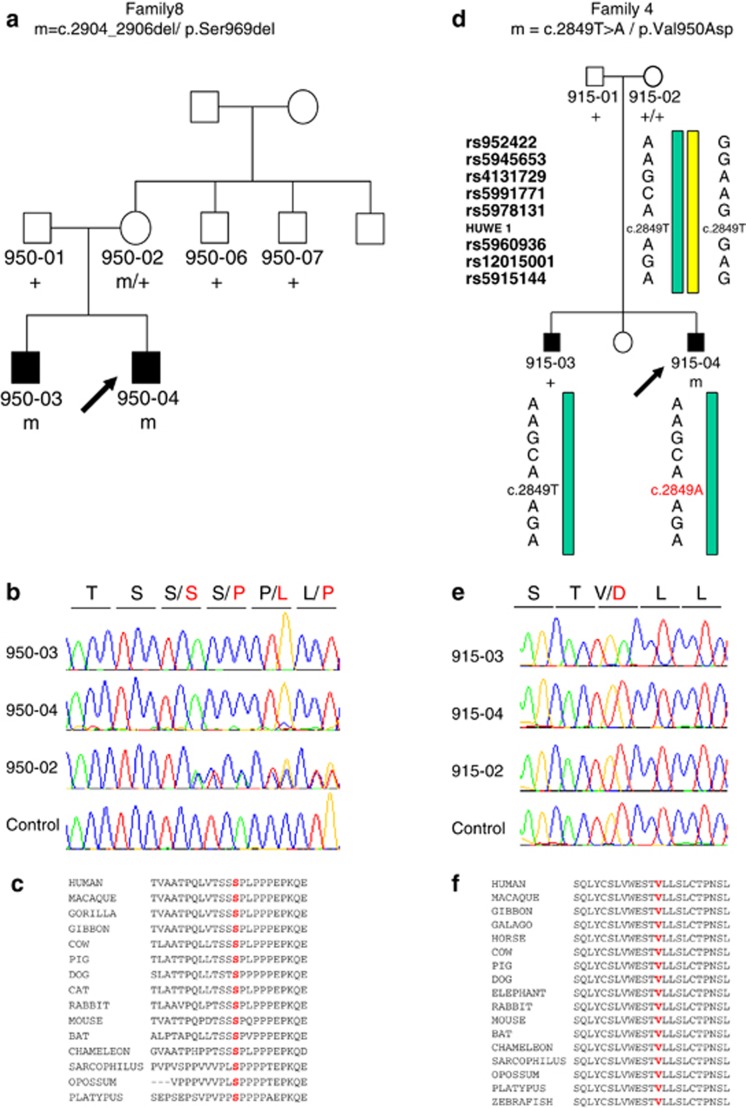
In two families, the variants were in genes previously implicated in ID.21, 22, 23, 24 Both variants, c.2904_2906del/p.Ser969del in PHF8 and c.2849 T>A/p.Val950Asp in HUWE1, affect amino-acids that were highly conserved during evolution and were not found in a large control population or reported in Hapmap, 1000 Genomes and the Exome Variant Server (Figure 2). Although, mutations in PHF8 causing a loss-of-function were previously identified in patients with ID and cleft lip/palate,21, 22, 23 the p.Ser969del variant segregated with high-functioning autism without other clinical features in family 8. In the index case of family 4, p.Val950Asp in HUWE1 was predicted to be deleterious by SIFT, Polyphen-2 and Mutpred algorithms. Surprisingly, this variant was not found in the proband's brother, who was less severely affected, and turned out to have occurred de novo in the proband. We hypothesized that Val950Asp in HUWE1 contributed to a genetically complex disorder by increasing the severity of the phenotype. Alternatively, the phenotype of the brother could have had a different etiology. These results illustrate the complexity of inheritance in ASD, in which a combination of rare inherited and de novo events can contribute to the disorder.6, 7, 8, 9, 10
Figure 2.
Identification of variants in PHF8 and HUWE1 in families 8 and 4. (a) Pedigree of family 8 and segregation analysis of the p.Ser969del variant in PHF8. The arrow indicates the index case. (b) Sequence electropherograms showing the presence of the p.Ser969del variant at the hemizygous state in the two affected brothers and at the heterozygous state in their mother. (c) Alignment of the region flanking the variant in orthologous proteins, showing the high conservation of Serine 969. (d) Pedigree of family 4 and haplotypes reconstructed from eight informative single nucleotide polymorphisms (SNPs) adjacent to HUWE1 (genotypes of these SNPs were obtained from Illumina cytoSNP-12 arrays analysis), showing that the same maternal haplotype was transmitted to the affected brothers with and without the p.Val950Asp mutation. The arrow indicates the index case. (e) Sequence electropherograms showing the presence of p.Val950Asp in the index case and its absence in the affected brother and in the mother. These results are consistent with the de novo occurrence of p.Val950Asp in the index case. (f) Alignment of the region flanking the variant in orthologous proteins, showing the high conservation of valine 950.
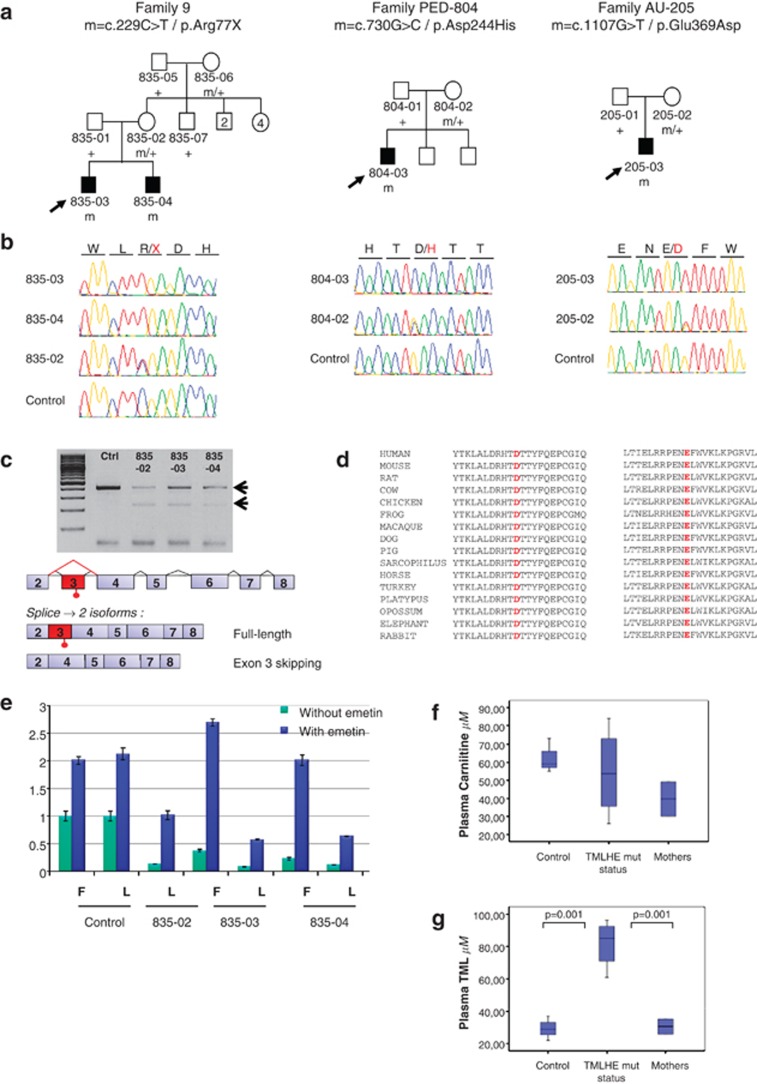
Among the remaining variants, a nonsense mutation (c.229C>T/p.Arg77X) in TMLHE, encoding ɛ-N-trimethyllysine hydroxylase, the enzyme catalyzing the first step of carnitine biosynthesis from TML, segregated with autism and moderate ID in family 9 (Figures 3a and b). The p.Arg77X mutation was absent from 508 healthy male controls and databases.
Figure 3.
Identification of TMLHE mutations in three families. (a) Pedigrees and segregation analysis of the TMLHE mutations in families 9, PED-804 and AU-205. The arrows indicate the index cases. (b) Sequence electropherograms of the mutations at the hemizygous state in the index cases (835–03 in family 9, 804–03 and 205–03) and the affected brother of family 9 (835–04), and at the heterozygous state in the mothers (835–02, 804–02 and 205–02). (c) Analysis of TMLHE mRNA in lymphoblasts from members of family 9 and schematic representations of the splicing isoforms detected in subjects with the p.Arg77X mutation in exon 3. Reverse transcriptase–PCR products using primer pairs in exons 2 and 4, run on 2% agarose gels, showed two mRNA isoforms in the index case (835–03), his affected brother (835–04) and his mother (835–02) and a single isoform in a control subject (c). Sequence analysis confirmed that the long isoform contains the premature termination codon in exon 3 and that exon 3 was skipped in the short isoform, probably as a consequence of nonsense-associated alternative splicing. (d) Alignment of the region flanking the two missense variants in orthologous proteins showing the conservation of the altered amino acids. (e) Quantification of TMLHE mRNA expression in fibroblasts (F) and lymphoblasts (L) from members of family 9 by quantitative real-time PCR, using primer pairs in exons 7 and 8. TMLHE mRNA was expressed 10 times less in patients compared with healthy controls (green bars). Overnight treatment with 10 μg ml−1 emetin (blue bars), an inhibitor of nonsense-mediated decay, restored the expression of the TMLHE mRNA. (f) Assay of free carnitine by UPLC (ultra performance liquid chromatography) chromatographic and TQD (tandem quadrupole detector) mass spectrometry in the plasma of patients. (g) Assay of trimethyllysine (TML) by UPLC chromatographic and TQD mass spectrometry showing a 2–3-fold increase in the plasma of patients.
This study identified several additional rare variants that might contribute to ASD such as c.521C>A/p.Ala174Asp (family 1) in ODZ1, which encodes teneurin-1, a transmembrane protein expressed in the developing central nervous system that might have a role in neuronal connectivity.25 In addition, 11 variants located in introns or UTRs of genes expressed in the brain and predicted to have a possible effect on gene expression were found in five families. Five of these genes are involved in axon guidance (PLXNA3, PLXNB3, KAL1) or neurotransmission (SYN1, GABRE), four regulate transcription, splicing or translation (TXLNG, TSPYL2, AFF2, involved in brain development, and RBM3, involved in RNA processing, regulation of translation and production of miRNA);26 the remaining genes are involved in ubiquitination (KLHL13) or in protein transport (BCAP31).
Finally, no rare variants meeting the criteria defined in Material and methods were found in three families (families 3, 7 and 10), suggesting that genetic factors in these families are possibly located in unexplored regions of the X chromosome, on autosomes, or were present with a frequency ⩾1%.
Screening of TMLHE and functional consequences of mutations
To investigate the effect of the p.Arg77X mutation at the mRNA level, we performed quantitative reverse transcriptase–PCR analysis in lymphoblasts and fibroblasts from the two affected brothers of family 9 and their mother. The mutated mRNA was significantly downregulated in the cells of the affected sibs. Pretreatment of the cells with emetin restored the mRNA levels, indicating that this downregulation corresponded to the degradation of the mutated mRNA by nonsense-mediated decay (Figure 3e). Interestingly, two mRNA isoforms were detectable in the patients and their heterozygous mother, one with a premature termination codon in exon 3 and one missing exon 3, which restored the reading frame (Figure 3c). Exon 3 skipping could have resulted from nonsense-associated altered splicing, a mechanism alternative to nonsense-mediated decay.27
To confirm that mutations in TMLHE are associated with ASD, we screened its coding sequence in 501 male probands with ASD. Two missense substitutions (c.730G>C/p.Asp244His and c.1107G>T/p.Glu369Asp) were found in two additional unrelated patients with ASD but not in 330 controls and not reported in databases. Aspartic acid 244 and glutamic acid 369 are highly conserved in other species (Figure 3d). Aspartic acid 244 is predicted to bind the 2-oxoglutarate cofactor and constitutes one of the three key residues of the catalytic core,28 suggesting a complete loss-of-function of the protein with this substitution.
To further analyze the consequences of the TMLHE mutations, we assayed carnitine and TML, the precursor of carnitine biosynthesis, in plasma and urine from the brothers with the p.Arg77X mutation and the patient with the p.Asp244His variant for whom biological samples were available. Carnitine was slightly but not significantly decreased in the plasma of the patients; the values remained in the normal range (Figure 3f). By contrast, mass spectrometry revealed a significant 2–3-fold increase in the TML precursor in the plasma of all the three patients with the Arg77X and Asp244His mutations compared with controls (Figure 3g). Carnitine intake, estimated from a 3-day dietary recall questionnaire, was normal in all patients (estimated at 55, 80 and 96 mg j−1, respectively).
Very few variants in TMLHE predicted to have deleterious effects are present in genetic databases (HapMap, 1000 Genomes); in particular, no nonsense mutations have been identified and only 18 non-synonymous variants have been reported on >8700 X chromosomes in the Exome Variant Server. Furthermore, non-synonymous variants are far more frequent in females than in males (n=22/3381 versus n=3/1998, if we exclude the Asn235Thr variant specific of the African population), indicating that variations in TMLHE are not well tolerated and are liable to be pathogenic. Mutations in TMLHE were found in patients with autism and ID but not in patients with Asperger syndrome. If we exclude patients with Asperger syndrome, the difference between point mutations in TMLHE in male patients with autism and mental retardation and male individuals from Exome variant server is significant (P=0.05).
Interestingly, Celestino-Soper et al.29 have recently reported that the deletion of exon 2 of TMLHE is a CNV present in ∼1/350 males and associated with ASD with a low penetrance. To compare the frequency of this CNV in healthy individuals and patients with ASD, we specifically assessed its presence in 896 unrelated healthy male controls and 691 patients with ASD by quantitative multiplex assay or Illumina Human 1M-single BeadChip arrays (see Material and methods). This study revealed the presence of one deletion of exon 2 of TMLHE in a single subject out of 896 male controls, whereas it was present in 3 out of 691 male patients with ASD (P=0.3). Interestingly, one of the three patients with the deletion of exon 2 had an affected brother who did not carry the deletion. In addition, a duplication encompassing exon 1 of TMLHE, absent from 525 healthy controls, was detected by Illumina Human 1M-single BeadChip arrays in an additional patient with ASD (P=0.4). Altogether, these results suggest that deficits in TMLHE could be rarer than previously reported. These deficits seem to be more frequent in patients with ASD, suggesting that they constitute susceptibility factors for autism, although the difference did not reach significance for microrearrangements in TMLHE.
Discussion
This study focused on rare variants on chromosome X in multiplex families with ASD compatible with X-linked inheritance. Altogether, this study identified 36 possibly deleterious variants in 33 genes, including PHF8, HUWE1 and TMLHE. Variants in genes common to at least two families were exceptional, confirming that ASD is genetically highly heterogeneous.
The X chromosome contains the largest number of genes expressed in the brain.30 For this reason, mutations causing monogenic forms of ID have been identified in numerous genes on chromosome X.31 Interestingly, almost all genes involved in ASD, such as NLGN3/4X on chromosome X or SHANK3 on chromosome 22qter, are also mutated in patients with ID without autistic features.11, 31 In this study, variants in two genes were previously implicated in ID: PHF8, which encodes a histone lysine demethylase that regulates rRNA synthesis32 and retinoic acid-induced neuronal differentiation,33 and HUWE1, which encodes an E3 ubiquitin-protein ligase that controls neural differentiation and proliferation by catalyzing the polyubiquitination and degradation of the N-Myc oncoprotein.34, 35 Missense mutations and microduplications encompassing HUWE1 were identified in a few large families with moderate-to-severe ID,24 whereas nonsense, truncating and one missense mutations in PHF8 were previously reported in patients with ID and cleft lip/palate.21, 22, 23 Interestingly, in a previous report, two brothers with a deletion encompassing PHF8 and two nearby genes (FAM120C and WNK3) also had autistic features.36 In our study, both variants alter highly conserved amino acids in the proteins and are predicted to be deleterious; the p.Ser969del variant in PHF8 segregated in the two affected brothers from family 8 and the p.Val950Asp in HUWE1 occurred de novo in the index case of family 4 who is more severely affected than his brother. The p.Glu441Lys in PPP1R3F was also present in the two affected brothers of family 4 but was inherited from their unaffected maternal grandfather. Interestingly, another missense variant (p.Phe245Leu) in PPP1R3F was reported in a patient with Asperger syndrome.17 This observation suggests that the variants identified in family 4 act as risk factors for ASD when associated with other deleterious variants on the X chromosome or autosomes. Further studies are needed to confirm the roles of these genes in ASD.
Among the variants detected in this study, there was a single nonsense mutation in TMLHE that segregated with ASD in the two affected brothers of family 9. TMLHE is located at the far end of the long arm of chromosome X (Xq28) and encodes ɛ-N-trimethyllysine hydroxylase, the enzyme that catalyzes the first of the four steps of endogenous carnitine biosynthesis. Screening of additional male patients with ASD identified two missense variants predicted to be deleterious in unrelated sporadic patients. The p.Arg77X and p.Asp244His mutations were associated with a significant increase of TML, the substrate of TMLHE and precursor of carnitine biosynthesis, in the plasma of the patients, confirming that they lead to loss of TMLHE function and deficit of endogenous carnitine biosynthesis. However, although carnitine was mildly decreased, it remained in the normal range in the plasma and urine of the patients. This result is not surprising as, in humans, the carnitine pool mainly comes from food intake. A small pool (∼25%) of carnitine is also synthesized in liver, kidney and brain, but the precise role of endogenous synthesis in these tissues remains unknown.28 Carnitine is an essential metabolite in all animal species as well as in numerous microorganisms and plants. In mammalian cells, carnitine is present as free carnitine and acylcarnitines, including acetylcarnitine.37, 38 The main role of carnitine is to transport activated long-chain fatty acids across the inner mitochondrial membrane for β-oxidation. But additional neuroprotective, neuromodulatory and neurotrophic roles have also been suggested.39, 40, 41, 42 In particular, carnitine is an antioxidant and that might protect mitochondria from oxidative stress.
Another recent study supports the observation that the loss-of-function of TMLHE is associated with ASD. The deletion of exon 2 of TMLHE, originally identified in ASD male patients,43 was shown to be a CNV that is present in male controls at a frequency of ∼1/350. The first coding exon of TMLHE, exon 2, encodes the signal peptide necessary to address the protein to mitochondria. Deletion of this exon causes enzyme deficiency and impairs endogenous carnitine biosynthesis, as observed for p.Arg77X and p.Asp244His mutations. Remarkably, 6 out of 7 sib pairs with ASD were concordant for the deletion of exon 2, whereas the frequency of this CNV was only slightly, not significantly, increased in sporadic ASD patient. The authors concluded that the deletion of exon 2 of TMLHE is a risk factor for nondysmorphic ASD, with a penetrance estimated at 2–4%.29 In this study, the deletion of exon 2 of TMLHE was present in three male patients with ASD out of 691 and only in one male control out of 896, indicating that this CNV could even be rarer than ∼1/350. Although CNVs altering TMLHE tend to be more frequent in patients with ASD than in controls, the difference of frequencies between patients and controls was not significant. Yet, this finding is concordant with the results of the previous study in which significance between patients and controls was not achieved either.29 Interestingly, in our case, the only sib pair with ASD was discordant for the deletion of exon 2 of TMLHE and all the other patients were sporadic cases. However, association studies are not appropriate for rare variants, especially in the case of a high genetic heterogeneity such as in ASD. The arguments suggesting that a loss-of-function of TMLHE could contribute to autism are: (i) the demonstration that mutations have functional biological impact; (ii) the segregation of the mutations with the disorder in the families; and (iii) the identification of different rare functional mutations in the same gene in unrelated individuals.44 Taking these arguments into account, our results confirm that the deficiency of TMLHE likely contributes to ASD, probably in association with other genetic or non-genetic abnormalities, and reinforces the view that point mutations could also be identified in patients with ASD. The penetrance of the point mutations remains to be determined but might be higher than previously anticipated for deletion of exon 2.
The mechanism by which TMLHE deficiency leads to ASD remains unclear. The increase in TML could be toxic at some stage during brain development or interfere with the establishment of normal neuronal networks. Alternatively, the deficiency of carnitine itself or one of the three intermediates of endogenous carnitine biosynthesis (HTML, TMABA or γ-BB, Supplementary Figure S4) might be deleterious for brain development, alone or combined with a deficit in carnitine intake. To test the hypothesis that patients with p.Arg77X and p.Asp244His had a deficiency of carnitine intake in addition of the TMLHE mutation, we assessed their dietary intake over several days. The calculated carnitine intake was normal in all the three patients but reflects only current carnitine intake; a lack of carnitine during specific antenatal or neonatal periods cannot be ruled out. A retrospective questionnaire revealed that patient PED-804–03 (with the p.Asp244His mutation) refused to eat meat around 12 months of age. However, young children frequently refuse meat, particularly those with ASD. Interestingly, low levels of carnitine and acetylcarnitines and altered brain fatty acid metabolism were reported in subjects with ASD.45, 46, 47 However, if a systemic carnitine deficiency constitutes a risk factor for ASD, infants with low meat intake would have a higher risk of developing ASD. This is unlikely as the prevalence of autism is not notably increased in vegetarians or populations with low meat intake. In addition, carnitine and related metabolites are often abnormal in patients without ASD. These arguments suggest that low levels of carnitine and acetylcarnitines in ASD patients could reflect a more complex deficit. Further studies are, therefore, needed to decipher the real role of carnitine and TMLHE deficiency in ASD.
Recent studies have emphasized the role of carnitine in promoting social interactions in animal models. Desert (Schistocerca gregaria) and migratory locusts (Locusta Migratoria) reversibly change between two phenotypes (solitarious and gregarious) that differ in bodily appearance, physiology, brain size and organization, and behavior. At low population density, locusts in the solitarious phase avoid their congeners; when the population increases, locusts become gregarious and aggregate in migratory swarms. The genomes of the two forms of locust are equivalent. The transformation is driven only by epigenetic regulation.48 Interestingly, carnitine was recently shown to constitute a key regulatory metabolite in the phase transition in the migratory locust.49 Remarkably, the only other metabolite that is known to regulate phase transition in desert locusts is serotonin.50 The link between carnitine and serotonin is unclear, but carnitine has been proposed to act as a neuromodulator in the animal central nervous system; in addition, acylcarnitines could promote the biosynthesis and release of neurotransmitters, including dopamine and melatonin.51 Although these results were obtained in species very distant from humans, they support the hypothesis that carnitine has a conserved role in socialization during evolution and offers potentially novel insights into the complex role of carnitine and its derivatives acetylcarnitine and acylcarnitines in the brain.
Finally, TMLHE could also have other yet unknown functions. An isoform has been reported in which exon 2 is spliced out and an alternative start codon in exon 3 is used.42 This isoform lacks the mitochondrial targeting signal and probably localizes in another cellular compartment. Contrary to the deletion of exon 2, which affects only the mitochondrial isoform,29 the mutations reported in this study are predicted to affect other isoforms as well. Interestingly, the TMLHE protein was reported to interact with nuclear complex p130/RBL2 that regulates gene expression, supporting the hypothesis that TMLHE has another cellular localization and function.52 The mechanism by which the TMLHE deficiency leads to ASD remains to be further characterized.
Altogether, our results confirm that a TMLHE deficiency is associated with ASD and support the hypothesis that rare variants on the X chromosome are involved in the etiology of ASD and contribute to the sex-ratio disequilibrium characteristic of these disorders.
Acknowledgments
We thank the patients for their participation in the study, the P3S platform and the genotyping and sequencing platform of the ICM for technical assistance and the DNA and cell bank of CRICM for DNA extraction and cell culture. We also thank Dr Merle Ruberg for critical reading of the manuscript and Pr André Mégarbané for kindly providing DNA of controls individuals from Lebanon. This study was financially supported by GIS-Maladies Rares, Fondation de France, ERA-NET NEURON EUHFAUTISM, INSERM and AP-HP.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Freitag CM, Staal W, Klauck SM, Duketis E, Waltes R. Genetics of autistic disorders: review and clinical implications. Eur Child Adolesc Psychiatry. 2010;19:169–178. doi: 10.1007/s00787-009-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Carlstrom E, Rastam M, Gillberg C, Anckarsater H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry. 2010;167:1357–1363. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Moreno-De-Luca D, Heron D, Bouteiller D, Gennetier A, Delorme R, et al. Screening for genomic rearrangements and methylation abnormalities of the 15q11-q13 region in autism spectrum disorders. Biol Psychiatry. 2009;66:349–359. doi: 10.1016/j.biopsych.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Sato D, Lionel AC, Leblond CS, Prasad A, Pinto D, Walker S, et al. SHANK1 deletions in males with autism spectrum disorder. Am J Human Genet. 2012;90:879–887. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G, et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 2012;8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillberg C, Cederlund M, Lamberg K, Zeijlon L. Brief report: ‘the autism epidemic'. The registered prevalence of autism in a Swedish urban area. J Autism Dev Disord. 2006;36:429–435. doi: 10.1007/s10803-006-0081-6. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton A, Gauthier J, Hamdan FF, Lafreniere RG, Yang Y, Henrion E, et al. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2011;16:867–880. doi: 10.1038/mp.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozonoff S, Young GS, Carter A, Messinger D, Yirmiya N, Zwaigenbaum L, et al. Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics. 2011;128:e488–495. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F, Holbert S, Ronce N, Faravelli F, Lenzner S, Schwartz CE, et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet. 2005;42:780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abidi FE, Miano MG, Murray JC, Schwartz CE. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin Genet. 2007;72:19–22. doi: 10.1111/j.1399-0004.2007.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivisto AM, Ala-Mello S, Lemmela S, Komu HA, Rautio J, Jarvela I. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin Genet. 2007;72:145–149. doi: 10.1111/j.1399-0004.2007.00836.x. [DOI] [PubMed] [Google Scholar]
- Froyen G, Corbett M, Vandewalle J, Jarvela I, Lawrence O, Meldrum C, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Human Gene. 2008;82:432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann D, Chiquet-Ehrismann R, Leachman NT, Tucker RP. Teneurin-1 is expressed in interconnected regions of the developing brain and is processed in vivo. BMC Dev Biol. 2008;8:30. doi: 10.1186/1471-213X-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilotte J, Dupont-Versteegden EE, Vanderklish PW. Widespread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein, RBM3. PLoS ONE. 2011;6:e28446. doi: 10.1371/journal.pone.0028446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Monfregola J, Cevenini A, Terracciano A, van Vlies N, Arbucci S, Wanders RJ, et al. Functional analysis of TMLH variants and definition of domains required for catalytic activity and mitochondrial targeting. J Cell Physiol. 2005;204:839–847. doi: 10.1002/jcp.20332. [DOI] [PubMed] [Google Scholar]
- Celestino-Soper PB, Violante S, Crawford EL, Luo R, Lionel AC, Delaby E, et al. A common X-linked inborn error of carnitine biosynthesis may be a risk factor for nondysmorphic autism. Proc Natl Acad Sci USA. 2012;109:7974–7981. doi: 10.1073/pnas.1120210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. High expression of the mammalian X chromosome in brain. Brain Res. 2006;1126:46–49. doi: 10.1016/j.brainres.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Raymond FL. X linked mental retardation: a clinical guide. J Med Genet. 2006;43:193–200. doi: 10.1136/jmg.2005.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Wang Y, Li X, Wang Y, Xu L, Wang X, et al. PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res. 2010;20:794–801. doi: 10.1038/cr.2010.75. [DOI] [PubMed] [Google Scholar]
- Qiu J, Shi G, Jia Y, Li J, Wu M, Li J, et al. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res. 2010;20:908–918. doi: 10.1038/cr.2010.81. [DOI] [PubMed] [Google Scholar]
- Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arca D, Zhao X, Xu W, Ramirez-Martinez NC, Iavarone A, Lasorella A. Huwe1 ubiquitin ligase is essential to synchronize neuronal and glial differentiation in the developing cerebellum. Proc Natl Acad Sci USA. 2010;107:5875–5880. doi: 10.1073/pnas.0912874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Liu X, Harvard C, Hildebrand MJ, Rajcan-Separovic E, Holden JJ, et al. Autism-associated familial microdeletion of Xp11.22. Clin Genet. 2008;74:134–144. doi: 10.1111/j.1399-0004.2008.01028.x. [DOI] [PubMed] [Google Scholar]
- Vaz FM, Wanders RJ. Carnitine biosynthesis in mammals. Biochem J. 2002;361 (Pt 3:417–429. doi: 10.1042/0264-6021:3610417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LL, McDonald DA, Borum PR. Acylcarnitines: role in brain. Prog Lipid Res. 2010;49:61–75. doi: 10.1016/j.plipres.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Virmani A, Binienda Z. Role of carnitine esters in brain neuropathology. Mol Aspects Med. 2004;25:533–549. doi: 10.1016/j.mam.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Nalecz KA, Miecz D, Berezowski V, Cecchelli R. Carnitine: transport and physiological functions in the brain. Mol Aspects Med. 2004;25:551–567. doi: 10.1016/j.mam.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Nalecz KA, Nalecz MJ. Carnitine--a known compound, a novel function in neural cells. Acta Neurobiol Exp. 1996;56:597–609. doi: 10.55782/ane-1996-1165. [DOI] [PubMed] [Google Scholar]
- Monfregola J, Napolitano G, Conte I, Cevenini A, Migliaccio C, D'Urso M, et al. Functional characterization of the TMLH gene: promoter analysis, in situ hybridization, identification and mapping of alternative splicing variants. Gene. 2007;395:86–97. doi: 10.1016/j.gene.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Celestino-Soper PB, Shaw CA, Sanders SJ, Li J, Murtha MT, Ercan-Sencicek AG, et al. Use of array CGH to detect exonic copy number variants throughout the genome in autism families detects a novel deletion in TMLHE. Human Mol Genet. 2011;20:4360–4370. doi: 10.1093/hmg/ddr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- Clark-Taylor T, Clark-Taylor BE. Is autism a disorder of fatty acid metabolism? Possible dysfunction of mitochondrial beta-oxidation by long chain acyl-CoA dehydrogenase. Med Hypotheses. 2004;62:970–975. doi: 10.1016/j.mehy.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Filipek PA, Juranek J, Nguyen MT, Cummings C, Gargus JJ. Relative carnitine deficiency in autism. J Autism Dev Disord. 2004;34:615–623. doi: 10.1007/s10803-004-5283-1. [DOI] [PubMed] [Google Scholar]
- Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17:290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows M, Rogers SM, Ott SR. Epigenetic remodelling of brain, body and behaviour during phase change in locusts. Neural Syst Circuits. 2011;1:11. doi: 10.1186/2042-1001-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Wu Z, Wang X, Yang P, Yu D, Zhao C, et al. Metabolomic analysis reveals that carnitines are key regulatory metabolites in phase transition of the locusts. Proc Natl Acad Sci USA. 2012;109:3259–3263. doi: 10.1073/pnas.1119155109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstey ML, Rogers SM, Ott SR, Burrows M, Simpson SJ. Serotonin mediates behavioral gregarization underlying swarm formation in desert locusts. Science (New York, NY. 2009;323:627–630. doi: 10.1126/science.1165939. [DOI] [PubMed] [Google Scholar]
- Scafidi S, Fiskum G, Lindauer SL, Bamford P, Shi D, Hopkins I, et al. Metabolism of acetyl-L-carnitine for energy and neurotransmitter synthesis in the immature rat brain. J Neurochem. 2010;114:820–831. doi: 10.1111/j.1471-4159.2010.06807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.